Abstract
Atmospheric pressure imaging mass spectrometry is a rapidly expanding field that offers advantages in the ability to study biological systems in their native condition, simplified sample preparation, and high-throughput experiments. In laser ablation electrospray ionization (LAE-SI), the native water molecules in biological tissues facilitate sampling by a focused mid-infrared laser beam. The ionization of the ablated material is accomplished by electrospray postionization. In this work, we demonstrate that the imaging variant of LAESI simultaneously provides lateral distributions for small metabolites and lipids directly in rat brain sections. To cope with the fragile nature and potential dehydration of the brain tissue due to drying in the ambient environment as well as to minimize analyte redistribution, a Peltier cooling stage is integrated into the LAESI imaging system. We demonstrate the utility of high-resolution (m/Δm > 6000) time-of-flight mass spectrometry with LAESI to deconvolute spatial distributions of different chemical species with identical nominal mass. To help with the evaluation of the massive data sets, Pearson colocalization maps are calculated for selected small metabolites and lipids. We show that this approach reveals biologically meaningful correlations between these two classes of biomolecules.
Continued developments in imaging mass spectrometry (MS) have provided a variety of techniques to report on the distribution of endogenous molecules and xenobiotics in biological tissues. The biological sciences and drug discovery have benefited from imaging MS methods,1 such as matrix-assisted laser desorption ionization (MALDI) and secondary ion mass spectrometry (SIMS), and most recently, nanostructure initiator mass spectrometry (NIMS). These techniques have demonstrated exceptional capabilities in molecular imaging of animal tissue and whole-body ections under vacuum conditions.2–9 To probe biochemical processes under native conditions, similar imaging capabilities are needed for the atmospheric pressure environment.
Molecular imaging with MS under atmospheric pressure conditions is based on emerging techniques that offer simplified sample preparation and a capability for high-throughput analysis.10–12 Although the imaging time for a ~1 cm2 area can be measured in hours, the acquired data set contains information on the distribution of hundreds of chemical species. This compares favorably with the time requirement for, e.g., optical methods that require separate tagging or staining for every species of interest. Among the direct ionization MS techniques, the spatial organization of small biomolecules and xenobiotics in biological tissues has been studied with the imaging variant of desorption electrospray ionization (DESI),13–16 atmospheric pressure infrared matrix-assisted laser desorption ionization (AP IR-MALDI)17,18 laser ablation electrospray ionization (LAESI),19–21 and most recently, probe electrospray ionization (PESI).22
LAESI MS is particularly tailored for biological samples with appreciable water content. In this technique, the sample is kept at atmospheric pressure and a focused mid-IR laser beam of 2940 nm wavelength excites the OH vibrations in its water molecules. As rapid microscale ablation driven by phase explosion sets in, a mixture of molecules, clusters, and particulate matter is ejected from the sample.19,23–25 The catapulted biomolecules then coalesce with charged droplets, produced by an electrospray, and a fraction of them is converted into gas-phase ions. Imaging by LAESI MS is realized by rastering the tissue surface with the laser beam across a selected area, while the generated ions are mass-analyzed and mass spectra are recorded. Using conventional focusing techniques, LAESI MS has shown success in uncovering the lateral and cross-sectional distribution of primary and secondary metabolites in plant leaves with 350 μm lateral and 40 μm depth resolution, respectively.20,26 Recently, based on focusing the mid-infrared radiation through a sharpened optical fiber tip, LAESI MS of single plant and animal cells has also been demonstrated.27 Typical LAESI mass spectra exhibit 200–300 ions related to metabolites and lipids in plant and animal tissues. Experiments on untreated fish tissues have demonstrated the feasibility of detecting numerous small metabolites and lipids.28
Small metabolites (m/z < 500) constitute a diverse group composed of building blocks for biosynthesis and products of degradation, species related to energy production, and signaling and defense molecules, among others. The interaction of small metabolites and lipids often modulates the role of the latter in the cell membranes and as signaling molecules. Imaging of intact small metabolites and lipids has been demonstrated by MALDI MS in vacuum.29 Simultaneous imaging of these two classes of biochemicals is yet to be demonstrated at atmospheric pressure.
The present study reports on the mass spectrometric imaging of rat brain sections with the LAESI technique in the atmospheric pressure environment. In comparison to the plant tissues stiffened by rigid cell walls in previous LAESI MS experiments, the brain tissue sections are softer and exhibit a much lower tensile strength, calling for special arrangements in the imaging experiments. For example, tissue handling, temperature control over an extended period of time (> 3 h), moisture condensation during ambient analysis, and a need for higher spatial resolution are apparent challenges. The simultaneous imaging of small metabolites and lipids in rat brain tissues by imaging LAESI MS reveals biologically meaningful correlations between these two classes of biomolecules. The importance of high mass resolving power is demonstrated for the imaging different chemical species with identical nominal mass. Colocalization maps are introduced to help identify anatomical areas in the brain tissue where biochemical correlations are observed.
EXPERIMENTAL SECTION
Laser Ablation Electrospray Ionization
The LAESI source was similar to the one we have recently described.19,20 The experimental parameters are summarized in Supporting Information Table S1. A coronal brain section was thaw-mounted on a microscope slide attached to a three-axis translation stage and positioned 10 mm below the electrospray axis. A Nd:YAG laser-driven optical parametric oscillator (Vibrant IR, Opotek Inc., Carlsbad, CA) provided mid-infrared pulses at 2940 nm wavelength and 20 Hz repetition rate. This laser beam was used to ablate samples ~5 mm downstream from the spray emitter tip at right angle. The average output energy of a laser pulse was ~100 μJ, which after focusing with a plano–convex ZnSe lens (Infrared Optical Products, Farmingdale, NY), translated into a fluence of ~0.3 J/cm2 at the sample surface.
Mass Spectrometry
The ablated material was intercepted by the electrospray plume, and the resulting ions were analyzed by an orthogonal acceleration time-of-flight mass spectrometer (Q-TOF Premier, Waters Co., Milford, MA) with a 0.5 s/spectrum integration time. The sampling cone of the mass spectrometer was located on axis with the spray emitter and 12 mm away from its tip. Electrospray solvent spectra were recorded without the laser ablation on and subtracted from the LAESI spectra. The mass spectra were externally calibrated with the collision-activated dissociation (CAD) fragments of the doubly protonated human [Glu1]-fibrinopeptide B peptide (Sigma-Aldrich), resulting in a mass accuracy of ~5 mDa and a mass resolution of mΔ/m >6000 between m/z 50 and 1000 in all experiments. In the CAD experiments, fragmentation was achieved in argon collision gas at 4 × 10−3 mbar pressure with the collision energy set between 10 and 30 eV.
Molecular Imaging with LAESI MS
A three-axis translation stage with motorized actuators (LTA-HS, Newport Corp., Irvine, CA) scanned the sample surface on a two-dimensional grid. The actuators had a travel range of 50 mm and a minimum incremental motion of 0.1 μm. In the imaging experiment, the sample plate, e.g., the microscope slide that holds the brain section, is moved in the focal plain of the laser in X and Y directions with lateral step sizes larger than or equal to the spot size of the laser. Thus, the ultimate resolution was determined by the focusing of the incident laser beam to ~200 μm diameter ablation craters. Under these conditions, lateral imaging of a 12.5 × 10.5 mm2 tissue area required a total imaging time of ~3 h. At each coordinate, the generated ions were analyzed and recorded by the mass spectrometer. Molecular images were then constructed by correlating the signal intensity of a particular ion with the absolute coordinate of the analysis. Software was written in-house (LabView 8.0, National Instruments Co., Austin, TX) to position the translation stage and render the analysis times to the corresponding coordinates. The exported data sets for particular ions were presented as contour plot images with a scientific visualization package (Origin 7.0, OriginLab Co., Northampton, MA).
The molecular imaging experiments were performed under conditions to maintain optimal signal-to-noise ratios. Table S1 in the Supporting Information lists the critical experimental parameters and their values. Because the imaging experiments took place at atmospheric pressure, a number of preventive steps were taken to minimize environmental effects. To prevent molecular migration in the sample, a Peltier cooling stage, equipped with a heat sink and fans, kept the tissue section frozen during the experiment. Additionally, a miniature enclosure surrounded the sample stage to minimize the emission of airborne particles into the environment. To eliminate condensation on the sample, the enclosure was filled with dry nitrogen gas.
Chemicals
Glacial acetic acid (TraceSelect grade), gradient grade methanol and water, adenosine, 2′-deoxyguanosine, N-acetylneuraminic acid, and human [Glu1]-fibrinopeptide B (>96% purity with HPLC) were obtained from Sigma-Aldrich and were used as received.
Preparation of Rat Brain Sections
All animal use and handling in this work followed the Guide for the Care and Use of Laboratory Animals (NIH). A male Sprague–Dawley rat (Harlan Industries, Indianapolis, IN) between 300 and 420 g was used. The rat was euthanized with intraperitoneal injection of sodium pentobarbital and decapitated upon cessation of respiration. The brain was quickly removed from the skull and frozen in dry ice-chilled isopentane for 15 s, prior to storage at −80 °C. For cryosectioning the frozen rat brain was transferred from −80 °C to the cryostat chamber (CM 3050 S; Leica Microsystems Nussloch GmbH, Nussloch, Germany) at −20 °C, where it was allowed to thermally equilibrate for 45 min. The tissue sample was attached to the cryostat sample stage using ice slush made from distilled water. The ice slush only came in contact with the tissue blocks at the surface opposing the sample stage and was frozen into a thin layer of ice within 5 s. This procedure was described in detail for MALDI MS imaging applications.30
The brain was cut into 100 μm sections. Serial brain sections were alternately collected onto clean microscope slides kept on dry ice. The slides were stored at −80 °C until LAESI analysis. Selected tissue sections on the microscope slides were stained with cresyl violet and a rat brain atlas31 was used for the assignment of the brain regions analyzed by MS.
RESULTS AND DISCUSSION
Direct Analysis of Rat Brain Tissue
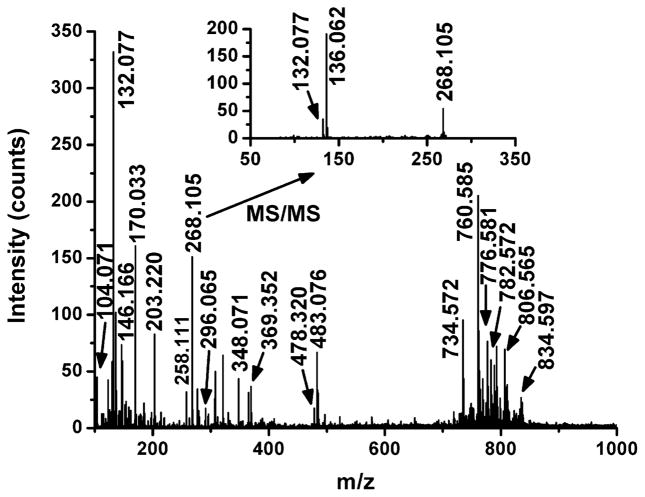
Initially the ions detected by LAESI MS from the rat brain tissue section were evaluated. To evaluate the role of potential molecular degradation upon laser exposure, we tested the shot-to-shot stability of the ion signal for individual m/z values. Similar to our experience with plant tissues,21 the outcome of these studies clearly indicated that consecutive laser pulses showed modest fluctuation in ion intensities but no noticeable trend in the average. A positive-ion mass spectrum recorded for the brain section is shown in Figure 1. A survey of the spectra indicated the presence of more than 200 sample-related ionic species. Some of the detected ions were assigned to particular metabolites in a multistep approach (see the Supporting Information for the protocol), and the assignments were confirmed by high-accuracy tandem MS. For example, for the m/z 268.105 ion, the database search returned the adenosine and its structural isomer deoxyguanosine (m/z 268.104, theoretical) along with neuraminic acid (m/z 268.103, theoretical) with 3 and 9 ppm mass accuracy, respectively. The inset of Figure 1 shows that CAD of the m/z 268.105 ion produced an abundant fragment at m/z 136.062. Fragmentation patterns induced by CAD for the related standards (see Figure S1 in the Supporting Information) showed that only the behavior of the adenosine was consistent with the CAD mass spectrum recorded for m/z 268.105 directly from the rat brain tissue. Formation of the m/z 136.062 fragment was not observed for the deoxyguanosine or neuraminic acid standards. A list of assignments for 27 ions is presented in Table S2 of the Supporting Information.
Figure 1.
Positive-ion LAESI mass spectrum from a selected pixel of a coronal rat brain section. The inset shows the CAD-induced tandem mass spectrum of the m/z 268.105 ion. The fragment ion at m/z 136.062 is consistent with protonated adenine, [C5H5N5 + H]+, (m/z 136.062, theoretical) within 1 ppm mass accuracy.
The low-mass region of the spectrum (m/z < 500) included several metabolite ions, such as the neurotransmitter molecules γ-aminobutyric acid and choline, and the polyamines spermidine and spermine. Essential metabolites for chemical energy transfer, specifically, the adenosine and adenosine monophosphate, were also observed. The ion measured at m/z 369.352 was assigned to the species produced by water loss from the cholesterol molecule, also observed by MALDI and SIMS methods.32–36 Indeed, in a control experiment, a standard solution of cholesterol gave abundant ions in the form of [M − H2O + H+] rather than [M + H]+. Ethanolamine (m/z 84.043) was also present with appreciable counts in the spectrum.
In the high-mass region (m/z > 500), the spectrum was dominated by lipid species. Lipid assignments were facilitated by tandem MS, and the observed fragmentation channels were evaluated in light of literature results.35 In earlier CAD studies, production of the phosphocholine headgroup at m/z 184.07 and its fragment with m/z 86.10 was considered diagnostic of glycerophosphocholines (PC), whereas decomposition of the glycerophosphoethanolamines (PE) characteristically occurred through the neutral loss of the phosphoethanolamine unit (see Figure S2 in the Supporting Information). This enabled us to differentiate between two major structural isomer groups, the PC(n:m) and PE((n + 3):m), where n is the combined length of the acyl chains and m is the number of double bonds. Due to the large variety of additional structural isomers, however, unambiguous identification of a particular lipid usually requires extensive studies, including chromatographic techniques and NMR. Our assignments are made with the understanding that no single technique can resolve all possible structural isomers. Nevertheless, tentative assignments are listed for 12 selected lipid ions in Table S2 of the Supporting Information. Their accurate masses are consistent with the measured values within 10 mDa. Structure alternatives between PC(n:m) and PE((n + 3):m) were resolved based on the tandem mass spectra.
The m/z 768.553 ion presented an interesting case because its fragmentation behavior was consistent with that of both PC(35:4) and PE(38:4). These lipids are structural isomers; thus, their unambiguous assignment is usually aided by chromatography. The tandem MS data acquired in our experiments suggested that both lipid species were present in the tissue.
Above m/z 600, a series of multiply charged ions also emerged in the spectrum. Deconvolution of the charge state distribution provided singly protonated ion masses with m/z 15197.3 and 15848.3, respectively, in good agreement with the primary amino acid sequence for the α1 and β1 subunit of rat hemoglobin that predicted37 protonated masses of 15197.8 and 15849.2, respectively. The m/z 616.176 ion was assigned to the heme prosthetic group of hemoglobin, consistent with the presence of the hemoglobin-related multiply charged ions.
Molecular Imaging of Brain Tissue by LAESI MS
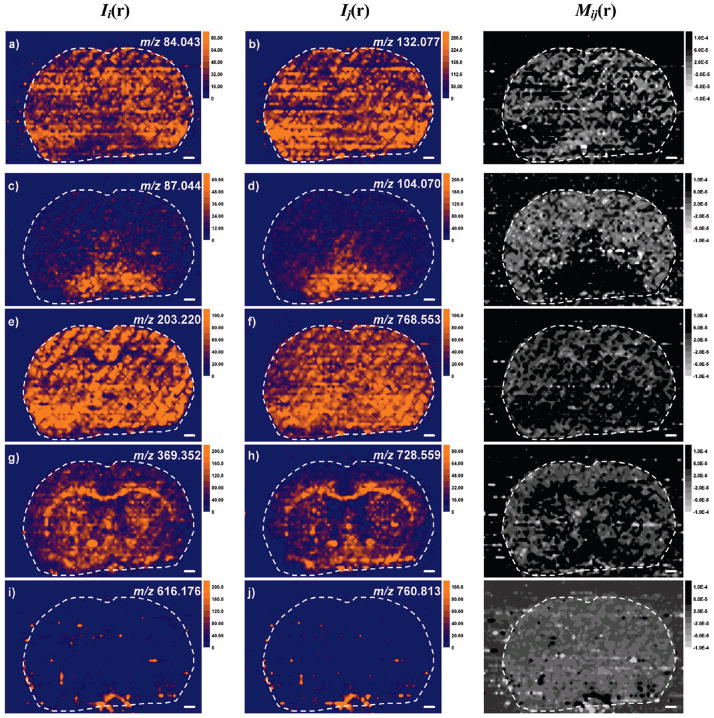
The imaging LAESI MS experiments yielded a large set of ion distributions for evaluation. Spatial distributions of selected ions recorded in the brain section are shown in Figure 2. The optical image of a stained tissue section was compared to rat brain atlases31,38 to identify the major anatomical features (see Figure S3 in the Supporting Information). The cerebral cortex, corpus callosum, caudate putamen, intergeniculate leaf, anterior commissure, and cingulum regions are marked in Supporting Information Figure S3 for reference.
Figure 2.
Atmospheric pressure imaging of positively charged molecular ions in a coronal section of the rat brain with LAESI MS reveals pairs of metabolite ions with highly correlated (ρij > 0.7) spatial distributions: (a) ethanolamine with (b) creatine; (c) γ-butyrolactone with (d) γ–aminobutyric acid and/or 2-methylalanine; (e) spermine with (f) PC(35:4) and PE(38:4); (g) cholesterol–H2O with (h) plasmalogens PC(O-33:3) and/or PE(O-36:3); and (i) heme with (j) α chain of hemoglobin. To the right of each pair is the corresponding colocalization map, Mij(r). See the definition in the text. Scale bars in white correspond to 1 mm.
The molecular ions exhibited a large variety of distributions in the tissue section. For example, Figure 2c reveals that γ-butyrolactone is observed at higher intensities in the septal complex and in the vicinity of the anterior commissure. Distinctively different ion distribution was noted for the spermine ion (Figure 2e) that showed high intensities throughout the entire section except for the corpus callosum. The cerebral cortex exhibited the adenosine ion with very high intensities (see Figure S4b in the Supporting Information). The cholesterol fragment (Figure 2g) and the plasmalogens PC(O-33:3) and/or PE(O-36:3) (Figure 2h) were detected with high abundance in the corpus callosum and in areas around the anterior commissure. These plasmalogens and the m/z 792.561 ion assigned to PC(37:6) and PE(40:6) (see Figure S4d in the Supporting Information) exhibited complementary presence. For the latter, abundant ion signal was registered across the entire tissue including the cerebral cortex and the caudate putamen, except for the corpus callosum region where the ion signal became very low. Close to homogeneous localization was noticed for the ion m/z 768.553 (Figure 2f), corresponding to the PC(35:4) and PE(38:4). The heme group was observed in regions (Figure 2i) that also appeared to be stained with blood.
Correlation and Colocalization Analysis
To explore the spatial correlation of metabolite distributions simple Pearson product-moment cross-correlation coefficients, ρij, were calculated between all possible pairs of ion intensity distributions, Ii(r) and Ij(r):
where cov is the covariance of the two ion intensities in the image and σi and σj stand for their standard deviations, respectively. This number is used as a measure of overall correlation between two images. For a more detailed assessment of the correlation, Pearson colocalization maps, Mij(r), were calculated:
where 〈Ii〉 and 〈Ij〉 are the average values for the corresponding ion intensity distributions. Similar colocalization analysis has been performed on protein distributions in confocal microscope images of neuronal tissues produced by immunostaining39 and on MALDI mass spectrometric images.40 In some cases, artifacts, such as ion suppression effects, may bias the correlation coefficients. Validation of potential biochemical pathways based on colocalization analysis requires the use of independent techniques.
Table S3 in the Supporting Information section lists the cross-correlation coefficient values calculated among 25 selected ions. Some highly correlated pairs (ρij > 0.7) and the corresponding colocalization maps are shown side by side in Figure 2. Inspecting the pairs in parts c and d of Figure 2 (γ-butyrolactone vs γ-aminobutyric acid and/or 2-methylalanine), parts g and h of Figure 2 [cholesterol–H2O vs plasmalogens PC(O-33:3) and/or PE(O-36:3)], and parts i and j of Figure 2 (heme vs the α chain of hemoglobin), the similarities are apparent. Indeed, the related colocalization maps in Figure 2 show high-intensity (dark) areas. For example, cholesterol and the plasmalogens colocalize in the corpus callosum, the anterior commissure, and its vicinity.
For the distributions in parts a and b of Figure 2 (ethanolamine vs creatine) and parts e and f of Figure 2 [spermine vs PC(35:4) and PE(38:4)] the parallels are more subtle but discernible by inspecting the colocalization maps. There is also strong correlation between the distributions of certain lipids, e.g., PC(36:1) and PC(38:4) with ρij = 0.85 (see Supporting Information Tables S2 and S3).
For some of the highly correlated pairs, the reason behind the spatial tracking is related to a common precursor. For example, the spatial distribution of the ions produced from heme and the α-1 chain of hemoglobin exhibited strong correlation likely because both formed from hemoglobin and, thus, followed its localization in the tissue. In other cases, understanding the high correlation might come from the underlying metabolic or other biochemical relationships between the chemical species involved. For example, the spatial correlation of spermine in Figure 2e with the lipids in Figure 2f (ρij = 0.75) as well as with other lipids [see Supporting Information Tables S2 and S3 for PC(35:4) and PE(38:4) (ρij = 0.75), PC(38:6) (ρij= 0.71) and PC(40:6) (ρij = 0.71)], can be rationalized based on the known role of polyamines in electrostatically shielding repulsive forces in lipid membranes.41 Polyamines are also implicated in membrane fusion and in the modulation of enzyme activity in phospholipid synthesis. Another highly correlated pair, ethanolamine and creatine (ρij = 0.71) (see Figure 2, parts a and b), is involved in phosphate metabolism in the brain.42
Parts g and h of Figure 2 and their colocalization map demonstrate that strong correlation is also found between cholesterol and the plasmalogens PC(O-33:3) and/or PE(O-36:3) (ρij= 0.76). Studies show that ethanolamine plasmalogens are necessary for cellular cholesterol transport.43 The tight correlation between γ-butyrolactone and γ-aminobutyric acid in parts c and d of Figures 2 (ρij = 0.81) probably reflects that the latter is the metabolic precursor of the former. In the brain γ-butyrolactone is converted into γ-hydroxybutyric acid by lactamase that, in turn, is converted into γ-aminobutyric acid by GABA transaminase.44
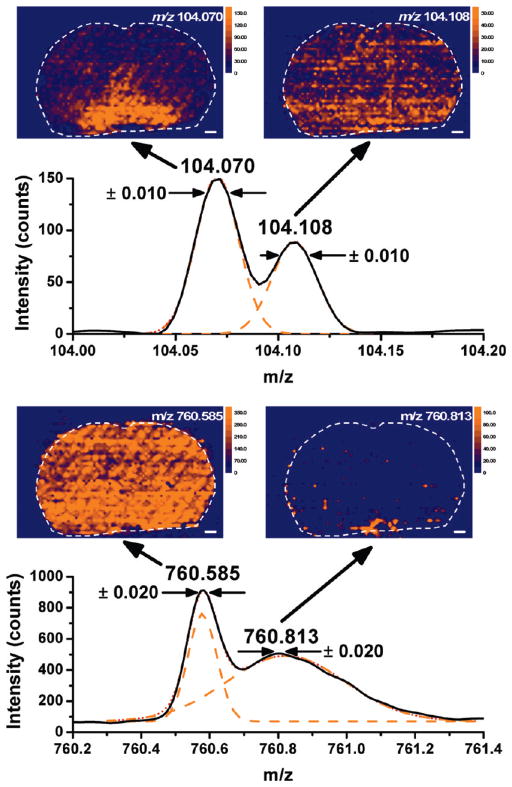
The spatial distributions for molecular ions with identical nominal mass were investigated in LAESI imaging experiments with high mass resolving power. Transmitting ions in a 10 mDa window around the centroids of the component peaks (see Figure 3) enabled the successful separation of the protonated γ-aminobutyric acid and choline+. Shown in the insets of Figure 3, the deconvoluted distributions exhibited large qualitative differences. Without sufficient mass resolving power these two components would have been lumped together resulting in a flawed overall distribution. The accurate masses of PC(34:1) and the 20-times protonated α-1 chain of rat hemoglobin differ by 228 mDa. Although the multiply charged hemoglobin peak is much broader, the Gaussian deconvolution shown in Figure 3 confirmed that a 20 mDa window provided acceptable separation between these ions. The recovered distributions were similar to the tissue accumulation of other lipids for PC(34:1) and to the heme unit for the hemoglobin.
Figure 3.
High mass resolving power enabled the separate imaging of positively charged species with identical nominal mass. The m/z values for the γ-aminobutyric acid (m/z 104.070) and choline (m/z 104.108) ions differed by only 38 mDa, whereas the PC(34:1) (m/z 760.585) and the multiply charged α-1 chain of the rat hemoglobin (m/z 760.813) were separated by 228 mDa. Deconvolution of these peaks (solid black line) enabled us to render the separated species (orange lines) into distinct images.
The results obtained with LAESI MS imaging were in qualitative agreement with studies that applied independent methodologies. For example, MALDI, cluster ion SIMS, and DESI imaging have indicated high abundance of cholesterol in the corpus callosum and the anterior commissure of rodents, areas that are rich in myelinated axons.6,33,34,36,45,46 Strategies based on dissection, extraction, and spectrofluorometric quantitation of brain metabolites found regional differences between spermidine and spermine47,48 similar to our findings (see Figure 2e and Supporting Information Figure S4e for the spermine and spermidine distributions, respectively). Regarding the lipid content, the PC(36:1) image in Supporting Information Figure S4c correlates particularly well with recent MALDI images.49
CONCLUSIONS
We presented several technical and methodological developments that allowed the extension of direct LAESI MS imaging from plant tissues to rat brain sections. Maintaining the native water content of the brain sections for successful mid-IR laser ablation presented nontrivial technical challenges associated with the potential dehydration of the sample in the ambient environment, its very low tensile strength, as well as the possibility of molecular migration upon tissue melting. A Peltier cooling stage with optimized heat transport kept the brain sections frozen for the duration of the analysis. Additionally, dry nitrogen environment prevented moisture condensation on the sample surface.
The results presented here demonstrate the feasibility of LAESI imaging MS on chemically untreated sections of rat brain at atmospheric pressure. High m/z precision and resolving power as well as tandem mass capabilities facilitated metabolite identification. Although a 200 μm spatial step size provided informative molecular images across the tissue sections, further improvement is needed in lateral resolution. In order to maintain the duration of the experiments within practical limits, improved spatial resolution requires faster surface scanning and higher laser pulse repetition rates. In the present study, neither of these parameters is at their technical limits, so further improvement in spatial resolution by at least a factor of 2 is feasible.
It is also essential to refine our understanding of analyte response over a wide set of molecular classes in the LAESI MS experiments. Although a large variety of primary and secondary metabolites as well as lipids are detected with LAESI, the coverage for other components remains to be determined. To improve the response for other molecular classes, in-plume reactions can be induced by introducing ionizing reactants into the electrospray or the gas environment.
The positive-ion mode molecular images provided by LAESI were consistent with literature results obtained by MALDI, SIMS, and DESI imaging methods. Although LAESI enables the generation of negative ions directly from tissues for local analysis, spray stability must be improved for imaging applications in this ion mode.
We demonstrated the utility of high-resolution MS to deconvolute spatial distributions of different chemical species with identical nominal mass. These species could not be distinguished in low-resolution spectra, resulting in a skewed distribution. This example shows the importance of mass resolving power in imaging and calls for continued improvement in this figure of merit.
The Pearson cross-correlation method and colocalization maps, adopted for finding synchronous variations in ion intensities, are informative tools for initial analysis in our imaging experiments. They, however, only measure linear relationships between the image data sets. More sophisticated methods of image analysis, such as Mander’s coefficients or Spearman’s rank correlation coefficient, can help to further explore the colocalization information in these large data sets. LAESI mass spectrometric imaging under ambient conditions offers a new approach to the mapping of biomolecules in animal tissues.
Supplementary Material
Acknowledgments
The authors are grateful for the financial support of this work by the U.S. National Science Foundation under Grant No. 0719232, by the U.S. Department of Energy (DEFG02-01ER15129), by the W. M. Keck Foundation (041904), and by Protea Biosciences, Inc. (Morgantown, WV) The help by J. D. Post and A. Delvolve in producing the rat brain sections is greatly appreciated.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Protocol for metabolite assignment with examples of tandem mass spectra for the chemical standards adenosine, 2′-deoxyguanosine, N-acetylneuraminic acid, and the lipid classes of glycerophosphocholines and glycerophosphoethanolamines (Figures S1 and S2, respectively), the optical image of a stained coronal rat brain section (Figure S3), complements of the molecular ion images shown in Figure 2 (Figure S4), optimized experimental parameters (Table S1), the tentative assignments for small metabolites and lipids (Table S2), and the Pearson product-momentum cross-correlation coefficients for selected pairs of ions (Table S3). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Rubakhin SS, Jurchen JC, Monroe EB, Sweedler JV. Drug Discovery Today. 2005;10:823–837. doi: 10.1016/S1359-6446(05)03458-6. [DOI] [PubMed] [Google Scholar]
- 2.Ostrowski SG, Van Bell CT, Winograd N, Ewing AG. Science. 2004;305:71–73. doi: 10.1126/science.1099791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cornett DS, Reyzer ML, Chaurand P, Caprioli RM. Nat Methods. 2007;4:828–833. doi: 10.1038/nmeth1094. [DOI] [PubMed] [Google Scholar]
- 4.Yanes O, Woo HK, Northen TR, Oppenheimer SR, Shriver L, Apon J, Estrada MN, Potchoiba MJ, Steenwyk R, Manchester M, Siuzdak G. Anal Chem. 2009;81:2969–2975. doi: 10.1021/ac802576q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khatib-Shahidi S, Andersson M, Herman JL, Gillespie TA, Caprioli RM. Anal Chem. 2006;78:6448–6456. doi: 10.1021/ac060788p. [DOI] [PubMed] [Google Scholar]
- 6.McDonnell LA, Heeren RMA. Mass Spectrom Rev. 2007;26:606–643. doi: 10.1002/mas.20124. [DOI] [PubMed] [Google Scholar]
- 7.Zhang H, Cha SW, Yeung ES. Anal Chem. 2007;79:6575–6584. doi: 10.1021/ac0706170. [DOI] [PubMed] [Google Scholar]
- 8.Andersson M, Groseclose MR, Deutch AY, Caprioli RM. Nat Methods. 2008;5:101–108. doi: 10.1038/nmeth1145. [DOI] [PubMed] [Google Scholar]
- 9.Northen TR, Yanes O, Northen MT, Marrinucci D, Uritboonthai W, Apon J, Golledge SL, Nordstrom A, Siuzdak G. Nature. 2007;449:1033–U1033. doi: 10.1038/nature06195. [DOI] [PubMed] [Google Scholar]
- 10.Takats Z, Wiseman JM, Gologan B, Cooks RG. Science. 2004;306:471–473. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- 11.Cooks RG, Ouyang Z, Takats Z, Wiseman JM. Science. 2006;311:1566–1570. doi: 10.1126/science.1119426. [DOI] [PubMed] [Google Scholar]
- 12.Cody RB, Laramee JA, Durst HD. Anal Chem. 2005;77:2297–2302. doi: 10.1021/ac050162j. [DOI] [PubMed] [Google Scholar]
- 13.Wiseman JM, Ifa DR, Song QY, Cooks RG. Angew Chem, Int Ed. 2006;45:7188–7192. doi: 10.1002/anie.200602449. [DOI] [PubMed] [Google Scholar]
- 14.Wiseman JM, Ifa DR, Cooks RG, Venter A. Nat Protoc. 2008;3:517–524. doi: 10.1038/nprot.2008.11. [DOI] [PubMed] [Google Scholar]
- 15.Wiseman JM, Ifa DR, Zhu YX, Kissinger CB, Manicke NE, Kissinger PT, Cooks RG. Proc Natl Acad Sci USA. 2008;105:18120–18125. doi: 10.1073/pnas.0801066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esquenazi E, Dorrestein PC, Gerwick WH. Proc Natl Acad Sci USA. 2009;106:7269–7270. doi: 10.1073/pnas.0902840106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Shrestha B, Vertes A. Anal Chem. 2007;79:523–532. doi: 10.1021/ac061577n. [DOI] [PubMed] [Google Scholar]
- 18.Li Y, Shrestha B, Vertes A. Anal Chem. 2008;80:407–420. doi: 10.1021/ac701703f. [DOI] [PubMed] [Google Scholar]
- 19.Nemes P, Vertes A. Anal Chem. 2007;79:8098–8106. doi: 10.1021/ac071181r. [DOI] [PubMed] [Google Scholar]
- 20.Nemes P, Barton AA, Li Y, Vertes A. Anal Chem. 2008;80:4575–4582. doi: 10.1021/ac8004082. [DOI] [PubMed] [Google Scholar]
- 21.Nemes P, Barton AA, Vertes A. Anal Chem. 2009;81:6668–6675. doi: 10.1021/ac900745e. [DOI] [PubMed] [Google Scholar]
- 22.Chen LC, Yoshimura K, Yu Z, Iwata R, Ito H, Suzuki H, Mori K, Ariyada O, Takeda S, Kubota T, Hiraoka K. J Mass Spectrom. 2009;44:1469–1477. doi: 10.1002/jms.1632. [DOI] [PubMed] [Google Scholar]
- 23.Vertes A, Nemes P, Shrestha B, Barton AA, Chen ZY, Li Y. Appl Phys A: Mater Sci Process. 2008;93:885–891. [Google Scholar]
- 24.Chen ZY, Bogaerts A, Vertes A. Appl Phys Lett. 2006:89. [Google Scholar]
- 25.Chen ZY, Vertes A. Phys Rev E. 2008:77. [Google Scholar]
- 26.Nemes P, Barton AA, Vertes A. Anal Chem. 2009;81:6668–6675. doi: 10.1021/ac900745e. [DOI] [PubMed] [Google Scholar]
- 27.Shrestha B, Vertes A. Anal Chem. 2009;81:8265–8271. doi: 10.1021/ac901525g. [DOI] [PubMed] [Google Scholar]
- 28.Sripadi P, Nazarian J, Hathout Y, Hoffman EP, Vertes A. Metabolomics. 2009;5:263–276. [Google Scholar]
- 29.Benabdellah F, Touboul D, Brunelle A, Laprevote O. Anal Chem. 2009;81:5557–5560. doi: 10.1021/ac9005364. [DOI] [PubMed] [Google Scholar]
- 30.Jackson SN, Wang HYJ, Woods AS. J Am Soc Mass Spectrom. 2005;16:133–138. doi: 10.1016/j.jasms.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 31.Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates. Academic Press; New York: 1997. [Google Scholar]
- 32.Altelaar AFM, van Minnen J, Jimenez CR, Heeren RMA, Piersma SR. Anal Chem. 2005;77:735–741. doi: 10.1021/ac048329g. [DOI] [PubMed] [Google Scholar]
- 33.Brunelle A, Laprevote O. Anal Bioanal Chem. 2009;393:31–35. doi: 10.1007/s00216-008-2367-3. [DOI] [PubMed] [Google Scholar]
- 34.Jones EA, Lockyer NP, Vickerman JC. Int J Mass Spectrom. 2007;260:146–157. [Google Scholar]
- 35.Murphy RC, Hankin JA, Barkley RM. J Lipid Res. 2009;50:S317–S322. doi: 10.1194/jlr.R800051-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Touboul D, Halgand F, Brunelle A, Kersting R, Tallarek E, Hagenhoff B, Laprevote O. Anal Chem. 2004;76:1550–1559. doi: 10.1021/ac035243z. [DOI] [PubMed] [Google Scholar]
- 37. [accessed June 23, 2009];NCBI Entrez Protein Database. at http://www.ncbi.nlm.nih.gov/sites/entrez.
- 38. [accessed July 1, 2009];Brainmaps.org Brain Atlas Database. at http://brainmaps.org.
- 39.Li Q, Lau A, Morris TJ, Guo L, Fordyce CB, Stanley EF. J Neurosci. 2004;24:4070–4081. doi: 10.1523/JNEUROSCI.0346-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McDonnell LA, van Remoortere A, van Zeijl RJM, Deelder AM. J Proteome Res. 2008;7:3619–3627. doi: 10.1021/pr800214d. [DOI] [PubMed] [Google Scholar]
- 41.Schuber F. Biochem J. 1989;260:1–10. doi: 10.1042/bj2600001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin S, Cohen MM. J Neurochem. 1966;13:85–92. doi: 10.1111/j.1471-4159.1966.tb03335.x. [DOI] [PubMed] [Google Scholar]
- 43.Munn NJ, Arnio E, Liu DL, Zoeller RA, Liscum L. J Lipid Res. 2003;44:182–192. doi: 10.1194/jlr.m200363-jlr200. [DOI] [PubMed] [Google Scholar]
- 44. [accessed December 15, 2009];Human Metabolome Database. at http://www.hmdb.ca/metabolites/HMDB00549.
- 45.Wu CP, Ifa DR, Manicke NE, Cooks RG. Anal Chem. 2009;81:7618–7624. doi: 10.1021/ac901003u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sjovall P, Lausmaa J, Johansson B. Anal Chem. 2004;76:4271–4278. doi: 10.1021/ac049389p. [DOI] [PubMed] [Google Scholar]
- 47.Shaw GG, Pateman AJ. J Neurochem. 1973;20:1225–1230. doi: 10.1111/j.1471-4159.1973.tb00091.x. [DOI] [PubMed] [Google Scholar]
- 48.Seiler N, Schmidt-Glenewinkel T. J Neurochem. 1975;24:791–795. [PubMed] [Google Scholar]
- 49.Mikawa S, Suzuki M, Fujimoto C, Sato K. Neurosci Lett. 2009;451:45–49. doi: 10.1016/j.neulet.2008.12.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.