Abstract
Background
It is well established in E. coli and Vibrio cholerae that strains harboring mutations in the ferric uptake regulator gene (fur) are unable to utilize tricarboxylic acid (TCA) compounds, due to the down-regulation of key TCA cycle enzymes, such as AcnA and SdhABCD. This down-regulation is mediated by a Fur-regulated small regulatory RNA named RyhB. It is unclear in the γ-proteobacterium S. oneidensis whether TCA is also regulated by Fur and RyhB.
Results
In the present study, we showed that a fur deletion mutant of S. oneidensis could utilize TCA compounds. Consistently, expression of the TCA cycle genes acnA and sdhA was not down-regulated in the mutant. To explore this observation further, we identified a ryhB gene in Shewanella species and experimentally demonstrated the gene expression. Further experiments suggested that RyhB was up-regulated in fur mutant, but that AcnA and SdhA were not controlled by RyhB.
Conclusions
These cumulative results delineate an important difference of the Fur-RyhB regulatory cycle between S. oneidensis and other γ-proteobacteria. This work represents a step forward for understanding the unique regulation in S. oneidensis.
Background
Fur (Ferric uptake regulator) is a global transcription factor that regulates a diversity of biological processes such as iron homeostasis, TCA cycle metabolism, acid resistance, oxidative stress response, chemotaxis and pathogenesis (reviewed in [1]). The active, DNA-binding form of this regulator is as a Fur homodimer complexed with ferrous iron. The DNA target recognized by Fe2+-Fur is a 19-bp inverted repeat sequence called a "Fur box" (GATAATGATAATCATTATC) [2]. The binding of Fe2+-Fur to a "Fur box" in the promoter regions of target genes effectively prevents the recruitment of the RNA polymerase holoenzyme, and thus represses transcription [3,4].
Although Fur typically acts as a transcriptional repressor, it also appears to positively regulate certain genes in E. coli [5,6]. This paradox was understood only recently, with the discovery of a 90-nt small RNA named RyhB [7]. RyhB negatively regulates a number of target genes by base pairing with their mRNAs and recruiting RNaseE, thus causing degradation of the mRNAs [7,8]. The ryhB gene itself is repressed by Fur via a "Fur box" in its promoter; thus, Fur repression of the negative regulator RyhB manifests as indirect positive regulation by Fur. The targets of RyhB include genes encoding iron-storage protein (Bfr) and enzymes of the TCA cycle (SdhABCD and AcnA) and oxidative stress response (SodB) [7]. The RyhB-mediated regulation of TCA cycle genes explains the inability of E. coli fur mutants to grow on succinate or fumarate [9].
S. oneidensis is a γ-proteobacterium with a striking capacity to reduce organic compounds and heavy metals, making it a potential bioremediator of environmental contaminants. The S. oneidensis Fur exhibits clear homology to its E. coli ortholog (73% amino acid identity). Physiological, transcriptomics and proteomics studies have shown that S. oneidensis Fur regulates genes involved in iron homeostasis and acid resistance [10-13]. Consistently, many of these target genes have a recognizable "Fur box" in their promoters. In the present study, we further characterize a fur null mutant of S. oneidensis with regard to its ability to utilize succinate and fumarate. Unexpectedly, HPLC analysis showed that the fur mutant was able to metabolize succinate and fumarate, and the growth of the mutant was enhanced in the presence of succinate and fumarate, indicating that the mutant can utilize these compounds. In addition, the expression of the TCA cycle genes acnA and sdhA was not down-regulated in the mutant. These differences between S. oneidensis and E. coli were traced to the small RNA gene ryhB, which we identified in several Shewanella species. Although S. oneidensis RyhB was up-regulated in the fur mutant, the TCA cycle genes did not appear to be regulated by RyhB. These results delineate differences in the gene regulation and physiological consequences of RyhB between S. oneidensis and E. coli.
Results
TCA cycle activity and regulation in the fur mutant
We showed recently that S. oneidensis harboring a fur deletion in the genome was sensitive to acidic conditions and de-repressed genes encoding iron acquisition systems [11]. Similar observations have been made in E. coli [14,15], suggesting that the functional roles of Fur are conserved in these species. Since Fur acts as a pleiotropic transcription factor involved in multiple biological processes, we proceeded to examine its role in regulating TCA cycle enzymes. The involvement of Fur in this biological process has been established in E. coli and V. cholerae by observations that fur mutants are unable to grow in defined media with succinate or fumarate as a carbon source [9,16], and that genes encoding certain TCA cycle enzymes, such as succinate dehydrogenase (SdhABCD) and aconitase (AcnA), are significantly down-regulated in a fur mutant [7].
Our initial tests showed that neither succinate nor fumarate, when provided as the sole carbon source in M1 defined media, could support detectable growth of S. oneidensis type strain MR-1 (data not shown), making it unlikely to analyze the growth of MR-1 and fur null mutant. However, the complete set of TCA genes is present in S. oneidensis genome, and recent studies have shown that the bacterium is capable of metabolizing succinate and fumarate [17,18]. To compare the metabolizing rates of the carbonates between MR-1 and the fur mutant, both strains were grown to mid-log phase with 10 mM lactate as the carbon source. Then equal numbers of cells (5 × 109) were washed and resuspended in fresh M1 medium with 10 mM lactate, succinate or fumarate as the sole carbon source. Unexpectedly, HPLC analyses showed that both succinate and fumarate were reduced to similar levels in MR-1 and the fur mutant after 36 and 54 hours' incubation at 30°C (Figure 1A and Figure 1B), suggesting that fur mutant is not deficient in metabolizing succinate and fumarate. In contrast, more lactate was consumed in MR-1 than in the fur mutant (Figure 1C). This could be explained by the observation that there were more MR-1 cells after 36 hours' incubation (data not shown), as the MR-1 grew faster than the fur mutant when lactate was provided as carbon source (Figure 2). To determine whether the ability of the fur mutant in metabolizing succinate and fumarate affects cell growth, we grew MR-1 and the fur mutant in M1 medium with 10 mM lactate plus succinate or fumarate. Addition of succinate or fumarate significantly enhanced the growth of the fur mutant (Figure 2). Together, succinate and fumarate can indeed be similarly metabolized by MR-1 and the fur mutant of S. oneidensis and be used to support the cell growth when combined with lactate, though they are unable to support the cell growth as the sole carbon source.
Figure 1.
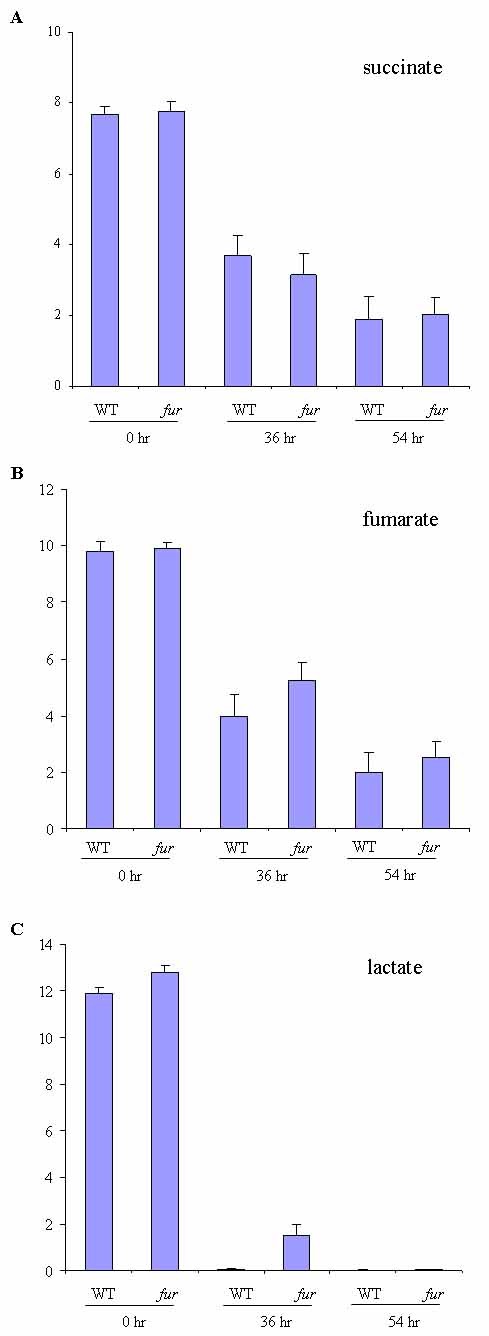
Comparison of MR-1 and the fur mutant for their ability to metabolize carbonate: (A) succinate, (B) fumarate and (C) lactate. 5 × 109 cells were incubated with 10 mM carbonate for 0, 36 and 54 hours. HPLC was used for carbonate measurements. Y-axis: the concentration of carbon source.
Figure 2.
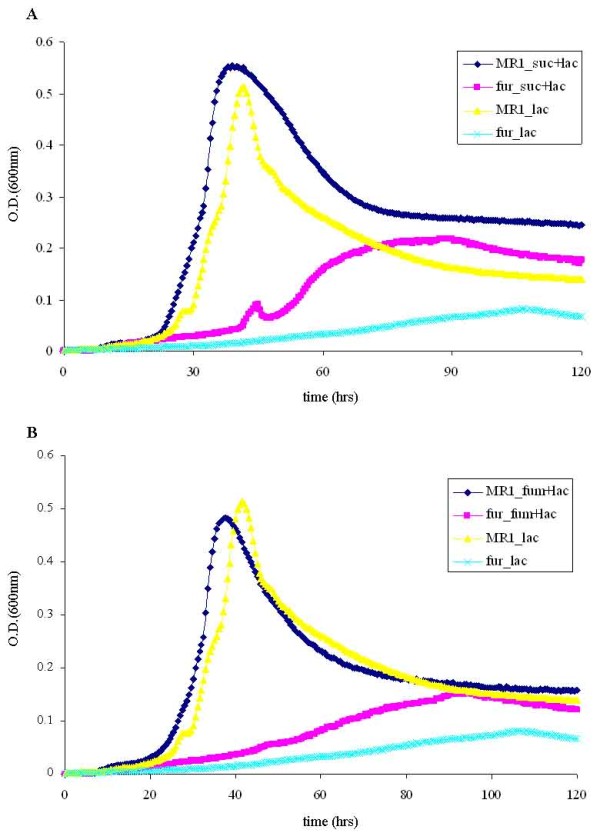
The growth of wild-type (MR-1) and fur mutant in the presence of 10 mM lactate (lac) and (A) succinate (suc) or (B) fumarate (fum), which were supplied as carbon sources in defined medium. Cell density was measured at OD600 every thirty minutes for five days. Data were averaged over triplicate samples.
A recent microarray study comparing the gene expression profile of the fur mutant to that of MR-1 showed that neither the sdhCDAB operon nor the acnA gene was down-regulated [11], which was unlike the observations in E. coli. To confirm this, quantitative RT-PCR was carried out on acnA and sdhA, a gene of the SdhCDAB operon. The housekeeping gene RecA was used as the internal standard to normalize the gene expression levels. The levels of SdhA and AcnA relative to RecA in MR-1 are 0.14 and 0.06, respectively. Both genes exhibited little change in expression in the fur mutant relative to MR-1 (Table 1). Therefore, the utilization of succinate or fumarate by the fur mutant (Figure 1) may be attributable to the persistent expression of TCA cycle genes. Notably, An putative iron uptake gene SO3032, which was expressed at the level of 0.04 relative to RecA in MR-1, was up-regulated in the S. oneidensis fur mutant. In contrast, the Fe-dependent superoxide dismutase encoded by sodB, a gene known to be regulated by Fur in E. coli [7], was repressed in the fur mutant (Table 1). This result agrees with previous observations that the transcript and protein expression levels of SodB are repressed in the fur mutant of S. oneidensis [10].
Table 1.
Quantitative RT-PCR results.
| Gene | fur mutant compared to MR-1 | pFur/fur mutant compared to MR-1 | pRyhB/fur mutant compared to vector/fur mutant | pRyhB/MR-1 compared to vector/MR-1 |
|---|---|---|---|---|
| RyhB | 20.1 (0.0006) | 0.46 (0.07) | 65.2 (0.0002) | 61.4 (0.0001) |
| SdhA | 1.06 (0.3) | 0.89 (0.81) | 1.07 (0.42) | 1.56 (0.25) |
| AcnA | 1.1 (0.42) | 1.29 (0.63) | 0.78 (0.44) | 1.05 (0.47) |
| SodB | 0.12 (0.03) | 0.89 (0.57) | 0.06 (0.01) | 0.06 (0.008) |
| SO3032 | 16.7 (0.04) | 2.32 (0.06) | N/A | N/A |
The numbers in the cells are ratios of gene expression changes and the numbers in the parenthesis are p values of two-sided t-test. 0.05 is used as threshold to determine the significance of the changes.
Identification of the small RNA RyhB in Shewanella species
In E. coli, TCA cycle genes are controlled by a Fur-regulated small RNA named RyhB [7,19]. However, its homolog in S. oneidensis was not identified by homology to the E. coli RyhB using BLAST [20] or by searches using the ryhB sequence alignment and covariance model from Rfam [21]. Therefore, we examined the S. oneidensis MR-1 genome sequence in the region syntenic with the V. cholerae genomic region encoding RyhB. Specifically, the V. cholerae ryhB gene is located downstream of the gene VC0106 [22,23], which is orthologous (by reciprocal best-hit criteria) to the S. oneidensis gene SO4716. We identified a region downstream of SO4716 that exhibited homology with a region that was well-conserved among enterobacterial ryhB sequences (Figure 3A). This "core" region encompasses the sequence believed to base-pair with E. coli sodB mRNA and the binding site for the RNA chaperone Hfq [24].
Figure 3.

Bioinformatics analyses of RyhB in S. oneidensis. (A) Muscle multiple sequence alignment [39]showing homology of the identified region of the S. oneidensis genome with the "core" region of ryhB from E. coli and V. cholerae. Genome coordinates for the sequences are from NC_000913 (E. coli), NC_002505 (V. cholerae), and NC004347 (S. oneidensis). The sequence shown in green is predicted to base pair with the E. coli SodB mRNA. The Hfq binding site is shown in red. (B) Muscle multiple sequence alignment of putative ryhB sequences from eleven species of Shewanella. The box indicates the conserved Fur binding site, the red stars are the start and end positions of the putative promoter, the bent arrow indicates the transcription start site for S. oneidensis, and the region highlighted in yellow is the region of RyhB shown in (A).
RT-PCR was performed to detect the expression of the putative RyhB transcript from this region of the S. oneidensis genome. Total RNA was prepared from wild type S. oneidensis MR-1 strain grown to mid-logarithmic phase and then used for reverse transcription-PCR. A PCR product with expected size of 119 bp was generated using ryhB-specific primers (Figure 4). This PCR product was absent when a PCR reaction was performed on RNA samples without reverse transcription, indicating that the RNA sample was free of genomic DNA contamination. This conclusion was also supported by the absence of PCR products when RT-PCR experiments were used to amplify six other intergenic regions (two examples are shown in Figure 4).
Figure 4.
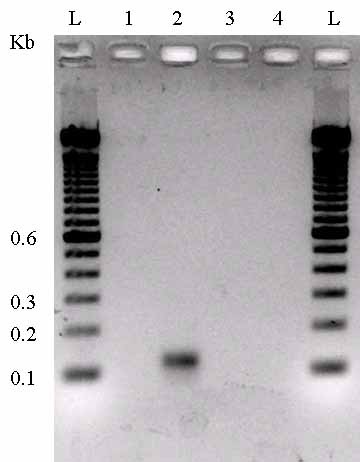
Verification of the expression of small RNA RyhB by RT-PCR. L: DNA ladder; 1. PCR amplification of S. oneidensis RNA without reverse transcription; 2. PCR amplification of sample after reverse transcription of RNA. The presence of the ~119 bp PCR product validates the expression of RyhB RNA. 3 and 4: PCR on two control intergenic regions (Chr. 367734-367820 and 796545-796665). The absence of PCR products indicates that genomic DNA has been completely removed from the RNA templates used for RT-PCR.
To determine the transcriptional boundaries of the RyhB transcript, 5'- and 3'-RACE experiments were carried out on the same sample used for RT-PCR, identifying a 168-nt transcript between nucleotides 4920234-4920401 of the S. oneidensis genome [25]. This transcript is longer than the 90-nt E. coli RyhB [19], but shorter than the 215-nt V. cholerae RyhB [22,23]. A "Fur box", matching 15 of the 19-base consensus sequence (GATAATGATAATCATTATC) [26], was predicted at positions -26 to -44 upstream of this gene (Figure 3B). Together, these results support the existence of a ryhB gene in S. oneidensis.
ryhB genes were subsequently identified in eleven other sequenced Shewanella species by BLASTN using the S. oneidensis ryhB sequence as the query. Extensive sequence conservation was observed (Figure 3B), including the "core" region identified as homologous with the enterobacterial ryhB. Two copies of ryhB were detected in the draft genome sequence of S. amazonensis, in a tandem arrangement similar to that observed for the P. aeruginosa ryhB [27]. The putative "Fur box" was also detected upstream of all of the ryhB homologs, suggesting that regulation of RyhB by Fur is a common feature among the Shewanella species.
Over-expression of RyhB has no impact on the expression of TCA cycle genes
In E. coli, RyhB is highly up-regulated in a fur mutant, which in turn inhibits the expression of AcnA and SdhABCD enzymes and thus the TCA cycle. Since the expression of AcnA and SdhA remained unchanged in the S. oneidensis fur mutant, two possibilities exist as either RyhB is not regulated by Fur or that acnA and sdhA expression is independent of RyhB. To test the possibility that RyhB is not regulated by Fur, quantitative RT-PCR was performed to examine RyhB expression. As shown in Table 1, RyhB was induced 20-fold in the fur mutant. When the fur deletion was complemented by exogenous expression of Fur on the expression vector pBBR1MCS5-1, the RhyB induction was abolished (Table 1). In addition, regulation of RyhB by Fur was also supported by the identification of a "Fur box" upstream of RyhB (Figure 3B). To test the possibility that the expression of acnA and sdhA is independent of RyhB, S. oneidensis was transformed with a RyhB expression plasmid and quantitative RT-PCR performed. RyhB was 60-fold over-expressed relative to endogenous levels in MR-1 and the fur mutant (Table 1). Notably, the expression of SdhA and AcnA mRNAs remained unchanged by RyhB over-expression. In contrast, expression of the superoxide dismutase encoded by sodB was repressed, suggesting that the S. oneidensis sodB was negatively regulated by RyhB. In addition, over-expression of RyhB did not change the growth pattern of MR-1 or the fur mutant in the presence of succinate or fumarate (data not shown). Together, these results suggest that negative regulation of RyhB by Fur exists in S. oneidensis, but sdhA and acnA are not part of Fur-RyhB regulon. Therefore, the TCA cycle in S. oneidensis is independent of Fur and RyhB control.
Discussion
It is of interest to note that succinate and fumarate cannot support the growth of MR-1. Genomics analysis indicates that MR-1 contain the complete gene set required for TCA cycle. However, a recent metabolic flux analysis [17] showed that the anaplerotic pathway (Pyr → Mal) and (Pyr → PEP) were unidirectional, indicating that succinate and fumarate could not be used to produce pyruvate and Acetyl-CoA. Since Acetyl-CoA is the precursor of critical biomass components such as lipids, the inability to convert succinate and fumarate into Acetyl-CoA leads to the growth inhibition of MR-1. In contrast, lactate could be metabolized into pyruvate as well as other central metabolites and thus supports the cell growth.
The inability of E. coli fur mutant to grow on succinate or fumarate has been attributed to the down-regulation of acnA and sdhCDAB by the Fur-regulated small RNA, RyhB [7]. However, this regulatory mechanism of TCA cycle is not present in the γ-proteobacterium S. oneidensis, as evidenced by three observations: (1) both microarray and quantitative RT-PCR experiments showed that expression of acnA and sdhA remained unchanged in the fur mutant; (2) MR-1 and the fur mutant showed similar reduction of succinate and fumarate; and (3) succinate or fumarate enhanced the growth of the fur mutant. To explain the observations, we showed that although S. oneidensis RyhB was up-regulated in the fur mutant, over-expressing RyhB caused little change in the expression of acnA and sdhA as well as the growth with succinate or fumarate. Therefore, acnA and sdhA are not part of the Fur-RyhB regulon in S. oneidensis.
Intriguingly, we found that over-expressing RyhB enhanced the growth of the fur mutant in LB medium containing iron chelator (unpublished data), suggesting that RyhB plays a role in iron response of S. oneidensis. However, additional work is needed to delineate the regulon of RyhB and its regulatory mechanism.
RyhB acts as a post-transcriptional regulator by base pairing with its target mRNAs [7]. Therefore, it is possible to predict its direct targets by surveying DNA sequences for possible base-pairing. A likely target is the SodB mRNA, as evidenced by the presence of sequences in the "core" region of Shewanella RyhB that could potentially base-pair with SodB mRNA [24] and the repression of sodB in strains over-expressing RyhB (Table 1). No likely base pairing between RyhB and sdhCDAB or acnA was noted by manual inspection or by computational programs designed to predict small RNA targets [28,29]. Worthy of mention is that a program called TargetRNA [29] identified possible base pairing between ryhB and Fur genes (Figure 5), implying the possibility of a regulatory feedback loop. Such a regulatory circuit has recently been verified in E. coli [30]. In addition, several genes involved in anaerobic respiration, such as those encoding alcohol dehydrogenase II (AdhB), anaerobic DMSO reductase (DmsA-1), NADH:ubiquinone oxidoreductase subunit (NqrC-2) and two c-type cytochromes (ScyA & SO1659), possess extensive complementary regions with ryhB (Figure 5). Although interesting, these predictions require experimental validation involving a ryhB null mutant. Nevertheless, we have not been able to generate the mutant despite of multiple attempts, which might be attributed to technical difficulties or the possibility that ryhB is an essential gene in S. oneidensis.
Figure 5.

Complementarity between RyhB and its potential targets. The alignment shows the predicted interaction between RyhB and the anti-sense strand of target genes. The numbers represent the start and end positions of the nucleotides. All of the base pairing is considered significant, as judged by p value less than 0.01 [29].
The differences we observed in the RyhB regulon, relative to that of E. coli, are perhaps not surprising in light of the low level of sequence conservation among ryhB genes in phylogenetically related bacteria, implying that ryhB evolves at a rapid pace. Thus far, the only persistent structural features among the known ryhB homologs are the presence of an upstream Fur binding site and a region complementary to the SodB mRNA. The former has been employed to identify ryhB in P. aeruginosa [27].
Accumulating evidence suggests that regulatory pathways in S. oneidensis are distinct from other γ-proteobacteria. For example, the E. coli cAMP receptor protein (CRP) controls the transcription of a number of catabolic genes, but its S. oneidensis homolog is involved in regulation of anaerobic respiration [31]. Also, a major regulator of anaerobic respiration in E. coil (FNR) shows little involvement in anaerobic respiration in S. oneidensis [32-34]. Furthermore, the regulons of the global regulators ArcA and Fur are clearly distinct from that in other bacteria despite significant overlap [10,35].
Conclusions
In accordance with current findings of distinct gene regulatory pathways in S. oneidensis, our study provides evidence to delineate the unique RhyB gene regulation in S. oneidensis.
Methods
Growth conditions and strain construction
M1 defined medium [36] was used. Cell growth was measured by a type FP-1100-C Bioscreen C machine (Thermo Labsystems) at 600 nm after growing cells to mid-logarithmic phase and diluting 1:100 into 300 μl fresh medium. Triplicate cultures were used to determine average and standard deviation. To prepare cultures for real time PCR (RT-PCR), strains were inoculated into 20-50 ml medium in 250 ml flasks. The cultures were incubated at 30°C with vigorous shaking (250 rpm). When the cultures reached mid-logarithmic phase, the cells were collected by centrifugation and flash frozen in liquid nitrogen. Cells were stored at -80°C prior to RNA extraction.
For exogenous expression of Fur and RyhB, the fur and ryhB open reading frames (ORFs) were PCR amplified with primers fur-F1 and fur-R1, and ryhB-F1 and ryhB-R1, respectively (Table 2). The PCR products were digested with SalI and EcoRI, and cloned into the broad-range expression vector pBBR1MCS5-1 (Kmr), placing the ORFs under the transcriptional control of a strong lac promoter. The resulting plasmids were verified by DNA sequencing and transferred into E. coli WM3064, which is a diaminopimelic acid (DAP) auxotroph with plasmid RK4 integrated in the chromosome to mobilize plasmid in trans during conjugation [37]. Conjugation was carried out by mating E. coli and S. oneidensis in 1:1 donor/recipient ratio for 8 hrs on a LB/DAP plate at 30°C followed by selection of S. oneidensis transconjugants on LB agar plates supplemented with 50 μg/ml kanamycin. The vector pBBR1MCS5-1 was also transformed into S. oneidensis for the purpose of comparison.
Table 2.
Oligonucleotide primers used in this study.
| Primer name | Sequence |
|---|---|
| strain construction | |
| fur-F1 | GGTCGACCAAGAGATTAGCAATGACAGATG |
| fur-R1 | GGAATTCGAGCAAGCTTATTCGTCGT |
| ryhB-F1 | GGTCGACAGGAGGAACTCTGATGACTGGTAATCTG |
| ryhB-R1 | GGAATTCAGTTAAATGTGGCGCAAAC |
| Reverse Transcription-PCR | |
| ryhB-F2 | TCTGACGTTGTTAAAGTGCTCC |
| ryhB-R2 | CCTAATGCGCCTATTCGCT |
| Control 1-F | TCAGGTTGTTTGGTATTGTGC |
| Control 1-R | CCATCAATCAAGGTTGTCG |
| Control 2-F | CTGTCAAATGGTGTGCTGC |
| Control 2-R | GTGTAACAGTGCTAAAGCCTGC |
| Control 3-F | TCTACTCAAATGACGAGCTGC |
| Control 3-R | GAAAAGCCGCCAAATGC |
| Control 4-F | TATGGTTTCCCGCTTTCG |
| Control 4-R | AACGCATCAGTGCTATTTGC |
| Control 5-F | TCACTCACAGAACGCTTCG |
| Control 5-R | GCAGCTACAGAATGTCACTACG |
| Control 6-F | TCTAGCAGGGATTAAATGAGC |
| Control 6-R | CCTTCGCCTTGTCTAAAGC |
| 5'- and 3'-RACE assays | |
| 5'- RNA adapter | GAUAUGCGCGAAUUCCUGUAGAACGAACACUAGAAGAAA |
| ryhB-R3 | AGAGTGTGTGAGCAATGTCG |
| 3'- RNA adapter | UUCACU GUUCUUAGCGGCCGCAUGCUC-idT |
| Quantitative RT-PCR | |
| RyhB-F | TCTGACGTTGTTAAAGTGCTCC |
| RyhB-R | CCTAATGCGCCTATTCGCT |
| SdhA-F | GAGCAGTTAAAAGCCATCC |
| SdhA-R | GTTGTCCAATTCTAAACACTCG |
| AcnA-F | ACCAACAAACGCTAGACTACC |
| AcnA-R | ATCATCGCTCCACAAACC |
| SodB-F | TCTACTGGAACTGCTTAGCACC |
| SodB-R | TGAATGCATCGAATGAACC |
| RecA-F | AACCCAGAAACCACAACG |
| RecA-R | ACCAACCACCTCATCACC |
Primer sequences were derived from the S. oneidensis MR-1 genome sequence [25]. F and R stand for forward and reverse primers, respectively.
HPLC analyses
S. oneidensis wild-type (strain MR-1) and the fur mutant were grown to mid-logarithmic phase in M1 medium with 10 mM lactate as the sole carbon source. Cell density was determined by cell counting under microscope and 5 × 109 cells were collected by centrifugation, washed three times with PBS, and inoculated into 3 ml of fresh M1 medium with 10 mM of one of the following carbon sources: lactate, succinate or fumarate. These cultures were incubated at 30°C with vigorous shaking, and at time 0, 36 and 54 hrs, 1 ml culture was centrifuged. The supernatant was used for HPLC with an Elite LaChrom system (Hitachi). The samples were filtered with PALL Life Science Acrodisc 13 mm syringe filters with 0.2 μm nylon membranes, and analyzed with 5 mM H2S04 mobile phase filtered with Gelman Sciences Nylaflo 47 mm 0.45 μm nylon membrane filter paper, degassed and at 0.5 mL/min flowrate for 35 mins with Biorad -Aminex HPX-87H column (300 × 7.8). The column temperature was maintained at 60°C, and the RI detector maintained at 50°C.
RNA isolation and Reverse Transcription-PCR
Total cellular RNA was isolated using the TRIzol reagent (Invitrogen) according to the manufacturer's instructions. RNA samples were treated with RNase-free DNase I (Ambion) to digest residual chromosomal DNA and purified with RNeasy Kit (Qiagen) prior to spectrophotometric quantification at 260 nm. For RT-PCR, 0.1 μg RNA template was used in a Superscript One-step RT-PCR kit (Invitrogen) as recommended by the manufacturer. The primers used were ryhB-F2 and R2, control 1-6 F and R (Table 2).
5'- and 3'-RACE assays
RACE (rapid amplification of cDNA ends) experiments were carried out essentially as described [19]. For 5' RACE, the 5'-triphosphates of 15 μg total RNAs were converted to monophosphates by 25 units of tobacco acid pyrophosphatase (Epicentre Technologies) at 37°C for 1 hr, followed by phenol/chloroform extraction and ethanol precipitation. Precipitated RNA was resuspended in water and ligated to 500 pmol 5'- RNA adapter (Table 2). The ligated product was purified by phenol/chloroform extraction and ethanol precipitation, and reverse transcribed with 2 pmol sRNA-specific primer RyhB-R3 using the Thermoscript RT system (Invitrogen). The product was amplified by PCR, cloned into a pCR2.1 TOPO vector (Invitrogen) and sequenced. 3'-RACE assays were performed similarly to 5'-RACE, except that total RNA was dephosphorylated by calf intestine alkaline phosphatase (New England Biolabs), ligated to a 3'-RNA adapter (Table 2) and reverse transcribed with 100 pmol of a single primer complementary to the 3'-RNA adapter.
Quantitative RT-PCR
The cDNA template for RT-PCR was synthesized in a 10 μl final reaction volume containing 3 μg of total RNA, 3 μg random primers (Invitrogen), 0.5 μM dNTPs, 10 mM DTT, 1 × first-strand buffer and 100 U of Superscript II reverse transcriptase (Invitrogen). After incubation at 42°C for 2 hours, the reaction was diluted five fold in H2O and stored at -80°C. Quantitative RT-PCR was carried out in an iCycler thermal cycler (Bio-Rad) in a 30 μl reaction mixture containing 15 μl iQ SYBR supermix (Molecular Probes), 1 μl cDNA template, and 160 nM forward and reverse primers. Primers were designed using the program Omiga 2.0 (Oxford Molecular) to yield a PCR product of ~100 bp in length (Table 2). Four technical replicates were performed for each of at least three biological replicates. The house-keeping gene recA was used as an internal control. That is, all results were normalized to the recA results obtained in parallel on the same sample to adjust for variation introduced during reverse transcription and RT-PCR. Specifically, the expression values were normalized by subtracting the mean of the recA expression values of the same samples. Different sources of variation (e.g. biological and technical replicates) were accounted for by linear mixed models [38]. The significance of the ratios between two samples was determined using a two-sided t-test, with a type 1 error of 0.05.
Authors' contributions
YY conceived the study, implemented experiments to identify ryhB and drafted the manuscript. LAM performed bioinformatics analyses and manuscript editing. ABP carried out quantitative RT-PCR and growth experiments and performed manuscript editing. SF performed statistical analyses. JZ coordinated the study and performed manuscript editing. All authors have read and approved the manuscript.
Contributor Information
Yunfeng Yang, Email: yangyf@tsinghua.edu.cn.
Lee Ann McCue, Email: leeann.mccue@pnl.gov.
Andrea B Parsons, Email: parsonsab@gmail.com.
Sheng Feng, Email: sheng.feng@duke.edu.
Jizhong Zhou, Email: jzhou@rccc.ou.edu.
Acknowledgements
We thank Choo Yieng Hamilton, Chris Hemme and Charles X. Guan for technical support. This work was supported by The United States Department of Energy's Office of Biological and Environmental Research under the Genomics:GTL Program through the Shewanella Federation, and the Microbial Genome Program. Oak Ridge National Laboratory is managed by University of Tennessee-Battelle LLC for the Department of Energy under contract DE-AC05-00OR22725. PNNL is operated by Battelle for the US Department of Energy under Contract DE-AC06-76RLO 1830.
References
- Escolar L, Perez-Martin J, de Lorenzo V. Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol. 1999;181(20):6223–6229. doi: 10.1128/jb.181.20.6223-6229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baichoo N, Helmann JD. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J Bacteriol. 2002;184(21):5826–5832. doi: 10.1128/JB.184.21.5826-5832.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagg A, Neilands JB. Ferric uptake regulation protein acts as a repressor, employing iron (II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry. 1987;26(17):5471–5477. doi: 10.1021/bi00391a039. [DOI] [PubMed] [Google Scholar]
- de Lorenzo V, Giovannini F, Herrero M, Neilands JB. Metal ion regulation of gene expression. Fur repressor-operator interaction at the promoter region of the aerobactin system of pColV-K30. J Mol Biol. 1988;203(4):875–884. doi: 10.1016/0022-2836(88)90113-1. [DOI] [PubMed] [Google Scholar]
- Niederhoffer EC, Naranjo CM, Bradley KL, Fee JA. Control of Escherichia coli superoxide dismutase (sodA and sodB) genes by the ferric uptake regulation (fur) locus. J Bacteriol. 1990;172(4):1930–1938. doi: 10.1128/jb.172.4.1930-1938.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrac S, Touati D. Fur positive regulation of iron superoxide dismutase in Escherichia coli: functional analysis of the sodB promoter. J Bacteriol. 2000;182(13):3802–3808. doi: 10.1128/JB.182.13.3802-3808.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci USA. 2002;99(7):4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17(19):2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantke K. Selection procedure for deregulated iron transport mutants (fur) in Escherichia coli K 12: fur not only affects iron metabolism. Mol Gen Genet. 1987;210(1):135–139. doi: 10.1007/BF00337769. [DOI] [PubMed] [Google Scholar]
- Wan XF, Verberkmoes NC, McCue LA, Stanek D, Connelly H, Hauser LJ, Wu L, Liu X, Yan T, Leaphart A. et al. Transcriptomic and proteomic characterization of the Fur modulon in the metal-reducing bacterium Shewanella oneidensis. J Bacteriol. 2004;186(24):8385–8400. doi: 10.1128/JB.186.24.8385-8400.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Harris DP, Luo F, Wu L, Parsons AB, Palumbo AV, Zhou J. Characterization of the Shewanella oneidensis Fur gene: roles in iron and acid tolerance response. BMC Genomics. 2008;9(Suppl 1):S11. doi: 10.1186/1471-2164-9-S1-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Zhu M, Wu L, Zhou J. Assessment of data processing to improve reliability of microarray experiments using genomic DNA reference. BMC Genomics. 2008;9(Suppl 2):S5. doi: 10.1186/1471-2164-9-S2-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Harris DP, Luo F, Xiong W, Joachimiak M, Wu L, Dehal P, Jacobsen J, Yang Z, Palumbo AV. et al. Snapshot of iron response in Shewanella oneidensis by gene network reconstruction. BMC Genomics. 2009;10:131. doi: 10.1186/1471-2164-10-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdul-Tehrani H, Hudson AJ, Chang YS, Timms AR, Hawkins C, Williams JM, Harrison PM, Guest JR, Andrews SC. Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. J Bacteriol. 1999;181(5):1415–1428. doi: 10.1128/jb.181.5.1415-1428.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Ngeleka M, Potter AA, Allan BJ. Effect of fur mutation on acid-tolerance response and in vivo virulence of avian septicemic Escherichia coli. Can J Microbiol. 2002;48(5):458–462. doi: 10.1139/w02-042. [DOI] [PubMed] [Google Scholar]
- Litwin CM, Calderwood SB. Analysis of the complexity of gene regulation by fur in Vibrio cholerae. J Bacteriol. 1994;176(1):240–248. doi: 10.1128/jb.176.1.240-248.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YJ, Hwang JS, Wemmer DE, Keasling JD. Shewanella oneidensis MR-1 fluxome under various oxygen conditions. Appl Environ Microbiol. 2007;73(3):718–729. doi: 10.1128/AEM.01532-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YJ, Martin HG, Deutschbauer A, Feng X, Huang R, Llora X, Arkin A, Keasling JD. Invariability of central metabolic flux distribution in Shewanella oneidensis MR-1 under environmental or genetic perturbations. Biotechnol Prog. 2009;25(5):1254–1259. doi: 10.1002/btpr.227. [DOI] [PubMed] [Google Scholar]
- Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol. 2001;11(12):941–950. doi: 10.1016/S0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005. pp. D121–124. [DOI] [PMC free article] [PubMed]
- Davis BM, Quinones M, Pratt J, Ding Y, Waldor MK. Characterization of the Small Untranslated RNA RyhB and Its Regulon in Vibrio cholerae. J Bacteriol. 2005;187(12):4005–4014. doi: 10.1128/JB.187.12.4005-4014.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mey AR, Craig SA, Payne SM. Characterization of Vibrio cholerae RyhB: the RyhB regulon and role of ryhB in biofilm formation. Infect Immun. 2005;73(9):5706–5719. doi: 10.1128/IAI.73.9.5706-5719.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissmann TA, Touati D. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. Embo J. 2004;23(2):396–405. doi: 10.1038/sj.emboj.7600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberg JF, Paulsen IT, Nelson KE, Gaidos EJ, Nelson WC, Read TD, Eisen JA, Seshadri R, Ward N, Methe B. et al. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat Biotechnol. 2002;20(11):1118–1123. doi: 10.1038/nbt749. [DOI] [PubMed] [Google Scholar]
- Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27(2-3):215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- Wilderman PJ, Sowa NA, FitzGerald DJ, FitzGerald PC, Gottesman S, Ochsner UA, Vasil ML. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc Natl Acad Sci USA. 2004;101(26):9792–9797. doi: 10.1073/pnas.0403423101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. miRU: an automated plant miRNA target prediction server. Nucleic Acids Res. 2005. pp. W701–704. [DOI] [PMC free article] [PubMed]
- Tjaden B, Goodwin SS, Opdyke JA, Guillier M, Fu DX, Gottesman S, Storz G. Target prediction for small, noncoding RNAs in bacteria. Nucleic Acids Res. 2006;34(9):2791–2802. doi: 10.1093/nar/gkl356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecerek B, Moll I, Blasi U. Control of Fur synthesis by the non-coding RNA RyhB and iron-responsive decoding. Embo J. 2007;26(4):965–975. doi: 10.1038/sj.emboj.7601553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffarini DA, Schultz R, Beliaev A. Involvement of cyclic AMP (cAMP) and cAMP receptor protein in anaerobic respiration of Shewanella oneidensis. J Bacteriol. 2003;185(12):3668–3671. doi: 10.1128/JB.185.12.3668-3671.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier TM, Myers CR. Isolation and characterization of a Shewanella putrefaciens MR-1 electron transport regulator etrA mutant: reassessment of the role of EtrA. J Bacteriol. 2001;183(16):4918–4926. doi: 10.1128/JB.183.16.4918-4926.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliaev AS, Thompson DK, Fields MW, Wu L, Lies DP, Nealson KH, Zhou J. Microarray transcription profiling of a Shewanella oneidensis etrA mutant. J Bacteriol. 2002;184(16):4612–4616. doi: 10.1128/JB.184.16.4612-4616.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Meier UT. Genetic interaction between a chaperone of small nucleolar ribonucleoprotein particles and cytosolic serine hydroxymethyltransferase. J Biol Chem. 2003;278(26):23553–23560. doi: 10.1074/jbc.M300695200. [DOI] [PubMed] [Google Scholar]
- Gralnick JA, Brown CT, Newman DK. Anaerobic regulation by an atypical Arc system in Shewanella oneidensis. Mol Microbiol. 2005;56(5):1347–1357. doi: 10.1111/j.1365-2958.2005.04628.x. [DOI] [PubMed] [Google Scholar]
- Myers CR, Nealson KH. Respiration-linked proton translocation coupled to anaerobic reduction of manganese(IV) and iron(III) in Shewanella putrefaciens MR-1. J Bacteriol. 1990;172(11):6232–6238. doi: 10.1128/jb.172.11.6232-6238.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltikov CW, Newman DK. Genetic identification of a respiratory arsenate reductase. Proc Natl Acad Sci USA. 2003;100(19):10983–10988. doi: 10.1073/pnas.1834303100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littell RC, Milliken GA, Stroup WW, Wolfinger RD. SAS system for mixed models. Cary, NC: SAS Institute; 1996. [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]


