Abstract
The U2AF35-related protein Urp has been implicated previously in splicing of the major class of U2-type introns. Here we show that Urp is also required for splicing of the minor class of U12-type introns. Urp is recruited in an ATP-dependent fashion to the U12-type intron 3′ splice site, where it promotes formation of spliceosomal complexes. Remarkably, Urp also contacts the 3′ splice site of a U2-type intron, but in this case is specifically required for the second step of splicing. Thus, through recognition of a common splicing element, Urp facilitates distinct steps of U2- and U12-type intron splicing.
Keywords: Urp, U2AF35, splicing, spliceosome assembly, U2-type intron, U12-type intron
Pre-mRNA splicing occurs in a dynamic ribonucleoprotein (RNP) complex termed the spliceosome, which is composed of numerous proteins and small nuclear RNP particles (snRNPs). For the major class of introns, called U2-type introns, splicing occurs in a spliceosome that contains four U snRNPs: U1, U2, U5, and U4/U6 snRNPs (Black 2003). A small subset of introns, called U12-type introns, is spliced through the conventional two-step pathway, but by a different spliceosome that contains U5, U11, and U12 snRNPs, and an alternative form of U4/U6 snRNP called U4atac/U6atac (Will and Luhrmann 2005). On U12-type introns, the 5′ and 3′ splice sites and branchpoint are highly conserved and differ from those of the conventional U2-type introns (Hall and Padgett 1994; Sharp and Burge 1997), and the characteristic polypyrimidine (Py) tract is typically absent (Burge et al. 1998).
Spliceosome assembly of U2-type introns is initiated by binding of U2AF to the Py tract/3′ splice site. U2AF is a heterodimer composed of a large (65-kDa) and a small (35-kDa) subunit (Zamore et al. 1992). The large subunit, U2AF65, binds specifically to the Py tract, whereas the small subunit, U2AF35, contacts the AG dinucleotide at the 3′ splice site (Merendino et al. 1999; Wu et al. 1999; Zorio and Blumenthal 1999). This contact results, at least in part, from a sequence-specific RNA-binding activity of U2AF35 that recognizes the 3′ splice site AG. For introns with weak Py tracts, the U2AF35–3′ splice site interaction is critical for U2AF binding and splicing.
Genome sequence analysis and expression studies have revealed the existence of several U2AF35-related proteins (U2AF35-RPs). Each U2AF35-RP contains a common core, consisting of a noncanonical RNA recognition motif called the U2AF homology motif (UHM) (Kielkopf et al. 2004) and two flanking zinc finger domains, but differs at the N and/or C termini. In mammals, there are at least three U2AF35-RPs: U2AF26 and two highly similar proteins, U2AF1-RS1 and U2AF1-RS2 (also called Urp) (Kitagawa et al. 1995; Tronchere et al. 1997). U2AF26 is nearly identical to U2AF35, but lacks the C-terminal arginine–serine-rich (RS) domain that is present in U2AF35 (Shepard et al. 2002). U2AF26 associates with U2AF65 and can functionally substitute for U2AF35 in both constitutive and enhancer-dependent splicing (Graveley et al. 2001; Shepard et al. 2002), suggesting that U2AF26 and U2AF35 have overlapping functions. In contrast, Urp does not appear to be functionally redundant with U2AF35, as biochemical complementation experiments have revealed that U2AF35 cannot substitute for Urp in U2-type intron splicing in Urp-depleted extracts (Tronchere et al. 1997). Like U2AF35, Urp contains a C-terminal RS domain, but also possesses an additional acidic N-terminal domain that is not found in U2AF35. In addition, like U2AF35, Urp can interact with U2AF65 (Tronchere et al. 1997). However, the precise role of Urp in pre-mRNA splicing is not known. Here we analyze the role of Urp in splicing of U2- and U12-type introns.
Results and Discussion
Urp is required for splicing of U12-type introns
We observed that there was a perfect phylogenetic relationship between organisms that have Urp and also U12-type splicing (Table 1), raising the possibility that Urp was a U12-type intron splicing factor. To perform functional experiments, we raised a polyclonal Urp antibody against a unique sequence in the N terminus. The anti-Urp antibody detected a 66-kDa polypeptide, consistent with the expected size of Urp, in HeLa cells (Supplemental Fig. 1). Moreover, this 66-kDa band was not detected following RNAi-mediated knockdown of Urp using two different Urp siRNAs.
Table 1.
Phylogenetic comparison of organisms for U2AF35, Urp, and U12-type splicing
The presence (●) or absence (—) of U12-type introns was assigned based on previous studies that had identified minor spliceosomal proteins, snRNAs, and/or introns in a variety of species (Russell et al. 2006). BLAST searches of the NCBI GenBank database were performed to identify novel Urp homologs (see the Materials and Methods for details).
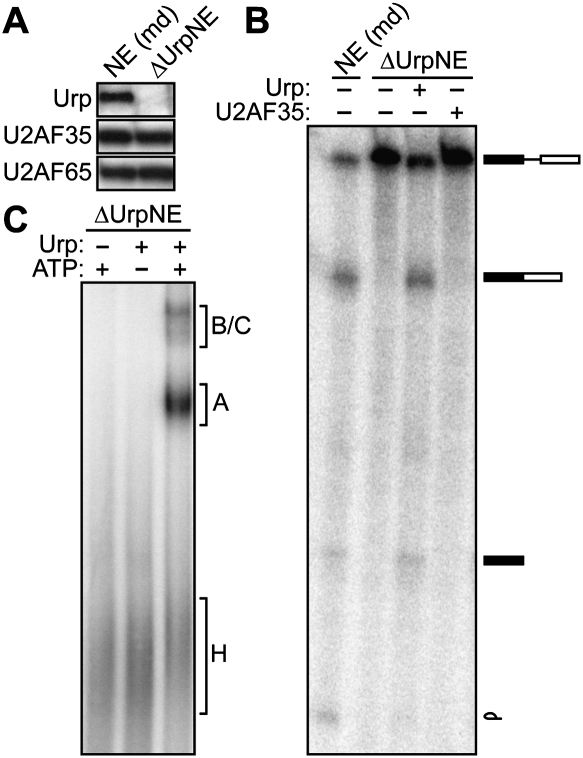
The anti-Urp antibody was used to prepare an Urp-depleted HeLa nuclear extract (ΔUrpNE). Figure 1A confirms that the ΔUrpNE lacks Urp but contains normal levels of U2AF35 and U2AF65. We then analyzed the ability of the ΔUrpNE to support splicing of the well-characterized P120 pre-mRNA substrate, which contains a U12-type intron. Figure 1B shows that P120 was spliced in the HeLa NE but not in the ΔUrpNE. Splicing of P120 was also not detectable in the ΔUrpNE following extended incubation (Supplemental Fig. 2). Addition of recombinant Urp to the ΔUrpNE restored its ability to support splicing of P120 (Fig. 1B). In contrast, addition of recombinant U2AF35—which, as described above, is present at normal levels in the ΔUrpNE—failed to restore P120 splicing, indicating that Urp and U2AF35 are not functionally redundant for U12-type intron splicing. Consistent with these in vitro splicing results, RNAi experiments indicate that both U2AF35 and Urp are required for cell viability (Supplemental Fig. 3), which provides further evidence that the two proteins are not functionally redundant. Finally, a U2AF35-depleted extract, which lacked U2AF35 but contained normal levels of Urp, supported P120 splicing (Supplemental Fig. 4). Thus, Urp is an essential factor for U12-type intron splicing.
Figure 1.
Urp is required for splicing and splicing complex assembly of a U12-type intron. (A) Immunoblot analysis showing Urp, U2AF35, and U2AF65 protein levels in mock-depleted HeLa NE [NE (md)] and in ΔUrpNE. (B) Conventional splicing assay. Urp and, as a control, U2AF35 were analyzed for their ability to complement splicing of the P120 pre-mRNA substrate in the ΔUrpNE. Splicing of P120 in NE (md) is shown as a control. Identities of spliced products are shown on the right. (C) Splicing complex assembly. Urp was analyzed for its ability to support assembly of the prespliceosome (complex A) and mature spliceosome (complexes B and C) on the P120 pre-mRNA substrate in the ΔUrpNE.
We next analyzed the role of Urp in the assembly of splicing complexes on the P120 pre-mRNA substrate using a native gel assay. Figure 1C shows that, in the ΔUrpNE, the P120 pre-mRNA was not assembled into either the prespliceosome (complex A) or spliceosome (complex B/C), and only the nonspecific H complex was detectable. Following addition of recombinant Urp to the ΔUrpNE, P120 was assembled into both prespliceosomal and spliceosomal complexes. Thus, Urp acts early to promote assembly of spliceosomal complexes.
Urp is a component of the U12-type spliceosome and specifically contacts the 3′ splice site
The requirement of Urp for P120 splicing and spliceosome assembly strongly suggested that Urp would be recruited into P120 splicing complexes. To confirm this possibility, a biotinylated P120 substrate was added to a HeLa NE under splicing conditions and affinity-purified on streptavidin-agarose, and the presence of Urp and other splicing factors was detected by immunoblotting. For comparison, a biotinylated adenovirus major late (Ad ML) pre-mRNA substrate, which contains a U2-type intron, was analyzed in parallel. Figure 2A shows, as expected, that the U11/U12 35-kDa snRNP protein, a U12-type splicing component, was present in splicing complexes formed on the P120 pre-mRNA but not the Ad ML pre-mRNA. Likewise, Urp was present in P120 but not Ad ML splicing complexes. Significantly, U2AF65 was not detected in P120 splicing complexes, which is consistent with our previous finding that U2AF65 is not required for U12-type intron splicing (Shen and Green 2007).
Figure 2.
Urp is a component of the U12-type spliceosome and selectively contacts the 3′ splice site. (A) Affinity purification of P120 splicing complexes. Biotin-labeled P120 and Ad ML pre-mRNAs were purified using streptavidin, and the association of Urp, U11/U12 snRNP 35K (U1SNRNPBP), and U2AF65 was monitored by immunoblot analysis. (B) UV-cross-linking analysis. The P120 substrate was uniformly labeled and incubated with NE (md) or ΔUrpNE under splicing conditions. Following cross-linking and RNase treatment, Urp was immunoprecipitated. The immunoprecipitate was analyzed by SDS-PAGE, and the presence of P120 was detected by PhosphorImager analysis. (C) UV-cross-linking analysis of the Urp–U12 intron interaction using the P120 substrate labeled with a single 32P at the 5′ splice site (5′ss), branchpoint (BP), 3′ splice site (3′ss), or exon 2. (D) UV-cross-linking analysis of the Urp–3′ splice site interaction in the absence of U11 or U12 snRNA. (E) UV-cross-linking analysis monitoring the ability of Urp to bind a series of P120 3′ splice site derivatives in which the AC dinucleotide was mutated to either AU, AA, AG, or UC.
To determine whether Urp contacted the P120 pre-mRNA substrate, we used a UV-cross-linking assay. In brief, a uniformly 32P-labeled P120 substrate was incubated in HeLa NE under splicing conditions, the reaction mixture was irradiated with UV light to induce RNA–protein cross-links and treated with RNase, and, following immunoprecipitation with the anti-Urp antibody, the presence of 32P-tagged Urp in the immunoprecipitate was detected by SDS-PAGE and PhosphorImager analysis. Figure 2B shows that Urp directly contacted the P120 pre-mRNA in the HeLa NE and in the ΔUrpNE complemented with recombinant Urp. Unexpectedly, however, we found that interaction of Urp with the P120 pre-mRNA was ATP-dependent.
The UV-cross-linking experiment described above indicates that Urp contacts the pre-mRNA, but does not reveal whether this interaction occurs at a specific splicing element. We therefore prepared a series of P120 derivatives that were site-specifically labeled at either the 5′ splice site, the branchpoint, the 3′ splice site, or exon 2. The UV-cross-linking assay of Figure 2C shows that Urp selectively contacted the P120 3′ splice site. As discussed above, splicing of U11 and U12-type introns requires U11 and U12 snRNAs. Figure 2D shows that, following oligonucleotide-directed RNase H degradation of either U11 or U12 snRNA, the Urp–P120 3′ splice site interaction did not occur.
Previous studies have shown that mutating the C of the 3′ splice site AC dinucleotide does not impair splicing of a U12-type intron, whereas mutation of the A is detrimental (Dietrich et al. 2001, 2005). To determine whether these splicing results correlated with binding of Urp to the 3′ splice site, we analyzed a series of P120 derivatives that have been characterized previously for splicing activity (Dietrich et al. 2001, 2005). The UV-cross-linking experiment of Figure 2E shows that mutation of the 3′ splice site AC dinucleotide to either AU, AA, or AG had no effect on binding of Urp. In contrast, mutation of the 3′ splice site AC dinucleotide to UC abolished Urp binding. These binding results correlate perfectly with the previously reported splicing activity of these P120 derivatives, further supporting the role of Urp in U12-type intron splicing.
Urp is required for the second step of U2-type splicing and selectively contacts the 3′ splice site
To determine whether Urp also has a role in splicing of U2-type introns, we analyzed the ability of the ΔUrpNE to support splicing of an Ad ML pre-mRNA substrate. Figure 3A shows, consistent with the original proposal of Tronchere et al. (1997), that the Ad ML pre-mRNA underwent the first step of splicing but was unable to undergo the second step, as evidenced by both an increased level of the intron–exon2 lariat intermediate and the absence of the fully spliced mRNA product. Ad ML second-step splicing products were also not detectable in the ΔUrpNE following extended incubation (Supplemental Fig. 5A). As expected, addition of recombinant Urp, but not U2AF35, restored the ability of the ΔUrpNE to support the second step of Ad ML pre-mRNA splicing (Fig. 3A). Moreover, Urp had no detectable effect on the Ad ML first-step splicing kinetics (Supplemental Fig. 5B). Consistent with these splicing results, splicing complex assembly on the Ad ML pre-mRNA occurred normally in the presence or absence of Urp (Fig. 3B).
Figure 3.
Urp is required for the second step of U2-type splicing. (A) Conventional splicing assay. Urp or, as a control, U2AF35 was analyzed for its ability to complement splicing of the Ad ML pre-mRNA substrate in the ΔUrpNE. Splicing of Ad ML in NE (md) is shown as a control. (B) Splicing complex assembly. Urp was analyzed for its ability to support assembly of the prespliceosome (complex A) and mature spliceosome (complexes B and C) on the Ad ML pre-RNA substrate in the ΔUrpNE. (C) Bimolecular ligation assay. Uniformly labeled 5′ RNA substrate was preincubated in the ΔUrpNE for 0 or 1 h. After 1 h of incubation, an excess of unlabeled 3′ RNA substrate was added in the presence or absence of Urp, followed by further incubation for 1 h. Samples were analyzed by denaturing PAGE.
To confirm that Urp is selectively required for the second step of U2-type intron splicing, we used a bimolecular exon ligation assay, which physically uncouples the first and second catalytic steps (Anderson and Moore 1997). The assay involves separate 5′ and 3′ RNA substrates. The 5′ RNA substrate contains an exon upstream of an intron lacking the 3′ splice site AG, and thus can undergo the first but not the second step of splicing. The 3′ RNA substrate comprises an intact 3′ splice site and a downstream exon.
In the experiment shown in Figure 3C, the 5′ RNA substrate was uniformly 32P-labeled and preincubated in the ΔUrpNE for 1 h under conditions that support splicing. As expected, in the absence of exogenous Urp, the 5′ RNA substrate underwent the first step of splicing, as evidenced by the appearance of the first exon and intron-containing lariat intermediates. Significantly, addition of Urp had no effect on the levels of these first-step splicing products, as expected if Urp is not involved in the first step of splicing. After 1 h of incubation, an excess of unlabeled 3′ RNA substrate was added in either the presence or absence of recombinant Urp. The results show that fully spliced product was detectable only when the 3′ RNA substrate was added with recombinant Urp.
To determine whether Urp selectively contacts a specific region of the Ad ML pre-mRNA, we performed a UV-cross-linking assay using a series of site-specifically labeled pre-mRNA substrates. Figure 4A shows that Urp selectively contacted the 3′ splice site of the Ad ML pre-mRNA.
Figure 4.
Urp selectively contacts the 3′ splice site of a U2-type intron. (A) UV-cross-linking assay using the Ad ML substrate labeled with a single 32P at the 5′ splice site (5′ss), branchpoint (BP), Py tract (PPT), 3′ splice site (3′ss), or exon 2. (B) Modified affinity purification assay. Biotin-labeled P120 and Ad ML pre-mRNAs were incubated with NE and then subjected to UV-cross-linking prior to streptavidin affinity purification. Association of Urp, U11/U12 snRNP 35K (U1SNRNPBP), and U2AF65 was monitored by immunoblot analysis. (C) UV-cross-linking assay using the Ad ML substrate, labeled as in A, in HeLa NE or heat-treated NE. (D) UV-cross-linking assay using an Ad ML substrate site-specifically labeled at the 3′ splice site following addition of untreated, recombinant Urp to a heat-treated NE (md) or ΔUrpNE. (E, top) Time-course analysis of splicing of the Ad ML pre-mRNA in the ΔUrpNE in the presence or absence of recombinant Urp. (Bottom) UV-cross-linking analysis monitoring binding of Urp, U2AF35, and U2AF65 to the Ad ML 3′ splice site.
The finding that Urp contacted the Ad ML 3′ splice site was somewhat unexpected in light of the apparent absence of Urp in affinity-purified spliceosomes formed on an Ad ML substrate (see Fig. 2A). A possible explanation for our ability to detect the association of Urp with the Ad ML pre-mRNA in a UV-cross-linking assay but not in an affinity purification assay is that Urp binding is relatively weak and therefore dissociates from the biotinylated pre-mRNA during the course of affinity purification. We therefore modified the affinity purification to include an initial UV-cross-linking step; in this modified affinity purification assay, the association of Urp with the Ad ML pre-mRNA was clearly detectable (Fig. 4B).
To investigate when during the splicing reaction Urp contacts the pre-mRNA, we used the UV-cross-linking assay to analyze the interaction between Urp and the Ad ML 3′ splice site in a heat-treated NE, which is unable to support the second step of splicing (Supplemental Figs. 5B, 6; Krainer and Maniatis 1985). Figure 4C shows that the Urp–3′ splice site interaction was readily detectable in the control but not the heat-treated NE. Thus, Urp does not interact with the 3′ splice site prior to the second step of splicing, the phase of the reaction at which Urp functions. To rule out the possibility that Urp itself is the heat-labile factor required for the second step of splicing, we repeated the cross-linking experiment following addition of untreated, recombinant Urp to a heat-treated NE or ΔUrpNE. In neither case was cross-linking of Urp to the 3′ splice site detected (Fig. 4D), indicating that Urp was not the heat-labile factor.
To further investigate the kinetics of U2-type intron 3′ splice site recognition, we performed UV-cross-linking time-course experiments in the ΔUrpNE in the presence or absence of Urp. In the absence of Urp, U2AF65 and U2AF35 remained bound to the Ad ML 3′ splice site throughout the entire time course (Fig. 4E, lanes 2–4). Following Urp addition, U2AF65 and U2AF35, but not Urp, were bound to the 3′ splice site during the first step of splicing (Fig. 4E, lane 6). Urp binding was first detected with the appearance of second-step products (Fig. 4E, lane 7), and, at a later time point, only binding of Urp, and not U2AF65 and U2AF35, was detected (Fig. 4E, lane 8). Collectively, the results of Figure 4 suggest a model in which the U2AF65–U2AF35 heterodimer, which is bound to the 3′ splice site during the first step of splicing, is replaced by Urp during the second step.
In this report, we showed that the U2AF35-related factor Urp is required for splicing of both U2- and U12-type introns and, in both instances, selectively contacts the 3′ splice site. Remarkably, however, Urp promotes different steps in U2 and U12 intron splicing. For U12-type introns, Urp is recruited to the pre-mRNA in an ATP-dependent manner and is required for assembly of the prespliceosome, a precursor to other spliceosomal complexes. Our results reveal for the first time how the 3′ splice site of a U12-type intron is recognized, and demonstrate that this involves a U2AF35-related factor.
Interestingly, U2AF65 is neither required for U12-type intron splicing (Shen and Green 2007) nor present in the U12-type spliceosome (Figs. 2A, 4B). Thus, the basis by which Urp is incorporated into the U12-type intron remains to be determined. An attractive possibility is that Urp is recruited by an ATP-dependent interaction with another U12-type spliceosomal component.
For U2-type introns, Urp is required only for the second step of splicing. However, as with U12-type introns, Urp selectively contacts the 3′ splice site. The 3′ splice sites of the U2-type Ad ML intron and U12-type P120 intron contain AG and AC, respectively. Notably, approximately two-thirds of human U12-type introns contain an AG at the 3′ splice site, and the remaining third contain an AC (Levine and Durbin 2001). These considerations suggest that the last nucleotide of the 3′ splice site is not a critical determinant for Urp recognition, which was directly confirmed by our UV-cross-linking results with P120 3′ splice site mutants. An implication of our findings is that the 3′ splice site of a U2-type intron is recognized multiple times: in the early complex (complex E) by U2AF35 to promote prespliceosome assembly, and after the first step of splicing by Urp to promote the second step. Recognition of the 3′ splice site at different stages of the splicing reaction may provide an additional mechanism to ensure splicing fidelity.
Materials and methods
Identification of Urp homologs
BLAST searches of the NCBI GenBank database were performed using either the human Urp sequence or an Urp consensus sequence derived from human, mouse, chicken, fish (Fugu rubipes and Danio rerio), and Drosophila Urp sequences that was generated using the CONS algorithm on a multiple sequence alignment: pekxsxKkyraalkkekrkkrrqxlarlrDxxxxxxeeEexsXxeexqxxxxxxxxxxxxexllexerqrlheEwllrexkaqeefRlkkekeEaarkrKeeqerKikxEweeQQrkereEeeqkrQekrereeavqkmldqaenelenxxxwxnpexpxxxxxxxxxmekDrancpfysktgacrfgDrcsrkhNfptssptlliksmfttfgmeqxxcrrDDyDxDasleysEeExyqqflDfyXDvlpefknvgkviqfkvscnlephlrgnvyvqyqseEecqaaxslfngrwyagrqlqcefcpvtrwkmaicglfEiqkcprgkhcnflhvfrnpNnefxEaNrDiymspDxtxXsxgknserrerxxhhDxyyxrxrxrxsxsxxXsxrXnxxxXrXxxXxrxxkxxxxxxxXxx.
All available eukaryotic sequences (genomic, cDNA/mRNA, EST, etc.) were queried to identify novel Urp homologs, which were defined as fitting the Urp consensus sequence and sharing its domain structure (i.e., a UHM motif [Kielkopf et al. 2004] flanked by two zinc finger motifs), but missing the well-conserved N-terminal block of amino acids that defines U2AF35.
Protein expression and purification
The complete human Urp ORF was PCR-amplified from HeLa cDNA and cloned into a derivative of pEF6V5-HisB (Invitrogen), which contains an N-terminal multiepitope (Flag, S-peptide, and epitope C) tag (X7), to generate a plasmid expressing X7-Urp (pX7-Urp). For protein expression, the Urp gene was PCR-amplified from pX7-Urp and cloned into a pET41a-based vector (Novagen) to generate GST-Urp; the construct was confirmed by DNA sequencing. GST-Urp was expressed in Escherichia coli and purified as described (Valcarcel et al. 1996). For U2AF35 expression, the coding region of U2AF35 was inserted into pET15b (Novagen). His-tagged U2AF35 was expressed in E. coli and purified under native conditions using Ni-NTA agarose (Qiagen).
In vitro splicing assays and spliceosome assembly reactions
ΔUrpNE was analyzed using a polyclonal α-Urp antibody (described in the Supplemental Material), or a monoclonal antibody to U2AF35 (provided by Juan Valcarcel, Center de Regulació Genòmica, Barcelona, Spain) or U2AF65 (MC3; Santa Cruz Biotechnology). For splicing reactions, 6 μL of Urp-depleted or mock-depleted NE was used per splicing reaction in a final volume of 10 μL, and splicing reactions were performed as described previously (Fleckner et al. 1997) using a plasmid-encoded Ad ML (Zillmann et al. 1988) or P120 (Hastings and Krainer 2001) substrate and GST-Urp (0.08 μM).
Spliceosome assembly reactions were performed as described previously (Kan and Green 1999). Spliceosomal complexes H, A, B, and C were resolved on nondenaturing 4% acrylamide:bisacrylamide (80:1)/0.5% low-melting agarose (LMA) in 50 mM Tris base/50 mM glycine buffer (Wu and Green 1997); spliceosomal complexes H and E were separated on a 1.5% LMA gel in 0.5× TBE (Das and Reed 1999). Signals were visualized by PhosphorImager (Fujifilm FLA-7000 imaging system). To deplete ATP, the ΔUrpNE was preincubated for 30 min at 30°C.
Affinity purification of splicing complexes
Biotinylated Ad ML or P120 pre-mRNA substrates (100 fmol) were incubated with HeLa NE (64 μL in a total volume of 100 μL) and GST-Urp (0.8 μM) for 20 min under conditions that allow spliceosome formation, after which heparin (200 ng/μL; Sigma) was added to stop the reaction. The reaction mixture was then incubated with streptavidin-agarose beads (Pierce) and washed five times (for 10 min each) in buffer R (20 mM Tris-HC1 at pH 7.8, 0.1% Triton X-100, 150 mM KC1, 2.5 mM EDTA). Affinity-purified complexes were digested with RNase A (Roche) to release the bound proteins, which were analyzed by immunoblotting using a polyclonal antibody to Urp (described above) or U11/U12 snRNP 35K (Abcam), or a monoclonal antibody to U2AF65.
Protein–RNA cross-linking assays
Protein–RNA cross-linking and site-specific labeling of pre-mRNA substrates were performed as described previously (Shen et al. 2004). Labeled products were visualized using a Fujifilm FLA-7000 PhosphorImager. Inactivation of U11 or U12 snRNA by RNase H-directed cleavage was performed as described previously (Shen et al. 2004) using a DNA oligonucleotide complementary to the 5′ end of U11 snRNA (5′-UCACGACAGAAGCCCUUUUU-3′) or the branchpoint base-pairing region of U12 snRNA (5′-AUUUUCCUUACUCAUAAG-3′). Previously described P120 mutants (Dietrich et al. 2005) were obtained from Richard Padgett (Lerner Research Institute). Mild heat treatment of NE or ΔUrpNE was performed by heating the extract for 20 min at 43°C (Krainer and Maniatis 1985; Wu and Green 1997).
Biomolecular exon ligation assays
The 5′ and 3′ RNA Ad ML substrates were transcribed from PCR products amplified from the plasmid MINX (Zillmann et al. 1988). The 5′ substrate was preincubated in HeLa NE for 1 h, and bimolecular exon ligation was initiated by the addition of the 3′ RNA substrate and GST-Urp. The final concentration of the 5′ substrate in the splicing reaction was 30 nM, the final concentration of the 3′ RNA substrate was 150 nM, and the final concentration of GST-Urp was 0.1 μM. Splicing reactions were performed at 30°C in 25% HeLa NE, 2 mM MgCl2, 1 mM ATP, and 5 mM creatine phosphate as described previously (Anderson and Moore 1997). RNAs were extracted and separated on 8% denaturing polyacrylamide gels.
Acknowledgments
We thank Juan Valcarcel and Richard Padgett for providing reagents, and Sara Evans for editorial assistance. This work was supported by the Mid-career Researcher Program through National Research Foundation (NRF) grant 2010-0000437 funded by the Ministry of Education, Science, and Technology (MEST), a Dasan Young Faculty Grant from Gwangju Institute of Science and Technology, and the Brain Korea 21 Project Research Foundation to H.S. and X.Z., and National Institutes of Health grant R01GM035490 to M.R.G. M.R.G. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1974810.
Supplemental material is available at http://www.genesdev.org.
References
- Anderson K, Moore MJ 1997. Bimolecular exon ligation by the human spliceosome. Science 276: 1712–1716 [DOI] [PubMed] [Google Scholar]
- Black DL 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72: 291–336 [DOI] [PubMed] [Google Scholar]
- Burge CB, Padgett RA, Sharp PA 1998. Evolutionary fates and origins of U12-type introns. Mol Cell 2: 773–785 [DOI] [PubMed] [Google Scholar]
- Das R, Reed R 1999. Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. RNA 5: 1504–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich RC, Peris MJ, Seyboldt AS, Padgett RA 2001. Role of the 3′ splice site in U12-dependent intron splicing. Mol Cell Biol 21: 1942–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich RC, Fuller JD, Padgett RA 2005. A mutational analysis of U12-dependent splice site dinucleotides. RNA 11: 1430–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleckner J, Zhang M, Valcarcel J, Green MR 1997. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP–branchpoint interaction. Genes Dev 11: 1864–1872 [DOI] [PubMed] [Google Scholar]
- Graveley BR, Hertel KJ, Maniatis T 2001. The role of U2AF35 and U2AF65 in enhancer-dependent splicing. RNA 7: 806–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall SL, Padgett RA 1994. Conserved sequences in a class of rare eukaryotic nuclear introns with non-consensus splice sites. J Mol Biol 239: 357–365 [DOI] [PubMed] [Google Scholar]
- Hastings ML, Krainer AR 2001. Functions of SR proteins in the U12-dependent AT-AC pre-mRNA splicing pathway. RNA 7: 471–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan JL, Green MR 1999. Pre-mRNA splicing of IgM exons M1 and M2 is directed by a juxtaposed splicing enhancer and inhibitor. Genes Dev 13: 462–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielkopf CL, Lucke S, Green MR 2004. U2AF homology motifs: Protein recognition in the RRM world. Genes Dev 18: 1513–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa K, Wang X, Hatada I, Yamaoka T, Nojima H, Inazawa J, Abe T, Mitsuya K, Oshimura M, Murata A, et al. 1995. Isolation and mapping of human homologues of an imprinted mouse gene U2af1-rs1. Genomics 30: 257–263 [DOI] [PubMed] [Google Scholar]
- Krainer AR, Maniatis T 1985. Multiple factors including the small nuclear ribonucleoproteins U1 and U2 are necessary for pre-mRNA splicing in vitro. Cell 42: 725–736 [DOI] [PubMed] [Google Scholar]
- Levine A, Durbin R 2001. A computational scan for U12-dependent introns in the human genome sequence. Nucleic Acids Res 29: 4006–4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merendino L, Guth S, Bilbao D, Martinez C, Valcarcel J 1999. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature 402: 838–841 [DOI] [PubMed] [Google Scholar]
- Russell AG, Charette JM, Spencer DF, Gray MW 2006. An early evolutionary origin for the minor spliceosome. Nature 443: 863–866 [DOI] [PubMed] [Google Scholar]
- Sharp PA, Burge CB 1997. Classification of introns: U2-type or U12-type. Cell 91: 875–879 [DOI] [PubMed] [Google Scholar]
- Shen H, Green MR 2007. RS domain-splicing signal interactions in splicing of U12-type and U2-type introns. Nat Struct Mol Biol 14: 597–603 [DOI] [PubMed] [Google Scholar]
- Shen H, Kan JL, Green MR 2004. Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol Cell 13: 367–376 [DOI] [PubMed] [Google Scholar]
- Shepard J, Reick M, Olson S, Graveley BR 2002. Characterization of U2AF(26), a splicing factor related to U2AF(35). Mol Cell Biol 22: 221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronchere H, Wang J, Fu XD 1997. A protein related to splicing factor U2AF35 that interacts with U2AF65 and SR proteins in splicing of pre-mRNA. Nature 388: 397–400 [DOI] [PubMed] [Google Scholar]
- Valcarcel J, Gaur RK, Singh R, Green MR 1996. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA. Science 273: 1706–1709 [DOI] [PubMed] [Google Scholar]
- Will CL, Luhrmann R 2005. Splicing of a rare class of introns by the U12-dependent spliceosome. Biol Chem 386: 713–724 [DOI] [PubMed] [Google Scholar]
- Wu S, Green MR 1997. Identification of a human protein that recognizes the 3′ splice site during the second step of pre-mRNA splicing. EMBO J 16: 4421–4432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Romfo CM, Nilsen TW, Green MR 1999. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402: 832–835 [DOI] [PubMed] [Google Scholar]
- Zamore PD, Patton JG, Green MR 1992. Cloning and domain structure of the mammalian splicing factor U2AF. Nature 355: 609–614 [DOI] [PubMed] [Google Scholar]
- Zillmann M, Zapp ML, Berget SM 1988. Gel electrophoretic isolation of splicing complexes containing U1 small nuclear ribonucleoprotein particles. Mol Cell Biol 8: 814–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio DA, Blumenthal T 1999. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature 402: 835–838 [DOI] [PubMed] [Google Scholar]