Abstract
Background
It has been proposed that anatomical differences in human and great ape guts arose in response to species-specific diets and energy demands. To investigate functional genomic consequences of these differences, we compared their physiological levels of phytanic acid, a branched chain fatty acid that can be derived from the microbial degradation of chlorophyll in ruminant guts. Humans who accumulate large stores of phytanic acid commonly develop cerebellar ataxia, peripheral polyneuropathy, and retinitis pigmentosa in addition to other medical conditions. Furthermore, phytanic acid is an activator of the PPAR-alpha transcription factor that influences the expression of genes relevant to lipid metabolism.
Results
Despite their trace dietary phytanic acid intake, all great ape species had elevated red blood cell (RBC) phytanic acid levels relative to humans on diverse diets. Unlike humans, chimpanzees showed sexual dimorphism in RBC phytanic acid levels, which were higher in males relative to females. Cultured skin fibroblasts from all species had a robust capacity to degrade phytanic acid. We provide indirect evidence that great apes, in contrast to humans, derive significant amounts of phytanic acid from the hindgut fermentation of plant materials. This would represent a novel reduction of metabolic activity in humans relative to the great apes.
Conclusion
We identified differences in the physiological levels of phytanic acid in humans and great apes and propose this is causally related to their gut anatomies and microbiomes. Phytanic acid levels could contribute to cross-species and sex-specific differences in human and great ape transcriptomes, especially those related to lipid metabolism. Based on the medical conditions caused by phytanic acid accumulation, we suggest that differences in phytanic acid metabolism could influence the functions of human and great ape nervous, cardiovascular, and skeletal systems.
Background
Humans and great apes (bonobos, chimpanzees, gorillas, and orangutans) share a common gut anatomy, consisting of a simple stomach, small intestine, small cecum terminating in an appendix, and a hindgut consisting of the large intestine, rectum, and anal canal [1]. Nevertheless, significant differences have been reported in their gut proportions [2-4]. While the large intestine represents the majority of the great ape gut volume, the majority of the modern human gut volume consists of the small intestine [2-4]. Initial surveys have also indicated modern humans have a smaller total gut volume to body mass ratio relative to the great apes [5-7]. However, this could be influenced by primate gut plasticity related to diet and genetic diversity [2,8].
It has been proposed that gut proportions changed at some point within the human lineage in response to higher quality foods which can be digested in the small intestine [2]. The diets of hominids and/or early human populations improved, in part, due to cooking [9] and the increased abundance of animal products obtained through scavenging, hunting, fishing, and dairy consumption [10-19]. In contrast, great ape species in the wild derive a significant amount of their total daily metabolic energy needs through the fermentation of lower quality plant materials in their hindguts [20-25]. Although hindgut fermentation also occurs in humans [26-28], there is evidence that wild great apes derive greater amount of total daily metabolic energy from this process than do humans on Western diets [20-22]. However, seasonal changes in great ape diets and the limited dietary diversity of the humans studied will influence the interpretation of these data sets.
To explore the systemic consequences of hindgut fermentation activities in humans and great apes, we evaluated their physiological levels and cellular metabolic activities of phytanic acid, a branched chain fatty acid that can bind to and/or activate PPAR-alpha [29-34] and RXR [35-38] transcription factors. Humans do not synthesize phytanic acid, but rather acquire it from the diet [39,40]. In ruminants, the gut fermentation of plant materials liberates phytol, a constituent of chlorophyll, which is then converted to phytanic acid and stored in fats [39,40]. While humans can covert free phytol into phytanic acid, they do not accumulate significant amounts of phytanic acid as a result of consuming plant materials [41,42]. However, they can obtain phytanic acid from ruminant fats, fish, and dairy products [41]. Humans with impaired phytanic acid catabolism can overaccumulate phytanic acid, which results in peripheral polyneuropathy, cerebellar ataxia, retinitis pigmentosa, anosmia, and hearing loss [43-45]. This can also result in cardiac arrhythmias, shortened metacarpals or metatarsals, and ichthyosis [43-45].
Despite the demonstrated importance of maintaining appropriate levels of phytanic acid in humans and mice [46-48], phytanic acid metabolism has not been studied in the great apes. Based on the biochemical evaluation of red blood cells and cultured primary skin fibroblasts, we uncovered candidate human-specific differences in the ability to obtain phytanic acid from dietary sources relative to the great apes. We also provide indirect evidence that these changes could affect physiological functions relevant to the evolution and health of these species.
Results
Red blood cell (RBC) phytanic acid levels
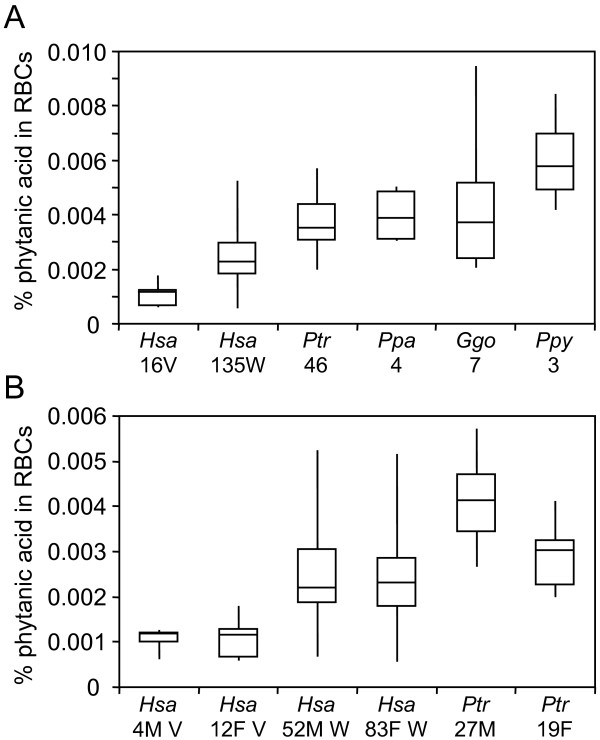
RBCs are commonly used to evaluate the physiological abundance of phytanic acid (Fig. 1) in clinical settings [49]. First, we measured RBC phytanic acid levels in a cohort of humans with Western diets (N = 135), chimpanzees (N = 46), bonobos (N = 4), lowland gorillas (N = 7), and Sumatran orangutans (N = 3) (Fig. 2A and Additional File 1). We found significant differences in RBC phytanic acid levels in humans with Western diets relative to the great apes (P < 2×10-13). Chimpanzee (1.5-fold, P < 3 × 10-10), bonobo (1.7-fold, P < 6 × 10-3), gorilla (1.7-fold, P < 7 × 10-4), and orangutan (2.6-fold, P < 4 × 10-5) RBC phytanic acid levels were all elevated relative to those of this human cohort. This is remarkable given the higher phytanic acid content of human Western diets, reportedly 50-100 mg per day [43], relative to the minimal (< 2 mg) daily amounts in the diets of our great ape cohort, even taking nutritional biscuits into consideration as described in the Materials and Methods section.
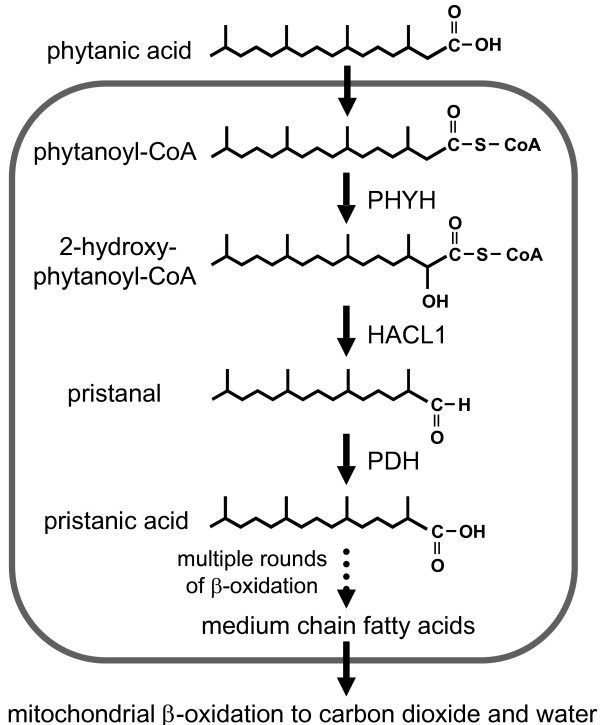
Figure 1.
Phytanic acid catabolism in mammals. Phytanic acid in ruminant fats is derived from phytol produced during the bacterial degradation of chlorophyll in their rumen (first stomach). After conversion to its CoA thioester, phytanic acid undergoes α-oxidation, yielding pristanic acid. This fatty acid then undergoes three subsequent rounds of β-oxidation in the peroxisome. The resulting medium chain fatty acid exits the peroxisome and translocates to the mitochondrion where the remaining carbon chain is degraded by β-oxidation. Abbreviations for the enzymes listed include: HACL1 (aka HPCL2) = 2-hydroxyphytanoyl-CoA lyase; PHYH = phytanoyl-CoA α-hydroxylase; PDH = pristanal dehydrogenase, whose gene is not yet known. We note that phytanic acid can also be degraded by β-oxidation; however, this activity of this pathway is relatively minor [40,44].
Figure 2.
Phytanic acid levels in human and great ape red blood cells. Box plots representing the percentage of phytanic acid relative to total fatty acids from red blood cells are provided. Median, quartile 1, quartile 3, minimum, and maximum values are provided. In Panels A and B, the species (Hsa: human; Ptr: chimpanzee; Ppa: bonobo; Ggo: gorilla; Ppy: orangutan), human diet (V: vegan, W: western), and number of individuals successfully analyzed is provided on the X-axes. Blood donor sex (M: male, F: female) is provided in Panel B.
To begin to address these dietary differences, we compared RBC phytanic acid levels in great apes and humans on phytanic acid-deficient diets (Fig. 2A). The latter are based on a cohort of 16 humans on vegan diets for over one year (Additional File 1). The absolute amount of phytanic acid intake in this population is unknown; however, it should be minimal (< 10 mg), based on typical components of vegan diets [41,50]. Phytanic acid RBC levels were lower in vegans relative to individuals on Western diets (2.3-fold, P < 1 × 10-13). We also found robust differences in vegan RBC phytanic acid levels relative to the great apes (P < 3 × 10-25). RBC phytanic acid levels in chimpanzees (3.5-fold, P < 6 × 10-20), bonobos (3.9-fold, P < 6 × 10-10), gorillas (3.8-fold, P < 2 × 10-12), and orangutans (5.9-fold, P < 5 × 10-12) were all dramatically higher relative to vegans.
Given the composition of our cohort, we had adequate statistical power to screen for possible sexual dimorphism in human (Western diet) and chimpanzee RBC phytanic acid levels. Although no sex-specific differences were found in humans on Western diets, male chimpanzees showed 1.4-fold higher (P < 2 × 10-6) RBC phytanic acid levels relative to females (Fig. 2B). This occurs despite the fact that our cohort of male and female chimpanzees had equivalent dietary exposure to phytanic acid and its precursor, phytol. Although our power is limited due to the numbers of vegan males in our cohort, there was also no evidence of sexual dimorphism in vegan RBC phytanic acid levels (Fig. 2B).
Phytanic acid catabolism in cell culture
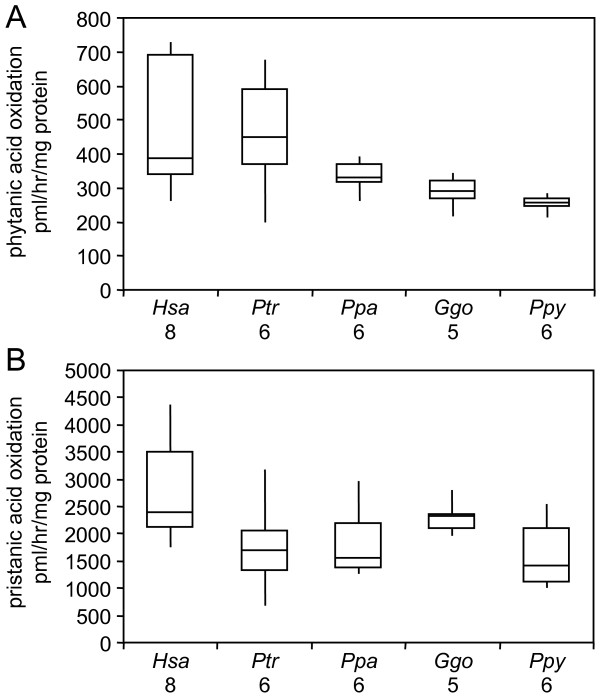
To evaluate cellular rates of phytanic acid metabolism, we analyzed cultured human and great ape dermal fibroblasts (Additional File 2), which are routinely used for the clinical diagnoses of phytanic acid and other peroxisomal disorders [43]. All fibroblast cultures showed robust catabolism of phytanic acid and its metabolite pristanic acid. Nevertheless, the oxidation rates of phytanic acid (P = 0.010) and pristanic acid (P = 0.048) differed in human relative to great ape cells (Fig. 3A-B). Human cells showed elevated phytanic acid oxidation rates relative to gorilla (1.6-fold, P = 0.017) and orangutan (1.8-fold, P = 0.002) cells (Fig. 3A). Humans cells also showed elevated pristanic acid oxidation rates compared to chimpanzee (1.7-fold, P = 0.018), bonobo (1.5-fold, P = 0.05), and orangutan (1.8-fold, P = 0.011) cells (Fig. 3B).
Figure 3.
Phytanic acid catabolism in human and great ape cultured fibroblasts. Box plots representing the relative rates of (A) phytanic acid and (B) pristanic acid oxidation in cultured fibroblasts are provided. Median, quartile 1, quartile 3, minimum, and maximum values are provided. The species and number of samples successfully analyzed is provided on the X-axis of both panels.
Comparative transcriptome analysis of peroxisomal α- and β-oxidation pathways
To evaluate potential in vivo consequences of cross-species differences in lipid metabolism, we reanalyzed oligonucleotide microarray gene expression data from 5 male chimpanzee and 6 male human livers, brains, kidneys, heart, and testes [51]. We only considered data from probes predicted to be perfectly matched to both genomes and calculated gene expression scores only for probe sets containing at least four probes after masking [52]. We focused on genes involved in peroxisomal phytanic acid α-oxidation, β-oxidation, and PPAR-responsive genes relevant to phytanic acid metabolism (Fig. 4, Additional File 3). Our criteria for differential gene expression are provided in the legend of Figure 4.
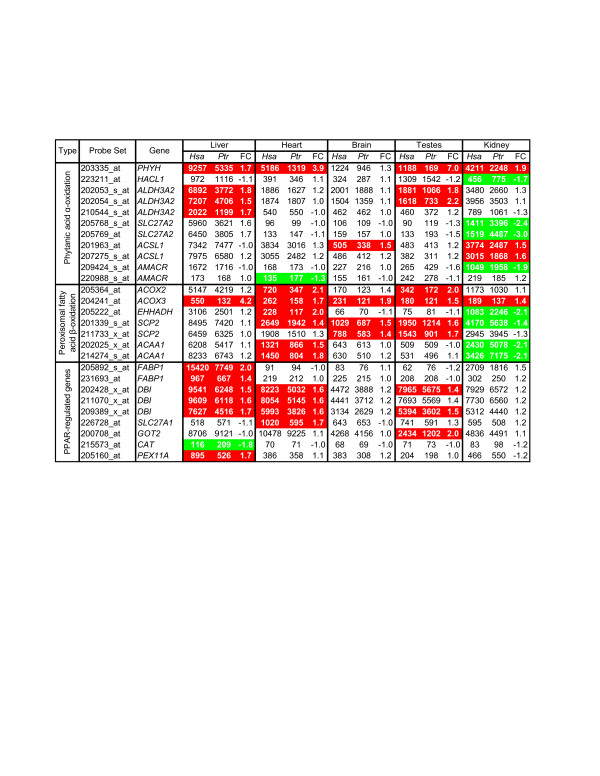
Figure 4.
Differential expression of genes related to peroxisomal lipid metabolism. We reanalyzed of Affymetrix GeneChip U133v2.0 expression profiles of human and chimpanzee tissues [51] using the masking strategy stated in the text. We used standard F-tests (FDR-adjusted using the Benjamini and Hochberg approach) to test for differences in the distributions by species for the 5 tissues. The fold change (FC) of human (Hsa) versus chimpanzee (Ptr) geometric mean gene expression scores are provided. Differentially expressed genes (≥1.2 FC in either direction with a Student's t-test, two-tailed P-value ≤0.05 after Bonferroni correction) for a given tissue are highlighted in red (higher in human) or green (higher in chimpanzee). Probe sets with (i) F-tests yielding a ≤5% FDR and (ii) differential expression in at least one tissue are shown. All data are provided in Additional File 3.
We found that cross-species differential expression of phytanic acid α-oxidation and peroxisomal β-oxidation genes in the liver, brain, and testes always reflected higher transcript levels in humans (Fig. 4). Genes in these categories also showed cross-species differential expression in heart and kidney; however, some transcripts were more abundant in humans (7 for heart and 4 for kidneys) while others were more abundant in chimpanzees (1 for heart and 8 for kidneys). Strikingly, ACOX3 transcript levels were higher in humans relative to chimpanzees for all five tissues examined. In 4 tissues (liver, heart, testes, and kidney), PHYH transcript levels were also higher in human relative to chimpanzee. To further explore this latter observation, we measured PHYH levels in human, chimpanzee, bonobo, and gorilla fibroblast cultures (6 different donors per species) via quantitative PCR (qPCR) using primers designed against a conserved region of this transcript. PHYH levels were at least 2-fold more abundant in human relative to chimpanzee, bonobo, and gorilla fibroblasts (P < 3 × 10-4 for each individual comparison, ANOVA P < 2 × 10-5 for human versus African great apes).
Lastly, we examined a group of five genes (FABP1, DBI, SLC27A1, GOT2, and CAT) previously used to evaluate PPAR-activity in mice with phytanic acid disorders [53] and PEX11A, a direct transcriptional target of PPAR-alpha [54]. All differentially expressed transcripts in heart (DBI and SLC27A1) and testes (DBI and GOT2) were more abundant in humans. FABP1, DBI, and PEX11A transcript levels were higher in human liver; however, CAT transcripts were more abundant in chimpanzee liver. None of these six genes showed cross-species differential expression in brain or kidney.
Discussion
Microbial fermentation in human [26-28] and great ape [20-25] hindguts are responsible for the break down of complex carbohydrates not processed and absorbed in the small intestine. The major end products of hindgut fermentation are short chain fatty acids, which provide energy-yielding substrates for colonic mucosa that regulate its growth and blood flow and promotes sodium and water absorption [26,27]. It has been estimated that wild chimpanzees derive 21-33% and wild orangutans derive 7-57% of their total daily metabolic energy from fiber fermentation [22]. This could be as high as 30-60% for wild Western gorillas [21]. In contrast, humans on Western diets are estimated to obtain no more than 10% of their daily energy needs through hindgut fermentation; however, this is likely higher in populations with lower quality diets and instances of small intestinal malabsorption [28].
Although we contrast human and the great ape dietary strategies, it is important to consider diversity within the great apes. For example, gorillas and orangutans can and often do subsist on lower quality foods (e.g. mature foliage and unripe fruits) relative to chimpanzees and bonobos, which can consume ample amounts of succulent ripe fruits [2]. Furthermore, chimpanzees hunt colobus monkeys and other small mammals; however, this comprises a minor component of their diets and varies among individuals and communities [55-59]. Thus, we propose that it is likely that the human, chimpanzee, bonobo, gorilla and orangutan lineages have all acquired molecular adaptations relevant to their diets.
In the studies presented here, we began to evaluate the broader systemic consequences of hindgut fermentation and related lipid metabolic activity in humans and great apes. In principle, RBC phytanic acid can be an effective biomarker of hindgut fermentation of plant materials as long as phytanic acid or its precursor phytol is not appreciably present in the human or great ape diets. The phytanic acid content of Western diets (50-100 mg daily intake) [43] is estimated to be at least 10 × greater than its free phytol content [41,60]. Since phytanic acid is primarily found in ruminant fats (organic beef fat has >325 mg/100 g), dairy products (45% fat cream cheese has >125 mg/100 g), and fish (canned salmon has >250 mg/100 g) [41], human vegans are appropriate for our studies. In this regard, a survey of fresh vegetables and fruits all showed minimal phytol levels (≤2 mg phytol/100 g) [41]. We also note a prior study demonstrating that an individual who ingested 3.5-kg of boiled spinach leaves within a 60 hour period as a supplement to an uncontrolled mixed diet later showed serum phytanic acid levels within normal limits [42]. Our captive great ape cohort should be exposed to similar low levels of dietary phytanic acid (< 2 mg daily intake) and phytol [41].
We discovered elevated RBC phytanic acid levels in all great apes relative to humans on both high- (Western) and low-phytanic acid (vegan) diets (Fig. 2). In principle, these could be influenced by differences in phytanic acid retention and/or biosynthesis. Relevant to the retention hypothesis, both humans and great apes showed robust cellular rates of phytanic and pristanic acid oxidation (Fig. 3). In fact, there were no differences in phytanic acid oxidation rates in human relative to chimpanzee or bonobo fibroblasts (Fig. 3A). Although it is possible that the lower phytanic acid oxidation rates in gorilla and orangutan relative to human fibroblasts (Fig. 3A) relate to the dietary strategies of these species [2], these observations need to be validated in larger-scale studies prior to commentary. We also cannot exclude the possibility that the cultured fibroblasts do not fully reflect in vivo activities; however, they are invaluable for the clinical diagnosis of phytanic acid oxidation disorders [61]. Likewise, we cannot exclude the possibility that great apes, unlike humans, show robust biological retention of trace dietary phytanic acid. However, laboratory mice do not accumulate significant physiological levels of phytanic acid when fed standard diets not supplemented with additional phytol or phytanic acid [53]. In addition, rats on standard diets can rapidly catabolize artificially elevated stores of phytanic acid in their tissues [62].
Our preferred hypothesis is that all the great apes, unlike humans [45,63], are capable of accumulating significant amounts of phytanic acid from the hindgut fermentation of plant materials. We propose that the great apes have maintained the ancestral condition and that humans diverged since their most recent common ancestor with chimpanzees and bonobos and now have a derived metabolism. We believe that changes in gut anatomy provide a likely basis for the proposed reduction of hindgut fermentation activity in the human lineage. In this regard, it has been proposed that improved dietary quality contributed to hindgut reduction in modern humans relative to great apes (i.e. the hindgut comprises ~20% of the total human gut volume, but >50% of the total great ape gut volume) [2,3] and the smaller total gut volume to body mass ratio in modern humans relative to great apes [6,7,64,65]. However, we also recognize that the human and great ape gut microbiomes, whose composition are influenced by the diets of these species and other environmental interactions [66,67], could also contribute to the proposed differences in hindgut fermentation activity. Thus, in principle, the improved dietary quality in the human lineage could have influenced the major factors (gut anatomy and microbial activities) that we propose are most likely to be causally responsible for the observed differences in human and great ape RBC phytanic acid levels.
Given that sexual dimorphism in human lipid metabolism is well-documented [68,69], we screened for sex-specific differences in RBC phytanic acid levels. Here, we found no evidence of sexual dimorphism in RBC levels in humans on Western or vegan diets. However, we observed 1.4-fold elevated RBC phytanic acid levels in male relative to female chimpanzees (Fig. 2B). The evolutionary implications and physiological origin of this sexual dimorphism are unclear. Phytanic acid profiling in additional males and females from other non-human primates could shed light on the extent of this phenomenon. As discussed above, phytanic acid levels are affected by multiple factors, including gut anatomies, microbiomes, and gene expression, which provide candidate mechanisms for the observed sexual dimorphism in chimpanzees.
We note that sexual dimorphism in phytanic acid metabolism has been documented in rodents. C57BL/6J laboratory mice show sexually dimorphic phytanic acid accumulation in response to phytol-enriched diets; however, unlike chimpanzees, females showed higher phytanic acid levels than males [53,70]. There is no evidence as to whether this is a metabolic adaptation to diet or a non-specific result of inbreeding. It is thought this sexual dimorphism is influenced by the higher liver expression of sterol carrier protein-x (SCP-x) in males relative to females [53]. SCP-x is required for branched chain fatty acid catabolism through its keto-acyl CoA thiolase activity [53,70]. Sexual dimorphism in liver SCP-x expression has also been observed in FVB mice [53], BALB/c mice [71], and laboratory rats [72]. This suggests that sexual dimorphism in phytanic acid metabolism could be present in additional rodent species. To date, SCP-x expression has not been measured in human and chimpanzee livers, even in a recent study involving both sexes from each species [73].
We also investigated possible relationships between human and chimpanzee RBC lipid profiles and their transcriptomes. While there is no known physiological role for phytanic acid, it can bind to and/or activate PPAR-alpha [29-34], RXR [35-38], and likely other transcription factors [29]. Experiments involving laboratory mice have concluded that phytanic acid is a physiological ligand for PPAR-alpha [29-31]. While the estimated phytanic acid concentration in normal human serum (6 μM total [42], 2 μM free [35]) was at the threshold needed for RXR stimulation of one cell culture study, the authors concluded that phytol metabolites, such as phytanic acid, are compelling candidate physiological effectors of RXR [35].
Here, we found that multiple genes reported to be influenced by PPAR-alpha activity, including those involved in peroxisomal α- and β-oxidation of fatty acids, show cross-species expression differences in the chimpanzee and human tissues surveyed (Fig. 4). Such genes were primarily more highly expressed in humans relative to chimpanzee for all tissues except kidney, where higher expression in chimpanzees was frequently found. Exploratory work, not involving non-human primates, highlighted the lower fatty acid α-oxidation capacity in the kidneys, but not livers, of carnivorous relative to herbivorous animals [74]. Also of interest, others have provided gene expression-based evidence that PPAR-signaling is higher in human relative to chimpanzee liver [75]. The elevated FABP1 transcript levels in human liver suggests an increased capacity for humans to transport phytanic acid to peroxisomes for degradation [53]. Nevertheless, we recognize phytanic acid is just one of several activators of PPAR transcription factors and that the dietary information and RBC lipid profiles of the humans and chimpanzees in the gene expression profiling studies are unknown.
We have previously noted that the toxicity associated with abnormal phytanic acid accumulation in humans could provide selective pressure to maintain or enhance its degradation [76]. For example, Refsum disease is an autosomal recessive disorder in humans that can result from mutations in the PHYH gene and subsequent accumulation of phytanic acid in tissues [46,47]. This typically affects the peripheral nerves (i.e. polyperipheral neuropathy), movement (i.e. cerebellar ataxia), and senses (e.g. retinitis pigmentosa, anosmia, and hearing loss) [43-45]. These individuals can also show cardiac arrhythmias, skeletal malformations (e.g. shortened metacarpals or metatarsals), and skin changes (i.e. ichthyosis) [43-45]. Given that cardiac arrhythmias are suspected to be responsible for the sudden deaths of Refsum disease patients [43], the commonly observed sudden cardiac death, presumed secondary to fatal arrhythmias, in captive male chimpanzees [77] could be influenced by their physiological levels of phytanic acid. However, the RBC phytanic acid levels in our cohort of male chimpanzees are far below those of Refsum disease patients, which was approximately 100 times higher than normal in one case we evaluated. Nevertheless, we raise the possibility that phytanic acid metabolism could influence physiological differences and medical conditions amongst human and non-human primates.
Conclusions
The candidate differences in human and great ape phytanic acid metabolism are consistent with prior reports that the regulation of genes involved in lipid metabolism likely evolved under directional selection in humans [75] and that the promoter regions of nutrition-related genes are under positive selection in humans [78]. It is also relevant that the effects human diets have on mouse liver transcriptomes can be used as a model for expression differences between humans and chimpanzees [79]. We note that even if transcriptome data of humans and chimpanzees on matching diets were obtained, differences in how each species digests their food and extract regulators of transcription would have a major impact on the observed cross-species and sex-specific differences.
Our study also relates to hypotheses that historic diets and/or changes in lipid metabolism influenced the evolution of numerous human traits [2,12,18,55,64,65,80-85]. To further explore such hypotheses, it will be necessary to determine if the RBC phytanic acid profiles in our cohort extend to other cell types. The comparative analyses of the lipid composition of human and great ape nervous and cardiovascular systems will be of special interest given the clinical phenotypes observed in humans with phytanic acid metabolic [43-45] and other peroxisomal disorders [86]. In addition, prostate tissue could be of interest given the differences of phytanic acid metabolic activities in normal and cancerous tissues [87-89] and male reproductive differences across human and non-human primates [90]. Lastly, we propose that larger-scale lipid profiling studies will likely identify additional candidate metabolic adaptations to human and non-human diets and that investigations into the microbial contributions toward specific metabolic differences is warranted.
Methods
Cohort for red blood cell lipid profiling
Bloods from adult humans with Western diets were collected from healthy individuals attending an international conference. Bloods from adult humans on vegan diets for over one year were collected in conjunction with a blood donor center. Appropriate Institutional Review Board (IRB) approval from the University of Southern California and Johns Hopkins Medicine was obtained for all human subjects research. Chimpanzee blood was collected at the Alamogordo Primate Facility. All chimpanzees took part in daily enrichment activities to maintain psychological well-being. Animals were maintained in accordance with the Guide for the Care and Use of Animals (U.S. Dept. of Health and Human Services, Public Health Service, Bethesda, MD., 1996). The APF and its program were fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care, International (AAALAC). Other great ape bloods were collected at the Zoological Society of San Diego. The gender and ages of blood donors are provided in Additional File 2. One gorilla blood donor, described in reference [91], carries a deletion of the distal q arm of chromosome 3 (the homolog of human chromosome 4) as indicated in Additional File 4. Great ape diets contain fresh fruits, vegetables, and nutritional biscuits. Capillary gas chromatography (GC) - electron-capture negative-ion mass spectrometry [92] analysis of nutritional biscuits indicated the great apes were exposed to <2 mg of daily phytanic acid intake.
Red blood cell lipid profiling
Whole blood samples were collected from fasting individuals and stored in EDTA blood collection tubes. RBCs were collected by centrifugation, washed twice with physiological saline, transferred to freezer vials, flushed with nitrogen, and stored at -80°C until analysis. RBCs were thawed briefly before 100 μl aliquots were taken for analysis of the total lipid fatty acid content by capillary GC - electron-capture negative-ion mass spectrometry [92]. Processed data are provided in Additional File 4.
Primary fibroblast cultures
Great ape dermal fibroblasts were obtained from the Zoological Society of San Diego while human dermal fibroblasts were obtained from the Coriell Institute for Medical Research or the Kennedy Krieger Institute. All individuals are thought to be unrelated. The gender, age, and biopsy site of all fibroblast donors and corresponding biochemical analyses are provided in Additional File 2. Fibroblasts were cultured as previously described [76].
Phytanic and pristanic acid oxidation in cultured fibroblasts
Fatty acids were dried under nitrogen and solubilized with β-cyclodextrin (Sigma; 10 mg/ml in 10 mM Tris, pH 7.5) by warming to 37°C and sonication. Fibroblasts were grown to approximately 80% confluence in 12-well culture plates under standard conditions. Afterwards, the media was removed and replaced with 0.6 mL of serum-free culture media supplemented with 10 μM [1-14C]-phytanic acid (~40,000 dpm/nmol) or 10 μM [1-14C]-pristanic acid (~20,000 dpm/nmol). The cells were incubated for two hours (pristanic) or four hours (phytanic) at 37°C in 5% CO2. Reactions were terminated by addition of 0.12 ml of 2.6 N HClO4. Water-soluble reaction products were separated from the labeled substrates and quantified by liquid scintillation counting. The specific activity of phytanic acid or pristanic acid oxidation (pmoles/hour/mg protein) were calculated based on the amount of water-soluble radioactivity in the test sample minus the blank per mg of protein, determined using the Lowry method [93]. All primary data are provided in Additional File 5.
Statistical considerations
We analyzed all data on the log2 scale. We used analysis of variance (ANOVA) to compare average blood lipid data across humans and great apes. Heterogeneity P-values are reported for the test that the mean level is different in at least one of the great apes groups, and Wald P-values for tests comparing the average level in individual non-human primate groups to humans. Under ANOVA, statistical tests use an estimate of within-group variation from all samples. Due to the unbalanced group sizes, this estimate is driven by the variation in humans and in chimpanzees. For gene expression studies, we tested for differential expression (Fig. 4) using ANOVA and standard F-tests. To account for testing differential expression across multiple genes, we adjusted p-values using the Benjamini and Hochberg approach for controlling the false-discovery rate (FDR). All analyses were done using the R programming language.
Authors' contributions
PAW, ABM, CBT, and SJS carried out the biochemical analyses in this project. PAW, ABM, and HWM were involved in the design and conception of the peroxisome components of this project. MWK and KR maintained and conducted genetic and biochemical analysis on human and great ape cells. KDS conducted statistical analyses of all biochemical and gene expression data. RL and JJE provided characterized chimpanzee blood samples and diet information. OAR provided characterized great ape cells, blood samples, and diet information. OAR, JJE and RL were involved in the design and conception of the great ape components of the project. JGH was involved in the overall design and conception of the project, statistical analysis of all data sets, and wrote the manuscript with the help of all authors.
Supplementary Material
Composition of blood donor cohort. A summary of the numbers, ages, and sex of blood donors is provided.
Skin fibroblast cultures used for phytanic and pristanic acid biochemical analysis. A summary of donor sex and age and anatomical source of the skin fibroblasts is provided.
Detailed summary of gene expression data for cross-species comparisons. More complete gene expression data summary statistics relevant to Figure 4 are provided.
Phytanic acid levels relative to total fatty acids in red blood cells from individual donors. Relative phytanic acid levels for all RBC donors are provided.
Rates of phytanic and pristanic acid oxidation in cultured dermal fibroblasts. The rates of phytanic and pristanic acid oxidation from all individual fibroblast cultures are provided.
Contributor Information
Paul A Watkins, Email: watkins@kennedykrieger.org.
Ann B Moser, Email: mosera@kennedykrieger.org.
Cicely B Toomer, Email: toomer@kennedykrieger.org.
Steven J Steinberg, Email: Steven.Steinberg@kennedykrieger.org.
Hugo W Moser, Email: mosera@kennedykrieger.org.
Mazen W Karaman, Email: mazenkaraman@gmail.com.
Krishna Ramaswamy, Email: rkrishna@usc.edu.
Kimberly D Siegmund, Email: kims@usc.edu.
D Rick Lee, Email: rick.lee@crl.com.
John J Ely, Email: John.Ely@crl.com.
Oliver A Ryder, Email: oryder@ucsd.edu.
Joseph G Hacia, Email: hacia@hsc.usc.edu.
Acknowledgements
We thank C. Finch, C. Stanford, K. Benirschke, M. Goodman, P. Gagneux, and N. Braverman for thoughtful discussion, M. Houck and L. Chemnick for technical assistance, and D. Johnson for the synthesis of [1-14C] pristanic acid. This study was funded by the National Institutes of Health (GM072447 to J.G.H. and HD24061 to P.A.W. and A.B.M).
References
- Mitchell P. C. V. On the intestinal tract of mammals. Trans Zool Soc Lond. 1905;XVII:437–536. [Google Scholar]
- Milton K. A hypothesis to explain the role of meat-eating in human evolution. Evolutionary Anthropology. 1999;8(1):11–21. doi: 10.1002/(SICI)1520-6505(1999)8:1<11::AID-EVAN6>3.0.CO;2-M. [DOI] [Google Scholar]
- Milton K. The critical role played by animal source foods in human (Homo) evolution. J Nutr. 2003;133(11 Suppl 2):3886S–3892S. doi: 10.1093/jn/133.11.3886S. [DOI] [PubMed] [Google Scholar]
- Leonard WR, Snodgrass JJ, Robertson ML. Effects of brain evolution on human nutrition and metabolism. Annu Rev Nutr. 2007;27:311–327. doi: 10.1146/annurev.nutr.27.061406.093659. [DOI] [PubMed] [Google Scholar]
- Martin RD, Chivers DJ, Maclarnon AM, Hladik CM. In: Size and Scaling in Primate Biology. Jungers WJ, editor. Plenum: Plenum Press; 1985. Gastrointestinal allometry in primates and other mammals; pp. 61–139. [Google Scholar]
- Milton K, Demment MW. Digestion and passage kinetics of chimpanzees fed high and low fiber diets and comparison with human data. J Nutr. 1988;118(9):1082–1088. doi: 10.1093/jn/118.9.1082. [DOI] [PubMed] [Google Scholar]
- Hladik CM, Chivers DJ, Pasquet P. On Diet and Gut Size in Non-human Primates and Humans: Is There a Relationship to Brain Size? Curr Anthropol. 1999;40(5):695–697. doi: 10.1086/300099. [DOI] [PubMed] [Google Scholar]
- Leonard WR, Robertson ML, Aiello LC, Wheeler P. On diet, energy metabolism, and brain size in human evolution. Current Anthropology. 1996;37:125–129. doi: 10.1086/204476. [DOI] [Google Scholar]
- Carmody RN, Wrangham RW. The energetic significance of cooking. J Hum Evol. 2009;57(4):379–391. doi: 10.1016/j.jhevol.2009.02.011. [DOI] [PubMed] [Google Scholar]
- Schoeninger MJ, Bunn HT, Murray S, Pickering T, Moore J. In: Meat-eating and Human Evolution. Stanford CB, Bunn HT, editor. Oxford University Press. New York; 2001. Meat-eating by the fourth African ape; pp. 179–195. [Google Scholar]
- Bunn HT, Kroll EM. Systematic butchery by Plio/Pleistocene hominids at Oldvai Gorge, Tanzania. Current Anthropology. 1986;27:431–452. doi: 10.1086/203467. [DOI] [Google Scholar]
- Bunn HT. In: Evolution of the human diet: The known, the unknown, and the unknowable. Ungar PS, editor. Oxford: Oxford University Press; 2006. Meat made us human; pp. 191–211. [Google Scholar]
- Marean CW, Bar-Matthews M, Bernatchez J, Fisher E, Goldberg P, Herries AI, Jacobs Z, Jerardino A, Karkanas P, Minichillo T. et al. Early human use of marine resources and pigment in South Africa during the Middle Pleistocene. Nature. 2007;449(7164):905–908. doi: 10.1038/nature06204. [DOI] [PubMed] [Google Scholar]
- Plummer T. Flaked stones and old bones: biological and cultural evolution at the dawn of technology. Am J Phys Anthropol. 2004. pp. 118–164. [DOI] [PubMed]
- Tishkoff SA, Reed FA, Ranciaro A, Voight BF, Babbitt CC, Silverman JS, Powell K, Mortensen HM, Hirbo JB, Osman M. et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet. 2007;39(1):31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rick TC, Erlandson JM. Anthropology. Coastal exploitation. Science. 2009;325(5943):952–953. doi: 10.1126/science.1178539. [DOI] [PubMed] [Google Scholar]
- Richards MP, Jacobi R, Cook J, Pettitt PB, Stringer CB. Isotope evidence for the intensive use of marine foods by Late Upper Palaeolithic humans. J Hum Evol. 2005;49(3):390–394. doi: 10.1016/j.jhevol.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Broadhurst CL, Wang Y, Crawford MA, Cunnane SC, Parkington JE, Schmidt WF. Brain-specific lipids from marine, lacustrine, or terrestrial food resources: potential impact on early African Homo sapiens. Comp Biochem Physiol B Biochem Mol Biol. 2002;131(4):653–673. doi: 10.1016/S1096-4959(02)00002-7. [DOI] [PubMed] [Google Scholar]
- Shipman P. Scavenging or hunting in early hominids: theoretical framework and tests. American Anthropologist. 1986;88:27–43. doi: 10.1525/aa.1986.88.1.02a00020. [DOI] [Google Scholar]
- Rothman JM, Dierenfeld ES, Hintz HF, Pell AN. Nutritional quality of gorilla diets: consequences of age, sex, and season. Oecologia. 2008;155(1):111–122. doi: 10.1007/s00442-007-0901-1. [DOI] [PubMed] [Google Scholar]
- Popovich DG, Jenkins DJ, Kendall CW, Dierenfeld ES, Carroll RW, Tariq N, Vidgen E. The western lowland gorilla diet has implications for the health of humans and other hominoids. J Nutr. 1997;127(10):2000–2005. doi: 10.1093/jn/127.10.2000. [DOI] [PubMed] [Google Scholar]
- Conklin-Brittain NL, Knott CD, Wrangham RW. In: Feeding Ecology in Apes and Other Primates: Ecological, Physical, and Behavioral Aspects. Hohmann G, Robbins MM, Boesch C, editor. Cambridge: Cambridge University Press; 2006. Energy intake by wild chimpanzees and orangutans: methodological considerations and a preliminary comparison; pp. 445–472. [Google Scholar]
- Remis MJ. Initial studies on the contributions of body size and gastrointestinal passage rates to dietary flexibility among gorillas. Am J Phys Anthropol. 2000;112(2):171–180. doi: 10.1002/(SICI)1096-8644(2000)112:2<171::AID-AJPA4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Remis MJ, Dierenfeld ES. Digesta passage, digestibility, and behavior in captive gorillas under two dietary regimens. International Journal of Primatology. 2004;25:825–845. doi: 10.1023/B:IJOP.0000029124.04610.c7. [DOI] [Google Scholar]
- Schmidt DA, Kerley MS, Dempsey JL, Porton IJ, Porter JH, Griffin ME, Ellersieck MR, Sadler WC. Fiber digestibility by the orangutan (Pongo abelii): in vitro and in vivo. J Zoo Wildl Med. 2005;36(4):571–580. doi: 10.1638/04-103.1. [DOI] [PubMed] [Google Scholar]
- Cummings JH, Englyst HN. Fermentation in the human large intestine and the available substrates. Am J Clin Nutr. 1987;45(5 Suppl):1243–1255. doi: 10.1093/ajcn/45.5.1243. [DOI] [PubMed] [Google Scholar]
- Mortensen PB, Clausen MR. Short-chain fatty acids in the human colon: relation to gastrointestinal health and disease. Scand J Gastroenterol Suppl. 1996;216:132–148. doi: 10.3109/00365529609094568. [DOI] [PubMed] [Google Scholar]
- McNeil NI. The contribution of the large intestine to energy supplies in man. Am J Clin Nutr. 1984;39(2):338–342. doi: 10.1093/ajcn/39.2.338. [DOI] [PubMed] [Google Scholar]
- Gloerich J, van Vlies N, Jansen GA, Denis S, Ruiter JP, van Werkhoven MA, Duran M, Vaz FM, Wanders RJ, Ferdinandusse S. A phytol-enriched diet induces changes in fatty acid metabolism in mice both via PPARalpha-dependent and -independent pathways. J Lipid Res. 2005;46(4):716–726. doi: 10.1194/jlr.M400337-JLR200. [DOI] [PubMed] [Google Scholar]
- Wolfrum C, Ellinghaus P, Fobker M, Seedorf U, Assmann G, Borchers T, Spener F. Phytanic acid is ligand and transcriptional activator of murine liver fatty acid binding protein. J Lipid Res. 1999;40(4):708–714. [PubMed] [Google Scholar]
- Ellinghaus P, Wolfrum C, Assmann G, Spener F, Seedorf U. Phytanic acid activates the peroxisome proliferator-activated receptor alpha (PPARalpha) in sterol carrier protein 2-/sterol carrier protein x-deficient mice. J Biol Chem. 1999;274(5):2766–2772. doi: 10.1074/jbc.274.5.2766. [DOI] [PubMed] [Google Scholar]
- Goto T, Takahashi N, Kato S, Egawa K, Ebisu S, Moriyama T, Fushiki T, Kawada T. Phytol directly activates peroxisome proliferator-activated receptor alpha (PPARalpha) and regulates gene expression involved in lipid metabolism in PPARalpha-expressing HepG2 hepatocytes. Biochem Biophys Res Commun. 2005;337(2):440–445. doi: 10.1016/j.bbrc.2005.09.077. [DOI] [PubMed] [Google Scholar]
- Heim M, Johnson J, Boess F, Bendik I, Weber P, Hunziker W, Fluhmann B. Phytanic acid, a natural peroxisome proliferator-activated receptor (PPAR) agonist, regulates glucose metabolism in rat primary hepatocytes. Faseb J. 2002;16(7):718–720. doi: 10.1096/fj.01-0816fje. [DOI] [PubMed] [Google Scholar]
- Lampen A, Meyer S, Nau H. Phytanic acid and docosahexaenoic acid increase the metabolism of all-trans-retinoic acid and CYP26 gene expression in intestinal cells. Biochim Biophys Acta. 2001;1521(1-3):97–106. doi: 10.1016/s0167-4781(01)00305-0. [DOI] [PubMed] [Google Scholar]
- Kitareewan S, Burka LT, Tomer KB, Parker CE, Deterding LJ, Stevens RD, Forman BM, Mais DE, Heyman RA, McMorris T. et al. Phytol metabolites are circulating dietary factors that activate the nuclear receptor RXR. Mol Biol Cell. 1996;7(8):1153–1166. doi: 10.1091/mbc.7.8.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter A, Barbera MJ, Iglesias R, Giralt M, Villarroya F. Phytanic acid, a novel activator of uncoupling protein-1 gene transcription and brown adipocyte differentiation. Biochem J. 2002;362(Pt 1):61–69. doi: 10.1042/0264-6021:3620061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemotte PK, Keidel S, Apfel CM. Phytanic acid is a retinoid × receptor ligand. Eur J Biochem. 1996;236(1):328–333. doi: 10.1111/j.1432-1033.1996.00328.x. [DOI] [PubMed] [Google Scholar]
- Radominska-Pandya A, Chen G. Photoaffinity labeling of human retinoid × receptor beta (RXRbeta) with 9-cis-retinoic acid: identification of phytanic acid, docosahexaenoic acid, and lithocholic acid as ligands for RXRbeta. Biochemistry. 2002;41(15):4883–4890. doi: 10.1021/bi0121151. [DOI] [PubMed] [Google Scholar]
- van den Brink DM, Wanders RJ. Phytanic acid: production from phytol, its breakdown and role in human disease. Cell Mol Life Sci. 2006;63(15):1752–1765. doi: 10.1007/s00018-005-5463-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanders RJ, Komen JC. Peroxisomes, Refsum's disease and the alpha- and omega-oxidation of phytanic acid. Biochem Soc Trans. 2007;35(Pt 5):865–869. doi: 10.1042/BST0350865. [DOI] [PubMed] [Google Scholar]
- Brown PJ, Mei G, Gibberd FB, Burston D, Mayne PD, McClinchy JE, Sidey M. Diet and Refsum's disease. The determination of phytanic acid and phytol in certain foods and the application of this knowledge to the choice of suitable convenience foods for patients with Refsum's disease. Journal of Human Nutrition and Dietetics. 1993;6:295–305. doi: 10.1111/j.1365-277X.1993.tb00375.x. [DOI] [Google Scholar]
- Avigan J. The presence of phytanic acid in normal human and animal plasma. Biochim Biophys Acta. 1966;116(2):391–394. doi: 10.1016/0005-2760(66)90022-1. [DOI] [PubMed] [Google Scholar]
- Steinberg D. In: Metabolic Basis of Inherited Disease. 5. Stanbury JB, Wyngarden JB, Fredericksen DS, Goldstein JL, Brown MS, editor. New York: McGraw Hill; 1983. Phytanic acid storage disease (Refsum's disease) pp. 731–747. [Google Scholar]
- Wierzbicki AS. Peroxisomal disorders affecting phytanic acid alpha-oxidation: a review. Biochem Soc Trans. 2007;35(Pt 5):881–886. doi: 10.1042/BST0350881. [DOI] [PubMed] [Google Scholar]
- Verhoeven NM, Wanders RJ, Poll-The BT, Saudubray JM, Jakobs C. The metabolism of phytanic acid and pristanic acid in man: a review. J Inherit Metab Dis. 1998;21(7):697–728. doi: 10.1023/A:1005476631419. [DOI] [PubMed] [Google Scholar]
- Jansen GA, Ofman R, Ferdinandusse S, Ijlst L, Muijsers AO, Skjeldal OH, Stokke O, Jakobs C, Besley GT, Wraith JE. et al. Refsum disease is caused by mutations in the phytanoyl-CoA hydroxylase gene. Nat Genet. 1997;17(2):190–193. doi: 10.1038/ng1097-190. [DOI] [PubMed] [Google Scholar]
- Mihalik SJ, Morrell JC, Kim D, Sacksteder KA, Watkins PA, Gould SJ. Identification of PAHX, a Refsum disease gene. Nat Genet. 1997;17(2):185–189. doi: 10.1038/ng1097-185. [DOI] [PubMed] [Google Scholar]
- Ferdinandusse S, Zomer AW, Komen JC, van den Brink CE, Thanos M, Hamers FP, Wanders RJ, van der Saag PT, Poll-The BT, Brites P. Ataxia with loss of Purkinje cells in a mouse model for Refsum disease. Proc Natl Acad Sci USA. 2008;105(46):17712–17717. doi: 10.1073/pnas.0806066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser AB, Jones DS, Raymond GV, Moser HW. Plasma and red blood cell fatty acids in peroxisomal disorders. Neurochem Res. 1999;24(2):187–197. doi: 10.1023/A:1022549618333. [DOI] [PubMed] [Google Scholar]
- Coppack SW, Evans R, Gibberd FB, Clemens ME, Billimoria JD. Can patients with Refsum's disease safely eat green vegetables? Br Med J (Clin Res Ed) 1988;296(6625):828. doi: 10.1136/bmj.296.6625.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, Hellmann I, Enard W, Nowick K, Leinweber M, Franz H, Weiss G, Lachmann M, Paabo S. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309(5742):1850–1854. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- Toleno DM, Renaud G, Wolfsberg TG, Islam M, Wildman DE, Siegmund KD, Hacia JG. Development and evaluation of new mask protocols for gene expression profiling in humans and chimpanzees. BMC Bioinformatics. 2009;10(1):77.. doi: 10.1186/1471-2105-10-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atshaves BP, McIntosh AL, Landrock D, Payne HR, Mackie JT, Maeda N, Ball J, Schroeder F, Kier AB. Effect of SCP-x gene ablation on branched-chain fatty acid metabolism. Am J Physiol Gastrointest Liver Physiol. 2007;292(3):G939–951. doi: 10.1152/ajpgi.00308.2006. [DOI] [PubMed] [Google Scholar]
- Anderson SP, Howroyd P, Liu J, Qian X, Bahnemann R, Swanson C, Kwak MK, Kensler TW, Corton JC. The transcriptional response to a peroxisome proliferator-activated receptor alpha agonist includes increased expression of proteome maintenance genes. J Biol Chem. 2004;279(50):52390–52398. doi: 10.1074/jbc.M409347200. [DOI] [PubMed] [Google Scholar]
- Finch CE, Stanford CB. Meat-adaptive genes and the evolution of slower aging in humans. Q Rev Biol. 2004;79(1):3–50. doi: 10.1086/381662. [DOI] [PubMed] [Google Scholar]
- Goodall J. The chimpanzees of Gombe: patterns of behavior. Cambridge, Mass.: Belknap Press of Harvard University Press; 1986. [Google Scholar]
- Stanford CB. Chimpanzee and red colobus: the ecology of predator and prey. Cambridge, Mass.: Harvard University Press; 1998. [Google Scholar]
- Stanford CB, Wallis J, Matama H, Goodall J. Patterns of predation by chimpanzees on red colobus monkeys in Gombe National Park, 1982-1991. Am J Phys Anthropol. 1994;94(2):213–228. doi: 10.1002/ajpa.1330940206. [DOI] [PubMed] [Google Scholar]
- Boesch C, Boesch H. Hunting behavior of wild chimpanzees in the Tai National Park. Am J Phys Anthropol. 1989;78(4):547–573. doi: 10.1002/ajpa.1330780410. [DOI] [PubMed] [Google Scholar]
- Steinberg D, Mize CE, Herndon JH, Fales HM, Engel WK, Vroom FQ. Phytanic acid in patients with Refsum's syndrome and response to dietary treatment. Arch Intern Med. 1970;125(1):75–87. doi: 10.1001/archinte.125.1.75. [DOI] [PubMed] [Google Scholar]
- Gootjes J, Mooijer PA, Dekker C, Barth PG, Poll-The BT, Waterham HR, Wanders RJ. Biochemical markers predicting survival in peroxisome biogenesis disorders. Neurology. 2002;59(11):1746–1749. doi: 10.1212/01.wnl.0000036609.14203.70. [DOI] [PubMed] [Google Scholar]
- Steinberg D, Avigan J, Mize CE, Baxter JH, Cammermeyer J, Fales HM, Highet PF. Effects of dietary phytol and phytanic acid in animals. J Lipid Res. 1966;7(5):684–691. [PubMed] [Google Scholar]
- Allen NE, Grace PB, Ginn A, Travis RC, Roddam AW, Appleby PN, Key T. Phytanic acid: measurement of plasma concentrations by gas-liquid chromatography-mass spectrometry analysis and associations with diet and other plasma fatty acids. Br J Nutr. 2008;99(3):653–659. doi: 10.1017/S000711450782407X. [DOI] [PubMed] [Google Scholar]
- Aiello LC. Guts and brains. Roebroeks W: Amsterdam University Press; 2007. Notes on the implications of the expensive tissue hypothesis for human biological and social evolution; pp. 17–28. [Google Scholar]
- Aiello LC, Wheeler P. The expensive-tissue hypothesis: The brain and the digestive system in human and primate evolution. Current Anthropology. 1995;36(2):199–221. doi: 10.1086/204350. [DOI] [Google Scholar]
- Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6(10):776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser MJ, Falkow S. What are the consequences of the disappearing human microbiota? Nat Rev Microbiol. 2009;7(12):887–894. doi: 10.1038/nrmicro2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittendorfer B. Sexual dimorphism in human lipid metabolism. J Nutr. 2005;135(4):681–686. doi: 10.1093/jn/135.4.681. [DOI] [PubMed] [Google Scholar]
- Kitson AP, Stroud CK, Stark KD. Elevated production of docosahexaenoic acid in females: potential molecular mechanisms. Lipids. 2010;45(3):209–224. doi: 10.1007/s11745-010-3391-6. [DOI] [PubMed] [Google Scholar]
- Atshaves BP, Payne HR, McIntosh AL, Tichy SE, Russell D, Kier AB, Schroeder F. Sexually dimorphic metabolism of branched-chain lipids in C57BL/6J mice. J Lipid Res. 2004;45(5):812–830. doi: 10.1194/jlr.M300408-JLR200. [DOI] [PubMed] [Google Scholar]
- Roff CF, Pastuszyn A, Strauss JF, Billheimer JT, Vanier MT, Brady RO, Scallen TJ, Pentchev PG. Deficiencies in sex-regulated expression and levels of two hepatic sterol carrier proteins in a murine model of Niemann-Pick type C disease. J Biol Chem. 1992;267(22):15902–15908. [PubMed] [Google Scholar]
- McLean MP, Billheimer JT, Warden KJ, Irby RB. Differential expression of hepatic sterol carrier proteins in the streptozotocin-treated diabetic rat. Endocrinology. 1995;136(8):3360–3368. doi: 10.1210/en.136.8.3360. [DOI] [PubMed] [Google Scholar]
- Blekhman R, Marioni JC, Zumbo P, Stephens M, Gilad Y. Sex-specific and lineage-specific alternative splicing in primates. Genome Res. 2010;20(2):180–189. doi: 10.1101/gr.099226.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokke O. Alpha-oxidation of fatty acids in various mammals, and a phytanic acid feeding experiment in an animal with a low alpha-oxidation capacity. Scand J Clin Lab Invest. 1967;20:305–312. doi: 10.3109/00365516709076960. [DOI] [Google Scholar]
- Blekhman R, Oshlack A, Chabot AE, Smyth GK, Gilad Y. Gene regulation in primates evolves under tissue-specific selection pressures. PLoS Genet. 2008;4(11):e1000271. doi: 10.1371/journal.pgen.1000271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaman MW, Houck ML, Chemnick LG, Nagpal S, Chawannakul D, Sudano D, Pike BL, Ho VV, Ryder OA, Hacia JG. Comparative analysis of gene-expression patterns in human and african great ape cultured fibroblasts. Genome Res. 2003;13(7):1619–1630. doi: 10.1101/gr.1289803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammey ML, Lee DR, Ely JJ, Sleeper MM. Sudden cardiac death in 13 captive chimpanzees (Pan troglodytes) J Med Primatol. 2008;37(Suppl 1):39–43. doi: 10.1111/j.1600-0684.2007.00260.x. [DOI] [PubMed] [Google Scholar]
- Haygood R, Fedrigo O, Hanson B, Yokoyama KD, Wray GA. Promoter regions of many neural- and nutrition-related genes have experienced positive selection during human evolution. Nat Genet. 2007;39(9):1140–1144. doi: 10.1038/ng2104. [DOI] [PubMed] [Google Scholar]
- Somel M, Creely H, Franz H, Mueller U, Lachmann M, Khaitovich P, Paabo S. Human and chimpanzee gene expression differences replicated in mice fed different diets. PLoS ONE. 2008;3(1):e1504.. doi: 10.1371/journal.pone.0001504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell MJ, McInerney JO. Adaptive evolution of the human fatty acid synthase gene: support for the cancer selection and fat utilization hypotheses? Gene. 2005;360(2):151–159. doi: 10.1016/j.gene.2005.06.020. [DOI] [PubMed] [Google Scholar]
- Horrobin DF. Lipid metabolism, human evolution and schizophrenia. Prostaglandins Leukot Essent Fatty Acids. 1999;60(5-6):431–437. doi: 10.1016/S0952-3278(99)80024-6. [DOI] [PubMed] [Google Scholar]
- O'Connell JF, Hawkes K, Blurton Jones NG. Grandmothering and the evolution of homo erectus. J Hum Evol. 1999;36(5):461–485. doi: 10.1006/jhev.1998.0285. [DOI] [PubMed] [Google Scholar]
- Erren TC, Erren M. Can fat explain the human brain's big bang evolution?-Horrobin's leads for comparative and functional genomics. Prostaglandins Leukot Essent Fatty Acids. 2004;70(4):345–347. doi: 10.1016/j.plefa.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Varki A. Multiple changes in sialic acid biology during human evolution. Glycoconj J. 2009;26(3):231–245. doi: 10.1007/s10719-008-9183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch CE. Evolution in health and medicine Sackler colloquium: Evolution of the human lifespan and diseases of aging: roles of infection, inflammation, and nutrition. Proc Natl Acad Sci USA. 2010;107(Suppl 1):1718–1724. doi: 10.1073/pnas.0909606106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yik WY, Steinberg SJ, Moser AB, Moser HW, Hacia JG. Identification of novel mutations and sequence variation in the Zellweger syndrome spectrum of peroxisome biogenesis disorders. Hum Mutat. 2009;30(3):E467–480. doi: 10.1002/humu.20932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Thornburg T, Turner AR, Vitolins M, Case D, Shadle J, Hinson L, Sun J, Liu W, Chang B. et al. Serum levels of phytanic acid are associated with prostate cancer risk. Prostate. 2005;63(3):209–214. doi: 10.1002/pros.20233. [DOI] [PubMed] [Google Scholar]
- Zha S, Ferdinandusse S, Hicks JL, Denis S, Dunn TA, Wanders RJ, Luo J, De Marzo AM, Isaacs WB. Peroxisomal branched chain fatty acid beta-oxidation pathway is upregulated in prostate cancer. Prostate. 2005;63(4):316–323. doi: 10.1002/pros.20177. [DOI] [PubMed] [Google Scholar]
- Magda D, Lecane P, Prescott J, Thiemann P, Ma X, Dranchak PK, Toleno DM, Ramaswamy K, Siegmund KD, Hacia JG. mtDNA depletion confers specific gene expression profiles in human cells grown in culture and in xenograft. BMC Genomics. 2008;9:521. doi: 10.1186/1471-2164-9-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixson AF. Primate sexuality: comparative studies of the prosimians, monkeys, apes, and human beings. Oxford; New York: Oxford University Press; 1998. [Google Scholar]
- Lear TL, Houck ML, Zhang YW, Debnar LA, Sutherland-Smith MR, Young L, Jones KL, Benirschke K. Trisomy 17 in a bonobo (Pan paniscus) and deletion of 3q in a lowland gorilla (Gorilla gorilla gorilla): comparison with human trisomy 18 and human deletion 4q syndrome. Cytogenet Cell Genet. 2001;95(3-4):228–233. doi: 10.1159/000059350. [DOI] [PubMed] [Google Scholar]
- Lagerstedt SA, Hinrichs DR, Batt SM, Magera MJ, Rinaldo P, McConnell JP. Quantitative determination of plasma c8-c26 total fatty acids for the biochemical diagnosis of nutritional and metabolic disorders. Mol Genet Metab. 2001;73(1):38–45. doi: 10.1006/mgme.2001.3170. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193(1):265–275. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Composition of blood donor cohort. A summary of the numbers, ages, and sex of blood donors is provided.
Skin fibroblast cultures used for phytanic and pristanic acid biochemical analysis. A summary of donor sex and age and anatomical source of the skin fibroblasts is provided.
Detailed summary of gene expression data for cross-species comparisons. More complete gene expression data summary statistics relevant to Figure 4 are provided.
Phytanic acid levels relative to total fatty acids in red blood cells from individual donors. Relative phytanic acid levels for all RBC donors are provided.
Rates of phytanic and pristanic acid oxidation in cultured dermal fibroblasts. The rates of phytanic and pristanic acid oxidation from all individual fibroblast cultures are provided.