Abstract
The unfolded protein response (UPR), a cellular defense mechanism against misfolded protein accumulation in the endoplasmic reticulum (ER), is associated with many human diseases such as aging, cancer and diabetes. X-box binding protein 1 (XBP1), a key transcription factor of the UPR, is critical in maintaining ER homeostasis. Nonetheless, the mechanism by which XBP1 transcriptional activity is regulated remains unexplored. Here we show that the active form of XBP1 protein, XBP1s, is SUMOylated mainly by the protein inhibitors of activated STAT 2 (PIAS2) at two lysine residues located at the C-terminal transactivation domain. Ablation of these SUMOylation events significantly enhances the transcriptional activity of XBP1s towards UPR target genes. Thus, our results reveal a previously unexpected role for SUMO in the regulation of UPR activation and ER homeostasis.
Keywords: UPR, SUMO, post-translational modification, transcriptional regulation
INTRODUCTION
The inositol-requiring enzyme 1α (IRE1α)-XBP1 pathway, the most highly conserved signaling branch of the UPR [1–3], is important for ER biogenesis and the secretory capacity of cells. Once activated, mammalian IRE1α splices 26 nucleotides from the Xbp1u mRNA (u, unspliced), leading to a frameshift and the generation of an active b-ZIP transcription factor called XBP1s (s, spliced) that contains a C-terminal transactivation domain absent from XBP1u [4–9]. The XBP1s protein subsequently translocates to the nucleus and activates the transcription of genes involved in lipid biosynthesis, protein folding, degradation and trafficking [6, 7, 10, 11].
Studies in animal models have revealed that the IRE1α-XBP1 pathway is essential for the development and function of the liver and heart as well as professional secretory cells such as pancreatic and plasma cells [12–16]. XBP1s has also been shown to regulate hepatic lipogenesis via a UPR-independent manner [17]. We recently showed that the IRE1α-XBP1 pathway is indispensable for adipocyte differentiation via XBP1s-mediated regulation of a key adipogenic factor C/EBPα [18]. Despite its physiological significance, the mechanism by which the transcriptional activity of XBP1s protein is regulated remains largely unknown.
The post-translational modification of proteins by the small ubiquitin-like modifier (SUMO) is an important transient regulatory mechanism in many cellular processes, most notably transcriptional regulation, DNA damage, and signal transduction [19, 20]. SUMO proteins have three isoforms (SUMO1-3), with 50% identity between SUMO1 and SUMO2/3. Similar to the process of ubiquitination, SUMO is first activated by the E1 enzyme Aos1/Uba2, then transferred to the E2 enzyme Ubc9, and subsequently conjugated to target proteins via the activity of an E3 ligase such as PIAS, Ran binding protein 2 (RanBP2) and polycomb group protein Pc2 [19, 20]. Here we show that the transcriptional activity of XBP1s is negatively modulated by SUMOylation. Our results reveal a previously unexpected role for SUMO in the regulation of UPR activation.
MATERIALS AND METHODS
Plasmids
Full length XBP1s, XBP1u, ATF6 and active form of ATF6N expression constructs were cloned from liver cDNA library and subcloned into either pcDNA3/Flag- or HA- or myc-vectors. The PCR product with the same XBP1u sequence but lacked 26 nucleotides was termed truncated XBP1s. XBP1s-GFP fusion constructs were made by subclone XBP1s PCR fragment into the pEGFP-N1 vector (Clontech). To construct the plasmid encoding luciferase gene driven by four XBP1 binding sites, the synthetic oligo containing four XBP1 binding sites (italicized), aagctagccgcgTGGATGACGTGTACATGGATGACGTGTACATGGATGACGTGTACATGGATGACGTGTACAaagctttt (underlined sequences are Nhe I and Hind III sites), were digested with the Nhe I/Hind III and then ligated with the Nhe I/Hind III fragment of the pGL3 vector (Promega). mPIAS1/2 was cloned using PCR of mouse liver cDNA library and subcloned into pcDNA3/Flag vector using the Not I and Xba I sites. Plasmids encoding hPIAS2a and hPIAS2b were gifts from Dr. Jorma Palvimo (University of Kuopio, Finland). Plasmids encoding Flag-HDAC1, 2, 4, 5 and 6 on the pBJ5 vector and Flag-HDAC3 on the pcDNA3 vector were gifts from Dr. Marc Montminy (Salk Institute). The pCMV10/3xFlag-hPIAS4, myc-Ubc9 and HA-SUMO1 plasmids were gifts from Dr. Ling Cai (UCSD). HA-SUMO2 and HA-SUMO3 were gifts from Katherine Kieckhafer (Cornell University). The same amount of plasmids was used in all transfection experiments.
Cells and reagents
XBP1−/− MEFs (a gift from Laurie Glimcher, Harvard Medical School), macrophage RAW264.7 and HEK293T cells were maintained in DMEM supplemented with 10% FBS (Hyclone). ER stress inducer thapsigargin (Tg, EMD Calbiochem) was dissolved in DMSO and used at 300 nM.
Quick-change mutagenesis
Site-directed mutagenesis to generate XBP1s mutants was carried out using pcDNA3/Flag-XBP1s as the template. The PCR reaction included the mutagenic primers as well as the pfu polymerase (Stratagene) with the following cycles: 95°C 30 sec, 55°C 30 sec, 72°C 5 min for a total of 13 cycles. After Dpn I digestion for 2–3 h, the PCR reaction was transformed into XL-Blue competent cells. All mutagenesis were confirmed by sequencing. All primers used in mutagenesis are listed in Supplemental Table 1.
Fluorescent microscopy
HEK293T cells were split onto sterile cover glasses coated with poly-L-lysine (Sigma). Cells were transfected with plasmids encoding wildtype or lysine mutant XBP1s-GFP fusion proteins. Cover glasses were removed from dishes and mounted onto slides with VECTASHIED with DAPI. The fluorescent pictures when taken with a Leica DM5000 B microscope using a 63× oil objective.
Immunoprecipitation
HEK293T cells split onto 10 cm dishes the day before were transfected with plasmids using polyethylenimiene. Transfected cells were lysed directly on the plates. 10 mM N-ethylmaleimide (NEM, Sigma) was added to lysis buffer when detecting SUMOylation. Anti-Flag agarose beads were added to the lysates and rocked gently at 4°C overnight. Following three washes, the beads were boiled and then supernatants were loaded onto a SDS-PAGE.
Western blot
Cells were lysed in Tris-based lysis buffer containing 1% NP-40. Western blot was performed using 15–30 µg of total cell lysates as described in [18]. Antibodies used in this study: Flag- and HA-HRP (Sigma), XBP1 (rabbit, Santa Cruz) and goat anti-rabbit IgG HRP (Biorad).
Luciferase reporter assay
After overnight transfection, cells on a 24-well plate were lysed in 100 µl extraction buffer containing Gly-Gly buffer (FISHER), 1% Triton X-100, 1 mM DTT. 50 µl of lysates were transferred to a 96-well plate containing 50 µl of assay mix (Gly-Gly buffer, 16 mM K2HPO4, 2 mM DTT and 2.5 mM ATP). 100 µl of 0.4 mM luciferin mix was added to each well through injector and readings were collected using the Synergy 2 plate reader (Bio-tek). Luciferase activity was normalized to activity from cotransfected CMV promoter driven-β-galactosidase expression plasmid.
RNA extraction and Q-PCR
Total RNA was extracted using Trizol per supplier’s protocol (Molecular Research Center) and reverse transcribed using Superscript III kit (Invitrogen). cDNA were analyzed using the SYBR Green PCR system on the Roche LightCycler 480 Q-PCR machine (Roche). All Q-PCR data were normalized to ribosomal l32 gene. Primer sequences are listed in Supplemental Table 1.
Statistical analysis
Results are expressed as mean ± s.e.m. Comparisons between groups were made by unpaired two-tailed Student’s t-test. P<0.05 was considered as statistically significant. All experiments were repeated at least twice and representative data are shown.
RESULTS
XBP1s of the UPR is SUMOylated
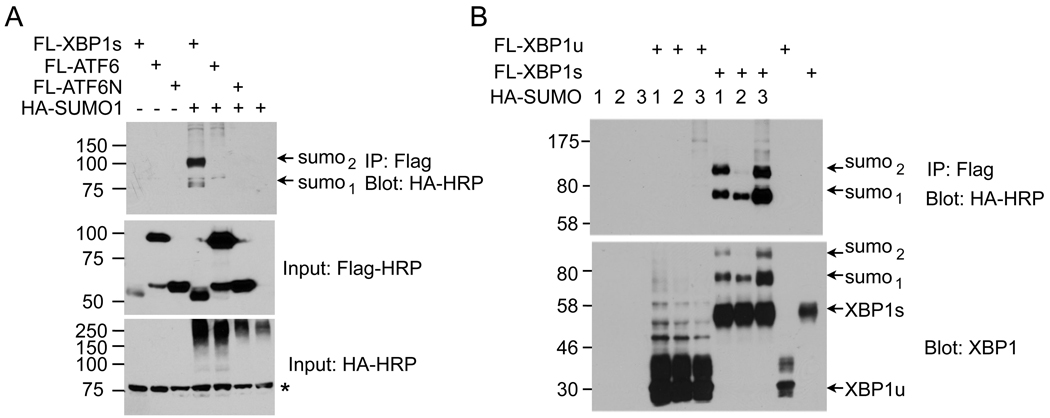
To identify potential UPR transcription factors that could be SUMOylated, we transfected HEK293T cells with vectors encoding XBP1s, ATF6 and the nuclear-active form of ATF6 (ATF6N) plus the E2 enzyme Ubc9 and SUMO1. Among the three factors, XBP1s protein was the only one undergoing SUMOylation (Figure 1A). Further confirming the specificity of SUMOylation in UPR, XBP1s, but not XBP1u, was SUMOylated by all three isoforms of SUMO, SUMO1 and SUMO2/3 (Figure 1B). Indeed, a recent genome-wide analysis of SUMO2 modification during heat shock response identified XBP1s as a SUMOylated protein by SUMO2 [21]. In line with previous reports [22–24], SUMOylation increased the molecular weight of XBP1s protein by ~20kDa. XBP1s SUMOylation was not cell-type specific as it was also detected in mouse embryonic fibroblasts (MEFs) and COS7 cells (not shown). Taken together, our data show that the XBP1s protein of the UPR is SUMOylated.
Figure 1. XBP1s is SUMOylated.
Immunoblots of HA-SUMO showing recovery of HA-SUMO from immunoprecipitates of Flag. (A) HEK293T cells were transfected with Flag-tagged XBP1, full length ATF6 and active form of ATF6 (ATF6N) together with HA-SUMO1. (B) HEK293T cells were transfected with Flag-XBP1s and XBP1u together with different SUMO isoforms (HA-SUMO1, 2, and 3). The n in sumon denotes the number of SUMOylation events. Inputs are shown. *, non-specific band.
XBP1s protein is SUMOylated at the C-terminal transactivation domain
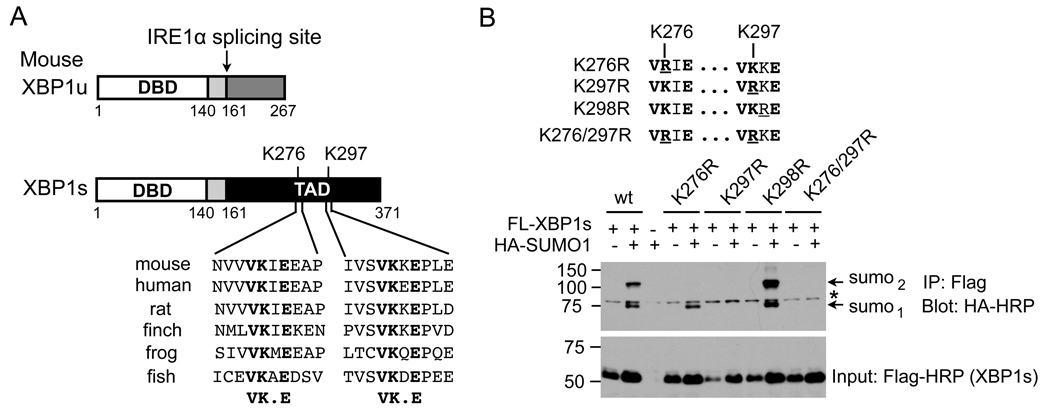
SUMOylation often occurs at the lysine (K) residue in the motif ΨKXE/D (Ψ is a hydrophobic residue; X is any residue; E/D, Aspartate/Glutamate) [25, 26]. Indeed, two highly-conserved SUMOylation motifs were identified at residues K276 and K297; both sites are located within the transactivation domain of the XBP1s protein, a region that is absent from the XBP1u protein (Figure 2A). Mutation of K276 to R (K276R) abolished one SUMOylation event (Figure 2B). Interestingly, mutation of K297 to R (K297R) completely abolished SUMOylation on XBP1s, suggesting that SUMOylation at K297 is required for SUMOylation at K276. Excluding the possibility of non-specific modification events, mutation of K298 to R (K298R), the X in the ΨKXE motif, had no effect on SUMOylation of XBP1s protein (Figure 2B). Thus, our results show that the XBP1s protein is SUMOylated at two lysine residues located at the C-terminal transactivation domain.
Figure 2. XBP1s is SUMOylated at K276 and 297 residues at the C-terminal transactivation domain.
(A) Conservation of two putative SUMOylation motifs in XBP1s. Schematic diagram shown for domains in both XBP1u and XBP1s – shared regions shown with the same color. DBD, DNA binding domain. TAD, transactivation domain. (B) (upper) Diagram of mutagenesis of lysine residues (K) to arginine (R) at the SUMO consensus motifs of mouse XBP1s protein. Mutated residues are underlined. (Lower) Immunoblots of HA-HRP (SUMO1) from immunoprecipitates of Flag in HEK293T cells upon transfection with different Flag-XBP1s constructs with or without HA-SUMO1. Input of XBP1s levels shown. Note that the same amounts of plasmids were added in all experiments and the variation between samples remains unclear. *, non-specific band.
XBP1s protein is SUMOylated endogenously
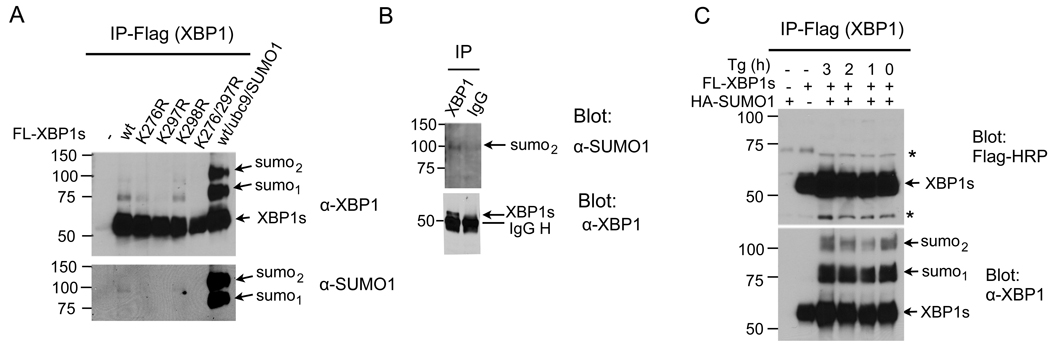
To determine whether endogenous SUMOylation machinery could mediate SUMOylation of XBP1s, cells were transfected with various XBP1s plasmids in the absence of exogenous Ubc9 and SUMO1. Indeed, both wildtype and K298R XBP1s were SUMOylated; the K276R mutant was modified to a lesser extent and no SUMOylation occurred in the K297R or K276/297R mutants (Figure 3A). The difference between the XBP1 and SUMO blots was highly reproducible and specific; therefore, we speculated that it was likely due to the specificity of the antibodies in that SUMOylation may reduce the recognition by the XBP1 antibody for the bi-SUMOylated XBP1s protein (top), which binds preferentially to the SUMO antibody (bottom). Furthermore, SUMOylated endogenous XBP1s protein could be detected following immunoprecipitation of XBP1 in macrophage cells treated with ER stress inducer Tg (Figure 3B). Nonetheless, ER stress itself did not increase the SUMOylation of XBP1s as Tg treatment failed to alter the amount of SUMO-XBP1s in HEK293T cells transfected with XBP1s (Figure 3C). Thus, our data suggest that the XBP1s protein may be constitutively SUMOylated during ER stress.
Figure 3. SUMOylation of endogenous XBP1s.
(A) Immunoblots of XBP1 (upper) and SUMO1 (lower) from immunoprecipitates (IP) of Flag-agarose prepared from transfected HEK293T cells. Cells were transfected with different XBP1s constructs without exogenous Ubc9/SUMO1. Cells transfected with wildtype (wt) XBP1s, E2 Ubc9 and SUMO1 were included in the last lane as a positive control. Note that exogenous SUMO is HA-tagged, thus larger than endogenous SUMO modification. (B) Immunoblots of SUMO1 (upper) and XBP1 (lower) from immunoprecipitates of XBP1 prepared from RAW264.7 macrophage cells treated with 300 nM Tg for 4 h. Immunoprecipitates of control rabbit IgG were included as a negative control. (C) Immunoblots of Flag-HRP (upper) and XBP1s (lower) from immunoprecipitates of Flag-agarose prepared from XBP1s- and SUMO- transfected HEK293T cells. Cells were treated with 300 nM Tg for 0, 1, 2, 3 h prior to IP. *, non-specific band.
SUMO E3 ligase PIAS2 interacts with and mediates SUMOylation of XBP1s
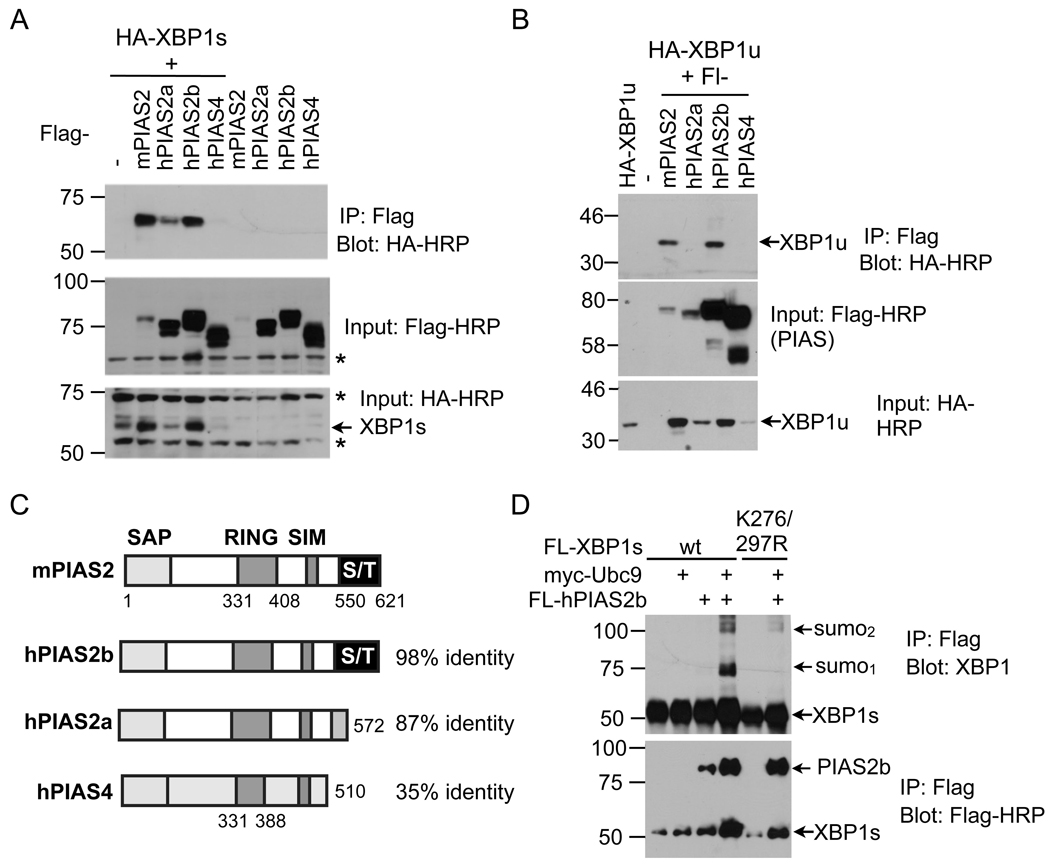
To identify the E3 ligase responsible for SUMOylation of XBP1s, we transfected HEK293T cells with XBP1s and various E3 ligase PIAS constructs (PIAS1, 2a/b, 3 and 4). Strikingly, XBP1s protein interacted specifically with PIAS2 proteins, either mouse PIAS2 or its human homolog PIAS2b. Interactions with other PIAS proteins, including PIAS1, PIAS3, and PIAS4 were either much weaker or not detected (Figure 4A and not shown). Similar observations were observed for XBP1u (Figure 4B), suggesting that the N-terminal region containing the DNA-binding domain shared by both XBP1u and XBP1s (Fig. 2A) may mediate the interaction between XBP1s and PIAS2. On the other hand, interaction of XBP1s or XBP1u protein with hPIAS2a was much weaker compared to that with mPIAS2 or hPIAS2b (Figure 4A–B). It is worth pointing out that the same amount of plasmids for XBP1s was used in all experiments and the difference in XBP1 protein levels (Fig. 4A–B) was likely due to the stabilization of XBP1s proteins by the interacting proteins.
Figure 4. SUMO E3 ligase PIAS2 interacts with and increases SUMOylation of XBP1s.
(A–B) Immunoblots showing recovery of HA-XBP1s (A) and HA-XBP1u (B) from immunoprecipitates of Flag (Flag-PIAS) prepared from transfected HEK293T cells. m, mouse; h, human. (C) Domain comparison of PIAS proteins and the identity shared with mouse PIAS2 protein. SAP, scaffold attachment factor A/B, acinus and PIAS; RING, RING zinc finger; SIM, SUMO interacting motif; S/T, serine/threonine-rich domain. Numbers indicate the position of residues. (D) Immunoblots of XBP1 from immunoprecipitates of Flag in HEK293T cells transfected with Flag-tagged XBP1, Flag-hPIAS2b and/or E2 myc-Ubc9. Note that the same amounts of plasmids were used in each condition in all experiments. *, non-specific band.
As splicing variants PIAS2a (PIASxα) and PIAS2b (PIASxβ) differ from each other at the C-terminal serine/threonine (S/T)-rich domain (Figure 4C), our data suggest that this domain of the PIAS protein may partially mediate its interaction with XBP1s. Further supporting its role in SUMOylation of XBP1s protein, overexpression of hPIAS2b increased SUMOylation of wildtype but not K276/297R XBP1s proteins (Figure 4D). Thus, our data suggest that mouse PIAS2 or human PIAS2b interacts with and mediates SUMOylation of XBP1s.
SUMOylation does not alter the nuclear localization of XBP1s
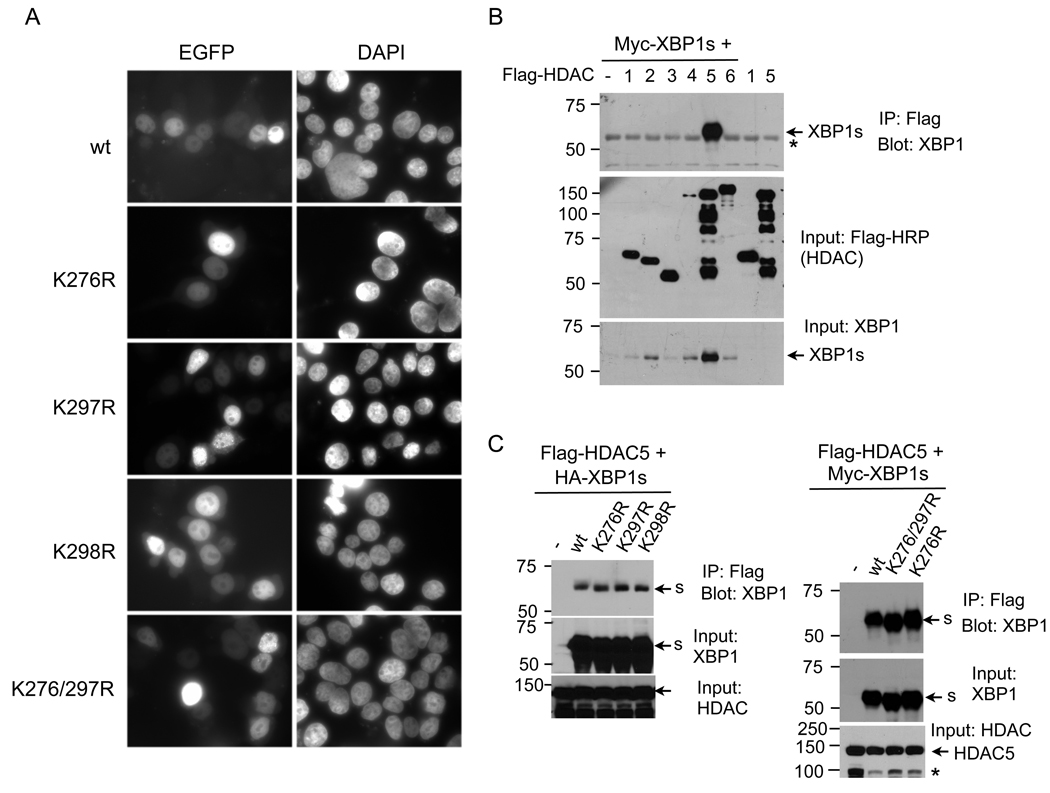
SUMOylation of transcription factors often leads to alterations in the intracellular localization of target proteins [19]. To address this, we expressed XBP1s-GFP fusion proteins for wildtype and mutant XBP1s in HEK293T cells. Comparable to wildtype XBP1s proteins, SUMOylation mutant proteins predominantly localized to the nucleus and did not exhibit altered intracellular distribution (Figure 5A). Thus, our data suggest that SUMOylation does not affect the nuclear localization of XBP1s protein.
Figure 5. SUMOylation of XBP1s does not affect its nuclear localization nor its interaction with HDAC5.
(A) Fluorescent microscopic images of XBP1s-EGFP fusion constructs upon 24 h transfection into HEK293T cells. Nuclei were counter-stained with DAPI. (B) Immunoblots of XBP1 from immunoprecipitates of Flag-agarose prepared from transfected HEK293T cells. Cells were transfected with myc-tagged XBP1s with or without different Flag-HDAC constructs (HDAC1 to HDAC6). Note that same amount of plasmids were transfected in each condition. (C) Immunoblots of XBP1 from immunoprecipitates of Flag-agarose prepared from transfected HEK293T cells. (Left) Cells were transfected with Flag-HDAC5 together with different HA-XBP1s constructs. (Right) Cells were transfected with Flag-HDAC5 together with different myc-XBP1s constructs. Inputs for XBP1s and HDAC5 shown. *, non-specific band.
SUMOylation does not affect the interaction between XBP1s and HDAC5
SUMOylation of transcription factors may also alter their interaction with transcriptional co-repressors such as histone deacetylases (HDAC) [27–29]. Indeed, out of all the HDACs (1 to 6), XBP1s strongly interacted with HDAC5 (Figure 5B). This interaction was not affected by mutations at the SUMOylation sites (Figure 5C). Thus, our data shows that XBP1s interacts with HDAC5 and this interaction is not affected by SUMOylation of XBP1s.
SUMOylation negatively regulates transcriptional activity of XBP1s
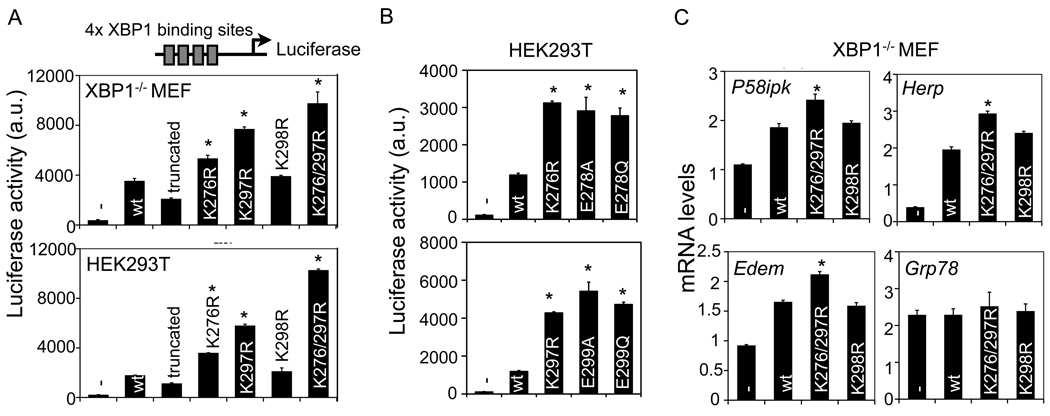
We next determined the physiological significance of SUMOylation on the transcriptional activity of XBP1s. First, we transfected wildtype or mutant XBP1s in XBP1−/− MEFs together with a luciferase reporter driven by a synthetic promoter containing four-tandem XBP1 binding sites TGACGT [30]. Wildtype XBP1s increased luciferase activity over 10 fold (Figure 6A). In line with the requirement of the C-terminal domain of XBP1s for transcriptional activation, loss of C-terminal 100 amino acids of XBP1s (i.e., a truncated form of XBP1s protein) reduced the activity by 50% (Figure 6A). Mutations at either K276 or K297 increased XBP1s transcriptional activity by 50 and 100% relative to wildtype XBP1s, respectively, while mutations at both residues further increased its transcriptional activity (Figure 6A). In contrast, mutation at K298 had no effect. Similar observations were obtained in 293T cells (Figure 6A). It remains unclear why K276/297R XBP1s has higher transcriptional activity compared to K276R XBP1s as both mutants abolish SUMOylation of XBP1s (Figure 2B). We speculate that the K276 residue in the K297R mutant may still be SUMOylated at low efficiency beyond the detection limit or modified via a SUMO-independent mechanism, which may negatively regulates its transcriptional activity.
Figure 6. SUMOylation represses transcriptional activity of XBP1s.
(A–B) Luciferase assay in transfected XBP1−/− MEF (upper A) and HEK293T cells (lower A and B) transfected with various XBP1 constructs. Luciferase gene expression is driven by a synthetic promoter consisted of four tandem XBP1 binding sites. Truncated, XBP1s with no C-terminal transactivation domain. -, GFP control; wt, wildtype. (C) Q-PCR analysis of UPR genes in XBP1−/− MEF transfected with control GFP (−) or different XBP1s constructs. Data are represented as mean ± s.e.m. *, P<0.05 using unpaired two-tailed Student’s t-test comparing mutant XBP1s to wildtype XBP1s.
To further exclude the possibility of SUMO-independent effects at the two lysines (K276 and K297), we mutated E in the SUMO motif ΨKXE to A or Q, i.e., E278A/Q and E299A/Q. Indeed, these mutants exhibited similar transcriptional activity as the lysine mutant located in the same motif (Figure 6B), pointing to a key role of both SUMO motifs in regulating XBP1s activity. Thus, these data demonstrate unequivocally that SUMOylation negatively regulates transcriptional activity of XBP1s.
We next determined the effect of SUMOylation on the expression of known XBP1s target genes. To this end, we transfected wildtype or mutant XBP1s into XBP1−/− MEFs and measured the expression of endogenous UPR genes. Mutations at K276/297, but not K298, strongly induced the expression of several well-known XBP1 target genes [10], including Edem, Herp and P58ipk, by nearly 30–50%, but not the ATF6 target gene Grp78 (Figure 6C). Taken together, our data show that SUMOylation of XBP1s downregulates its transcriptional activity.
DISCUSSION
Here we show that a key transcription factor of the UPR signaling pathway is subjected to covalent post-translational modifications via SUMOylation. Our data showed that XBP1s protein, generated upon activation of IRE1α RNase activity during ER stress response or UPR, is SUMOylated at two lysine residues at the C-terminal transactivation domain, and that these SUMOylation events have additive suppressive effects on XBP1s transcriptional activity. Thus, the transcriptional activity of XBP1s protein is regulated not only by the IRE1α-mediated splicing event, but also by post-translational modifications such as SUMOylation.
Our data adds XBP1s to a growing list of transcription factors whose activity is regulated by SUMOylation and suggests an unexpected role for SUMOylation in regulating ER homeostasis and UPR activity. As SUMOylation is a highly transient post-translational modification event, it may serve as a mechanism to further fine-tune the transcriptional activity of XBP1s and in turn, affect the outcome of UPR. Nonetheless, similar to the scenario for most other transcription factors that are regulated by SUMOylation [19, 20], the detailed molecular mechanism by which SUMOylation affects XBP1s transcriptional activity requires further investigation.
Previous studies have established the indispensible role of the IRE1α-XBP1 branch of the UPR in plasma cell and adipocyte differentiation [15, 16, 18, 31]. During plasma cell differentiation, accumulation of XBP1s protein induces a series of transcriptional events leading to enlarged cell size, expanded ER membrane/volume and increased secretory capacity [32]. On the other hand, during adipocyte differentiation, accumulation of XBP1s protein increases the expression of C/EBPα, a key adipogenic factor, which subsequently controls cellular adaptation to a mature adipocyte phenotype [18]. Further studies are required to determine whether SUMOylation of XBP1s plays a role in cellular differentiation processes.
UPR, a critical cellular defense mechanism against misfolded proteins and aggregates in the ER, has been implicated in many human diseases, collectively termed as “conformational diseases”, including diabetes, cancer and neurodegeneration [33]. When the UPR is overwhelmed or fails, it often leads to cellular dysfunction and cell death [11, 33]. As our knowledge of the role of UPR and XBP1s in human diseases [34, 35], the ability to modulate transcriptional activity of XBP1s may be an important first-step towards improving therapeutical strategies targeting dysfunctional UPR.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Laurie Glimcher, Jorma Palvimo, Marc Montminy, Ling Cai and Katherine Kieckhafer for cell lines and plasmids; Yin He for critical reading of the manuscript; and other members of the Qi laboratory for insightful comments and suggestions. L. Q. is the recipient of the Rosalinde and Arthur Foundation/American Federation for Aging Research New Investigator Award in Alzheimer’s Diseases and the American Diabetes Association (ADA) Junior Faculty Award. This study was funded by AFAR (RAG08061) and NIH NIDDK (R01DK082582).
Abbreviations
- ATF6
activating transcription factor 6
- ER
endoplasmic reticulum
- HDAC
histone deacetylase
- IRE1α
inositol-requiring enzyme 1α
- PIAS
protein inhibitors of activated STAT
- RanBP2
Ran binding protein 2
- SUMO
small ubiquitin-related modifier
- Tg
thapsigargin
- UPR
unfolded protein response
- XBP1
X-box binding protein 1
- XBP1u
unspliced form of XBP1 protein
- XBP1s
spliced form of XBP1 protein
Footnotes
AUTHOR CONTRIBUTION
H.C. and L.Q. conceived the projects. H.C. did the experiments and analyzed the data. L.Q. wrote the paper and H.C. edited the paper.
REFERENCES
- 1.Mori K, Ma W, Gething MJ, Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- 2.Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 3.Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 4.Sidrauski C, Cox JS, Walter P. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell. 1996;87:405–413. doi: 10.1016/s0092-8674(00)81361-6. [DOI] [PubMed] [Google Scholar]
- 5.Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- 6.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 7.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 8.Lee K, Tirasophon W, Shen X, Michalak M, Prywes R, Okada T, Yoshida H, Mori K, Kaufman RJ. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002;16:452–466. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen X, Ellis RE, Lee K, Liu CY, Yang K, Solomon A, Yoshida H, Morimoto R, Kurnit DM, Mori K, Kaufman RJ. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 2001;107:893–903. doi: 10.1016/s0092-8674(01)00612-2. [DOI] [PubMed] [Google Scholar]
- 10.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 12.Lee AH, Chu GC, Iwakoshi NN, Glimcher LH. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J. 2005;24:4368–4380. doi: 10.1038/sj.emboj.7600903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masaki T, Yoshida M, Noguchi S. Targeted disruption of CRE-binding factor TREB5 gene leads to cellular necrosis in cardiac myocytes at the embryonic stage. Biochem. Biophys. Res. Commun. 1999;261:350–356. doi: 10.1006/bbrc.1999.0972. [DOI] [PubMed] [Google Scholar]
- 14.Reimold AM, Etkin A, Clauss I, Perkins A, Zhang J, Horton HF, Scott A, Orkin SH, Byrne MC, Grusby MJ, Glimcher LH. An essential role in liver development for transcription factor XBP-1. Genes Dev. 2000;14:152–157. [PMC free article] [PubMed] [Google Scholar]
- 15.Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 16.Zhang K, Wong HN, Song B, Miller CN, Scheuner D, Kaufman RJ. The unfolded protein response sensor IRE1alpha is required at 2 distinct steps in B cell lymphopoiesis. J. Clin. Invest. 2005;115:268–281. doi: 10.1172/JCI21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sha H, He Y, Chen H, Wang C, Zenno A, Shi H, Yang X, Zhang X, Qi L. The IRE1alpha-XBP1 pathway of the unfolded protein response is required for adipogenesis. Cell Metab. 2009;9:556–564. doi: 10.1016/j.cmet.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hay RT. SUMO: a history of modification. Mol. Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 20.Rytinki MM, Kaikkonen S, Pehkonen P, Jaaskelainen T, Palvimo JJ. PIAS proteins: pleiotropic interactors associated with SUMO. Cell Mol. Life Sci. 2009;66:3029–3041. doi: 10.1007/s00018-009-0061-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay RT. System-wide changes to SUMO modifications in response to heat shock. Sci. Signal. 2009;2 doi: 10.1126/scisignal.2000282. ra24. [DOI] [PubMed] [Google Scholar]
- 22.Johnson ES. Protein modification by SUMO. Annu. Rev. Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 23.Oishi Y, Manabe I, Tobe K, Ohsugi M, Kubota T, Fujiu K, Maemura K, Kubota N, Kadowaki T, Nagai R. SUMOylation of Kruppel-like transcription factor 5 acts as a molecular switch in transcriptional programs of lipid metabolism involving PPAR-delta. Nat. Med. 2008;14:656–666. doi: 10.1038/nm1756. [DOI] [PubMed] [Google Scholar]
- 24.Stankovic-Valentin N, Deltour S, Seeler J, Pinte S, Vergoten G, Guerardel C, Dejean A, Leprince D. An acetylation/deacetylation-SUMOylation switch through a phylogenetically conserved psiKXEP motif in the tumor suppressor HIC1 regulates transcriptional repression activity. Mol. Cell Biol. 2007;27:2661–2675. doi: 10.1128/MCB.01098-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duprez E, Saurin AJ, Desterro JM, Lallemand-Breitenbach V, Howe K, Boddy MN, Solomon E, de The H, Hay RT, Freemont PS. SUMO-1 modification of the acute promyelocytic leukaemia protein PML: implications for nuclear localisation. J. Cell Sci. 1999;112(Pt 3):381–393. doi: 10.1242/jcs.112.3.381. [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 2001;276:12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- 27.Girdwood D, Bumpass D, Vaughan OA, Thain A, Anderson LA, Snowden AW, Garcia-Wilson E, Perkins ND, Hay RT. P300 transcriptional repression is mediated by SUMO modification. Mol. Cell. 2003;11:1043–1054. doi: 10.1016/s1097-2765(03)00141-2. [DOI] [PubMed] [Google Scholar]
- 28.Shiio Y, Eisenman RN. Histone sumoylation is associated with transcriptional repression. Proc. Natl. Acad. Sci. U S A. 2003;100:13225–13230. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang SH, Sharrocks AD. SUMO promotes HDAC-mediated transcriptional repression. Mol. Cell. 2004;13:611–617. doi: 10.1016/s1097-2765(04)00060-7. [DOI] [PubMed] [Google Scholar]
- 30.Clauss IM, Chu M, Zhao JL, Glimcher LH. The basic domain/leucine zipper protein hXBP-1 preferentially binds to and transactivates CRE-like sequences containing an ACGT core. Nucleic Acids Res. 1996;24:1855–1864. doi: 10.1093/nar/24.10.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iwakoshi NN, Lee AH, Vallabhajosyula P, Otipoby KL, Rajewsky K, Glimcher LH. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat. Immunol. 2003;4:321–329. doi: 10.1038/ni907. [DOI] [PubMed] [Google Scholar]
- 32.Shaffer AL, Shapiro-Shelef M, Iwakoshi NN, Lee AH, Qian SB, Zhao H, Yu X, Yang L, Tan BK, Rosenwald A, Hurt EM, Petroulakis E, Sonenberg N, Yewdell JW, Calame K, Glimcher LH, Staudt LM. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 33.Yoshida H. ER stress and diseases. Febs. J. 2007;274:630–658. doi: 10.1111/j.1742-4658.2007.05639.x. [DOI] [PubMed] [Google Scholar]
- 34.Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, Nieuwenhuis EE, Higgins DE, Schreiber S, Glimcher LH, Blumberg RS. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim I, Xu W, Reed JC. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat. Rev. Drug Discov. 2008;7:1013–1030. doi: 10.1038/nrd2755. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.