Abstract
Nonsense-mediated mRNA decay (NMD) is a quality control mechanism that detects and degrades mRNAs containing premature stop codons (PTCs). In vertebrates, PTCs trigger efficient NMD when located upstream of an exon junction complex (EJC). Degradation of PTC-containing mRNAs requires the endonucleolytic activity of SMG6, a conserved NMD factor; nevertheless, the precise role for the EJC in NMD and how the SMG6 endonuclease is recruited to NMD targets have been unclear. Here we show that SMG6 interacts directly with the EJC via two conserved EJC-binding motifs (EBMs). We further show that the SMG6–EJC interaction is required for NMD. Our results reveal an unprecedented role for the EJC in recruiting the SMG6 endonuclease to NMD targets. More generally, our findings identify the EBM as a protein motif present in a handful of proteins, and suggest that EJCs establish multiple and mutually exclusive interactions with various protein partners, providing a plausible explanation for the myriad functions performed by this complex in post-transcriptional mRNA regulation.
Keywords: mRNA decay, exon junction complex, NMD, UPF1
The nonsense-mediated mRNA decay (NMD) pathway triggers the rapid degradation of mRNAs containing premature stop codons (PTCs, or nonsense codons), thereby preventing the accumulation of potentially deleterious truncated proteins. In vertebrates, a stop codon is efficiently recognized as being premature if it lies at least 50–55 nucleotides (nt) upstream of an exon–exon boundary, which is marked by the exon junction complex (EJC) (Nagy and Maquat 1998; Le Hir et al. 2000). PTC recognition causes a “surveillance complex” to assemble on the mRNA; this complex marks the PTC-containing transcript for degradation (Isken and Maquat 2007; Rebbapragada and Lykke-Andersen 2009; Nicholson et al. 2010).
The surveillance complex consists of a well-defined set of NMD factors, including the evolutionarily conserved proteins UPF1, UPF2, and UPF3 (Chang et al. 2007; Isken and Maquat 2007; Rebbapragada and Lykke-Andersen 2009; Nicholson et al. 2010). In metazoa, additional NMD factors include the proteins SMG1 (a phosphatidylinositol 3-kinase-like kinase that phosphorylates UPF1) and SMG5, SMG6, and SMG7, which trigger UPF1 dephosphorylation (Cali et al. 1999; Page et al. 1999; Denning et al. 2001; Pal et al. 2001; Yamashita et al. 2001; Anders et al. 2003; Chiu et al. 2003; Ohnishi et al. 2003; Fukuhara et al. 2005).
Current models for NMD suggest that UPF1 and SMG1 are recruited by ribosomes terminating translation prematurely through interactions with the eukaryotic release factors eRF1 and eRF3 (Czaplinski et al. 1998; Kashima et al. 2006). In the presence of UPF2 and/or UPF3, presumably bound to downstream EJCs on the mRNA, SMG1 phosphorylates UPF1; this in turn recruits SMG5, SMG6, and SMG7 (Denning et al. 2001; Pal et al. 2001; Yamashita et al. 2001; Anders et al. 2003; Chiu et al. 2003; Ohnishi et al. 2003; Fukuhara et al. 2005).
SMG5, SMG6, and SMG7 are three related proteins that are thought to bind phosphorylated UPF1 through a common 14-3-3-like domain (Fukuhara et al. 2005). SMG5 and SMG7 interact with each other (Anders et al. 2003; Ohnishi et al. 2003) and may link the surveillance complex to the general mRNA decay machinery, thereby coupling premature translation termination to mRNA degradation (Unterholzner and Izaurralde 2004). SMG6 is characterized by a C-terminal PIN (PilT N terminus) domain, which exhibits nuclease activity (Glavan et al. 2006). Recent studies in Drosophila melanogaster, zebrafish, and human cells indicate that degradation of NMD targets requires the endonucleolytic activity of SMG6 (Gatfield and Izaurralde 2004; Huntzinger et al. 2008; Eberle et al. 2009; Wittkopp et al. 2009). Nevertheless, a number of key questions remain: For example, how is SMG6 recruited to NMD targets? And, aside from the endonucleolytic activity of the PIN domain, how do the other SMG6 domains function during NMD?
In this study, we addressed these questions by generating a series of SMG6 mutants and testing them in protein–protein interaction and functional assays. We show that, like the PIN domain, the N-terminal and 14-3-3-like domains of SMG6 are essential for NMD. Accordingly, the N-terminal domain is required for SMG6 to associate with other NMD factors, including SMG6 itself, suggesting this domain enables SMG6 to incorporate into assembled surveillance complexes. Unexpectedly, the SMG6 N-terminal domain also mediates binding to the EJC.
Sequence analyses revealed that the N-terminal domains of SMG6 orthologs from various species contain one or two conserved motifs that are related to a C-terminal motif present in UPF3b. The UPF3b C-terminal motif binds to the EJC (Gehring et al. 2003; Buchwald et al. 2010). Similarly, the conserved SMG6 N-terminal motifs confer direct binding to the EJC and were termed EJC-binding motifs (EBMs). Recently, Buchwald et al. (2010) defined the structure of the UPF3b EBM bound to the EJC. Using this as a guide, we designed point mutations in SMG6 EBMs that disrupt EJC binding. Remarkably, these mutations abrogated SMG6 association with mRNA targets and its ability to complement NMD in cells lacking endogenous SMG6. These findings reveal an unexpected role for the EJC in the recruitment of the SMG6 endonuclease, and thereby in the degradation of NMD targets. More generally, our findings define the EBM: a novel protein sequence motif that we show is present in a handful of metazoan proteins in addition to SMG6 and UPF3.
Results
The SMG6 N-terminal region, 14-3-3-like domain, and PIN domain are all required for NMD
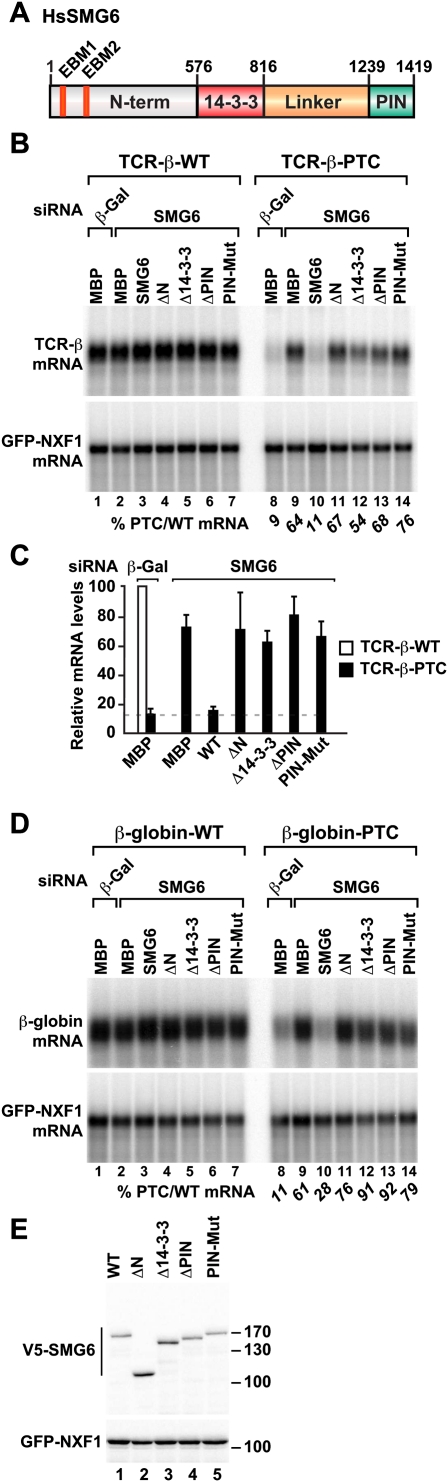
SMG6 contains two highly conserved, globular domains connected by a linker region. The globular domains consist of the C-terminal PIN domain, which exhibits endonuclease activity, and the 14-3-3-like domain, which is thought to interact with phosphorylated UPF1 (Fig. 1A; Fukuhara et al. 2005; Glavan et al. 2006). The N terminus, upstream of the 14-3-3-like domain, is predicted to be unstructured and is less conserved among SMG6 orthologs. To investigate which domains of SMG6 are important for NMD activity, we designed a series of deletion mutants lacking each domain individually and tested them in a complementation assay (described in Huntzinger et al. 2008). Briefly, an siRNA complementary to the ORF of the smg6 mRNA was used to deplete endogenous SMG6; this depletion inhibits NMD. In SMG6-depleted HeLa cells, SMG6 mutants were then tested for the ability to restore NMD. Transcripts encoding the recombinant proteins were resistant to the siRNA because we introduced mutations that disrupt the mRNA–siRNA base pair interactions but do not alter the protein sequence.
Figure 1.
The SMG6 N-terminal domain is required for NMD. (A) Domain organization of SMG6. (N-term) N-terminal region; (14-3-3) domain with a 14-3-3-like fold; (Linker) linker region; (PIN) PIN domain. (B–D) HeLa cells were transfected with the indicated siRNAs plus a mixture of two plasmids: one expressing the NMD reporters (β-globin or TCR-β with or without PTC), and another expressing GFP-NXF1 as transfection control. Plasmids expressing siRNA-resistant, wild-type, or mutant versions of human SMG6 were included in the transfection mixtures, as indicated. These proteins are fused to a V5 tag. V5-MBP served as a control. The levels of wild-type or PTC-containing reporters were analyzed by Northern blot and normalized to those of GFP-NXF1 mRNA. The numbers in italics below B and D represent the normalized levels of the PTC-containing reporter relative to those of the wild-type, which were set to 100 for each condition. (C shows mean values ± standard deviation obtained in three independent experiments using the TCR-β reporter. Note that the normalized values of the wild-type reporter were set to 100 for each condition (white bar), but are shown only for control cells expressing MBP and treated with β-Gal siRNA. (E) The expression of wild-type or mutant V5-SMG6 was analyzed by Western blotting using an anti-V5 antibody. Cotransfected GFP-NXF1 served as a transfection control.
We monitored NMD using two well-characterized NMD reporters: One was based on a TCR-β minigene (Wang et al. 2002), and a second was based on a β-globin gene (Thermann et al. 1998). As reported before, the levels of the PTC-containing TCR-β and β-globin mRNAs were reduced to ∼10% of the wild-type levels (Fig. 1B,D [lanes 8], C; Thermann et al. 1998; Wang et al. 2002). Depleting SMG6, however, caused the PTC-containing TCR-β and β-globin reporters to increase by sixfold (Fig. 1B,D [lanes 9], C). In contrast, the expression levels of the corresponding wild-type mRNAs did not change significantly (Fig. 1B,D, lanes 1–7).
In cells depleted of endogenous SMG6, expressing an siRNA-resistant version of wild-type SMG6 restored NMD (Fig. 1B,D, lanes 10). In contrast, the NMD activity was not restored if cells expressed an siRNA-resistant SMG6 that lacked either the N-terminal domain or the 14-3-3-like domain (Fig. 1B,D, lanes 11,12). SMG6 mutants lacking the PIN domain or carrying mutations in catalytic residues were also inactive in complementation assays, as shown before (Fig. 1B,D, lanes 13,14; Huntzinger et al. 2008). The effect of deleting the SMG6 linker region could not be tested because this mutant was expressed at low levels and was extensively degraded (data not shown). Since SMG6 mutants were expressed at equivalent or higher levels than wild-type SMG6 (Fig. 1E), we conclude that the SMG6 N-terminal region, the 14-3-3-like domain, and the PIN domain are all required for NMD.
The N-terminal domain is required for SMG6 incorporation into surveillance complexes
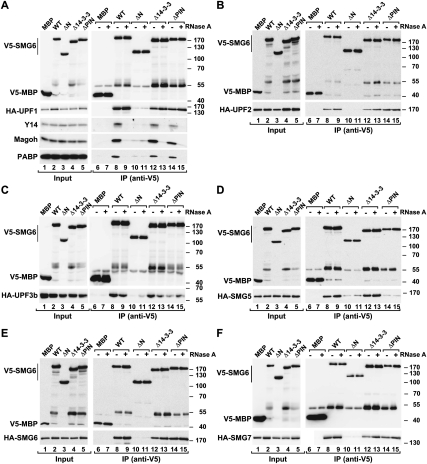
While previous work elucidated functions for the 14-3-3-like and PIN domains of SMG6 (namely, binding to phosphorylated UPF1 and promoting RNA endonucleolytic cleavage, respectively), no function for the N-terminal region of SMG6 is known. To test whether this region might be involved in binding NMD factors, we performed coimmunoprecipitation assays. We observed that SMG6 interacted with all NMD effectors, including SMG6 itself (Fig. 2A–F, lanes 8). Notably, these interactions were insensitive to RNase A treatment (Fig. 2A–F, lanes 9). In contrast, the interaction of SMG6 with the cytoplasmic poly(A)-binding protein PABP was sensitive to RNase A treatment, indicating that it is mediated by RNA (Fig. 2A, lanes 8,9).
Figure 2.
The N-terminal region of SMG6 confers binding to NMD factors and EJC components. (A–F) Interaction of V5-tagged SMG6 (wild type or deletion mutant) with HA-tagged NMD factors or endogenous Y14 and Magoh in human cells. Proteins were immunoprecipitated using anti-V5 antibodies. A V5-tagged MBP served as a negative control. The presence of endogenous Y14 and Magoh in the immunoprecipitates was analyzed by Western blotting using specific antibodies. In all panels, the protein bands observed at ∼55 kDa represent SMG6 degradation products comigrating with the immunoglobulin heavy chain that cross-react with the anti-V5 antibody.
Surprisingly, deleting the N-terminal region of SMG6 prevented it from interacting with any NMD factor (Fig. 2A–F, lanes 10). Also, deleting the N-terminal region abolished the interaction of SMG6 with PABP, suggesting a role for the N-terminal region in mediating SMG6 recruitment to target mRNAs (Fig. 2A, lane 10). Unexpectedly, deleting the 14-3-3-like domain did not affect the SMG6 interaction with UPF1 or the other NMD factors (Fig. 2A–F, lanes 12). Our experimental conditions, however, might not detect the interaction between the SMG6 14-3-3-like domain and phosphorylated UPF1, because either only a small fraction of UPF1 is phosphorylated in vivo and/or UPF1 is rapidly dephosphorylated in cell lysates (Ohnishi et al. 2003).
Interestingly, if the SMG6 PIN domain is deleted, then the association between SMG6 and SMG5 is disrupted (Fig. 2D, lane 14); this indicates that SMG6 requires both the N-terminal region and the PIN domain to associate with SMG5. The coimmunoprecipitation assays described above probably reflect an association between SMG6 and preassembled surveillance complexes on RNA fragments (which are resistant to the RNase A treatment), because it is unlikely that SMG6 interacts directly with all NMD factors. This association with surveillance complexes requires the SMG6 N-terminal domain.
The SMG6 N-terminal domain mediates direct binding to the EJC
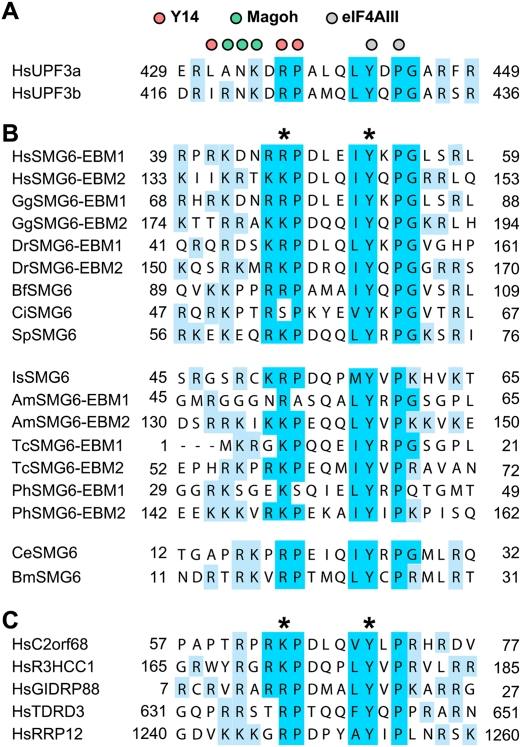
To obtain insight into how the N terminus of SMG6 might associate with NMD surveillance complexes and mRNA targets, we compared orthologous sequences for potential conserved features. BLAST searches revealed two conserved, related motifs in the N-terminal domains of SMG6 proteins from several species (Fig. 3A,B). Surprisingly, these motifs exhibited 23% and 38% identity (38% and 57% similarity) to a UPF3b C-terminal motif, which is known to mediate binding to the EJC (Fig. 3A,B; Gehring et al. 2003; Kunz et al. 2006; Buchwald et al. 2010). We therefore termed these motifs EBMs, or EBM1 and EBM2 (Fig. 3B). The EBMs are well-conserved in SMG6 orthologs, except in dipterans, even though the rest of the N-terminal domain evolves rapidly. SMG6 proteins contain two copies of the motif in vertebrates and insects, and one copy in all other animals.
Figure 3.
EBMs in UPF3, SMG6, and additional proteins. Alignment of EBMs in UPF3 (A), SMG6 homologs (B), and other human proteins (C). The organisms are Homo sapiens (Hs), Gallus gallus (Gg), Danio rerio (Dr), Branchiostoma floridae (Bf), Ciona intestinalis (Ci), Strongylocentrotus purpuratus (Sp), Ixodes scapularis (Is), Apis mellifera (Am), Tribolium castaneum (Tc), Pediculus humanus (Ph), Caenorhabditis elegans (Ce), and Brugia malayi (Bm). Conserved residues of the motif are highlighted in blue. Positively charged residues flanking the motif are highlighted in cyan. Colored circles above the UPF3 sequences indicate the residues of human UPF3b that interact with Y14 (magenta), Magoh (green), and eIF4AIII (gray), according to Buchwald et al. (2010). Asterisks indicate residues mutated in this study.
To test whether SMG6 interacts with the EJC, as suggested by the presence of the EBMs, we performed coimmunoprecipitation assays. We observed that full-length SMG6 did indeed associate with endogenous Y14 and Magoh, two EJC components (Fig. 2A, lane 8). The N-terminal region of SMG6 was both necessary (Fig. 2A, lane 10) and sufficient (see below) for EJC binding.
In contrast to the interaction with NMD factors, but similar to PABP association, SMG6 binding to Y14 and Magoh was disrupted when cell lysates were treated with RNase A (Fig. 2A, lane 9). Usually if a coimmunoprecipitation reaction is sensitive to RNase A, it is interpreted to mean that the protein interaction is indirect and mediated by RNA. This may not be the case for the SMG6 interaction with the EJC, because EJC assembly requires the presence of RNA. Therefore, the RNase A treatment may destabilize EJCs in cell lysates. Accordingly, although UPF3b is known to bind directly to EJC components (Buchwald et al. 2010), this interaction was also sensitive to RNase A treatment in cell lysates (Supplemental Fig. S1A).
To verify whether SMG6 associates directly with the EJC, we expressed the recombinant, purified proteins in Escherichia coli and tested them in binding assays in vitro. The core of the human EJC was reconstituted using recombinant eIF4AIII, Y14, Magoh, and the SELOR domain of Barentsz (BTZ), as described before (Ballut et al. 2005; Bono et al. 2006). These proteins assemble into a stable complex in the presence of a U15 RNA, AMPPNP, and magnesium ions (Ballut et al. 2005; Bono et al. 2006). We observed that EJC complexes could be pulled down by a GST-tagged, N-terminal fragment of SMG6 that contains the two EBMs (amino acids 1–207) (Fig. 4, lane 7). As expected, the EJC was precipitated by a GST-tagged UPF3b C-terminal fragment (Fig. 4, lane 6; Buchwald et al. 2010). The EJC could also be precipitated with a biotinylated peptide corresponding to the sequence of EBM2 (Supplemental Fig. S1B), indicating this sequence is a bona fide EBM. We conclude that the N-terminal region of SMG6 interacts directly with the EJC.
Figure 4.
SMG6 EBMs confer direct EJC binding. GST pull-downs of the preassembled EJC, with wild-type or mutant GST-SMG6 (amino acids 1–207). Inputs (20%) and bound fractions (100%) were analyzed on a 4%–12% gradient SDS-PAGE. GST-UPF3bΔN serves as a positive control for EJC binding. In lanes 1 and 5, the identical reaction, with EJC alone, shows the absence of nonspecific binding to the GST beads.
The two EBMs of SMG6 are partially redundant
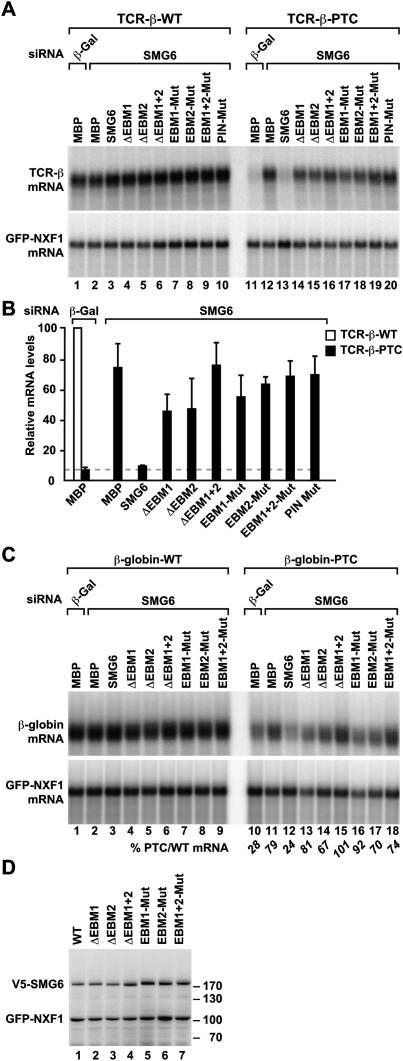
To verify that the EBMs of SMG6 interacted with the EJC, and to determine their relative contribution, we deleted these motifs either individually or in combination and performed coimmunoprecipitation assays with EJC components. Deleting either EBM1 or EBM2 strongly reduced the SMG6 interaction with Y14 and Magoh (Fig. 5A, lanes 11,12); however, some residual binding was observed with the mutant lacking EBM1 (Fig. 5A, lane 11), suggesting that EBM2 contributes the majority of the affinity for the EJC. Binding was abolished when both EBMs were deleted (Fig. 5A, lane 13).
Figure 5.
Mutations in the EBMs abolish EJC binding, but not the interaction with NMD factors. (A–G) Interaction of V5-tagged SMG6 (wild type or the indicated EBM mutants) with HA-tagged NMD factors or endogenous Y14 and Magoh in human cells. Proteins were immunoprecipitated using anti-V5 antibodies and analyzed as described in Figure 2.
Recently, the structure of the UPF3b EBM bound to the EJC was determined (Buchwald et al. 2010). Based on this structure, we introduced two amino acid substitutions in the SMG6 EBMs: We substituted a positively charged residue (which is invariantly arginine or lysine) and an invariant tyrosine with glutamic acid residues (see above; Fig. 3B, asterisks). When bound to the EJC, the equivalent residues in the UPF3b EBM contact Y14 and eIF4AIII, respectively. Substituting these two residues in either EBM1 or EBM2 had a similar effect as deleting the entire motif. Moreover, the substitutions in EBM1 reduced EJC binding (twofold reduction relative to wild type), but not as strongly as substituting the equivalent residues in EBM2 (fourfold reduction relative to wild type), reinforcing the idea that EBM2 provides the higher-affinity binding site (Fig. 5A, lanes 14,15). When both EBMs carried the substitutions, SMG6 binding to the EJC was abolished (Fig. 5A, lane 16). Together, these results indicate that the two SMG6 EBMs contribute to EJC binding in an additive or cooperative manner, although EBM2 appears to exhibit a higher affinity than EBM1.
In agreement with the coimmunoprecipitation results, a GST fusion of an SMG6 fragment (amino acids 1–207) carrying substitutions in both EBM motifs did not associate with the EJC in in vitro pull-down assays (Fig. 4, lane 8). Finally, in pull-down experiments, the EJC could not be precipitated with a biotinylated peptide corresponding to EBM2 that contained substitutions in the residues described above (Supplemental Fig. S1B). Interestingly, SMG6 mutants deficient in EJC binding retained the ability to interact with UPF1, UPF2, UPF3, SMG5, and SMG7 and to self-interact (Fig. 5B–G), indicating that SMG6 binds to NMD factors independently of the EJC and this binding is likely mediated by sequences in the SMG6 N-terminal region that are distinct from the EBM motifs. Consistently, SMG6 EBM mutants exhibited higher affinity for SMG5 and SMG7 than wild type (Fig. 5F,G). More importantly, SMG6 mutants that failed to interact with the EJC were also strongly impaired in PABP binding (Fig. 6A), suggesting that the SMG6–EJC interaction plays a crucial role in recruiting SMG6 to target mRNAs.
Figure 6.
Mutations in the EBMs strongly reduce the interaction of SMG6 with PABP. (A) V5-tagged SMG6 wild type or the indicated EBM mutants were expressed in human cells and immunoprecipitated using anti-V5 antibodies. The presence of endogenous PABP, Y14, and Magoh in the immunoprecipitates was analyzed by Western blotting. (B) UPF3b competes with SMG6 for binding to the EJC. GST pull-downs of the preassembled EJC with GST-UPF3bΔN or GST-SMG6 (1–207). (Lanes 1,2,5,6) To test for competition of EJC binding, the pull-downs with GST-UPF3bΔN were done in the absence or presence of untagged SMG6 (1–207). (Lanes 3,4,7,8) Conversely, the pull-downs with GST-SMG6 (1–207) were done in the absence or presence of MBP-UPF3bΔN. Samples were analyzed as described in Figure 4.
SMG6 and UPF3 bind at the same site on the EJC
As described above, substituting EBM equivalent residues in SMG6 and UPF3b abolishes EJC binding for both, implying that EBMs in these proteins interact with the EJC in a similar way. To test whether SMG6 and UPF3b bind to the same surface on the EJC, we performed competition experiments. In these studies, adding a sixfold molar excess of an untagged SMG6 N-terminal fragment to binding reactions did not affect the interaction of the EJC with UPF3b (Fig. 6B, cf. lanes 6 and 5). In contrast, a twofold molar excess of maltose-binding protein (MBP)-tagged UPF3b blocked SMG6 from binding to the EJC (Fig. 6B, cf. lanes 8 and 7). We conclude that UPF3b has a higher affinity for the EJC than SMG6, an idea supported by the observation that a higher concentration of SMG6 is required to pull down equivalent amounts of the EJC (Fig. 6B). Together with the observation that substituting residues located at equivalent positions in the EBMs of both SMG6 and UPF3b abolish EJC binding, these results indicate that SMG6 and UPF3b compete for the same surface on the EJC.
SMG6 interaction with the EJC is required for NMD
To evaluate whether the interaction of SMG6 with the EJC contributes to NMD, we tested whether SMG6 mutants that are impaired in EJC binding could complement NMD in cells lacking endogenous SMG6. We observed that, independently of the reporter, SMG6 mutants lacking either EBM1 or EBM2 or carrying amino acid substitution in either of the two EBMs were strongly impaired in complementation assays (Fig. 7A–C). NMD activity was abolished when both EBMs were deleted or mutated simultaneously (Fig. 7A–C). All proteins were expressed at comparable levels (Fig. 7D). We conclude that, in human cells, the interaction of SMG6 with the EJC is required for NMD.
Figure 7.
SMG6 interaction with the EJC is required for NMD. (A–D) Complementation assays were performed with wild-type SMG6 and various SMG6 mutants and the indicated NMD reporters, as described in Figure 1. An SMG6 protein carrying mutations in catalytic residues of the PIN domain and MBP served as controls. A and C show Northern blot analysis of representative RNA samples. Wild-type and PTC-containing mRNA levels were normalized to those of the GFP-NXF1 mRNA. For each condition, the normalized values of the wild-type reporter were set to 100 (white bar in B). Bshows mean values and standard deviation obtained in three independent experiments using the TCR-β reporter. The corresponding values for the β-globin reporter are shown in italics below C. (D) Western blotting showing the expression levels of recombinant proteins.
EBM motifs are present in additional proteins
Since we found that the EBMs in SMG6 and UPF3b are related in sequence and mediate direct binding to the EJC, we searched for EBM motifs in other proteins using iterative hidden Markov model (HHM) searches with HHpred (see the Materials and Methods). In addition to the statistical significance of the matches, two criteria served to validate the identified occurrences of the motif: They had to be part of a predicted unstructured region, and they had to be conserved in homologs.
This analysis led to the identification of EBMs in the human proteins RRP12 (Oeffinger et al. 2004) and TDRD3 (Tudor domain-containing protein 3) (Goulet et al. 2008; Linder et al. 2008), the uncharacterized protein C2orf68, and the paralog proteins R3HCC1 (R3H and coiled-coil-containing protein 1) and GIDRP88 (growth inhibition and differentiation-related protein 88) (Fig. 3C). We also found EBMs in the uncharacterized D. melanogaster proteins CG2691, CG2162, and CG13472, which are orthologous to RRP12, R3HCC1/GIDRP88, and TDRD3, respectively.
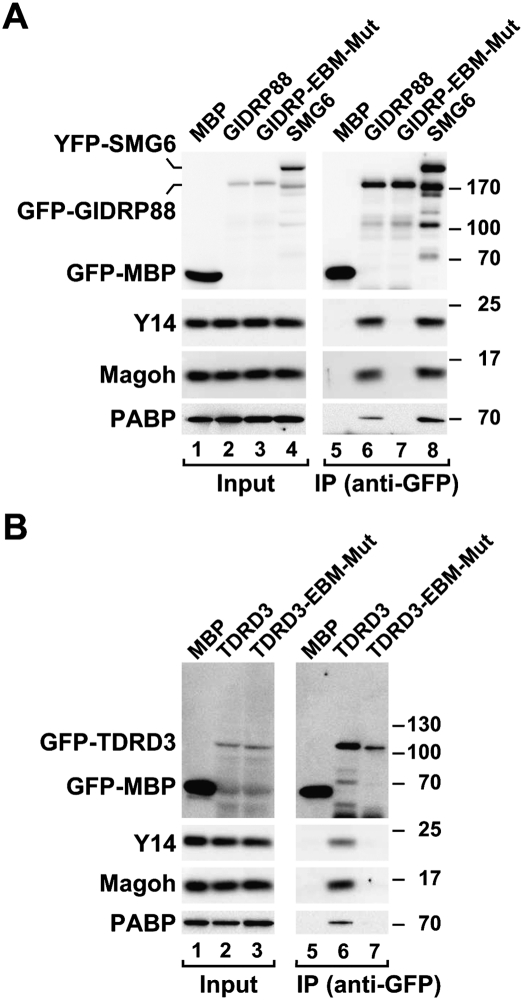
To validate the predicted EBMs experimentally, we expressed GIDRP88, TDRD3, and C2orf68 in human cells and performed coimmunoprecipitation assays as described above. We observed that GIDRP88 and TDRD3 coimmunoprecipitated endogenous Y14 and Magoh, as well as PABP (Fig. 8A,B, lanes 6). These interactions were mediated by the respective EBMs. Indeed, substituting the conserved K/R and Y residues in the predicted EBMs (see Fig. 3C, asterisks) abolished GIDRP88 and TDRD3 association with Y14 and Magoh (Fig. 8A,B, lanes 7). Remarkably, as in SMG6, the EBMs of GIDRP88 and TDRD3 were also required for PABP binding, suggesting a role for these motifs in the recruitment of GIDRP88 and TDRD3 to mRNAs. C2orf68, in contrast, did not coimmunoprecipitate endogenous Magoh or Y14 (data not shown). Together, these findings suggest that the EJC may serve as a binding platform for various EBM-containing proteins, and each may perform distinct post-transcriptional functions after hooking onto mRNAs via the EJC.
Figure 8.
Interaction of GFP-tagged GIDRP88 and TDRD3 with endogenous Y14, Magoh, and PABP in human cells. (A,B) Proteins were immunoprecipitated using anti-GFP antibodies and analyzed as described in Figure 2. GFP-MBP and YFP-SMG6 served as negative and positive controls, respectively.
Discussion
It is now well established in D. melanogaster and human cells that SMG6 mediates degradation of NMD targets (Gatfield and Izaurralde 2004; Huntzinger et al. 2008; Eberle et al. 2009). Previous work showed SMG6 contains a C-terminal PIN domain that provides endonucleolytic activity and a 14-3-3-like domain that likely binds phosphorylated UPF1 and eventually promotes its dephosphorylation (Denning et al. 2001; Pal et al. 2001; Yamashita et al. 2001; Anders et al. 2003; Chiu et al. 2003; Ohnishi et al. 2003; Fukuhara et al. 2005; Wittmann et al. 2006). To further elucidate the choreography of interactions centered on SMG6, we generated a new series of SMG6 mutants and tested them in protein–protein interaction and functional assays.
We observed that the SMG6 N-terminal region is required for NMD and associates with NMD factors in vivo. These results were unexpected because, after first examining orthologs from various species, the SMG6 N-terminal region did not appear to be well conserved. Closer examination, however, revealed that this region does contain two short sequence motifs that are conserved. These motifs are related to the EBM found in the C terminus of the NMD factors UPF3a and UPF3b (Gehring et al. 2003; Buchwald et al. 2010).
We show that, indeed, the EBMs in SMG6 interact directly with the EJC. Mutating or deleting the SMG6 EBMs disrupts EJC binding and also the ability of the protein to rescue NMD in cells lacking endogenous SMG6. Collectively, these results indicate that the interaction of SMG6 with the EJC is required for NMD. Interestingly, the SMG6 mutants that no longer interact with the EJC (i.e., those carrying mutations or deletions of the EBM motifs) still interact with other NMD factors (Fig. 5), indicating that the N-terminal region must contain separate sequence motifs that associate with NMD factors.
EBMs are conserved in SMG6 orthologs except in dipterans. Therefore, it will be of interest to determine whether the presence of EBMs in SMG6 correlates with a role for the EJC in NMD.
Role of the SMG6–EJC interaction in NMD
What role can the SMG6–EJC interaction play in NMD? A possible function of this interaction could be to facilitate the recruitment of SMG6 to mRNA targets (Fig. 9). Supporting this interpretation is the observation that the interaction of SMG6 with PABP, which is mediated by RNA, is strongly reduced by mutating or deleting the EBMs. This suggests that SMG6 EBM mutants, which no longer bind to the EJC, also lose interaction with mRNAs. Alternatively, the interaction with the EJC may be required to activate the SMG6 PIN domain (which in vitro exhibits poor nuclease activity) (Glavan et al. 2006) or promote mRNP remodeling steps during NMD. Independently of the precise role of the SMG6–EJC interaction in NMD, future studies should elucidate how SMG6 is recruited or activated in both organisms that lack the EBMs (e.g., D. melanogaster) and organisms and/or targets in which NMD is EJC-independent.
Figure 9.
Model illustrating SMG6 interactions with NMD factors. NMD occurs on mRNAs containing PTCs. In vertebrates, PTCs trigger efficient NMD when located upstream of an EJC. The core EJC components are Y14, Magoh, BTZ, and eIF4AIII (not shown). In a prevailing model of NMD, UPF1 and SMG1 are recruited by ribosomes terminating translation prematurely through interactions with the release factors eRF1 and eRF3. In the presence of UPF2 and UPF3, presumably bound to downstream EJCs on the mRNA, SMG1 phosphorylates UPF1; this in turn recruits SMG5, SMG6, and SMG7. SMG5, SMG6, and SMG7 are thought to bind phosphorylated UPF1 through a common 14-3-3-like domain. SMG5 and SMG7 interact with each other, and may recruit components of the general mRNA decay machinery. In this study, we show that SMG6 is recruited to surveillance complexes through interactions mediated by its N-terminal domain. This domain contains two EBMs that mediate direct interactions with the EJC, and additional sequences that confer binding to the additional NMD factors. Moreover, SMG6 contains a C-terminal PIN domain, which exhibits endonuclease activity and cleaves the mRNA in the proximity of the PTC. Because SMG6 and UPF3 compete for binding to the EJC, UPF3 must be released from the EJC to allow SMG6 binding. The step of the pathway at which these interactions take place is not known.
When does the SMG6–EJC interaction occur?
Our mutational analysis and competition studies revealed that UPF3b and SMG6 compete for binding to the EJC, with UPF3b having a higher affinity for the EJC. This suggests that UPF3b must dissociate from the EJC to make way for SMG6 to bind. Accordingly, Ohnishi et al. (2003) reported that UPF1 phosphorylation triggers a rearrangement of NMD complexes and the release of a long isoform of UPF3, which is replaced by a short isoform with higher affinity for SMG5 and SMG7. Both UPF3 isoforms contain a C-terminal EBM, but the shorter isoform may exhibit lower affinity for the EJC, enabling SMG6 to compete for binding.
At which step of the NMD process does SMG6 interact with the EJC? Although we do not have an answer to this question, we envisage that UPF3b may block SMG6 from being recruited to “normal” mRNAs. The assembly of the surveillance complex together with UPF1 phosphorylation on PTC-containing mRNAs may trigger the release of UPF3b, or its replacement by a shorter isoform with lower affinity for the EJC. This rearrangement could allow SMG6 to bind the EJC only in mRNAs that terminate translation prematurely (Fig. 9). Alternatively, SMG6 may interact with the subset of EJCs lacking UPF3. As a corollary of this scenario, not all NMD targets would be cleaved by SMG6; and those that are not may depend partly on the SMG5–SMG7 complex for degradation. Finally, an alternative, UPF3-independent branch of the NMD pathway has been described (Chan et al. 2007), suggesting that UPF3 may not always compete with SMG6 for EJC binding.
In conclusion, we demonstrated that SMG6 interacts directly with the EJC, and that this interaction is required for NMD. These results establish a new role for the EJC in mediating multiple interactions with NMD factors and facilitating their function. More generally, our findings uncover a novel protein sequence motif: the EBM. We further show that EBMs are present in a handful of metazoan proteins in addition to SMG6 and UPF3. EBMs may allow these various proteins to hook onto mRNAs via the EJC, which in turn may provide a mechanistic explanation for the myriad functions performed by this protein complex in post-transcriptional processes such as mRNA export, localization, and translation, and NMD.
Materials and methods
DNA constructs
Plasmids expressing wild-type SMG6, SMG6-PIN-Mut, SMG6ΔPIN, and MBP were described before (Huntzinger et al. 2008). A plasmid expressing V5-tagged SMG6 was obtained by inserting the corresponding cDNA into the MluI and XhoI sites of pCIneo-V5-SBP. SMG6 mutants were generated by site-directed mutagenesis using pCIneo-V5-SBP-SMG6 as template, the QuikChange mutagenesis kit (Stratagene), and the appropriate oligonucleotide sequences. SMG6ΔN, Δ14-3-3, ΔPIN, ΔEBM1, and ΔEBM2 carry deletions of the following amino acids, respectively: 1–576, 577–816, 1239–1419, 42–58, and 136–152. Plasmids for expression of HA-tagged NMD factors or MBP were constructed using the mammalian expression vector pCI-neo-λN (Gehring et al. 2003) following standard procedures.
To express the SMG6 N-terminal fragment (residues 1–207) in E. coli, the corresponding cDNA was cloned into the EcoRI and XhoI restriction sites of the pGEX6P1 vector (GE Healthcare), resulting in an N-terminal GST fusion. Mutations were introduced using the QuikChange mutagenesis kit (Stratagene). GST-UPF3bΔN was obtained by inserting a UPF3b cDNA encoding residues 145–467 into the BamHI and XhoI sites of pGEX6P-1 (GE Healthcare). For the MBP-tagged UPF3bΔN construct, the cDNA was excised from the pGEX6P1-UPF3bΔN plasmid using restriction enzymes BamHI and NotI and cloned into the respective restriction sites of the pETM41 vector, resulting in an N-terminal fusion with MBP.
Plasmids for expression of GFP-tagged GIDRP88 and TDRD3 (amino acids 94–774) were obtained by inserting the corresponding cDNAs into the XhoI and EcoRI sites of pEGFC1 vector.
Coimmunoprecipitation assays and Western blotting
For coimmunoprecipitation assays, HEK-293 cells were grown in 10-cm plates and transfected using the calcium phosphate method. To express V5-tagged proteins, cells were transfected with 10 μg of plasmid and, 2 d later, lysed for 15 min on ice in lysis buffer (50 mM HEPES at pH 7.4, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 2.5 mM MgCl2, 0.5 mM DTT) supplemented with protease inhibitor (Roche). Cell lysates were spun at 18,000g for 15 min at 4°C. When indicated, the cleared lysates were treated with 200 μg/mL RNase A (Qiagen) for 30 min and respun at 18,000g for 10 min at 4°C. Mouse monoclonal anti-V5 (Invitrogen) or rabbit polyclonal anti-GFP antibodies was added to the supernatants (1:1000 dilution). Samples were incubated for 1 h at 4°C. Then, 25 μL of GammaBind G Sepharose (GE Healthcare) was added and the mixtures were rotated for an additional hour at 4°C. Beads were washed four times with lysis buffer lacking sodium deoxycholate. Bound proteins were eluted with 100 μL of protein sample buffer and analyzed by Western blotting.
V5-tagged proteins were detected with anti-V5 antibodies (1:5000 dilution; Invitrogen). Horseradish peroxidase-linked anti-HA high-affinity antibodies and mouse monoclonal antibodies were purchased from Roche. Y14 and Magoh were detected using rabbit polyclonal antibodies. Secondary horseradish peroxidase-linked anti-mouse IgG antibodies or horseradish peroxidase-linked protein A were purchased from GE Healthcare. Endogenous PABP was detected using anti-PABP antibody (1:3000 dilution; ab21060-100, Abcam). All Western blot experiments were developed with the ECL Western blotting detection system (GE Healthcare) as recommended by the manufacturer.
NMD assays in HeLa cells
Human NMD reporters have been described before (Thermann et al. 1998; Wang et al. 2002). The plasmid pEGFPC1-NXF1 (Unterholzner and Izaurralde 2004) served as a transfection control. Transfections of human HeLa cells were performed in six-well plates using the Lipofectamine 2000 transfection reagent. The siRNAs were transfected at a final concentration of 70 nM, together with a mixture of three plasmids: one expressing the reporters (0.25 μg of β-globin or TCR-β reporters with or without PTC), one expressing the transfection control (0.25 μg, pEGFPC1-NXF1), and a third (0.4 μg) expressing recombinant proteins or the corresponding empty vector. The following target sequences were used: SMG6 (5′-AAGCCAGTGATACAGCGAATT-3′), and the control β-Gal (5′-CTACACAAATCAGCGATTT-3′) (Dharmacon). Cells were harvested 3 d after transfection. Total RNA was isolated using TriFast (Peqlab Biotechnologies) and analyzed as described before (Unterholzner and Izaurralde 2004).
Protein expression and purification
GST-SMG6 (amino acids 1–207; wild type and mutants) were expressed for 3 h at 37°C in BL21 Star (DE3) cells (Invitrogen) in LB medium. Cells were resuspended in lysis buffer (50 mM Tris-HCl at pH 8.0, 500 mM NaCl, 1 mM DTT) supplemented with an EDTA-free protease inhibitor (Roche), 1 mg/mL lysozyme, and 5 μg/mL DNaseI, and were lysed by sonication. The lysate was cleared by ultracentrifugation at 41,000 rpm for 1 h and applied to 5 mL of pre-equilibrated glutathione agarose beads (Macherey-Nagel). The beads were washed with 2 column volumes (CV) of lysis buffer and the protein was eluted by incubating with 2 CV of lysis buffer containing 25 mM L-glutathione. Subsequently, proteins were dialyzed into low-salt buffer (50 mM Tris-HCl at pH 8.0, 50 mM NaCl, 1 mM DTT) and purified over a MonoQ column (GE Healthcare), eluting with a salt gradient >25 CV to 300 mM NaCl. To prepare untagged SMG6 (1–207), the GST tag was cleaved with Prescission protease after affinity purification. GST was removed by rebinding to glutathione beads before dialysis and anion exchange chromatography. Protein-containing fractions were pooled, concentrated, and loaded onto a Superdex 200 column (GE Healthcare) that was run in gel filtration buffer (50 mM Tris-HCl at pH 8.0, 250 mM NaCl, 1 mM DTT).
GST-tagged UPF3bΔN was expressed in BL21 (DE3) pLysS cells (Promega) grown in ZY autoinduction medium (Studier 2005) for 12 h at 20°C. Lysis was performed in buffer A (50 mM Tris-HCl at pH 7.5, 200 mM NaCl, 10% glycerol, 2 mM DTT) with protease inhibitor, lysonase, and DNaseI, using an emulsiflex. Glutathione affinity purification was performed as described above, using buffer A with 500 mM NaCl for washing and 25 mM L-glutathione for elution. The protein was then loaded directly onto a HiTrap heparin column (GE Healthcare) and eluted with a salt gradient >10 CV to 1.2 M NaCl. Protein fractions were pooled, concentrated, and purified over a Superdex 200 column in buffer A without glycerol. UPF3bΔN proteins were stored at −80°C in buffer containing 100 mM Tris-HCl (pH 7.5), 300 mM NaCl, 3 mM DTT, 1 mM MgCl2, and 20% (v/v) glycerol.
MBP-tagged UPF3bΔN was also expressed in BL21 (DE3) pLysS cells but in LB medium for 3 h at 37°C. Cells were lysed in lysis buffer at pH 7.5 and the protein was purified using an MBPTrap column (GE Healthcare). After purification over the heparin column, the protein was released from contaminating E. coli chaperones by incubation with 5 mM MgCl2 and 5 mM ATP for 1 h at room temperature, and was purified by gel filtration chromatography as described above. MBP-UPF3bΔN proteins were stored at −80°C in buffer containing 100 mM Tris-HCl (pH 7.5), 300 mM NaCl, 3 mM DTT, 1 mM MgCl2, and 20% (v/v) glycerol.
EJC pull-down assays
EJCs were assembled as described before (Buchwald et al. 2010). For binding assays, 2 μM preassembled EJC (in a total volume of 30 μL of buffer containing 25 mM Tris-HCl at pH 7.5, 250 mM NaCl, 2.5 mM magnesium acetate, 12.5% [v/v] gylcerol, 0.15% [v/v] NP-40, 2 mM DTT) was incubated with 7 μM wild-type or mutant GST-SMG6 1–207 or 1.1 μM GST-UPF3bΔN, plus 40 μL of GST beads (50% slurry) in binding buffer (20 mM Tris-HCl at pH 7.5, 250 mM NaCl, 2 mM magnesium acetate, 10% [v/v] glycerol, 0.15% [v/v] NP-40, 1 mM DTT) for 1 h at 4°C. The beads were washed three times with 500 μL of binding buffer and loaded onto filter-spin columns (MoBiTec). The proteins were eluted with 30 μL of elution buffer (20 mM Tris-HCl at pH 7.5, 200 mM NaCl, 2 mM magnesium acetate, 2.5% [v/v] gylcerol, 0.1% [v/v] NP-40, 1 mM DTT, 25 mM glutathione). After TCA precipitation, 10 μL of SDS loading buffer was added and samples were separated on a 4%–12% bis-tricine gradient gel (Invitrogen).
Sequence analysis
The repetition of a short sequence motif in the N-terminal domain of SMG6 proteins was detected during alignment in MACAW (Schuler et al. 1991). All subsequent bioinformatic analyses were performed in the MPI Bioinformatics Toolkit (http://toolkit.tuebingen.mpg.de) (Biegert et al. 2006). Consensus secondary structure prediction showed that the motif is part of an extended natively unstructured region, and that it has no pronounced secondary structure propensity. Equivalence of this motif to the EBM in UPF3 was established by HMM searches with profiles of the two motifs over an HMM database of all human proteins using HHpred (http://toolkit.tuebingen.mpg.de/hhpred), which showed that the two motifs are reciprocal best matches. Further occurrences of the motif in human proteins were identified by iterative HMM searches with HHpred and pooling of the identified sequences after every round. In addition to the statistical significance of the matches, two criteria served to validate the identified occurrences: They had to be part of an extended unstructured region, and they had to be conserved in homologs.
Acknowledgments
We are grateful to Maria Fauser and Sigrun Helms for excellent technical assistance, and Eric Huntzinger and Fulvia Bono for helpful discussions. This study was supported by the Max Planck Society, and by grants from the Deutsche Forschungsgemeinschaft (DFG, FOR855, and the Gottfried Wilhelm Leibniz Program awarded to E.I.).
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.604610.
Supplemental material is available at http://www.genesdev.org.
References
- Anders KR, Grimson A, Anderson P 2003. SMG-5, required for C. elegans nonsense-mediated mRNA decay, associates with SMG-2 and protein phosphatase 2A. EMBO J 22: 641–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballut L, Marchadier B, Baguet A, Tomasetto C, Séraphin B, Le Hir H 2005. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol 12: 861–869 [DOI] [PubMed] [Google Scholar]
- Biegert A, Mayer C, Remmert M, Söding J, Lupas AN 2006. The MPI Bioinformatics Toolkit for protein sequence analysis. Nucleic Acids Res 34: W335–W339 doi: 10.1093/nar/gkl217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono F, Ebert J, Lorentzen E, Conti E 2006. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell 126: 713–725 [DOI] [PubMed] [Google Scholar]
- Buchwald G, Ebert J, Basquin C, Sauliere J, Jayachandran U, Bono F, Le Hir H, Conti E 2010. Insights into the recruitment of the NMD machinery from the crystal structure of a core EJC–UPF3b complex. Proc Natl Acad Sci 107: 10050–10055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cali BM, Kuchma SL, Latham J, Anderson P 1999. smg-7 is required for mRNA surveillance in Caenorhabditis elegans. Genetics 151: 605–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan WK, Huang L, Gudikote JP, Chang YF, Imam JS, MacLean JA II, Wilkinson MF 2007. An alternative branch of the nonsense-mediated decay pathway. EMBO J 26: 1820–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Imam JS, Wilkinson MF 2007. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem 76: 51–74 [DOI] [PubMed] [Google Scholar]
- Chiu SY, Serin G, Ohara O, Maquat LE 2003. Characterization of human Smg5/7a, a protein with similarities to Caenorhabditis elegans SMG5 and SMG7 that functions in the dephosphorylation of Upf1. RNA 9: 77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV, Han X, Weng Y, Perlick HA, Dietz HC, Ter-Avanesyan MD, Peltz SW 1998. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes & Dev 12: 1665–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning G, Jamieson L, Maquat LE, Thompson EA, Fields AP 2001. Cloning of a novel phosphatidylinositol kinase-related kinase: Characterization of the human SMG-1 RNA surveillance protein. J Biol Chem 276: 22709–22714 [DOI] [PubMed] [Google Scholar]
- Eberle AB, Lykke-Andersen S, Mühlemann O, Jensen TH 2009. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol 16: 49–55 [DOI] [PubMed] [Google Scholar]
- Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E 2005. SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell 18: 537–547 [DOI] [PubMed] [Google Scholar]
- Gatfield D, Izaurralde E 2004. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature 429: 575–578 [DOI] [PubMed] [Google Scholar]
- Gehring NH, Neu-Yilik G, Schell T, Hentze MW, Kulozik AE 2003. Y14 and hUpf3b form an NMD-activating complex. Mol Cell 11: 939–949 [DOI] [PubMed] [Google Scholar]
- Glavan F, Behm-Ansmant I, Izaurralde E, Conti E 2006. Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. EMBO J 25: 5117–5125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulet I, Boisvenue S, Mokas S, Mazroui R, Côté J 2008. TDRD3, a novel Tudor domain-containing protein, localizes to cytoplasmic stress granules. Hum Mol Genet 17: 3055–3074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Kashima I, Fauser M, Saulière J, Izaurralde E 2008. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoa. RNA 14: 2609–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Maquat LE 2007. Quality control of eukaryotic mRNA: Safeguarding cells from abnormal mRNA function. Genes & Dev 21: 1833–1856 [DOI] [PubMed] [Google Scholar]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S 2006. Binding of a novel SMG-1–Upf1–eRF1–eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes & Dev 20: 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz JB, Neu-Yilik G, Hentze MW, Kulozik AE, Gehring NH 2006. Functions of hUpf3a and hUpf3b in nonsense-mediated mRNA decay and translation. RNA 12: 1015–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ 2000. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J 19: 6860–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder B, Plöttner O, Kroiss M, Hartmann E, Laggerbauer B, Meister G, Keidel E, Fischer U 2008. Tdrd3 is a novel stress granule-associated protein interacting with the Fragile-X syndrome protein FMRP. Hum Mol Genet 17: 3236–3246 [DOI] [PubMed] [Google Scholar]
- Nagy E, Maquat LE 1998. A rule for termination-codon position within intron-containing genes: When nonsense affects RNA abundance. Trends Biochem Sci 23: 198–199 [DOI] [PubMed] [Google Scholar]
- Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Mühlemann O 2010. Nonsense-mediated mRNA decay in human cells: Mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell Mol Life Sci 67: 677–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M, Dlakic M, Tollervey D 2004. A pre-ribosome-associated HEAT-repeat protein is required for export of both ribosomal subunits. Genes & Dev 18: 196–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T, Yamashita A, Kashima I, Schell T, Anders KR, Grimson A, Hachiya T, Hentze MW, Anderson P, Ohno S 2003. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell 12: 1187–1200 [DOI] [PubMed] [Google Scholar]
- Page MF, Carr B, Anders KR, Grimson A, Anderson P 1999. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol Cell Biol 19: 5943–5951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal M, Ishigaki Y, Nagy E, Maquat LE 2001. Evidence that phosphorylation of human Upf1 protein varies with intracellular location and is mediated by a wortmannin-sensitive and rapamycin-sensitive PI 3-kinase-related kinase signaling pathway. RNA 7: 5–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbapragada I, Lykke-Andersen J 2009. Execution of nonsense-mediated mRNA decay: What defines a substrate. Curr Opin Cell Biol 21: 394–402 [DOI] [PubMed] [Google Scholar]
- Schuler GD, Altschul SF, Lipman DJ 1991. A workbench for multiple alignment construction and analysis. Proteins 9: 180–190 [DOI] [PubMed] [Google Scholar]
- Studier WF 2005. Protein production by auto-induction in high-density shaking cultures. Protein Expr Purif 1: 207–234 [DOI] [PubMed] [Google Scholar]
- Thermann R, Neu-Yilik G, Deters A, Frede U, Wehr K, Hagemeier C, Hentze MW, Kulozik AE 1998. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J 17: 3484–3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Izaurralde E 2004. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell 16: 587–596 [DOI] [PubMed] [Google Scholar]
- Wang J, Vock VM, Li S, Olivas OR, Wilkinson MF 2002. A quality control pathway that down-regulates aberrant T-cell receptor (TCR) transcripts by a mechanism requiring UPF2 and translation. J Biol Chem 277: 18489–18493 [DOI] [PubMed] [Google Scholar]
- Wittkopp N, Huntzinger E, Weiler C, Saulière J, Schmidt S, Sonawane M, Izaurralde E 2009. Nonsense-mediated mRNA decay effectors are essential for zebrafish embryonic development and survival. Mol Cell Biol 29: 3517–3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann J, Hol EM, Jäck H-M 2006. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol Cell Biol 26: 1272–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Ohnishi T, Kashima I, Taya Y, Ohno S 2001. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes & Dev 15: 2215–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]