Abstract
Alternative splicing of mRNA precursors is a nearly ubiquitous and extremely flexible point of gene control in humans. It provides cells with the opportunity to create protein isoforms of differing, even opposing, functions from a single gene. Cancer cells often take advantage of this flexibility to produce proteins that promote growth and survival. Many of the isoforms produced in this manner are developmentally regulated and are preferentially re-expressed in tumors. Emerging insights into this process indicate that pathways that are frequently deregulated in cancer often play important roles in promoting aberrant splicing, which in turn contributes to all aspects of tumor biology.
Keywords: Alternative splicing, cancer, proliferation, RNA-binding proteins, translation
Each regulatory point in the control of gene expression (which includes chromatin structure, splicing and polyadenylation of mRNA precursors, translation, and mRNA and protein stability) is subject to profound alterations during the development of most, if not all, cancers (Venables 2006; Mayr and Bartel 2009; Chi et al. 2010; Silvera et al. 2010). Of all of these points in the gene expression pathway, none provides the potential for more diverse outcomes than alternative splicing (AS). AS, the alternative selection of splice sites present within a pre-mRNA, leads to the production of multiple mRNAs from a single gene. AS thus has the capacity to radically alter the composition and function of the encoded protein. For example, a frequent outcome of AS is the production of proteins with opposing functions, a phenomenon illustrated perhaps most dramatically by the fact that a large majority of genes encoding proteins that function in apoptotic cell death pathways give rise to either pro- or anti-apoptotic isoforms by AS (Schwerk and Schulze-Osthoff 2005).
The plasticity offered by AS to remodel the proteome means that this process is rich with opportunities for cancer cells to subvert the process to produce proteins that suit the needs of the growing and spreading tumor. All areas of tumor biology appear to be affected by changes in AS, including metabolism, apoptosis, cell cycle control, invasion, and metastasis, as well as angiogenesis (Venables 2004; Ghigna et al. 2008). Many of these events appear to represent a return to isoforms normally expressed in a tightly controlled manner during development, but down-regulated in most adult cells. Therefore, as in many other areas of tumor biology, the regulation of these AS events in cancer (often by a recurring cast of splicing factors, as discussed below) can be understood as a consequence of the deregulation of important developmental pathways.
Genome-wide approaches have revealed that tumorigenesis often involves large-scale alterations in AS (Venables et al. 2009). Such approaches have been valuable in providing insight into the regulation of splicing in cancer, and have even proven useful in the classification of tumors (Venables 2006; Skotheim and Nees 2007; Omenn et al. 2010). While the number of AS events observed to differ in cancer has grown quickly with the use of such methods, relatively few have been demonstrated to be functionally important. Most such events instead have been discovered, often serendipitously, by investigators working in various areas of cancer biology. After a brief discussion of general splicing regulatory strategies, this review focuses on AS events for which a functional significance has been established in processes relevant to cancer biology, and for which the underlying regulatory mechanisms have been investigated. Several excellent reviews that deal with related topics have been published recently, and the reader is referred to these for additional insight into the importance of AS in development and disease (Skotheim and Nees 2007; Ghigna et al. 2008; Grosso et al. 2008; Cooper et al. 2009).
Regulation of AS by RNA-binding proteins (RBPs)
Pre-mRNA splicing, the joining of two exons accompanied by the removal of intronic sequence, requires a very large ribonucleoprotein (RNP) complex termed the spliceosome (Wahl et al. 2009). The spliceosome must be guided to the correct splice sites, so what determines which sequences will be included in the mRNA, and which will be excised and degraded in the nucleus? The answer to this question has turned out to be quite complex, especially in light of recent revelations that almost all genes produce transcripts that undergo AS, with a great deal of cell type variation in what sequences are defined as exons and included in the final mRNA (ET Wang et al. 2008). Most of the information necessary to decide which sections of pre-mRNA will be included when and where is present in the sequence of the pre-mRNA (Barash et al. 2010). This information is “read” by RBPs that bind to RNA with varying degrees of sequence specificity and dictate the fate of the surrounding RNA sequences (Chen and Manley 2009; Nilsen and Graveley 2010). Alterations in the levels and activity of these RBPs thus provide the primary means of AS regulation.
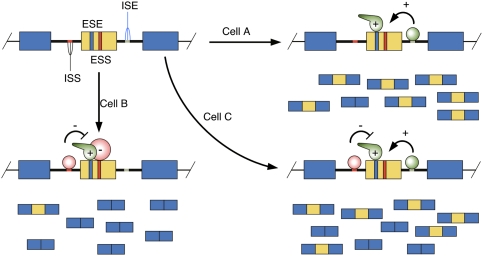
A number of RBPs that function to control AS have been well studied. The classical regulators of splice site choice are serine/arginine-rich (SR) proteins, which, when bound to exonic sequences known as exonic splicing enhancers (ESEs), tend to promote exon inclusion, and heterogeneous RNPs (hnRNPs), which frequently bring about exon exclusion when bound to exonic splicing silencers (ESSs) and/or intronic splicing silencers (ISSs) (Fig. 1). These proteins are joined by several dozen additional RBPs, some with more restricted cell type expression patterns that play important roles in a more limited number of tissue-specific AS events (for review, see Chen and Manley 2009). Many RBPs can act positively or negatively on exon inclusion, depending on the location of their binding sites relative to the regulated exon. This principle was demonstrated systematically by Darnell and colleagues (Ule et al. 2006), who showed that the brain-specific Nova proteins inhibit exon inclusion when their binding sites are located within the exon, while Nova-binding sites in the adjacent intron tend to promote exon inclusion.
Figure 1.
Combinatorial control of splicing by RBPs. In this schematic, a regulated exon (yellow) is flanked by two constitutively spliced exons (blue). Regulatory elements lying within the exon are known as ESEs and ESSs, while intronic regulatory elements are referred to as intronic splicing enhancers (ISEs) and ISSs. The trans-acting factors that bind to these elements can be regulated by changes in intracellular levels, as well as post-translational modifications that affect their cellular localization or activity. The balance of positive- and negative-acting factors present in a given cell determine the extent of regulated exon inclusion.
In light of their crucial role in regulating AS, it follows that aberrant expression and regulation of RBPs likely results in the deregulation of splicing observed in cancer. As discussed below, the production of critical AS isoforms can be a matter of life or death for cancer cells, meaning that there is strong selection for the expression of certain variants. However, as is also discussed below, most of the RBPs that regulate these important AS events have pleiotropic effects on splicing and other processes (especially translation), meaning that the critical changes in AS come as part of a wider program of RBP-mediated changes in gene expression. Most of the additional activities of these RBPs are generally consonant with the outcomes of the functionally important splicing events they govern. This indicates that cancer-associated RBPs are normally regulated as part of developmental pathways that are dysregulated at various stages of tumorigenesis. While the regulation of these RBPs is still not well understood, some new insights are available, as discussed below.
Splicing and apoptosis
Apoptosis, or programmed cell death, occurs through activation of one of several pathways present in normal cells. Because cancer cells display behavior that would normally elicit apoptosis, these cells must in some way or another suppress this process (Letai 2008). As mentioned above, transcripts from a significant number of genes involved in apoptosis are alternatively spliced, often resulting in isoforms with opposing roles in promoting or preventing cell death (Schwerk and Schulze-Osthoff 2005). Apoptotic signaling pathways have been shown to alter the balance of some of these isoforms in favor of proapoptotic isoforms, indicating that altering the balance of pro/anti-apoptotic isoforms is a normal part of programmed cell death (see below).
Given the apparent importance of particular splicing decisions in regulating apoptosis, it is not surprising that the levels of certain RBPs that control these events are a determinant of whether a cell undergoes apoptotic death; the biology of some proteins implicated in the control apoptotic AS events is discussed at the end of the section. While the number of genes involved in apoptosis that are alternatively spliced is large (see Schwerk and Schulze-Osthoff 2005), here we focus on a few well-studied examples for which insight into the relevant splicing regulatory mechanisms is available.
Bcl-x
One of the earliest discovered examples of AS creating opposing isoforms in apoptosis is Bcl-x. The Bcl-x pre-mRNA can be alternatively spliced to produce two isoforms: Bcl-x(L), which has anti-apoptotic effects, and Bcl-x(s), which promotes apoptosis (Boise et al. 1993). The two isoforms arise from AS at two competing 5′ splice sites in exon 2, the first coding exon of the Bcl-x transcript (Fig. 2A). Expression of Bcl-x(L), like Bcl-2, was found to prevent cell death after growth factors were removed from the medium (Boise et al. 1993). The shorter isoform antagonized the protective effects of both Bcl-2 and Bcl-x(L), and the expression of Bcl-x(s) alone was sufficient to induce apoptotic cell death in a wide range of cancer cell types (Boise et al. 1993; Clarke et al. 1995; Minn et al. 1996).
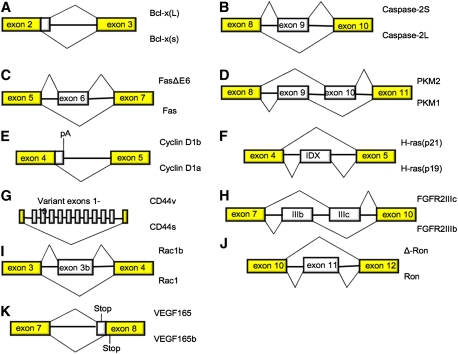
Figure 2.
Schematic representation of the AS events discussed in this review. In each case, isoforms that are up-regulated in cancer or that are otherwise shown to have positive effects on growth, survival, or invasive behavior are shown at the top of each diagram.
High Bcl-x(L)/Bcl-x(s) ratios are observed in a variety of cancer types, consistent with an important role for the long isoform in cancer cell survival (Xerri et al. 1996; Olopade et al. 1997; Takehara et al. 2001). This likely reflects a reduction in Bcl-x(s) in cancer, as an examination of endometrial carcinomas showed a down-regulation of Bcl-x(s) mRNA compared with normal endometrial tissue, with the extent of Bcl-x(s) down-regulation correlated with clinical staging (Ma et al. 2010). Illustrating the importance of this AS event to cancer cells, an antisense oligonucleotide complementary to the Bcl-x(L) isoform 5′ splice site shifted splicing of Bcl-x to the Bcl-x(s) isoform, and was sufficient to induce apoptosis in a prostate cancer (PCa) cell line (Mercatante et al. 2002).
A variety of signals and effectors that regulate Bcl-x AS have been identified. Prompted by reports that Sam68 overexpression can result in apoptosis in NIH-3T3 cells (Babic et al. 2006), and their observation that Sam68 interacts with the Bcl-x mRNA, Sette and colleagues (Paronetto et al. 2007) investigated a possible role for this protein in Bcl-x splicing. Overexpression of Sam68 in 293 cells resulted in an increase in Bcl-x(s) isoform, consistent with the proapoptotic effects observed upon Sam68 overexpression (Taylor et al. 2004; Paronetto et al. 2007). Interestingly, Sam68 phosphorylation by the Src-like tyrosine kinase Fyn reversed the effects of Sam68 overexpression, switching splicing of Bcl-x back to the long isoform. This result indicates that, in the presence of Sam68, growth factors or other signals that activate Fyn or other tyrosine kinases are necessary to maintain expression of Bcl-x(L), providing an additional connection between mitogenic signaling pathways and regulation of apoptosis.
While mitogenic signaling pathways have been implicated in maintaining high levels of Bcl-x(L), a proapoptotic pathway initiated by the sphingolipid ceramide has been suggested to promote Bcl-x(s) splicing (Chalfant et al. 2002; Pettus et al. 2002). Ceramide activates the serine/threonine phosphatases PP1 and PP2A, and treatment of cells with an inhibitor of PP1 negated the effects of ceramide on Bcl-x splicing (Chalfant et al. 2002). Ceramide-induced activation of PP1 has been shown to result in widespread dephosphorylation of SR proteins, although no direct connection between this event and Bcl-x splicing has been established (Chalfant et al. 2001). The RBP SAP155, best known as a member of the SF3b complex that associates with the U2 snRNP, has been shown to bind to a ceramide-responsive element present in the Bcl-x pre-mRNA and is necessary for the effects of ceramide on Bcl-x splicing (Massiello et al. 2006). Incidentally, SAP155 is a known target of PP1/PP2A prior to the second step of splicing (Shi et al. 2006). It is tempting to speculate that dephosphorylation of SAP155 by PP1 has a role in regulating Bcl-x splicing. In a separate pathway, expression of the transcription factor E2F1, which can promote apoptosis, resulted in an increase in Bcl-x(s) (Merdzhanova et al. 2008). Depletion of the SR protein SRSF2 (formerly SC35) (Manley and Krainer 2010), which is specifically induced by E2F1, reversed this effect, implicating SRSF2 in regulation of Bcl-x splicing.
Caspase-2
Caspase-2 is a highly conserved cysteine protease first identified as a mammalian homolog of the CED-3 caspase in Caenorhabditis elegans (Wang et al. 1994). While it was first implicated in apoptosis on the basis of its similarity to CED-3, it has since been shown to act as a tumor suppressor that participates in a wide variety of cellular processes (Ho et al. 2009; Kumar 2009). Caspase-2 mRNA is alternatively processed to produce multiple isoforms (Fig. 2B). The predominant form in most tissues, caspase-2L, produces a full-length protein with proapoptotic properties (Wang et al. 1994). However, in certain differentiated tissues such as brain and skeletal muscle, an additional mRNA isoform containing an additional 61-nucleotide (nt) exon (exon 9) was detected (Wang et al. 1994). Inclusion of this exon results in a frameshift leading to the introduction of a premature termination codon (PTC) in the mRNA, creating a short-lived nonsense-mediated decay (NMD) substrate (Solier et al. 2005). It is unclear under what circumstances caspase-2S mRNA might be stabilized, and conclusive evidence that it is translated is still lacking (Kitevska et al. 2009). In any event, when expressed from cDNA, the truncated product of the caspase-2S mRNA was shown to protect against cell death in some contexts (Wang et al. 1994; Droin et al. 2001).
While the existence of the caspase-2S protein remains to be demonstrated conclusively (Kitevska et al. 2009), alignment of caspase-2 gene sequences from multiple organisms reveals that E9 and the sequences flanking it are, in fact, highly conserved throughout vertebrates (in fact, E9 is among the most highly conserved portions of the gene) (Fig. 3A). This leaves little doubt that E9 inclusion plays an important role at some point in development. One clue to a potential function of E9 inclusion comes from a recent study that identified a class of AS event in which increased inclusion in differentiated cells of a PTC-inducing exon acts as a means of post-transcriptional gene control, resulting in NMD-mediated down-regulation of the gene product (Barash et al. 2010). Caspase-2 expression has been shown to be reduced in the developing retina, where the 2L isoform is down-regulated with a concomitant increase in a small amount of detectable 2S isoform, indicating that E9 inclusion may be a mechanism for developmental control (Kojima et al. 1998).
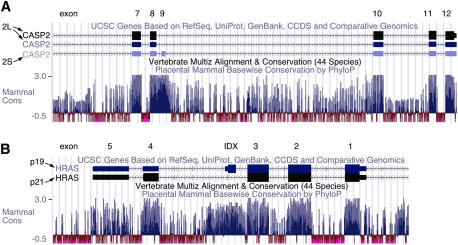
Figure 3.
Caspase-2 and H-ras alternative exons lie in highly conserved regions. Images are taken from the University of California at Santa Cruz Genome Browser, using the March 2006 assembly, available at http://genome.ucsc.edu (Kent et al. 2002), with the exon/intron structure of each gene indicated at that top and the mammalian conservation of the corresponding regions indicated below. Conserved regions are indicated by blue bars. (A) For caspase-2, the alternatively spliced exon 9 is highly conserved, and is also flanked by conserved regions, a hallmark of regulated alternatively spliced exons. (B) The IDX exon of H-ras lies within a similarly highly conserved region.
Whether exon 9 inclusion in caspase-2 results in the production of an anti-apoptotic protein or simply results in reduction of caspase-2 mRNA levels through NMD, it is clear that production of the 2S isoform would favor survival of cancer cells, making its regulation of interest. Early experiments showed that overexpression of hnRNP A1 promoted inclusion of E9 in a minigene construct, while overexpression of SRSF2 resulted in the opposite effect (Jiang et al. 1998). The RNA cis-elements responsible for the effects of hnRNP A1 and SRSF2 were not identified, but an element in intron 9 (I9) termed In100 was shown to contain binding sites for another hnRNP protein, polypyrimidine tract-binding protein (PTB), from which it repressed E9 inclusion (Cote et al. 2001b). In100 also contains a “decoy” 3′ splice site capable of forming nonproductive spliceosome-like complexes with the E9 5′ splice site (Cote et al. 2001a,b). E9 inclusion is promoted by the RBP RBM5. RBM5 binds to an element in I9 to promote E9 exclusion, producing the full-length proapoptotic isoform of caspase-2 (Fushimi et al. 2008).
In addition to RBPs, promoter choice appears to affect caspase-2 splicing. Corcos and colleagues (Logette et al. 2003) demonstrated that caspase-2S and caspase-2L mRNAs derive from different transcription start sites and contain different 5′ untranslated exons. It is known that alternative promoters can have a profound impact on AS patterns (Kornblihtt 2005), and caspase-2 splicing provides another example of this phenomenon. The importance of this phenomenon in regulating caspase-2 splicing in cancer, as well as the mechanism of promoter-dependent differential E9 inclusion, have yet to be explored.
Fas
AS of the Fas receptor pre-mRNA provides a potentially important means by which tumors cells can escape elimination by the immune system. The Fas protein (also known as CD95) is a widely expressed cell surface receptor that, when bound to Fas ligand (FasL) expressed on cytotoxic T cells, can initiate a cascade that eventually leads to cell death (Bouillet and O'Reilly 2009). In addition to producing the full-length mRNA, the Fas pre-mRNA can be alternatively spliced to produce a number of shorter products (Fig. 2C). The most abundant of these is an isoform in which the 63-nt exon 6 (E6) is skipped, deleting the transmembrane domain (Cheng et al. 1994; Cascino et al. 1995). The protein produced by the ΔE6 Fas isoform is soluble and capable of inhibiting Fas-mediated cell death, presumably by binding to FasL and preventing the interaction of FasL with membrane-bound Fas.
Elevated production of soluble Fas (sFas) has been observed in a wide range of cancers, as determined by Fas serum concentrations, which show a strong correlation with tumor staging (e.g., Sheen-Chen et al. 2003; Kondera-Anasz et al. 2005). Consistent with a role for AS in producing the soluble variants, examination of Fas mRNA isoforms in peripheral blood mononuclear cells (PBMCs) from patients with large granular lymphocyte leukemia revealed a large increase in the ΔE6 isoform compared with PBMCs from healthy individuals, consistent with increased serum concentrations of sFas in these patients (Liu et al. 2002). Studies examining the expression of Fas mRNA isoforms in other forms of cancer will be important to provide support for the idea that changes in Fas AS (as opposed to mechanisms such as proteolytic cleavage) indeed underlie the widespread appearance of sFas in cancer.
In light of the potential for Fas AS to play a role in the suppression of the anti-tumor immune response, the regulation of Fas AS has been extensively investigated. Several RBPs have been shown to be involved in promoting the production of full-length Fas mRNA. T-cell intracellular antigen-1 (TIA-1) and TIA-1-related protein (TIAR), two closely related RNA recognition motif (RRM)-containing proteins involved in apoptosis (see below), bind to U-rich sequences downstream from Fas E6 and promote its inclusion in the mRNA (Izquierdo et al. 2005). TIA-1 binding downstream from E6 results in increased U1 snRNP recruitment, presumably through an interaction with the U1 snRNP protein U1C (Forch et al. 2002; Izquierdo et al. 2005). Interestingly, Fas receptor activation influences the splicing of its own pre-mRNA through the activation of the Fas-associated S/T kinase (FAST K). FAST K can phosphorylate TIA-1/TIAR, potentiating their ability to activate E6 inclusion by increasing their ability to recruit U1 snRNPs to the pre-mRNA (Fig. 4; Izquierdo and Valcarcel 2007).
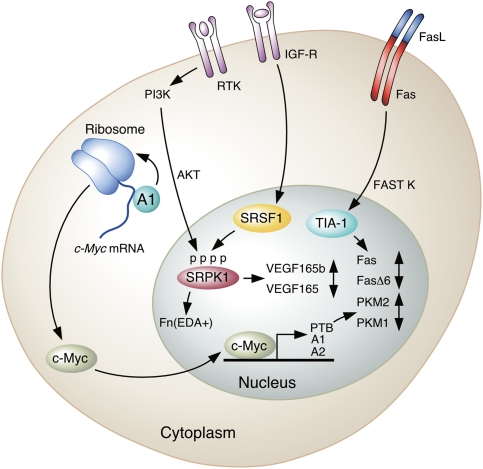
Figure 4.
Selected signal transduction pathways that affect AS of transcripts of genes important in cancer. (From top left,) hnRNP A1 promotes translation of c-Myc mRNA. Translated c-Myc protein can promote the transcription of PTB, hnRNP A1, and hnRNP A2, which in turn promote the production of PKM2. The PI3K pathway is activated downstream from ligand binding by receptor tyrosine kinases (RTKs), activating AKT, which phosphorylates SR proteins such as SRSF1, leading to altered splicing of Fn transcripts. IGF1 binding to the IGF receptor (IGF-R) results in activation of the SR protein kinases SRPK1/2, which phosphorylate SRSF1 and alter VEGF165 splicing in favor of the proangiogenesis isoform. Finally, Fas receptor activation by binding to FasL on another cell results in FAST K activation, which in turn results in TIA-1 phosphorylation and inclusion of exon 6 in the Fas transcript.
A number of repressors of E6 inclusion have also been identified. Valcarcel and colleagues (Izquierdo et al. 2005) found that PTB inhibits E6 inclusion by interfering with binding of the general splicing factor U2AF to the polypyrimidine tract upstream of E6. RBM5 was also identified as an inhibitor of Fas E6 inclusion. RBM5, unlike PTB, does not disrupt early events in the recognition of E6 by the splicing machinery, but rather inhibits the pairing between spliceosomal complexes assembled on E6 and those on the neighboring exons (Bonnal et al. 2008). Another RBP that promotes the expression of the sFas isoform is HuR, which binds to inhibitory sequences in E6 to promote its exclusion (Izquierdo 2008).
Biological functions of apoptosis-regulating RBPs
As just discussed, a number of RBPs that affect the splicing of apoptosis-related transcripts have been identified, highlighting the possibility that these proteins are, in fact, regulators of apoptosis. Below we discuss the function and biology of some of these proteins in normal development and cancer.
TIA-1/TIAR
TIA-1 and TIAR are highly conserved and widely expressed in mammals (Beck et al. 1996). The two proteins function in similar processes and often exhibit partial redundancy (e.g., in Fas splicing, as mentioned above). However, they are not fully redundant, as knockout of either results in embryonic lethality (Beck et al. 1998; Piecyk et al. 2000). In addition, TIAR appears to have a unique role in germ cell development (Beck et al. 1998). Among the earliest functions attributed to TIA-1 and TIAR was the ability to bind mRNAs and recruit them to cytoplasmic bodies (known as stress granules) in response to cellular stress such as heat shock (Kedersha et al. 1999). Both proteins have also been found to bind AU-rich elements (AREs) and act as translational repressors (Piecyk et al. 2000; Kawai et al. 2006; Kim et al. 2007). TIA-1 and TIAR have been shown to be regulators of the inflammatory response that function by silencing translation of key mediators of inflammation, such as TNF-α and COX-2 (Piecyk et al. 2000; Lopez de Silanes et al. 2005a).
Following the realization that TIA-1/TIAR show similarity to the Saccharomyces cerevisiae U1 snRNP-associated protein Nam8, it was demonstrated that the two proteins can function as regulators of pre-mRNA splicing (Gottschalk et al. 1998; Del Gatto-Konczak et al. 2000; Forch et al. 2000). Notable splicing substrates for TIA-1/TIAR are Fas (discussed above) and the epithelial-specific exon IIIb in the fibroblast growth factor receptor 2 (FGFR2) pre-mRNA, which is frequently excluded during cancer progression (Del Gatto-Konczak et al. 2000; see below). Consistent with the fact that TIA-1/TIAR can promote production of the proapoptotic form of Fas, introduction of TIA-1/TIAR into cells promotes apoptosis (Tian et al. 1991; Iseni et al. 2002). In addition to their proapoptotic effects, TIA-1/TIAR depletion in HeLa cells was shown to result in increased proliferation (Reyes et al. 2009).
Given that TIA-1 and TIAR have functions associated with tumor suppressor genes (i.e., promoting apoptosis and inhibiting proliferation), alterations in the expression or regulation of these proteins might be expected to contribute to tumorigenesis in some contexts. However, there is currently little evidence for widespread down-regulation of TIA-1/TIAR in cancer. That being said, TIA-1 expression in tumors correlates strongly with responsiveness to immunotherapy for melanoma patients (Wang et al. 2002). This result supports the idea that down-regulation of TIA-1 is indeed a means by which tumors can evade the immune system. In light of its role in processes important to the growth and survival of cancer cells, an examination of changes in TIA-1/TIAR expression and regulation during tumorigenesis is warranted.
RNA-binding motif 5 (RBM5)
RBM5 is a putative tumor suppressor that contains a large number of motifs, including a pair of RRMs, two Zn finger motifs, an RS domain, and an octamer repeat (OCRE) motif, a newly identified domain now implicated in splicing regulation (Bonnal et al. 2008). RBM5, along with the less extensively studied proteins RBM6 and RBM10, comprises a small family of highly similar RBPs that, in at least some activities, can function redundantly (Bonnal et al. 2008).
Evidence exists supporting the idea that RBM5 is a tumor suppressor (Sutherland et al. 2010). An early suggestion for this came from the presence of the RBM5 gene on a piece of chromosome 3 (3p21.3) that is frequently deleted in lung cancer. Loss of heterozygosity at this locus occurs in 95% of small-cell lung cancer (SCLC), as well as 70% of non-SCLC (Sutherland et al. 2010). Consistent with a role as a tumor suppressor, RBM5 (also known as LUCA-15 and H37) was later shown to be down-regulated in a high proportion of lung cancers (Oh et al. 2002). RBM5 was also one of nine genes down-regulated as part of a “metastatic signature” identified by microarray (Ramaswamy et al. 2003). In stark contrast to observations that RBM5 functions as a tumor suppressor, RBM5 is consistently overexpressed in breast cancer (Rintala-Maki et al. 2007). Together, these results suggest an important, but likely complex, role for RBM5 in regulating genes important in several cancers.
Given the association with cancer, what are the biological functions of RBM5? A 326-base-pair (bp) fragment of cDNA from the RBM5 locus was identified as a suppressor of Fas-mediated apoptosis in a cDNA screen (Sutherland et al. 2000). It was soon realized that this encoded an inhibitory RNA complementary to the 3′ untranslated region (UTR) of the RBM5 mRNA, and that full-length, sense-oriented RBM5 actually potentiates apoptosis initiated by Fas (Rintala-Maki and Sutherland 2004). In addition to its role in apoptosis, expression of RBM5 has been found to inhibit proliferation when transfected into several different cell lines (Edamatsu et al. 2000; Oh et al. 2006). RBM5 also appears to stimulate p53 transcription and promote higher levels of p53 transcripts through an unknown mechanism (Kobayashi et al. 2010).
While the results described above all support an important role for RBM5 in apoptosis and cell cycle regulation, apart from its role in Fas and caspase-2 splicing, very little is known about its biochemical functions. Judging from its large number of domains, these are no doubt diverse. In addition, while the effect of RBM5 on caspase-2 splicing (promoting the 2L isoform) appears to be in line with its role in promoting apoptosis, the inhibition of Fas E6 inclusion would be predicted to protect against Fas-mediated apoptosis, which runs counter to its demonstrated role in promoting Fas-mediated apoptosis. Unraveling this apparent paradox, which may underlie the differential regulation of RBM5 in distinct cancer types, as well as identifying additional targets and functions of RBM5 (and of RBM6 and RBM10), will be of considerable future interest.
HuR
HuR is another multifunctional RRM-containing protein that is frequently up-regulated in cancer (Blaxall et al. 2000; Lopez de Silanes et al. 2003; Denkert et al. 2004). HuR (unlike the related proteins HuB, HuC, and HuD) is ubiquitously expressed and shuttles between the nucleus and the cytoplasm. In synchronized cells, the presence of HuR in the cytoplasm is cell cycle-dependent, while a fraction of HuR is constitutively nuclear (Wang et al. 2000). HuR has a relatively well-characterized role in the cytoplasm, where it binds to mRNAs containing AREs and stabilizes, as well as affects translation of, a large number of mRNAs relevant to proliferation and apoptosis (Lopez de Silanes et al. 2005b; Hinman and Lou 2008). HuR is subject to regulation by phosphorylation as a result of a number of different signaling pathways, which result in changes in its shuttling ability, as well as its RNA binding (Doller et al. 2008). HuR promotes cell proliferation and is one of only a few RBPs for which an oncogenic function has been demonstrated. This was shown in a study in which HuR-overexpressing RKO colon cancer cells, when injected into nude mice, produced much larger tumors than control cells (Lopez de Silanes et al. 2003).
As mentioned above, HuR functions as a suppressor of apoptosis in cancer cells. HuR knockdown in HeLa cells resulted in apoptosis, while HuR overexpression promoted survival of UV-irradiated HeLa cells (Lal et al. 2005). HuR binds the mRNA encoding prothymosin α, an inhibitor of apoptosis, as well as that of Bcl-2, stabilizing both transcripts (Lal et al. 2005; Ishimaru et al. 2009). While the nuclear role for HuR is much more poorly understood than its cytoplasmic function, a role for HuR in regulating splicing and polyadenylation is beginning to emerge (Hinman and Lou 2008). The identification of Fas exon 6 as a target of HuR repression is in general agreement with observations that HuR is a repressor of apoptosis (Izquierdo 2008). This finding may portend a more widespread role for HuR in the regulation of post-transcriptional processing of genes involved in apoptosis and proliferation.
AS regulation of metabolism—pyruvate kinase M
AS also plays an important role in the control of metabolism in cancer through the regulation of a key metabolic gene, pyruvate kinase M (PKM). One of the earliest observations of the molecular differences between cancer cells and noncancerous tissue was that cancer cells consume large amounts of glucose and produce prodigious amounts of lactate, even in the presence of oxygen, a process referred to as aerobic glycolysis (also known as the Warburg effect, named for its discoverer) (Warburg 1956). This effect was later shown to be shared by noncancerous cells induced to proliferate (Wang et al. 1976). From an energy production standpoint, the use of glucose in proliferating cells appears wasteful, as glycolytic conversion of glucose to lactate produces only two molecules of ATP per glucose, while oxidative phosphorylation is capable of producing 36 molecules of ATP per glucose molecule (Vander Heiden et al. 2009). Why do tumor cells forsake the efficient extraction of energy from glucose, instead opting for high glucose consumption and high glycolytic flux? One explanation is that this allows proliferating cells to use carbon derived from glucose for biosynthetic processes necessary for proliferation (Mazurek et al. 2005; Jones and Thompson 2009; Vander Heiden et al. 2009). Multiple glycolytic intermediates represent precursors for the production of nucleotides, lipids, and amino acids, so high glycolytic flux may mean an increased supply of intermediates necessary for growth.
How do proliferating cells reprogram their metabolism to engage in aerobic glycolysis? The Warburg effect requires the shunting of pyruvate from the mitochondria—where, in differentiated cells, it provides the substrate for oxidative phosphorylation—to cytoplasmic conversion to lactate, catalyzed by lactate dehydrogenase (LDH). One important part of this process is increased production of LDH-A, necessary for the final step in aerobic glycolysis, the production of lactate from pyruvate (Fantin et al. 2006). The oncogenic transcription factor c-Myc is known to promote the up-regulation of LDH-A, as well as of several other glycolytic enzymes, an activity that appears to underlie, in part, the effect of Myc on aerobic glycolysis (Shim et al. 1997; Kim et al. 2004; Dang et al. 2009).
AS of the pre-mRNA encoding the enzyme that produces pyruvate, PK, is also an important determinant of how glucose is used in cancerous versus differentiated cells. PKM is the PK gene expressed in all mammalian tissues except liver and erythrocytes (Mazurek et al. 2005). It is subject to mutually exclusive AS, with exon 9 (E9) or exon 10 (E10) included to produce either the adult isoform, PKM1 (E9), or the embryonic version, PKM2 (E10) (Fig. 2D). Cantley and colleagues (Christofk et al. 2008a) found that, in tumors (which uniformly express PKM2), replacing PKM2 with PKM1 reduced the production of lactate and increased oxidative phosphorylation, implicating the PKM2 isoform as a promoter of the Warburg effect. Importantly, and consistent with the idea that aerobic glycolysis is vital for cell growth, replacing PKM2 with PKM1 in cancer cells resulted in impaired growth and reduced the ability to form tumors when injected into nude mice. An additional property specific to PKM2 is its ability to bind phosphotyrosine residues, which frequently result from the activation of mitogenic signaling cascades. This results in the release of its allosteric activator, fructose 1-6 bisphosphate, and the transient inhibition of PK activity, which is proposed to promote accumulation of glycolytic intermediates that can then be used for biosynthetic processes (Christofk et al. 2008b).
The universal switching of tumors to PKM2, along with its functional importance to tumor cells, makes the regulation of this AS event of great interest, because it likely reflects an alteration in the splicing regulatory machinery shared by all proliferating cells. Three hnRNP proteins that often act as splicing repressors—PTB, hnRNP A1, and hnRNP A2—were shown recently to promote the formation of the PKM2 isoform by binding to sequences upstream of and downstream from E9 (David et al. 2010). siRNA-mediated depletion of these proteins in a variety of cancer cells resulted in switching of the PKM splicing pattern to the production of PKM1, indicating that, in the absence of the repressive hnRNP proteins, the derepressed E9 is able to outcompete E10 for splicing to E11, resulting in the default production of PKM1 (Clower et al. 2010). Supporting the idea that these three RBPs play an important role in the production of PKM2 in tumors, up-regulation of all three proteins correlated perfectly with PKM2 expression in a panel of human gliomas (David et al. 2010). Consistent with the effect on PKM splicing, depletion of hnRNP A1/A2 or PTB also resulted in decreased lactate production in a glioblastoma cell line (Clower et al. 2010). The reduction in lactate production appeared greater than would be expected based solely on the effect of knockdown on PKM splicing, indicating that these proteins likely have additional targets relevant to aerobic glycolysis. One possibility is that knockdown of hnRNP A1/A2 and PTB results in impaired translation of c-Myc, a process promoted by both hnRNP A1 and PTB (Mitchell et al. 2005; Jo et al. 2008).
PKM2 expression appears to be universal in tumors, and likewise the up-regulation of hnRNP A1/A2 and PTB is very widely observed in cancer (see below), suggesting that pathways shared by many tumor types promote the overexpression of these RBPs. Chromatin immunoprecipitation (ChIP) and sequencing (Chip-seq) data revealed binding of c-Myc to the promoters of all three genes (Chen et al. 2008), suggesting that c-Myc may activate their transcription. Indeed, shRNA-mediated knockdown of c-Myc in NIH-3T3 cells resulted in decreased levels of hnRNP A1/A2 and PTB, and switched AS to favor accumulation of the PKM1 isoform (David et al. 2010). These data suggest that, in addition to its role in up-regulating glycolytic enzymes, c-Myc contributes to the Warburg effect by indirectly regulating PKM splicing (Chen et al. 2010). However, the effect of c-Myc knockdown on hnRNP protein levels was not observed in all cells tested (David et al. 2010), meaning that additional proliferation-associated transcriptional pathways may play a larger role in inducing overexpression of hnRNP A1/A2 and PTB in other contexts.
Regulation of proto-oncogenes by AS
Activation of proto-oncogenes is a classic initiating event in cancer. While this often happens through mutation, it is now clear that changes in AS that are unconnected to mutations can profoundly affect the activity of some oncogenes. Here we discuss two examples of proto-oncogenes for which AS yields products with distinct functions.
Cyclin D1
Cyclin D1 was first identified as a regulator of cell cycle progression through its association with cyclin-dependent kinase 4 or 6 (CDK4/6) (for review, see Knudsen et al. 2006). One role of the CDK4/6/cyclin D1 complex is to phosphorylate the transcription factor and tumor suppressor RB, thus relieving RB repression of E2F target genes and promoting cell cycle progression. In addition to its association with CDKs, cyclin D1 was later shown to have CDK-independent nuclear functions as a transcriptional regulator (Knudsen et al. 2006).
It is now known that cyclin D1 is subject to AS/polyadenylation that results in a more oncogenic protein. Cyclin D1a, the more common full-length variant, contains five exons, whereas a variant isoform known as cyclin D1b is polyadenylated at a site in intron 4 (Fig. 2E; Betticher et al. 1995). Cyclin D1b can be detected in noncancerous cells, but is up-regulated in some cancers such as breast and prostate (Burd et al. 2006; Y Wang et al. 2008). The D1b isoform, like D1a, associates with CDK4 and appears to regulate CDK activity similarly to D1a (Lu et al. 2003). Instead, a major difference between D1a and D1b is that, while D1a shuttles between the nucleus and cytoplasm in a cell cycle-dependent manner, cyclin D1b is constitutively nuclear, likely due to loss of a glycogen synthase kinase 3β phosphorylation site present at the C terminus of D1a (Lu et al. 2003; Solomon et al. 2003). Phosphorylation at this site results in export from the nucleus and proteolytic degradation, while mutation of this residue results in constitutive nuclear localization and a more oncogenic protein (Alt et al. 2000). Consistent with the greater transforming potential of the constitutively nuclear D1a mutant, D1b transforms NIH-3T3 cells with much greater efficiency than does D1a (Solomon et al. 2003). D1b has been shown to associate with the androgen receptor (AR), but, unlike D1a, D1b fails to inhibit AR transcriptional activity, a phenomenon of particular relevance to PCa, which shows frequent up-regulation of cyclin D1b (Burd et al. 2006; Comstock et al. 2009).
The choice between D1a and D1b production, unlike the other examples described thus far, represents a competition between the splicing of intron 4 and use of a polyadenylation site within the intron. Some insights into factors affecting this alternative processing event are available. A common polymorphism that occurs near the 5′ splice site of exon 4 influences the extent of cyclin D1b produced. The G870A polymorphism, with a reported allele frequency of 42% in one European population, has been associated with production of D1b (Betticher et al. 1995; Comstock et al. 2009). The polymorphism occurs at the very last nucleotide of exon 4 and may affect the recognition of exon 4 by the splicing machinery. A simple model for how G870A favors D1b production is that the G-to-A alteration results in impaired recognition of E4, resulting in slower kinetics of intron 4 splicing, thus favoring polyadenylation at the intronic site, resulting in production of D1b. Significantly, the G870A allele, which does not alter the encoded protein, is associated with increased risks for multiple cancers, supporting a role for D1b in tumorigenesis (Knudsen et al. 2006).
While the G870A polymorphism appears to affect the production of D1b in normal tissues, in cancer cells production of D1b occurs regardless of the identity of the final E4 nucleotide (Olshavsky et al. 2010). Consistent with this, RBPs that influence the choice between D1a and D1b have also been identified. Sam68, described above for its role in Bcl-x splicing, has been implicated in favoring the production of D1b (Paronetto et al. 2010). Binding sites for Sam68 were identified in intron 4, where it either inhibits E4 recognition or promotes polyadenylation in intron 4. A strong correlation between Sam68 expression levels and D1b production was also observed, underscoring the biological significance of these findings. The SR protein SRSF1 (formerly ASF/SF2) has also been shown to bind preferentially to the D1b transcript and promote the production of D1b, and SRSF1 levels were also found to correlate with D1b production (Olshavsky et al. 2010). The effects of SRSF1 on D1b production were enhanced with the 870G allele, potentially providing an explanation for the fact that, in cancer, both alleles result in similar D1b production. Finally, it has also been reported that an oncogenic fusion between the multifunctional RBP EWS and the transcription factor FLI1, which is found in Ewing's sarcoma, can influence production of D1b. While EWS itself was shown to promote production of full-length D1a, the EWS-FLI1 fusion promoted production of D1b (Sanchez et al. 2008). This was proposed to occur due to impaired transcription elongation in the presence of the fusion protein, which would allow more time for polyadenylation to occur in intron 4 before transcription of E5.
H-Ras
The H-Ras gene appears to provide another example of an AS event that produces two proteins with entirely different activities with regard to proliferation. In the late 1980s, Levinson and colleagues (Cohen and Levinson 1988) identified an intronic mutation in H-Ras that significantly enhanced H-Ras expression, significantly increasing the transforming potential of the gene. The mutation was soon found to disrupt the 5′ splice site of a previously unknown alternatively spliced exon that Cohen et al. (1989) named IDX (Fig. 2F). The IDX exon and flanking intronic regions are highly conserved in mammals (Fig. 3B), a strong indication that IDX inclusion is a functionally important regulated AS event (Cohen et al. 1989; Sorek and Ast 2003).
What is the function of IDX inclusion? The fact that mutation of the IDX splice site results in greatly increased production of full-length H-Ras mRNA led to the initial proposal that one function of the IDX-containing isoform could be to modulate levels of H-Ras production. When included, an in-frame stop codon in IDX (the third-to-last exon if all downstream introns are removed; see below) results in a presumptive NMD product, which was first proposed to channel much of the H-Ras transcript into a degradation pathway (Cohen et al. 1989). This was confirmed by a later study (Barbier et al. 2007).
While IDX-containing H-Ras (p19) appears to be an NMD target, Guil et al. (2003a), using antibodies to the divergent C terminus of p19 H-Ras, found that p19 is indeed expressed in HeLa cells. Specific knockdown of p19 H-Ras resulted in increased proliferation of multiple cell lines (Guil et al. 2003a; Jang et al. 2010). p19, in contrast with other Ras proteins, is strongly localized in the nucleus, and was found to bind to p53 and p73 and to promote transcription by the p73 isoform p73β, possibly by preventing an inhibitory interaction between MDM2 and p73 (Jeong et al. 2006).
Because the functions of p19 H-Ras apparently differ greatly from full-length H-Ras, the regulation of IDX inclusion is likely important at some point in development, an idea reinforced by the fact that the IDX exon and its flanking introns are highly conserved in mammals (Fig. 3B). A number of RBPs have been implicated in the control of IDX inclusion. Using an in vitro splicing assay, Guil et al. (2003b) identified an ISS downstream from IDX that was found to bind hnRNP A1, hnRNP H, and the RNA helicase p68. hnRNP A1 and p68 were both found to negatively regulate IDX splicing, while hnRNP H was shown to promote the production of IDX-containing mRNA. Sequences downstream from IDX are predicted to form an extensive stem–loop structure, leading to speculation that p68 helicase activity plays a role in negatively regulating IDX inclusion, which was proposed to interfere with binding of hnRNP H to that region (Camats et al. 2008). In addition to investigating the contributions of the hnRNP proteins and helicase, Bach-Elias and colleagues (Guil et al. 2003b) also provided evidence, using in vitro assays and transient transfections, that the SR proteins SRSF2 and SRSF5 (SRp40) can also function in promoting IDX splicing.
Invasion and metastasis
AS regulates several genes that play important roles in promoting invasive behavior. In addition, a process that often plays a role in the acquisition of invasive behavior in cancer cells, the epithelial-to-mesenchymal transition (EMT), is accompanied by a reprogramming of AS (Thiery et al. 2009). Down-regulation of a pair of epithelial-specific splicing factors may underlie many of the changes in splicing that occur during EMT (see below). In the following paragraphs, we discuss several genes in which AS creates isoforms that are associated with metastasis and the acquisition of invasive properties.
CD44
The transmembrane protein CD44 was among the first genes for which specific splice variants were associated with metastasis. In a 1991 report, Herrlich and colleagues (Gunthert et al. 1991) demonstrated that CD44 molecules containing variant exons v4–7 and CD44 v6–7 were expressed specifically in a metastasizing pancreatic carcinoma cell line, but not in the parental tumor. Indeed, the expression of these variants in cells derived from the parental tumor was sufficient to render them metastatic. CD44 pre-mRNA is subject to complex AS involving 10 adjacent variant exons that can be included singly or in combination (Fig. 2G; Ponta et al. 2003). Later analyses demonstrated the presence of variant exon-containing CD44 molecules throughout adult mammals in a variety of contexts. For example, the variant exons 8–10 are frequently included in CD44 expressed in epithelial tissues (Galiana-Arnoux et al. 2003), while the variant exon 6-containing CD44 first observed to be expressed in metastatic cells was shown to be transiently expressed in B and T lymphocytes after antigenic stimulation, an event necessary for lymphocyte activation (Arch et al. 1992). In an insight into the metastatic properties of CD44 v6, Orian-Rousseau et al. (2002) showed that this molecule is required for the assembly on cell membranes of a ternary complex containing the receptor tyrosine kinase Met, its ligand hepatocyte growth factor (HGF), and CD44 v6 (Orian-Rousseau et al. 2002). Formation of this complex is necessary for the activation of Met by HGF, an event that is strongly implicated in the acquisition of metastatic properties by cancer cells (Cecchi et al. 2010). Additionally, siRNA-mediated knockdown of v5-containing CD44 in HeLa cells resulted in drastically reduced invasion in an in vitro assay, confirming the importance of v5 inclusion in metastatic behavior (Cheng and Sharp 2006).
Because of its long-known association with metastasis, the regulatory mechanisms that underlie the changes in CD44 splicing in cancer have been the subject of long-standing interest, particularly centering around variant exons most strongly associated with metastasis (v4–v7). CD44 was among the first genes for which AS was shown to be altered by signaling pathways associated with growth. For example, inclusion of both exon v5 and v6 can be induced by Ras signaling (Konig et al. 1998; Weg-Remers et al. 2001; Cheng et al. 2006). Activation of v5 inclusion by Ras was shown to require the Ras–Raf–MEK–ERK pathway, and it was later demonstrated that ERK directly phosphorylates Sam68, which was shown to bind to v5 (Weg-Remers et al. 2001; Matter et al. 2002). ERK phosphorylation was shown to be required for Sam68 to activate v5 inclusion (Matter et al. 2002), a finding that for the first time showed a direct connection between a mitogenic signaling pathway and splicing control.
FGFRs
Splicing of transcripts of FGFR genes, which encode four closely related receptor tyrosine kinases, is closely connected to EMT and is frequently observed to change during the course of tumorigenesis. The FGFR genes are highly conserved, and FGFR1–3 share a tightly regulated mutually exclusive AS event that determines the composition of the third extracellular Ig domain (Turner and Grose 2010). For FGFR2, this event, involving the choice between exons IIIb and IIIc, is controlled in an exquisitely cell type-specific manner, with the IIIb exon included in epithelial cells and the IIIc exon included in mesenchymal cells, with nearly complete switching occurring during EMT (Fig. 2H; Yan et al. 1993). EMT, as discussed above, is often a critical event during tumorigenesis, and likewise, switching between the FGFR2 IIIb/IIIc isoforms accompanies crucial changes in tumor behavior. One example of this occurs in PCa cells, which in early stages are dependent on androgen (androgen-sensitive) and can be controlled by therapies that reduce the level of circulating androgen. However, in most cases, the cancer will return in a form that is no longer sensitive to such androgen deprivation (androgen-insensitive). Using a rat PCa model that recapitulates the transition from androgen-sensitive to androgen-insensitive PCa, Garcia-Blanco and colleagues (Carstens et al. 1997) showed that androgen-sensitive tumors express almost exclusively the FGFR2 IIIb isoform, while androgen-insensitive cells exhibit complete switching to the IIIc isoform.
The switch-like nature of this AS event implies an important function in the two different cell types. The mutually exclusive exons compose part of the ligand-binding domain, and one functional consequence of AS is a change in ligand affinity. For example, the FGFR2 IIIb isoform has high affinity for FGF7, which is secreted by stromal cells, while FGFR2 IIIc does not (Yan et al. 1993). Because of the role of FGF7 in mediating both proliferation and differentiation of FGFR2 IIIb-expressing cells, it was proposed that loss of responsiveness to this ligand may be an important event in the development of androgen-insensitive PCa (Carstens et al. 1997). Reintroduction of FGFR2 IIIb into a highly malignant PCa cell line reduced tumor formation and resulted in increased differentiation and apoptosis, suggesting that the switch to FGFR2 IIIc may be important for maximal tumorigenicity (Yasumoto et al. 2004). However, a direct comparison of effects of the two isoforms in cancer must be performed before any conclusions can be made about the functional importance of FGFR2 AS in tumor progression.
While much remains to be learned about the functions of FGFR IIIb/c splicing in development and cancer, a great deal is known about how the switch is regulated, which in turn is instructive of changes in splicing that occur during EMT. Two hnRNP proteins frequently overexpressed in cancer—hnRNP A1 and PTB—have been implicated in silencing of the epithelial-specific IIIb exon, while hnRNP H/F proteins can contribute to IIIc silencing (Del Gatto-Konczak et al. 1999; Carstens et al. 2000; Mauger et al. 2008). There is currently no strong evidence to suggest that changes in the levels of these hnRNP proteins provide an important means of regulating switching from exon IIIb to IIIc. However, differences in recruitment of these proteins to genes offers an additional and, until recently, unexplored potential point of regulation. In a interesting report, Misteli and colleagues (Luco et al. 2010) showed that differences in H3K36 trimethylation can result in differential recruitment of PTB to genes through the histone H3K36 trimethyl-binding protein MRG15. ChIP assays revealed that, in mesenchymal stem cells that repress the IIIb exon, H3K36 trimethylation is increased when compared with epithelial cells in which IIIb is included, a phenomenon proposed to result in increased recruitment of PTB to FGFR2 IIIb in mesenchymal cells.
While the proteins discussed above may play a role in FGFR2 AS regulation, the most important regulators of the IIIb/IIIc AS during EMT appear to be a pair of recently discovered cell type-specific RBPs, ESRP1 and ESRP2 (Warzecha et al. 2009). These two RRM-containing proteins, identified in a cDNA screen for splicing factors that promote the epithelial isoform of FGFR2, are expressed exclusively in epithelial-type cells. The two proteins function redundantly, and simultaneous knockdown of the two in an epithelial cell line promoted a complete reversal of FGFR2 splicing to exon IIIc, while transfection of ESRP1 or ESRP2 expression vectors into cells that express the IIIc exon had the opposite effect. The complete nature of the IIIb/IIIc switching induced by ESRP1/2 knockdown in an epithelial cell line (from 95% IIIb to 96% IIIc), along with the perfect correlation of ESRP1/2 expression and IIIb inclusion, indicates that expression of these RBPs is the primary determinant of FGFR2 IIIb/IIIc inclusion.
To examine FGFR2 splicing in vivo, Garcia-Blanco and colleagues (Oltean et al. 2006) developed a minigene reporter system designed to express RFP in cells in which exon IIIc is not included. They injected mesenchymal AT3 prostate tumor cells expressing the minigene into mice and found that, after invading the lungs, the tumor cells frequently exhibited IIIc skipping, indicative of a mesenchymal-to-epithelial transition (MET), which was confirmed by the presence of the epithelial marker E-cadherin. This result indicates that, in addition to EMT, the reverse transition MET also occurs during tumor progression, and this process entails a reversion to epithelial splicing patterns. While it remains to be examined directly, much of the altered AS that occurs during MET is likely the result of re-expression of ESRP1/2. FGFR1 is subject to another AS event that is of possible relevance to cancer. The α-exon, which comprises one of the extracellular Ig-like domains, is normally included in the brain, but in a variety of gliomas it was found to be skipped, producing an isoform named FGFR1β (Yamaguchi et al. 1994). The skipping of the α-exon is most drastic in the most aggressive glioma, glioblastoma multiforme (GBM). The FGFR1β isoform has higher affinity for FGF1, a feature that might contribute to tumor growth (Wang et al. 1995). The inclusion of the α-exon was shown to be repressed by PTB, which is overexpressed in GBMs (Jin et al. 2003).
Rac1
Rac1 is another Ras superfamily GTPase that, as a result of AS, can exist as an alternative isoform with important functional consequences in cancer cells. Like Ras, Rac1 cycles between an active GTP-bound form and an inactive GDP-bound form. The best studied function of the canonical isoform of Rac1 is regulation of cell migration through its control of lamellipodial protrusion (Bosco et al. 2009). Additional roles in cell proliferation and the creation of reactive oxygen species have also been uncovered (Kheradmand et al. 1998; Bosco et al. 2009). Rac1 has been shown to activate transcription by NFκB as well as the AKT kinase, both important pathways in many cancers (Keely et al. 1997; Perona et al. 1997). Unlike Ras, however, activating mutations of Rac1 are found infrequently in cancer, although mutations analogous to those that activate Ras have been engineered into Rac1 and the mutant protein found to be capable of transforming NIH-3T3 fibroblasts (Qiu et al. 1995; Schnelzer et al. 2000).
While mutation of Rac1 might be rare, in 1999 it was found that Rac1 pre-mRNA is frequently alternatively spliced in colorectal cancer to produce a previously unknown isoform named Rac1b, which contains a new internal 57-nt exon, 3b (Fig. 2I; Jordan et al. 1999). This isoform showed a striking pattern of tumor-specific expression in a set of colorectal tumors when compared with neighboring noncancerous tissue, and was most up-regulated (with reference to the normal Rac1 isoform) in metastatic disease. This was soon followed by a report of the same isoform up-regulated in breast cancer (Schnelzer et al. 2000). Exon 3b and surrounding intronic sequences show considerable conservation in vertebrates, suggesting that 3b inclusion is an important event during development.
Inclusion of exon 3b maintains the reading frame and adds 19 amino acids to a region immediately C-terminal to the switch II region, which, along with switch I, is critical for relaying the GTP/GDP-binding status of Rac1 to downstream effectors (Fiegen et al. 2004). Importantly, biochemical assays performed by multiple groups showed that Rac1b exhibits reduced intrinsic GTPase activity, but increased GDP/GTP exchange, expected to result in a higher level of constitutive activation (Schnelzer et al. 2000; Fiegen et al. 2004; Singh et al. 2004). Consistent with this observation, Rac1b, unlike Rac1, was able to cooperate with an activated mutant of Raf-1 to transform NIH-3T3 cells (Singh et al. 2004). Expression of Rac1b is critical for cell viability in at least one colorectal cancer cell line: Caco-2 (Matos and Jordan 2008). An important insight into the biological function of Rac1b in cancer was provided by a study showing that induction of Rac1b splicing is critical in mediating malignant transformation mediated by matrix metalloproteinase-3 (MMP-3) (Radisky et al. 2005). Addition of MMP-3 to mammary epithelial cells causes them to undergo EMT, leading to increased invasiveness and to genomic instability. Significantly, this process also leads to the production of Rac1b, and isoform-specific knockdown of Rac1b both reversed the effects of MMP3 on cell motility and also led to reduced production of reactive oxygen species that were shown to be responsible for the observed genomic instability (Radisky et al. 2005).
The generation of Rac1b is thus closely connected to fundamental changes such as EMT that occur during tumorigenesis, making the regulation of the splicing event extremely interesting. To investigate the regulation of Rac1 splicing, Jordan and colleagues (Goncalves et al. 2009) transiently overexpressed a variety of well-known splicing factors and found that SRSF1 overexpression resulted in increased exon 3b inclusion, while SRSF3 (SRp20) and SRSF7 (9G8) had the opposite effect. These results were confirmed using siRNA knockdowns. Jordan and colleagues (Goncalves et al. 2009) went on to show that inhibition of the Wnt/β-catenin pathway resulted in decreased SRSF3 levels and increased Rac1b. In accordance with this, the transcriptional activity of β-catenin negatively correlated with Rac1b in colorectal cancer cell lines (Goncalves et al. 2008). Additionally, inhibition of the PI3K/AKT pathway was found to promote increased SRSF1 levels, as well as increased Rac1b. The fact that the Wnt/β-catenin pathway appears to inhibit Rac1b splicing in colorectal cancer cell lines appears to pose a paradox, since this pathway is up-regulated during MMP3-mediated EMT (Radisky et al. 2005). Additional work is necessary to unravel the complexity of Rac1 splicing regulation in the various contexts in which it has been observed.
Ron
Like Rac1, AS of Ron transcripts (which encode the receptor of the macrophage-stimulating protein [MSP]) in cancer cells is closely tied to the invasive phenotype. Ron is a heterodimeric transmembrane receptor tyrosine kinase composed of an α chain and a β chain, each derived from proteolytic cleavage of a common precursor (Lu et al. 2007). Under normal conditions, binding of MSP to Ron results in tyrosine autophosphorylation of the receptor, leading to activation of signaling pathways that result in increased motility and invasive growth (Ghigna et al. 2005; Wagh et al. 2008). In 1996, a Ron variant missing exon 11, termed Δ-Ron, was found to be expressed in the gastric carcinoma cell line KATO-III (Fig. 2J; Collesi et al. 1996). The Δ-Ron isoform and several additional AS-generated isoforms have been found in additional epithelial cancers, including colorectal and breast cancer, and expression of these isoforms correlates with metastasis (Zhou et al. 2003; Ghigna et al. 2005). E11 skipping results in an in-frame deletion of 49 amino acids from the membrane-proximal extracellular domain, leading to a failure to undergo proper proteolytic processing and constitutive activation of the protein (Collesi et al. 1996).While incapable of transforming NIH-3T3 cells, expression of Δ-Ron in two different cell types resulted in increased motility in the absence of MSP, indicating that Δ-Ron indeed functions as a constitutively active receptor in promoting invasive behavior.
Like the constitutively active Rac1b isoform described above, the production of Δ-Ron can be mediated by SRSF1 (Ghigna et al. 2005). SRSF1 was found to bind to an ESE present in E12 and to promote E11 skipping when overexpressed. The importance of SRSF1 for Δ-Ron production was confirmed by knockdown experiments that demonstrated conclusively that SRSF1 expression levels can be an important determinant of the ratio of full-length Ron to Δ-Ron. Consistent with its effect on Ron splicing, SRSF1 overexpression had a profound effect on cell motility and morphology. Perhaps most interestingly, SRSF1 overexpression resulted in the hallmark changes of EMT, including down-regulation of E-cadherin and changes in β-catenin localization. Active Ron is known to activate EMT, and, accordingly, the effect of SRSF1 overexpression was reversed by Δ-Ron-specific siRNAs, indicating that regulation of Ron splicing is the key event mediating the effect of SRSF1 on cell motility (Bardella et al. 2004; Wang et al. 2004).
Regulation of angiogenesis: VEGFA
Angiogenesis is another area of cancer biology in which AS plays an important regulatory role. VEGFA transcripts, which encode the key ligand secreted by tumors in response to hypoxia to promote the formation of new blood vessels, are extensively alternatively spliced (Harper and Bates 2008). Perhaps the alternate isoform with the most pertinence to cancer was discovered in 2002, when a variant differing only in the final six amino acids was identified (Bates et al. 2002; Harper and Bates 2008). This six-amino-acid change occurred due to the choice of a distal 3′ splice site in the final exon (Fig. 2K). The most abundant isoform resulting from use of this 3′ splice site, VEGF165b, was distributed throughout most adult tissues, in some cases representing the major VEGF isoform. While canonical VEGF was broadly up-regulated in kidney carcinomas, VEGF165b was undetectable in most tumors examined. The same pattern of VEGF165b down-regulation was observed in several other cancer types, including PCa and malignant melanoma (Woolard et al. 2004; Pritchard-Jones et al. 2007).
VEGF165b was quickly shown to be an inhibitor of angiogenesis, providing yet another example of an AS event that produces antagonistic isoforms (Bates et al. 2002; Woolard et al. 2004). How does what appears to be a minor modification in VEGF result in such drastically different properties? The canonical form of VEGFA binds to VEGF receptor 1 (VEGFR1) or VEGFR2, as well as coreceptors neurophilin 1 (NRP1) and heparan sulfate proteoglycan (HSPG) (Olsson et al. 2006). Binding to VEGFR2 expressed on endothelial precursors by VEGFA is a critical event in the formation of new blood vessels. VEGFA binding to VEGFR2 as well as NRP1 results in dimerization of VEGFR2 and activation of receptor tyrosine kinase activity, leading to autophosphorylation of key residues necessary for signaling events required for formation of new vasculature (Olsson et al. 2006). VEGF165b, unlike VEGF165, fails to engage NRP1 in a multimeric complex on the cell surface, required for full VEGFR-induced signal transduction (Kawamura et al. 2008). VEGF165b binding to VEGFR2 also fails to result in phosphorylation of the critical residue Y1054, which is necessary for full activation of VEGFR tyrosine kinase activity (Kawamura et al. 2008). VEGF165b was originally proposed to inhibit angiogenesis by acting as a competitive inhibitor of isoforms that promote activation of VEGFR2, although other possibilities, such as activation of alternate signal transduction cascades upon binding VEGFR, cannot be ruled out (Harper and Bates 2008).
Investigation into the regulation of VEGFA splicing has yielded some interesting insights into the pathways and splicing factors involved in regulating the choice between the proximal and distal sites in the final exon. Nowak et al. (2008) examined the effects of three growth factors known to up-regulate VEGFA expression on VEGFA splicing. They found that insulin-like growth factor 1 (IGF1) and TNFα promoted up-regulation of the canonical isoform of VEGFA and down-regulation of VEGF165b, while TGFβ1 also up-regulated VEGFA expression but had the opposite effect on splicing. Inhibition of the p38 MAPK, activated by TGFβ1 signaling, reversed the effect of TGFβ1 on VEGFA splice site selection. The increase in VEGFA expression, as well as the splicing switch in favor of the canonical isoform in IGF1-treated cells, was inhibited when cells were treated simultaneously with inhibitors of protein kinase C (PKC) or the SR protein kinases SRPK1/2 (Nowak et al. 2010).
That inhibition of SRPK1/2 affects splice site choice in the VEGFA transcript implies that SR proteins are involved in regulating the choice between the angiogenic and anti-angiogenic forms of VEGFA. Predicted binding sites for SRSF1 and SRSF5 were identified within exon 8a (included in the canonical isoform), upstream of the distal splice site (DSS), and overexpression of these two SR proteins indeed promoted an increase in VEGF165/VEGF165b ratio (Nowak et al. 2008). In addition, SRSF6 (SRp55)-binding sites were found downstream from the DSS in exon 8b, and SRSF6 overexpression strongly promoted the use of the DSS and expression of VEGF165b (Nowak et al. 2008). Furthermore, IGF1 treatment of cells resulted in an increase in SRSF1 nuclear localization, a phenomenon dependent on SRPK1/2. Although it would be important to confirm these results with appropriate siRNA knockdowns, these findings suggest a pathway that connects a growth factor that promotes vascularization (IGF1) with the alteration of an AS event by several SR proteins in favor of a proangiogenic form of VEGFA.
hnRNP and SR proteins in proliferation and cancer
The first proteins identified to regulate AS belong to the SR and hnRNP protein families (Dreyfuss et al. 1993; Fu 1995; Manley and Tacke 1996). As detailed above, several members of these two protein families have been shown recently to play important roles in proliferation and cancer. Here we examine the regulation and functions of these proteins.
hnRNP A/B family proteins
hnRNPs A1, A2, and A3 constitute the hnRNP A/B family of RBPs. With the exception of hnRNP A3, these proteins have been studied extensively and implicated in a variety of processes, and are expressed in a proliferation-associated manner. A connection between increased hnRNP A1 expression and proliferation was established well before the functions of A1 in regulation of gene expression were understood (LeStourgeon et al. 1978). It was later demonstrated that transcription of the hnRNP A1 gene could be induced by growth factor stimulation of cells, while differentiation of some cell types resulted in decreased A1 levels (Planck et al. 1988; Minoo et al. 1989; Biamonti et al. 1993). In healthy adult tissues, the expression of hnRNP A1/A2 appears confined to proliferating cells, such as those in the basal layer of the skin, although they are also expressed in some neurons (Patry et al. 2003). Extending the analysis of A1 levels to an in vivo model of mouse lung tumorigenesis, Zerbe et al. (2004) showed that nuclear A1 protein levels were dramatically increased in tumors compared with surrounding nonneoplastic cells (Zerbe et al. 2004). Furthermore, induction of nonneoplastic proliferation in the lung was sufficient to lead to increased hnRNP A1 levels, which were further increased in tumors. Importantly, hnRNP A1 has been shown to be up-regulated in a wide variety of cancers, including breast, colorectal, and lung, and, as described above, gliomas (Pino et al. 2003; Ushigome et al. 2005; Li et al. 2009; David et al. 2010). Likewise, hnRNP A2 is consistently overexpressed in a wide range of cancers (Zhou et al. 2001; Yan-Sanders et al. 2002; Wu et al. 2003). hnRNP A2 is expressed at high levels during mouse lung development, then down-regulated in the adult lung, indicating an important role during highly proliferative periods of normal development and confirming the connection between proliferation and hnRNP A2 up-regulation (Montuenga et al. 1998).
Increased expression of hnRNP A/B proteins has important functional consequences. For one, it was found to have a protective effect against apoptosis, possibly in part through an effect on AS of caspase-2 pre-mRNA (Jiang et al. 1998). In addition, depletion of hnRNP A/B proteins in Colo16 cells resulted in reduced proliferation, with hnRNP A2 playing a particularly important role (Jiang et al. 1998; He et al. 2005). Consistent with an important role in tumors, siRNA-mediated knockdown of hnRNP A1/A2 in cancer cells, but not normal cells, can result in apoptosis (Patry et al. 2003).
hnRNP A/B proteins are multifunctional, and many of these functions appear consistent with a role in proliferation. The regulation of pre-mRNA splicing was one of the earliest known functions of hnRNP A1, a function it often carries out by binding to ESS sequences and repressing exon inclusion (Mayeda and Krainer 1992; Del Gatto-Konczak et al. 1999). hnRNP A1 bound to ISS sequences can also promote exon exclusion, possibly through a mechanism that involves self-interaction of A1 molecules bound to distal sites and loop formation (Blanchette and Chabot 1999; Kashima et al. 2007). While best characterized as repressors of splicing, hnRNP A/B proteins can also activate inclusion of some exons (Martinez-Contreras et al. 2006; Venables et al. 2008). As observed in PKM splicing regulation (David et al. 2010), hnRNP A1 and A2 frequently function redundantly in splicing regulation (Kashima and Manley 2003; Licatalosi and Darnell 2010). However, a genome-wide approach indicated that hnRNP A1/A2 targets may, in fact, be quite divergent (Venables et al. 2008). Additional investigation is required to establish the extent of their redundancy, and whether any redundancy exists with a third family member, hnRNP A3, which shares more identity with hnRNP A1 than A2 does. A recent report demonstrated a unique function for hnRNP A2, connecting AS regulation with invasive tumor cell behavior (Moran-Jones et al. 2009). Moran-Jones et al. (2009) found that hnRNP A2 was required for two different cell lines to invade matrigel or to migrate on a cell-derived matrix. Using an exon array, they also showed that hnRNP A2 promotes inclusion of exon 2 in transcripts of the gene encoding a p53 target, TP53INP2, in invasive cells, and that inclusion of this exon is important for invasive behavior.
Despite its important role as a splicing repressor, there have been relatively few reports implicating changes in hnRNP A/B levels in regulation of AS during normal development. In one example, hnRNP A1 and A2 were shown to repress exon 16 inclusion in the protein 4.1R transcript. Differentiation of erythroblasts accompanied an AS switch to exon 16 inclusion, a process that coincided with down-regulation of hnRNP A1 and A2 (Hou et al. 2002). Because up-regulation of hnRNP A1/A2 appears to be shared by most if not all proliferating cells, it will be interesting to see if additional proliferation/differentiation-associated changes in AS are the result of changes in hnRNP A1/A2 levels.
In addition to its role in splicing, hnRNP A1 has numerous additional functions that involve nucleic acid binding. In another role that is likely significant in cancer, human A1 has been implicated in lengthening and maintaining telomeres, a function that is conserved in C. elegans and possibly yeast (Lin and Zakian 1994; LaBranche et al. 1998; Joeng et al. 2004). A1 binds to the single-stranded telomeric DNA repeat sequence TAGGGT (a sequence almost identical to the high-affinity SELEX-binding site identified for A1 using RNA), where it is capable of stimulating telomerase activity (Burd and Dreyfuss 1994; Zhang et al. 2006). A similar function has been shown for hnRNP A2 (Kamma et al. 2001). hnRNP A1 shuttles between the nucleus and cytoplasm, and also has a well-established role in regulating translation of mRNAs through its binding to internal ribosome entry sites (IRES), present in the 5′ UTR of many mRNAs. Binding to such sites in the cyclin D1 and c-Myc mRNAs results in increased translation, making hnRNP A1 an important positive regulator of critical proto-oncogene expression (Jo et al. 2008). The fact that hnRNP A1 is transcriptionally up-regulated by c-Myc and itself promotes the expression of c-Myc (see above) indicates that these two proteins may constitute a positive feedback loop in some circumstances. An additional recently discovered activity of hnRNP A1 is in microRNA (miRNA) biogenesis. A1 was shown to promote processing of miRNA-18a, a miRNA frequently overexpressed in cancer (Guil and Caceres 2007; Motoyama et al. 2009). In light of its newfound role in regulating expression of miRNAs, it will be interesting to see if changes in hnRNP A1 expression are responsible for some of the large-scale changes in miRNA expression observed in cancer.
PTB
PTB also has a myriad of functions in cancer. The splicing-repressive functions of PTB are best studied, although it is also clear that it can promote inclusion of some exons (Xue et al. 2009). Like hnRNP A/B proteins, PTB is expressed throughout development, then down-regulated in many adult tissues, most notably brain and muscle (Boutz et al. 2007a; Makeyev et al. 2007). In differentiating neurons, PTB is replaced by a related protein, nPTB, which binds to similar sequences but functions as a weaker splicing repressor than PTB (Markovtsov et al. 2000; Boutz et al. 2007b). PTB has been shown to be overexpressed in ovarian cancer as well as gliomas (Jin et al. 2003; He et al. 2007; David et al. 2010). Like hnRNP A2, PTB promotes invasive behavior in a number of cancer cell types (He et al. 2007; Cheung et al. 2009). In glioma-derived cells, the increased inclusion of exon 3 in the RTN4 transcript, which is inhibited by PTB, appears to underlie the adverse effects of PTB depletion on cell migration (Cheung et al. 2009). PTB also exerts an effect on cell migration by binding to mRNAs encoding vinculin and α-actinin 4 and localizing to focal adhesions upon cell adhesion (Babic et al. 2009). PTB depletion reduced the number of cell protrusions and reduced the amount of vinculin mRNA present at the cellular edge.
PTB also plays significant roles in translation. In another parallel with hnRNP A1, PTB has been found to bind to the c-Myc IRES sequence, where it up-regulates c-Myc translation synergistically with another RBP, YB-1 (Cobbold et al. 2010). Cobbold et al. (2010) found a striking correlation between c-Myc and PTB protein levels, both of which were elevated in multiple myeloma cell lines compared with B-cell lines. A similar correlation between c-Myc and PTB (as well as hnRNP A1/A2) levels was observed in gliomas (David et al. 2010). Like hnRNP A1/A2, the level of PTB in some cell types is up-regulated by c-Myc, again potentially forming the basis of a positive feedback loop. In addition to c-Myc, PTB was found to bind to most if not all putative IRES elements present in a large number of target mRNAs involved in proliferation and apoptosis (Mitchell et al. 2005). The translational activity of PTB was shown to be important for the up-regulation of translation of a subset of apoptotic mRNAs upon the induction of TRAIL-induced apoptosis (Sawicka et al. 2008). In addition, PTB binds to the IRES of the CDK inhibitor p27Kip1 and promotes its translation (Cho et al. 2005). Depletion of PTB by siRNA-mediated knockdown resulted in a shortened G1 phase, indicating that control of p27Kip1 may play an important role in cell cycle regulation. So, while PTB is frequently overexpressed in cancer, it can have proapoptotic and anti-proliferative effects through its effect on translation of specific mRNAs. cAMP-dependent PKA phosphorylation of PTB results in its redistribution to the cytoplasm, a phenomenon shown to result in PTB-mediated stabilization of insulin mRNA in response to cAMP signaling (Xie et al. 2003; Knoch et al. 2006). It will be of interest to determine in more detail how the various activities of PTB are regulated during development and dysregulated in cancer.
SRSF1
The SR protein with the best-demonstrated role in cancer is SRSF1. As discussed above, SRSF1 has been implicated in a number of cancer-associated changes in AS events related to cell motility (Rac1 and Ron) and proliferation (Cyclin D1). Consistent with this, Krainer and colleagues (Karni et al. 2007) demonstrated that the gene encoding SRSF1 can function as a proto-oncogene. SFSR1 expression was shown to be up-regulated frequently in cancer (sometimes by amplification of its chromosomal locus). In addition, overexpression of SRSF1 resulted in transformation of immortalized cell lines, while reduction of SRSF1 levels in transformed cells reversed the malignant phenotype. Karni et al. (2007) hypothesized that some of the effects of SRSF1 overexpression on the transformed phenotype were due to altered AS, and examined candidate genes in signaling pathways frequently deregulated in cancer for changes in AS. For one gene, encoding the translational regulator S6K1, AS/polyadenylation creates a truncated product, dubbed isoform 2. This mRNA and its truncated protein product were up-regulated in cells overexpressing SRSF1, and isoform 2 itself was found to be sufficient to transform cells. In addition, SRSF1 overexpression promoted AS of the mRNA encoding the tumor suppressor BIN1, producing an isoform with no tumor suppressor activity. SRSF1 also plays a more direct role in translation, promoting cap-dependent translation requiring eIF4E, the deregulation of which can have oncogenic consequences (Karni et al. 2008; Michlewski et al. 2008). This appears to occur in part due to the activation by SRSF1 of mTORC1 through an unknown mechanism (Karni et al. 2008). Like hnRNP A1, SFSR1 has been shown to have a splicing-independent role in promoting the maturation of several miRNAs, some of which are themselves up-regulated in cancer (Wu et al. 2010).
Control of SR protein phosphorylation by signaling pathways
Phosphorylation of SR proteins, most notably on serines in the SR-rich domains that give them their name, is an important determinant of their localization and activity (Shepard and Hertel 2009). Regulation of SR protein activity by phosphorylation has now been implicated in a cancer-associated AS event. Specifically, the EDA exon of the fibronectin (Fn) transcript, an extracellular matrix protein involved in cell adhesion and migration, is expressed at low levels in adult tissues, and its inclusion has been shown to be a highly specific marker of liver metastasis (Rybak et al. 2007). Importantly, the EDA-containing variant appears to promote cellular proliferation to a greater extent than the EDA-lacking variant, indicating a potential functional importance in cancer progression (Manabe et al. 1999). Srebrow and colleagues (Blaustein et al. 2004) demonstrated that growth factors can promote EDA inclusion, and went on to demonstrate that this required the activity of PI3K. Furthermore, activation of the AKT kinase, a key mediator of PI3K signaling in response to growth signals, was capable of activating EDA inclusion (Fig. 4; Blaustein et al. 2005). AKT was shown to be capable of phosphorylating SR proteins SRSF1 and SRSF7 in vitro (Blaustein et al. 2005). These two proteins had been shown to activate splicing of the EDA exon, suggesting the possibility that direct AKT phosphorylation of SR proteins might underlie the change in Fn splicing in response to growth factors. AKT phosphorylation of SRSF1 and SRSF7 occurred mainly in the RS domain, which contains multiple consensus AKT phosphorylation sites (RXRXXS/T). However, the effect of AKT activation of Fn splicing was opposite the effect of overexpression of either SRPK1 or another SR protein kinase, Clk/Sty (Colwill et al. 1996), and the intracellular distribution of the SR proteins was unchanged in response to AKT activation, in contrast to SRPK1 and Clk/Sty activation. This indicates that SR protein phosphorylation by AKT differs from that by SRPK1 and Clk/Sty. Providing additional support for the notion that AKT activation can affect splicing through SR protein phosphorylation, another study suggested that SRSF5 may similarly be regulated by AKT phosphorylation (Patel et al. 2005). Future studies will be necessary to show that the effects of AKT on AS are indeed mediated by direct SR protein phosphorylation, and, if so, how AKT phosphorylation of SR proteins affects their splicing activities.
Conclusions
The above examination of a limited number of important AS events in cancer provides only a small window into the regulation of splicing in cancer. However, a few themes are apparent from these examples. For one, some growth pathways are able to influence splicing through their control of splicing factor expression or post-translational modifications. c-Myc-mediated up-regulation of PTB, hnRNP A1, and hnRNP A2; AKT phosphorylation of SR proteins; and the regulation of Sam68 activity by ERK and Fyn provide examples of this. The dysregulation of these pathways in cancer thus contributes to large-scale changes in AS as well as to alterations in other areas of gene expression controlled by these proteins. Gaining a better understanding of the pathways that lead to up-regulation of key AS regulatory proteins will be of future interest. Because post-translational modifications often have profound effects on the activity of splicing factors, a full understanding of AS regulation in cancer will likely require additional discoveries in this challenging area. A second theme that emerges is that RBPs that regulate AS are all multifunctional, and many of their activities (with some exceptions) appear to be functionally aligned. Systematic analysis of cancer-associated RBPs through the use of genome-wide methods will identify additional targets for these proteins and provide further insight into the extent of “biological coherence” in the processes they regulate (Licatalosi and Darnell 2010), but it should also be apparent that additional mechanistic studies will be needed to understand fully the diverse functions of these proteins and how they contribute to control of cell proliferation in normal and transformed cells.
There is certainly much more to learn about splicing regulation in cancer. The list of splicing regulatory proteins that control AS in cancer is very likely to expand, as is the number of genes affected. Nonetheless, significant insights have been obtained from examination of the relatively small number of splicing events reviewed here, and it is now apparent that dysregulation of AS plays critical roles in numerous cancers and at multiple points in disease progression. It is hoped that future studies will not only provide a deeper understanding of the role of AS in cancer, but also suggest possible therapeutic approaches.
Acknowledgments
We thank Mo Chen and other members of the Manley laboratory for helpful discussions, and Jim Duffy for help with the figures. Relevant work in our laboratory is supported by grants from the NIH.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1973010.
References
- Alt JR, Cleveland JL, Hannink M, Diehl JA 2000. Phosphorylation-dependent regulation of cyclin D1 nuclear export and cyclin D1-dependent cellular transformation. Genes Dev 14: 3102–3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arch R, Wirth K, Hofmann M, Ponta H, Matzku S, Herrlich P, Zoller M 1992. Participation in normal immune responses of a metastasis-inducing splice variant of CD44. Science 257: 682–685 [DOI] [PubMed] [Google Scholar]
- Babic I, Cherry E, Fujita DJ 2006. SUMO modification of Sam68 enhances its ability to repress cyclin D1 expression and inhibits its ability to induce apoptosis. Oncogene 25: 4955–4964 [DOI] [PubMed] [Google Scholar]
- Babic I, Sharma S, Black DL 2009. A role for polypyrimidine tract binding protein in the establishment of focal adhesions. Mol Cell Biol 29: 5564–5577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ 2010. Deciphering the splicing code. Nature 465: 53–59 [DOI] [PubMed] [Google Scholar]
- Barbier J, Dutertre M, Bittencourt D, Sanchez G, Gratadou L, de la Grange P, Auboeuf D 2007. Regulation of H-ras splice variant expression by cross talk between the p53 and nonsense-mediated mRNA decay pathways. Mol Cell Biol 27: 7315–7333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardella C, Costa B, Maggiora P, Patane S, Olivero M, Ranzani GN, De Bortoli M, Comoglio PM, Di Renzo MF 2004. Truncated RON tyrosine kinase drives tumor cell progression and abrogates cell-cell adhesion through E-cadherin transcriptional repression. Cancer Res 64: 5154–5161 [DOI] [PubMed] [Google Scholar]
- Bates DO, Cui TG, Doughty JM, Winkler M, Sugiono M, Shields JD, Peat D, Gillatt D, Harper SJ 2002. VEGF165b, an inhibitory splice variant of vascular endothelial growth factor, is down-regulated in renal cell carcinoma. Cancer Res 62: 4123–4131 [PubMed] [Google Scholar]
- Beck AR, Medley QG, O'Brien S, Anderson P, Streuli M 1996. Structure, tissue distribution and genomic organization of the murine RRM-type RNA binding proteins TIA-1 and TIAR. Nucleic Acids Res 24: 3829–3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck AR, Miller IJ, Anderson P, Streuli M 1998. RNA-binding protein TIAR is essential for primordial germ cell development. Proc Natl Acad Sci 95: 2331–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betticher DC, Thatcher N, Altermatt HJ, Hoban P, Ryder WD, Heighway J 1995. Alternate splicing produces a novel cyclin D1 transcript. Oncogene 11: 1005–1011 [PubMed] [Google Scholar]
- Biamonti G, Bassi MT, Cartegni L, Mechta F, Buvoli M, Cobianchi F, Riva S 1993. Human hnRNP protein A1 gene expression. Structural and functional characterization of the promoter. J Mol Biol 230: 77–89 [DOI] [PubMed] [Google Scholar]
- Blanchette M, Chabot B 1999. Modulation of exon skipping by high-affinity hnRNP A1-binding sites and by intron elements that repress splice site utilization. EMBO J 18: 1939–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaustein M, Pelisch F, Coso OA, Bissell MJ, Kornblihtt AR, Srebrow A 2004. Mammary epithelial–mesenchymal interaction regulates fibronectin alternative splicing via phosphatidylinositol 3-kinase. J Biol Chem 279: 21029–21037 [DOI] [PubMed] [Google Scholar]
- Blaustein M, Pelisch F, Tanos T, Munoz MJ, Wengier D, Quadrana L, Sanford JR, Muschietti JP, Kornblihtt AR, Caceres JF, et al. 2005. Concerted regulation of nuclear and cytoplasmic activities of SR proteins by AKT. Nat Struct Mol Biol 12: 1037–1044 [DOI] [PubMed] [Google Scholar]
- Blaxall BC, Dwyer-Nield LD, Bauer AK, Bohlmeyer TJ, Malkinson AM, Port JD 2000. Differential expression and localization of the mRNA binding proteins, AU-rich element mRNA binding protein (AUF1) and Hu antigen R (HuR), in neoplastic lung tissue. Mol Carcinog 28: 76–83 [PubMed] [Google Scholar]
- Boise LH, Gonzalez-Garcia M, Postema CE, Ding L, Lindsten T, Turka LA, Mao X, Nunez G, Thompson CB 1993. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell 74: 597–608 [DOI] [PubMed] [Google Scholar]
- Bonnal S, Martinez C, Forch P, Bachi A, Wilm M, Valcarcel J 2008. RBM5/Luca-15/H37 regulates Fas alternative splice site pairing after exon definition. Mol Cell 32: 81–95 [DOI] [PubMed] [Google Scholar]
- Bosco EE, Mulloy JC, Zheng Y 2009. Rac1 GTPase: A ‘Rac’ of all trades. Cell Mol Life Sci 66: 370–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouillet P, O'Reilly LA 2009. CD95, BIM and T cell homeostasis. Nat Rev Immunol 9: 514–519 [DOI] [PubMed] [Google Scholar]
- Boutz PL, Chawla G, Stoilov P, Black DL 2007a. MicroRNAs regulate the expression of the alternative splicing factor nPTB during muscle development. Genes Dev 21: 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, Shiue L, Ares M Jr, Black DL 2007b. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev 21: 1636–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G 1994. RNA binding specificity of hnRNP A1: Significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J 13: 1197–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CJ, Petre CE, Morey LM, Wang Y, Revelo MP, Haiman CA, Lu S, Fenoglio-Preiser CM, Li J, Knudsen ES, et al. 2006. Cyclin D1b variant influences prostate cancer growth through aberrant androgen receptor regulation. Proc Natl Acad Sci 103: 2190–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camats M, Guil S, Kokolo M, Bach-Elias M 2008. P68 RNA helicase (DDX5) alters activity of cis- and trans-acting factors of the alternative splicing of H-Ras. PLoS ONE 3: e2926 doi: 10.1371/journal.pone.0002926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstens RP, Eaton JV, Krigman HR, Walther PJ, Garcia-Blanco MA 1997. Alternative splicing of fibroblast growth factor receptor 2 (FGF-R2) in human prostate cancer. Oncogene 15: 3059–3065 [DOI] [PubMed] [Google Scholar]
- Carstens RP, Wagner EJ, Garcia-Blanco MA 2000. An intronic splicing silencer causes skipping of the IIIb exon of fibroblast growth factor receptor 2 through involvement of polypyrimidine tract binding protein. Mol Cell Biol 20: 7388–7400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascino I, Fiucci G, Papoff G, Ruberti G 1995. Three functional soluble forms of the human apoptosis-inducing Fas molecule are produced by alternative splicing. J Immunol 154: 2706–2713 [PubMed] [Google Scholar]
- Cecchi F, Rabe DC, Bottaro DP 2010. Targeting the HGF/Met signalling pathway in cancer. Eur J Cancer 46: 1260–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfant CE, Ogretmen B, Galadari S, Kroesen BJ, Pettus BJ, Hannun YA 2001. FAS activation induces dephosphorylation of SR proteins; dependence on the de novo generation of ceramide and activation of protein phosphatase 1. J Biol Chem 276: 44848–44855 [DOI] [PubMed] [Google Scholar]
- Chalfant CE, Rathman K, Pinkerman RL, Wood RE, Obeid LM, Ogretmen B, Hannun YA 2002. De novo ceramide regulates the alternative splicing of caspase 9 and Bcl-x in A549 lung adenocarcinoma cells. Dependence on protein phosphatase-1. J Biol Chem 277: 12587–12595 [DOI] [PubMed] [Google Scholar]
- Chen M, Manley JL 2009. Mechanisms of alternative splicing regulation: Insights from molecular and genomics approaches. Nat Rev Mol Cell Biol 10: 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117 [DOI] [PubMed] [Google Scholar]
- Chen M, David CJ, Manley JL 2010. Tumor metabolism: hnRNP proteins get in on the act. Cell Cycle 9: 1863–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Sharp PA 2006. Regulation of CD44 alternative splicing by SRm160 and its potential role in tumor cell invasion. Mol Cell Biol 26: 362–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Zhou T, Liu C, Shapiro JP, Brauer MJ, Kiefer MC, Barr PJ, Mountz JD 1994. Protection from Fas-mediated apoptosis by a soluble form of the Fas molecule. Science 263: 1759–1762 [DOI] [PubMed] [Google Scholar]
- Cheng C, Yaffe MB, Sharp PA 2006. A positive feedback loop couples Ras activation and CD44 alternative splicing. Genes Dev 20: 1715–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung HC, Hai T, Zhu W, Baggerly KA, Tsavachidis S, Krahe R, Cote GJ 2009. Splicing factors PTBP1 and PTBP2 promote proliferation and migration of glioma cell lines. Brain 132: 2277–2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P, Allis CD, Wang GG 2010. Covalent histone modifications—Miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer 10: 457–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S, Kim JH, Back SH, Jang SK 2005. Polypyrimidine tract-binding protein enhances the internal ribosomal entry site-dependent translation of p27Kip1 mRNA and modulates transition from G1 to S phase. Mol Cell Biol 25: 1283–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC 2008a. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452: 230–233 [DOI] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC 2008b. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature 452: 181–186 [DOI] [PubMed] [Google Scholar]
- Clarke MF, Apel IJ, Benedict MA, Eipers PG, Sumantran V, Gonzalez-Garcia M, Doedens M, Fukunaga N, Davidson B, Dick JE, et al. 1995. A recombinant bcl-x s adenovirus selectively induces apoptosis in cancer cells but not in normal bone marrow cells. Proc Natl Acad Sci 92: 11024–11028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clower CV, Chatterjee D, Wang Z, Cantley LC, Vander Heiden MG, Krainer AR 2010. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc Natl Acad Sci 107: 1894–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbold LC, Wilson LA, Sawicka K, King HA, Kondrashov AV, Spriggs KA, Bushell M, Willis AE 2010. Upregulated c-myc expression in multiple myeloma by internal ribosome entry results from increased interactions with and expression of PTB-1 and YB-1. Oncogene 29: 2884–2891 [DOI] [PubMed] [Google Scholar]
- Cohen JB, Levinson AD 1988. A point mutation in the last intron responsible for increased expression and transforming activity of the c-Ha-ras oncogene. Nature 334: 119–124 [DOI] [PubMed] [Google Scholar]
- Cohen JB, Broz SD, Levinson AD 1989. Expression of the H-ras proto-oncogene is controlled by alternative splicing. Cell 58: 461–472 [DOI] [PubMed] [Google Scholar]
- Collesi C, Santoro MM, Gaudino G, Comoglio PM 1996. A splicing variant of the RON transcript induces constitutive tyrosine kinase activity and an invasive phenotype. Mol Cell Biol 16: 5518–5526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwill K, Pawson T, Andrews B, Prasad J, Manley JL, Bell JC, Duncan PI 1996. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J 15: 265–275 [PMC free article] [PubMed] [Google Scholar]
- Comstock CE, Augello MA, Benito RP, Karch J, Tran TH, Utama FE, Tindall EA, Wang Y, Burd CJ, Groh EM, et al. 2009. Cyclin D1 splice variants: Polymorphism, risk, and isoform-specific regulation in prostate cancer. Clin Cancer Res 15: 5338–5349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G 2009. RNA and disease. Cell 136: 777–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote J, Dupuis S, Jiang Z, Wu JY 2001a. Caspase-2 pre-mRNA alternative splicing: Identification of an intronic element containing a decoy 3′ acceptor site. Proc Natl Acad Sci 98: 938–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote J, Dupuis S, Wu JY 2001b. Polypyrimidine track-binding protein binding downstream of caspase-2 alternative exon 9 represses its inclusion. J Biol Chem 276: 8535–8543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV, Le A, Gao P 2009. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin Cancer Res 15: 6479–6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Chen M, Assanah M, Canoll P, Manley JL 2010. hnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 463: 364–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Gatto-Konczak F, Olive M, Gesnel MC, Breathnach R 1999. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol Cell Biol 19: 251–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Gatto-Konczak F, Bourgeois CF, Le Guiner C, Kister L, Gesnel MC, Stevenin J, Breathnach R 2000. The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol Cell Biol 20: 6287–6299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denkert C, Weichert W, Pest S, Koch I, Licht D, Kobel M, Reles A, Sehouli J, Dietel M, Hauptmann S 2004. Overexpression of the embryonic-lethal abnormal vision-like protein HuR in ovarian carcinoma is a prognostic factor and is associated with increased cyclooxygenase 2 expression. Cancer Res 64: 189–195 [DOI] [PubMed] [Google Scholar]
- Doller A, Pfeilschifter J, Eberhardt W 2008. Signalling pathways regulating nucleo-cytoplasmic shuttling of the mRNA-binding protein HuR. Cell Signal 20: 2165–2173 [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Matunis MJ, Pinol-Roma S, Burd CG 1993. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem 62: 289–321 [DOI] [PubMed] [Google Scholar]
- Droin N, Rebe C, Bichat F, Hammann A, Bertrand R, Solary E 2001. Modulation of apoptosis by procaspase-2 short isoform: Selective inhibition of chromatin condensation, apoptotic body formation and phosphatidylserine externalization. Oncogene 20: 260–269 [DOI] [PubMed] [Google Scholar]
- Edamatsu H, Kaziro Y, Itoh H 2000. LUCA15, a putative tumour suppressor gene encoding an RNA-binding nuclear protein, is down-regulated in ras-transformed Rat-1 cells. Genes Cells 5: 849–858 [DOI] [PubMed] [Google Scholar]
- Fantin VR, St-Pierre J, Leder P 2006. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell 9: 425–434 [DOI] [PubMed] [Google Scholar]
- Fiegen D, Haeusler LC, Blumenstein L, Herbrand U, Dvorsky R, Vetter IR, Ahmadian MR 2004. Alternative splicing of Rac1 generates Rac1b, a self-activating GTPase. J Biol Chem 279: 4743–4749 [DOI] [PubMed] [Google Scholar]
- Forch P, Puig O, Kedersha N, Martinez C, Granneman S, Seraphin B, Anderson P, Valcarcel J 2000. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol Cell 6: 1089–1098 [DOI] [PubMed] [Google Scholar]
- Forch P, Puig O, Martinez C, Seraphin B, Valcarcel J 2002. The splicing regulator TIA-1 interacts with U1-C to promote U1 snRNP recruitment to 5′ splice sites. EMBO J 21: 6882–6892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XD 1995. The superfamily of arginine/serine-rich splicing factors. RNA 1: 663–680 [PMC free article] [PubMed] [Google Scholar]
- Fushimi K, Ray P, Kar A, Wang L, Sutherland LC, Wu JY 2008. Up-regulation of the proapoptotic caspase 2 splicing isoform by a candidate tumor suppressor, RBM5. Proc Natl Acad Sci 105: 15708–15713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiana-Arnoux D, Lejeune F, Gesnel MC, Stevenin J, Breathnach R, Del Gatto-Konczak F 2003. The CD44 alternative v9 exon contains a splicing enhancer responsive to the SR proteins 9G8, ASF/SF2, and SRp20. J Biol Chem 278: 32943–32953 [DOI] [PubMed] [Google Scholar]
- Ghigna C, Giordano S, Shen H, Benvenuto F, Castiglioni F, Comoglio PM, Green MR, Riva S, Biamonti G 2005. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell 20: 881–890 [DOI] [PubMed] [Google Scholar]
- Ghigna C, Valacca C, Biamonti G 2008. Alternative splicing and tumor progression. Curr Genomics 9: 556–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves V, Matos P, Jordan P 2008. The β-catenin/TCF4 pathway modifies alternative splicing through modulation of SRp20 expression. RNA 14: 2538–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves V, Matos P, Jordan P 2009. Antagonistic SR proteins regulate alternative splicing of tumor-related Rac1b downstream of the PI3-kinase and Wnt pathways. Hum Mol Genet 18: 3696–3707 [DOI] [PubMed] [Google Scholar]
- Gottschalk A, Tang J, Puig O, Salgado J, Neubauer G, Colot HV, Mann M, Seraphin B, Rosbash M, Luhrmann R, et al. 1998. A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. RNA 4: 374–393 [PMC free article] [PubMed] [Google Scholar]
- Grosso AR, Martins S, Carmo-Fonseca M 2008. The emerging role of splicing factors in cancer. EMBO Rep 9: 1087–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guil S, Caceres JF 2007. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol 14: 591–596 [DOI] [PubMed] [Google Scholar]
- Guil S, de La Iglesia N, Fernandez-Larrea J, Cifuentes D, Ferrer JC, Guinovart JJ, Bach-Elias M 2003a. Alternative splicing of the human proto-oncogene c-H-ras renders a new Ras family protein that trafficks to cytoplasm and nucleus. Cancer Res 63: 5178–5187 [PubMed] [Google Scholar]
- Guil S, Gattoni R, Carrascal M, Abian J, Stevenin J, Bach-Elias M 2003b. Roles of hnRNP A1, SR proteins, and p68 helicase in c-H-ras alternative splicing regulation. Mol Cell Biol 23: 2927–2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunthert U, Hofmann M, Rudy W, Reber S, Zoller M, Haussmann I, Matzku S, Wenzel A, Ponta H, Herrlich P 1991. A new variant of glycoprotein CD44 confers metastatic potential to rat carcinoma cells. Cell 65: 13–24 [DOI] [PubMed] [Google Scholar]
- Harper SJ, Bates DO 2008. VEGF-A splicing: The key to anti-angiogenic therapeutics? Nat Rev Cancer 8: 880–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Brown MA, Rothnagel JA, Saunders NA, Smith R 2005. Roles of heterogeneous nuclear ribonucleoproteins A and B in cell proliferation. J Cell Sci 118: 3173–3183 [DOI] [PubMed] [Google Scholar]
- He X, Pool M, Darcy KM, Lim SB, Auersperg N, Coon JS, Beck WT 2007. Knockdown of polypyrimidine tract-binding protein suppresses ovarian tumor cell growth and invasiveness in vitro. Oncogene 26: 4961–4968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman MN, Lou H 2008. Diverse molecular functions of Hu proteins. Cell Mol Life Sci 65: 3168–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho LH, Taylor R, Dorstyn L, Cakouros D, Bouillet P, Kumar S 2009. A tumor suppressor function for caspase-2. Proc Natl Acad Sci 106: 5336–5341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou VC, Lersch R, Gee SL, Ponthier JL, Lo AJ, Wu M, Turck CW, Koury M, Krainer AR, Mayeda A, et al. 2002. Decrease in hnRNP A/B expression during erythropoiesis mediates a pre-mRNA splicing switch. EMBO J 21: 6195–6204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iseni F, Garcin D, Nishio M, Kedersha N, Anderson P, Kolakofsky D 2002. Sendai virus trailer RNA binds TIAR, a cellular protein involved in virus-induced apoptosis. EMBO J 21: 5141–5150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru D, Ramalingam S, Sengupta TK, Bandyopadhyay S, Dellis S, Tholanikunnel BG, Fernandes DJ, Spicer EK 2009. Regulation of Bcl-2 expression by HuR in HL60 leukemia cells and A431 carcinoma cells. Mol Cancer Res 7: 1354–1366 [DOI] [PubMed] [Google Scholar]
- Izquierdo JM 2008. Hu antigen R (HuR) functions as an alternative pre-mRNA splicing regulator of Fas apoptosis-promoting receptor on exon definition. J Biol Chem 283: 19077–19084 [DOI] [PubMed] [Google Scholar]
- Izquierdo JM, Valcarcel J 2007. Fas-activated serine/threonine kinase (FAST K) synergizes with TIA-1/TIAR proteins to regulate Fas alternative splicing. J Biol Chem 282: 1539–1543 [DOI] [PubMed] [Google Scholar]
- Izquierdo JM, Majos N, Bonnal S, Martinez C, Castelo R, Guigo R, Bilbao D, Valcarcel J 2005. Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol Cell 19: 475–484 [DOI] [PubMed] [Google Scholar]
- Jang SM, Kim JW, Kim CH, Kim D, Rhee S, Choi KH 2010. p19ras represses proliferation of non-small cell lung cancer possibly through interaction with neuron-specific enolase (NSE). Cancer Lett 289: 91–98 [DOI] [PubMed] [Google Scholar]
- Jeong MH, Bae J, Kim WH, Yoo SM, Kim JW, Song PI, Choi KH 2006. p19ras interacts with and activates p73 by involving the MDM2 protein. J Biol Chem 281: 8707–8715 [DOI] [PubMed] [Google Scholar]
- Jiang ZH, Zhang WJ, Rao Y, Wu JY 1998. Regulation of Ich-1 pre-mRNA alternative splicing and apoptosis by mammalian splicing factors. Proc Natl Acad Sci 95: 9155–9160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Bruno IG, Xie TX, Sanger LJ, Cote GJ 2003. Polypyrimidine tract-binding protein down-regulates fibroblast growth factor receptor 1 α-exon inclusion. Cancer Res 63: 6154–6157 [PubMed] [Google Scholar]
- Jo OD, Martin J, Bernath A, Masri J, Lichtenstein A, Gera J 2008. Heterogeneous nuclear ribonucleoprotein A1 regulates cyclin D1 and c-myc internal ribosome entry site function through Akt signaling. J Biol Chem 283: 23274–23287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joeng KS, Song EJ, Lee KJ, Lee J 2004. Long lifespan in worms with long telomeric DNA. Nat Genet 36: 607–611 [DOI] [PubMed] [Google Scholar]
- Jones RG, Thompson CB 2009. Tumor suppressors and cell metabolism: A recipe for cancer growth. Genes Dev 23: 537–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P, Brazao R, Boavida MG, Gespach C, Chastre E 1999. Cloning of a novel human Rac1b splice variant with increased expression in colorectal tumors. Oncogene 18: 6835–6839 [DOI] [PubMed] [Google Scholar]
- Kamma H, Fujimoto M, Fujiwara M, Matsui M, Horiguchi H, Hamasaki M, Satoh H 2001. Interaction of hnRNP A2/B1 isoforms with telomeric ssDNA and the in vitro function. Biochem Biophys Res Commun 280: 625–630 [DOI] [PubMed] [Google Scholar]
- Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR 2007. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol 14: 185–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karni R, Hippo Y, Lowe SW, Krainer AR 2008. The splicing-factor oncoprotein SF2/ASF activates mTORC1. Proc Natl Acad Sci 105: 15323–15327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima T, Manley JL 2003. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat Genet 34: 460–463 [DOI] [PubMed] [Google Scholar]
- Kashima T, Rao N, Manley JL 2007. An intronic element contributes to splicing repression in spinal muscular atrophy. Proc Natl Acad Sci 104: 3426–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Lal A, Yang X, Galban S, Mazan-Mamczarz K, Gorospe M 2006. Translational control of cytochrome c by RNA-binding proteins TIA-1 and HuR. Mol Cell Biol 26: 3295–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura H, Li X, Harper SJ, Bates DO, Claesson-Welsh L 2008. Vascular endothelial growth factor (VEGF)-A165b is a weak in vitro agonist for VEGF receptor-2 due to lack of coreceptor binding and deficient regulation of kinase activity. Cancer Res 68: 4683–4692 [DOI] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J Cell Biol 147: 1431–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keely PJ, Westwick JK, Whitehead IP, Der CJ, Parise LV 1997. Cdc42 and Rac1 induce integrin-mediated cell motility and invasiveness through PI(3)K. Nature 390: 632–636 [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D 2002. The Human Genome Browser at UCSC. Genome Res 12: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheradmand F, Werner E, Tremble P, Symons M, Werb Z 1998. Role of Rac1 and oxygen radicals in collagenase-1 expression induced by cell shape change. Science 280: 898–902 [DOI] [PubMed] [Google Scholar]
- Kim JW, Zeller KI, Wang Y, Jegga AG, Aronow BJ, O'Donnell KA, Dang CV 2004. Evaluation of myc E-box phylogenetic footprints in glycolytic genes by chromatin immunoprecipitation assays. Mol Cell Biol 24: 5923–5936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Kuwano Y, Zhan M, Pullmann R Jr, Mazan-Mamczarz K, Li H, Kedersha N, Anderson P, Wilce MC, Gorospe M, et al. 2007. Elucidation of a C-rich signature motif in target mRNAs of RNA-binding protein TIAR. Mol Cell Biol 27: 6806–6817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitevska T, Spencer DM, Hawkins CJ 2009. Caspase-2: Controversial killer or checkpoint controller? Apoptosis 14: 829–848 [DOI] [PubMed] [Google Scholar]
- Knoch KP, Meisterfeld R, Kersting S, Bergert H, Altkruger A, Wegbrod C, Jager M, Saeger HD, Solimena M 2006. cAMP-dependent phosphorylation of PTB1 promotes the expression of insulin secretory granule proteins in β cells. Cell Metab 3: 123–134 [DOI] [PubMed] [Google Scholar]
- Knudsen KE, Diehl JA, Haiman CA, Knudsen ES 2006. Cyclin D1: Polymorphism, aberrant splicing and cancer risk. Oncogene 25: 1620–1628 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Ishida J, Musashi M, Ota S, Yoshida T, Shimizu Y, Chuma M, Kawakami H, Asaka M, Tanaka J, et al. 2010. p53 transactivation is involved in the antiproliferative activity of the putative tumor suppressor RBM5. Int J Cancer doi: 10.1002/ijc.25345 [DOI] [PubMed] [Google Scholar]
- Kojima M, Asahi M, Kikuchi H, Hashimoto N, Noda M, Hoshimaru M 1998. Expression of Nedd2/ICH-1 (caspase-2) in the developing rat retina. Neurosci Res 31: 211–217 [DOI] [PubMed] [Google Scholar]
- Kondera-Anasz Z, Mielczarek-Palacz A, Sikora J 2005. Soluble Fas receptor and soluble Fas ligand in the serum of women with uterine tumors. Apoptosis 10: 1143–1149 [DOI] [PubMed] [Google Scholar]
- Konig H, Ponta H, Herrlich P 1998. Coupling of signal transduction to alternative pre-mRNA splicing by a composite splice regulator. EMBO J 17: 2904–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt AR 2005. Promoter usage and alternative splicing. Curr Opin Cell Biol 17: 262–268 [DOI] [PubMed] [Google Scholar]
- Kumar S 2009. Caspase 2 in apoptosis, the DNA damage response and tumour suppression: Enigma no more? Nat Rev Cancer 9: 897–903 [DOI] [PubMed] [Google Scholar]
- LaBranche H, Dupuis S, Ben-David Y, Bani MR, Wellinger RJ, Chabot B 1998. Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nat Genet 19: 199–202 [DOI] [PubMed] [Google Scholar]
- Lal A, Kawai T, Yang X, Mazan-Mamczarz K, Gorospe M 2005. Antiapoptotic function of RNA-binding protein HuR effected through prothymosin α. EMBO J 24: 1852–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeStourgeon WM, Beyer AL, Christensen ME, Walker BW, Poupore SM, Daniels LP 1978. The packaging proteins of core hnRNP particles and the maintenance of proliferative cell states. Cold Spring Harb Symp Quant Biol 42: 885–898 [DOI] [PubMed] [Google Scholar]
- Letai AG 2008. Diagnosing and exploiting cancer's addiction to blocks in apoptosis. Nat Rev Cancer 8: 121–132 [DOI] [PubMed] [Google Scholar]
- Li S, Xu H, Ding H, Huang Y, Cao X, Yang G, Li J, Xie Z, Meng Y, Li X, et al. 2009. Identification of an aptamer targeting hnRNP A1 by tissue slide-based SELEX. J Pathol 218: 327–336 [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB 2010. RNA processing and its regulation: Global insights into biological networks. Nat Rev Genet 11: 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JJ, Zakian VA 1994. Isolation and characterization of two Saccharomyces cerevisiae genes that encode proteins that bind to (TG1–3)n single strand telomeric DNA in vitro. Nucleic Acids Res 22: 4906–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JH, Wei S, Lamy T, Li Y, Epling-Burnette PK, Djeu JY, Loughran TP Jr 2002. Blockade of Fas-dependent apoptosis by soluble Fas in LGL leukemia. Blood 100: 1449–1453 [PubMed] [Google Scholar]
- Logette E, Wotawa A, Solier S, Desoche L, Solary E, Corcos L 2003. The human caspase-2 gene: Alternative promoters, pre-mRNA splicing and AUG usage direct isoform-specific expression. Oncogene 22: 935–946 [DOI] [PubMed] [Google Scholar]
- Lopez de Silanes I, Fan J, Yang X, Zonderman AB, Potapova O, Pizer ES, Gorospe M 2003. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene 22: 7146–7154 [DOI] [PubMed] [Google Scholar]
- Lopez de Silanes I, Galban S, Martindale JL, Yang X, Mazan-Mamczarz K, Indig FE, Falco G, Zhan M, Gorospe M 2005a. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol Cell Biol 25: 9520–9531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Silanes I, Lal A, Gorospe M 2005b. HuR: Post-transcriptional paths to malignancy. RNA Biol 2: 11–13 [DOI] [PubMed] [Google Scholar]
- Lu F, Gladden AB, Diehl JA 2003. An alternatively spliced cyclin D1 isoform, cyclin D1b, is a nuclear oncogene. Cancer Res 63: 7056–7061 [PubMed] [Google Scholar]
- Lu Y, Yao HP, Wang MH 2007. Multiple variants of the RON receptor tyrosine kinase: Biochemical properties, tumorigenic activities, and potential drug targets. Cancer Lett 257: 157–164 [DOI] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T 2010. Regulation of alternative splicing by histone modifications. Science 327: 996–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Zhao Y, Li Y, Lu H, He Y 2010. Relevance of Bcl-x expression in different types of endometrial tissues. J Exp Clin Cancer Res 29: 14 doi: 10.1186/1756-9966-29-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T 2007. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell 27: 435–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manabe R, Oh-e N, Sekiguchi K 1999. Alternatively spliced EDA segment regulates fibronectin-dependent cell cycle progression and mitogenic signal transduction. J Biol Chem 274: 5919–5924 [DOI] [PubMed] [Google Scholar]
- Manley JL, Krainer AR 2010. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev 24: 1073–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley JL, Tacke R 1996. SR proteins and splicing control. Genes Dev 10: 1569–1579 [DOI] [PubMed] [Google Scholar]
- Markovtsov V, Nikolic JM, Goldman JA, Turck CW, Chou MY, Black DL 2000. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol Cell Biol 20: 7463–7479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Contreras R, Fisette JF, Nasim FU, Madden R, Cordeau M, Chabot B 2006. Intronic binding sites for hnRNP A/B and hnRNP F/H proteins stimulate pre-mRNA splicing. PLoS Biol 4: e21 doi: 10.1371/journal.pbio.0040021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massiello A, Roesser JR, Chalfant CE 2006. SAP155 Binds to ceramide-responsive RNA cis-element 1 and regulates the alternative 5′ splice site selection of Bcl-x pre-mRNA. FASEB J 20: 1680–1682 [DOI] [PubMed] [Google Scholar]
- Matos P, Jordan P 2008. Increased Rac1b expression sustains colorectal tumor cell survival. Mol Cancer Res 6: 1178–1184 [DOI] [PubMed] [Google Scholar]
- Matter N, Herrlich P, Konig H 2002. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature 420: 691–695 [DOI] [PubMed] [Google Scholar]
- Mauger DM, Lin C, Garcia-Blanco MA 2008. hnRNP H and hnRNP F complex with Fox2 to silence fibroblast growth factor receptor 2 exon IIIc. Mol Cell Biol 28: 5403–5419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A, Krainer AR 1992. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell 68: 365–375 [DOI] [PubMed] [Google Scholar]
- Mayr C, Bartel DP 2009. Widespread shortening of 3′ UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurek S, Boschek CB, Hugo F, Eigenbrodt E 2005. Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin Cancer Biol 15: 300–308 [DOI] [PubMed] [Google Scholar]
- Mercatante DR, Mohler JL, Kole R 2002. Cellular response to an antisense-mediated shift of Bcl-x pre-mRNA splicing and antineoplastic agents. J Biol Chem 277: 49374–49382 [DOI] [PubMed] [Google Scholar]
- Merdzhanova G, Edmond V, De Seranno S, Van den Broeck A, Corcos L, Brambilla C, Brambilla E, Gazzeri S, Eymin B 2008. E2F1 controls alternative splicing pattern of genes involved in apoptosis through upregulation of the splicing factor SC35. Cell Death Differ 15: 1815–1823 [DOI] [PubMed] [Google Scholar]
- Michlewski G, Sanford JR, Caceres JF 2008. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol Cell 30: 179–189 [DOI] [PubMed] [Google Scholar]
- Minn AJ, Boise LH, Thompson CB 1996. Bcl-x(S) anatagonizes the protective effects of Bcl-x(L). J Biol Chem 271: 6306–6312 [DOI] [PubMed] [Google Scholar]
- Minoo P, Sullivan W, Solomon LR, Martin TE, Toft DO, Scott RE 1989. Loss of proliferative potential during terminal differentiation coincides with the decreased abundance of a subset of heterogeneous ribonuclear proteins. J Cell Biol 109: 1937–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell SA, Spriggs KA, Bushell M, Evans JR, Stoneley M, Le Quesne JP, Spriggs RV, Willis AE 2005. Identification of a motif that mediates polypyrimidine tract-binding protein-dependent internal ribosome entry. Genes Dev 19: 1556–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montuenga LM, Zhou J, Avis I, Vos M, Martinez A, Cuttitta F, Treston AM, Sunday M, Mulshine JL 1998. Expression of heterogeneous nuclear ribonucleoprotein A2/B1 changes with critical stages of mammalian lung development. Am J Respir Cell Mol Biol 19: 554–562 [DOI] [PubMed] [Google Scholar]
- Moran-Jones K, Grindlay J, Jones M, Smith R, Norman JC 2009. hnRNP A2 regulates alternative mRNA splicing of TP53INP2 to control invasive cell migration. Cancer Res 69: 9219–9227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motoyama K, Inoue H, Takatsuno Y, Tanaka F, Mimori K, Uetake H, Sugihara K, Mori M 2009. Over- and under-expressed microRNAs in human colorectal cancer. Int J Oncol 34: 1069–1075 [DOI] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR 2010. Expansion of the eukaryotic proteome by alternative splicing. Nature 463: 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak DG, Woolard J, Amin EM, Konopatskaya O, Saleem MA, Churchill AJ, Ladomery MR, Harper SJ, Bates DO 2008. Expression of pro- and anti-angiogenic isoforms of VEGF is differentially regulated by splicing and growth factors. J Cell Sci 121: 3487–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak DG, Amin EM, Rennel ES, Hoareau-Aveilla C, Gammons M, Damodoran G, Hagiwara M, Harper SJ, Woolard J, Ladomery MR, et al. 2010. Regulation of vascular endothelial growth factor (VEGF) splicing from pro-angiogenic to anti-angiogenic isoforms: A novel therapeutic strategy for angiogenesis. J Biol Chem 285: 5532–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh JJ, West AR, Fishbein MC, Slamon DJ 2002. A candidate tumor suppressor gene, H37, from the human lung cancer tumor suppressor locus 3p21.3. Cancer Res 62: 3207–3213 [PubMed] [Google Scholar]
- Oh JJ, Razfar A, Delgado I, Reed RA, Malkina A, Boctor B, Slamon DJ 2006. 3p21.3 tumor suppressor gene H37/Luca15/RBM5 inhibits growth of human lung cancer cells through cell cycle arrest and apoptosis. Cancer Res 66: 3419–3427 [DOI] [PubMed] [Google Scholar]
- Olopade OI, Adeyanju MO, Safa AR, Hagos F, Mick R, Thompson CB, Recant WM 1997. Overexpression of BCL-x protein in primary breast cancer is associated with high tumor grade and nodal metastases. Cancer J Sci Am 3: 230–237 [PubMed] [Google Scholar]
- Olshavsky NA, Comstock CE, Schiewer MJ, Augello MA, Hyslop T, Sette C, Zhang J, Parysek LM, Knudsen KE 2010. Identification of ASF/SF2 as a critical, allele-specific effector of the cyclin D1b oncogene. Cancer Res 70: 3975–3984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson AK, Dimberg A, Kreuger J, Claesson-Welsh L 2006. VEGF receptor signalling—In control of vascular function. Nat Rev Mol Cell Biol 7: 359–371 [DOI] [PubMed] [Google Scholar]
- Oltean S, Sorg BS, Albrecht T, Bonano VI, Brazas RM, Dewhirst MW, Garcia-Blanco MA 2006. Alternative inclusion of fibroblast growth factor receptor 2 exon IIIc in Dunning prostate tumors reveals unexpected epithelial mesenchymal plasticity. Proc Natl Acad Sci 103: 14116–14121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omenn GS, Yocum AK, Menon R 2010. Alternative splice variants, a new class of protein cancer biomarker candidates: Findings in pancreatic cancer and breast cancer with systems biology implications. Dis Markers 28: 241–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orian-Rousseau V, Chen L, Sleeman JP, Herrlich P, Ponta H 2002. CD44 is required for two consecutive steps in HGF/c-Met signaling. Genes Dev 16: 3074–3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paronetto MP, Achsel T, Massiello A, Chalfant CE, Sette C 2007. The RNA-binding protein Sam68 modulates the alternative splicing of Bcl-x. J Cell Biol 176: 929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paronetto MP, Cappellari M, Busa R, Pedrotti S, Vitali R, Comstock C, Hyslop T, Knudsen KE, Sette C 2010. Alternative splicing of the cyclin D1 proto-oncogene is regulated by the RNA-binding protein Sam68. Cancer Res 70: 229–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel NA, Kaneko S, Apostolatos HS, Bae SS, Watson JE, Davidowitz K, Chappell DS, Birnbaum MJ, Cheng JQ, Cooper DR 2005. Molecular and genetic studies imply Akt-mediated signaling promotes protein kinase CβII alternative splicing via phosphorylation of serine/arginine-rich splicing factor SRp40. J Biol Chem 280: 14302–14309 [DOI] [PubMed] [Google Scholar]
- Patry C, Bouchard L, Labrecque P, Gendron D, Lemieux B, Toutant J, Lapointe E, Wellinger R, Chabot B 2003. Small interfering RNA-mediated reduction in heterogeneous nuclear ribonucleoparticule A1/A2 proteins induces apoptosis in human cancer cells but not in normal mortal cell lines. Cancer Res 63: 7679–7688 [PubMed] [Google Scholar]
- Perona R, Montaner S, Saniger L, Sanchez-Perez I, Bravo R, Lacal JC 1997. Activation of the nuclear factor-κB by Rho, CDC42, and Rac-1 proteins. Genes Dev 11: 463–475 [DOI] [PubMed] [Google Scholar]
- Pettus BJ, Chalfant CE, Hannun YA 2002. Ceramide in apoptosis: An overview and current perspectives. Biochim Biophys Acta 1585: 114–125 [DOI] [PubMed] [Google Scholar]
- Piecyk M, Wax S, Beck AR, Kedersha N, Gupta M, Maritim B, Chen S, Gueydan C, Kruys V, Streuli M, et al. 2000. TIA-1 is a translational silencer that selectively regulates the expression of TNF-α. EMBO J 19: 4154–4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pino I, Pio R, Toledo G, Zabalegui N, Vicent S, Rey N, Lozano MD, Torre W, Garcia-Foncillas J, Montuenga LM 2003. Altered patterns of expression of members of the heterogeneous nuclear ribonucleoprotein (hnRNP) family in lung cancer. Lung Cancer 41: 131–143 [DOI] [PubMed] [Google Scholar]
- Planck SR, Listerud MD, Buckley SD 1988. Modulation of hnRNP A1 protein gene expression by epidermal growth factor in Rat-1 cells. Nucleic Acids Res 16: 11663–11673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponta H, Sherman L, Herrlich PA 2003. CD44: From adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol 4: 33–45 [DOI] [PubMed] [Google Scholar]
- Pritchard-Jones RO, Dunn DB, Qiu Y, Varey AH, Orlando A, Rigby H, Harper SJ, Bates DO 2007. Expression of VEGF(xxx)b, the inhibitory isoforms of VEGF, in malignant melanoma. Br J Cancer 97: 223–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu RG, Chen J, Kirn D, McCormick F, Symons M 1995. An essential role for Rac in Ras transformation. Nature 374: 457–459 [DOI] [PubMed] [Google Scholar]
- Radisky DC, Levy DD, Littlepage LE, Liu H, Nelson CM, Fata JE, Leake D, Godden EL, Albertson DG, Nieto MA, et al. 2005. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature 436: 123–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S, Ross KN, Lander ES, Golub TR 2003. A molecular signature of metastasis in primary solid tumors. Nat Genet 33: 49–54 [DOI] [PubMed] [Google Scholar]
- Reyes R, Alcalde J, Izquierdo JM 2009. Depletion of T-cell intracellular antigen proteins promotes cell proliferation. Genome Biol 10: R87 doi: 10.1186/gb-2009-10-8-r87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rintala-Maki ND, Sutherland LC 2004. LUCA-15/RBM5, a putative tumour suppressor, enhances multiple receptor-initiated death signals. Apoptosis 9: 475–484 [DOI] [PubMed] [Google Scholar]
- Rintala-Maki ND, Goard CA, Langdon CE, Wall VE, Traulsen KE, Morin CD, Bonin M, Sutherland LC 2007. Expression of RBM5-related factors in primary breast tissue. J Cell Biochem 100: 1440–1458 [DOI] [PubMed] [Google Scholar]
- Rybak JN, Roesli C, Kaspar M, Villa A, Neri D 2007. The extra-domain A of fibronectin is a vascular marker of solid tumors and metastases. Cancer Res 67: 10948–10957 [DOI] [PubMed] [Google Scholar]
- Sanchez G, Bittencourt D, Laud K, Barbier J, Delattre O, Auboeuf D, Dutertre M 2008. Alteration of cyclin D1 transcript elongation by a mutated transcription factor up-regulates the oncogenic D1b splice isoform in cancer. Proc Natl Acad Sci 105: 6004–6009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicka K, Bushell M, Spriggs KA, Willis AE 2008. Polypyrimidine-tract-binding protein: A multifunctional RNA-binding protein. Biochem Soc Trans 36: 641–647 [DOI] [PubMed] [Google Scholar]
- Schnelzer A, Prechtel D, Knaus U, Dehne K, Gerhard M, Graeff H, Harbeck N, Schmitt M, Lengyel E 2000. Rac1 in human breast cancer: Overexpression, mutation analysis, and characterization of a new isoform, Rac1b. Oncogene 19: 3013–3020 [DOI] [PubMed] [Google Scholar]
- Schwerk C, Schulze-Osthoff K 2005. Regulation of apoptosis by alternative pre-mRNA splicing. Mol Cell 19: 1–13 [DOI] [PubMed] [Google Scholar]
- Sheen-Chen SM, Chen HS, Eng HL, Chen WJ 2003. Circulating soluble Fas in patients with breast cancer. World J Surg 27: 10–13 [DOI] [PubMed] [Google Scholar]
- Shepard PJ, Hertel KJ 2009. The SR protein family. Genome Biol 10: 242 doi: 10.1186/gb-2009-10-10-242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Reddy B, Manley JL 2006. PP1/PP2A phosphatases are required for the second step of Pre-mRNA splicing and target specific snRNP proteins. Mol Cell 23: 819–829 [DOI] [PubMed] [Google Scholar]
- Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, Dalla-Favera R, Dang CV 1997. c-Myc transactivation of LDH-A: Implications for tumor metabolism and growth. Proc Natl Acad Sci 94: 6658–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvera D, Formenti SC, Schneider RJ 2010. Translational control in cancer. Nat Rev Cancer 10: 254–266 [DOI] [PubMed] [Google Scholar]
- Singh A, Karnoub AE, Palmby TR, Lengyel E, Sondek J, Der CJ 2004. Rac1b, a tumor associated, constitutively active Rac1 splice variant, promotes cellular transformation. Oncogene 23: 9369–9380 [DOI] [PubMed] [Google Scholar]
- Skotheim RI, Nees M 2007. Alternative splicing in cancer: Noise, functional, or systematic? Int J Biochem Cell Biol 39: 1432–1449 [DOI] [PubMed] [Google Scholar]
- Solier S, Logette E, Desoche L, Solary E, Corcos L 2005. Nonsense-mediated mRNA decay among human caspases: The caspase-2S putative protein is encoded by an extremely short-lived mRNA. Cell Death Differ 12: 687–689 [DOI] [PubMed] [Google Scholar]
- Solomon DA, Wang Y, Fox SR, Lambeck TC, Giesting S, Lan Z, Senderowicz AM, Conti CJ, Knudsen ES 2003. Cyclin D1 splice variants. Differential effects on localization, RB phosphorylation, and cellular transformation. J Biol Chem 278: 30339–30347 [DOI] [PubMed] [Google Scholar]
- Sorek R, Ast G 2003. Intronic sequences flanking alternatively spliced exons are conserved between human and mouse. Genome Res 13: 1631–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland LC, Edwards SE, Cable HC, Poirier GG, Miller BA, Cooper CS, Williams GT 2000. LUCA-15-encoded sequence variants regulate CD95-mediated apoptosis. Oncogene 19: 3774–3781 [DOI] [PubMed] [Google Scholar]
- Sutherland LC, Wang K, Robinson AG 2010. RBM5 as a putative tumor suppressor gene for lung cancer. J Thorac Oncol 5: 294–298 [DOI] [PubMed] [Google Scholar]
- Takehara T, Liu X, Fujimoto J, Friedman SL, Takahashi H 2001. Expression and role of Bcl-xL in human hepatocellular carcinomas. Hepatology 34: 55–61 [DOI] [PubMed] [Google Scholar]
- Taylor SJ, Resnick RJ, Shalloway D 2004. Sam68 exerts separable effects on cell cycle progression and apoptosis. BMC Cell Biol 5: 5 doi: 10.1186/1471-2121-5-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA 2009. Epithelial–mesenchymal transitions in development and disease. Cell 139: 871–890 [DOI] [PubMed] [Google Scholar]
- Tian Q, Streuli M, Saito H, Schlossman SF, Anderson P 1991. A polyadenylate binding protein localized to the granules of cytolytic lymphocytes induces DNA fragmentation in target cells. Cell 67: 629–639 [DOI] [PubMed] [Google Scholar]
- Turner N, Grose R 2010. Fibroblast growth factor signalling: From development to cancer. Nat Rev Cancer 10: 116–129 [DOI] [PubMed] [Google Scholar]
- Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB 2006. An RNA map predicting Nova-dependent splicing regulation. Nature 444: 580–586 [DOI] [PubMed] [Google Scholar]
- Ushigome M, Ubagai T, Fukuda H, Tsuchiya N, Sugimura T, Takatsuka J, Nakagama H 2005. Up-regulation of hnRNP A1 gene in sporadic human colorectal cancers. Int J Oncol 26: 635–640 [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB 2009. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 324: 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables JP 2004. Aberrant and alternative splicing in cancer. Cancer Res 64: 7647–7654 [DOI] [PubMed] [Google Scholar]
- Venables JP 2006. Unbalanced alternative splicing and its significance in cancer. Bioessays 28: 378–386 [DOI] [PubMed] [Google Scholar]
- Venables JP, Koh CS, Froehlich U, Lapointe E, Couture S, Inkel L, Bramard A, Paquet ER, Watier V, Durand M, et al. 2008. Multiple and specific mRNA processing targets for the major human hnRNP proteins. Mol Cell Biol 28: 6033–6043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables JP, Klinck R, Koh C, Gervais-Bird J, Bramard A, Inkel L, Durand M, Couture S, Froehlich U, Lapointe E, et al. 2009. Cancer-associated regulation of alternative splicing. Nat Struct Mol Biol 16: 670–676 [DOI] [PubMed] [Google Scholar]
- Wagh PK, Peace BE, Waltz SE 2008. Met-related receptor tyrosine kinase Ron in tumor growth and metastasis. Adv Cancer Res 100: 1–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R 2009. The spliceosome: Design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Wang T, Marquardt C, Foker J 1976. Aerobic glycolysis during lymphocyte proliferation. Nature 261: 702–705 [DOI] [PubMed] [Google Scholar]
- Wang L, Miura M, Bergeron L, Zhu H, Yuan J 1994. Ich-1, an Ice/ced-3-related gene, encodes both positive and negative regulators of programmed cell death. Cell 78: 739–750 [DOI] [PubMed] [Google Scholar]
- Wang F, Kan M, Yan G, Xu J, McKeehan WL 1995. Alternately spliced NH2-terminal immunoglobulin-like Loop I in the ectodomain of the fibroblast growth factor (FGF) receptor 1 lowers affinity for both heparin and FGF-1. J Biol Chem 270: 10231–10235 [DOI] [PubMed] [Google Scholar]
- Wang W, Caldwell MC, Lin S, Furneaux H, Gorospe M 2000. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J 19: 2340–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Miller LD, Ohnmacht GA, Mocellin S, Perez-Diez A, Petersen D, Zhao Y, Simon R, Powell JI, Asaki E, et al. 2002. Prospective molecular profiling of melanoma metastases suggests classifiers of immune responsiveness. Cancer Res 62: 3581–3586 [PMC free article] [PubMed] [Google Scholar]
- Wang D, Shen Q, Chen YQ, Wang MH 2004. Collaborative activities of macrophage-stimulating protein and transforming growth factor-β1 in induction of epithelial to mesenchymal transition: Roles of the RON receptor tyrosine kinase. Oncogene 23: 1668–1680 [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Dean JL, Millar EK, Tran TH, McNeil CM, Burd CJ, Henshall SM, Utama FE, Witkiewicz A, Rui H, et al. 2008. Cyclin D1b is aberrantly regulated in response to therapeutic challenge and promotes resistance to estrogen antagonists. Cancer Res 68: 5628–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O 1956. On the origin of cancer cells. Science 123: 309–314 [DOI] [PubMed] [Google Scholar]
- Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP 2009. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell 33: 591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weg-Remers S, Ponta H, Herrlich P, Konig H 2001. Regulation of alternative pre-mRNA splicing by the ERK MAP-kinase pathway. EMBO J 20: 4194–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolard J, Wang WY, Bevan HS, Qiu Y, Morbidelli L, Pritchard-Jones RO, Cui TG, Sugiono M, Waine E, Perrin R, et al. 2004. VEGF165b, an inhibitory vascular endothelial growth factor splice variant: Mechanism of action, in vivo effect on angiogenesis and endogenous protein expression. Cancer Res 64: 7822–7835 [DOI] [PubMed] [Google Scholar]
- Wu S, Sato M, Endo C, Sakurada A, Dong B, Aikawa H, Chen Y, Okada Y, Matsumura Y, Sueoka E, et al. 2003. hnRNP B1 protein may be a possible prognostic factor in squamous cell carcinoma of the lung. Lung Cancer 41: 179–186 [DOI] [PubMed] [Google Scholar]
- Wu H, Sun S, Tu K, Gao Y, Xie B, Krainer AR, Zhu J 2010. A splicing-independent function of SF2/ASF in microRNA processing. Mol Cell 38: 67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xerri L, Parc P, Brousset P, Schlaifer D, Hassoun J, Reed JC, Krajewski S, Birnbaum D 1996. Predominant expression of the long isoform of Bcl-x (Bcl-xL) in human lymphomas. Br J Haematol 92: 900–906 [DOI] [PubMed] [Google Scholar]
- Xie J, Lee JA, Kress TL, Mowry KL, Black DL 2003. Protein kinase A phosphorylation modulates transport of the polypyrimidine tract-binding protein. Proc Natl Acad Sci 100: 8776–8781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon YS, Zhang C, Yeo G, Black DL, Sun H, et al. 2009. Genome-wide analysis of PTB–RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell 36: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi F, Saya H, Bruner JM, Morrison RS 1994. Differential expression of two fibroblast growth factor-receptor genes is associated with malignant progression in human astrocytomas. Proc Natl Acad Sci 91: 484–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan G, Fukabori Y, McBride G, Nikolaropolous S, McKeehan WL 1993. Exon switching and activation of stromal and embryonic fibroblast growth factor (FGF)-FGF receptor genes in prostate epithelial cells accompany stromal independence and malignancy. Mol Cell Biol 13: 4513–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan-Sanders Y, Hammons GJ, Lyn-Cook BD 2002. Increased expression of heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNP) in pancreatic tissue from smokers and pancreatic tumor cells. Cancer Lett 183: 215–220 [DOI] [PubMed] [Google Scholar]
- Yasumoto H, Matsubara A, Mutaguchi K, Usui T, McKeehan WL 2004. Restoration of fibroblast growth factor receptor2 suppresses growth and tumorigenicity of malignant human prostate carcinoma PC-3 cells. Prostate 61: 236–242 [DOI] [PubMed] [Google Scholar]
- Zerbe LK, Pino I, Pio R, Cosper PF, Dwyer-Nield LD, Meyer AM, Port JD, Montuenga LM, Malkinson AM 2004. Relative amounts of antagonistic splicing factors, hnRNP A1 and ASF/SF2, change during neoplastic lung growth: Implications for pre-mRNA processing. Mol Carcinog 41: 187–196 [DOI] [PubMed] [Google Scholar]
- Zhang QS, Manche L, Xu RM, Krainer AR 2006. hnRNP A1 associates with telomere ends and stimulates telomerase activity. RNA 12: 1116–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Allred DC, Avis I, Martinez A, Vos MD, Smith L, Treston AM, Mulshine JL 2001. Differential expression of the early lung cancer detection marker, heterogeneous nuclear ribonucleoprotein-A2/B1 (hnRNP-A2/B1) in normal breast and neoplastic breast cancer. Breast Cancer Res Treat 66: 217–224 [DOI] [PubMed] [Google Scholar]
- Zhou YQ, He C, Chen YQ, Wang D, Wang MH 2003. Altered expression of the RON receptor tyrosine kinase in primary human colorectal adenocarcinomas: Generation of different splicing RON variants and their oncogenic potential. Oncogene 22: 186–197 [DOI] [PubMed] [Google Scholar]