Abstract
This study presents the first comparative BAC map of the gilthead sea bream (Sparus aurata), a highly valuated marine aquaculture fish species in the Mediterranean. High-throughput end sequencing of a BAC library yielded 92,468 reads (60.6 Mbp). Comparative mapping was achieved by anchoring BAC end sequences to the three-spined stickleback (Gasterosteus aculeatus) genome. BACs that were consistently ordered along the stickleback chromosomes accounted for 14,265 clones. A fraction of 5,249 BACs constituted a minimal tiling path that covers 73.5% of the stickleback chromosomes and 70.2% of the genes that have been annotated. The N50 size of 1,485 “BACtigs” consisting of redundant BACs is 337,253 bp. The largest BACtig covers 2.15 Mbp in the stickleback genome. According to the insert size distribution of mapped BACs the sea bream genome is 1.71-fold larger than the stickleback genome. These results represent a valuable tool to researchers in the field and may support future projects to elucidate the whole sea bream genome.
1. Introduction
Genome mapping has become an essential tool in order to identify and characterize the genetic basis of phenotypic traits of organisms as well as to detect single genes of interest (e.g. disease related genes) or to improve selective breeding. Genome mapping also offers the possibility of comparative mapping, which brings valuable knowledge production potential about genome structure, function, and evolution. Several mapping methods exist that have shaped the work of geneticists in the past decades. Genetic linkage maps or radiation hybrid maps give a global view of genetic markers mapped to groups that in a best case scenario correspond to whole chromosomes [1, 2]. While these approaches characterize a species genome with relatively low marker density (about one marker per Mbp genomic sequence), today mapping approaches that use large insert clone libraries such as BACs (bacterial artificial chromosomes) [3, 4] have a higher impact, not only offering very dense maps but also revealing a part of the genome sequence and gene content. In the “age of genomics” mapped clones are of special value for the analysis of distinct genomic regions by a variety of molecular techniques. Especially, the sequencing of large genomes takes profit from BAC maps, as they enable the reconstruction of chromosomal sized sequences from draft whole genome shotgun assemblies or sequencing strategies that apply a hierarchical “first map then sequence” strategy. Among the methodologies to construct BAC maps, fingerprinting by restriction analysis [5–7] or hybridization of labeled marker sequences to BAC colony filters [8], as well as PCR-based methods [9, 10], have been widely used for a variety of species to build “de novo” maps of overlapping BAC clones. Today, protocols for high-throughput sequencing of BAC end sequences (BAC-ES) enable comparative mapping strategies that rely on sequence alignment to closely related species, which have been sequenced to completeness [11, 12]. This strategy is based on automated pipelines for sequencing and bioinformatics that have been established worldwide during the last decade enabling the genome analysis of a wide range of species.
Genomic information of species spanning a wider range of evolutionary distance will contribute to the complex questions of duplication and divergence. Furthermore, comparison between “genomic-rich” species and “genomic-poor” species has been a powerful tool for unravelling genes as well as specific genome regions of interest. In addition, comparative genomics is broadly used to identify and characterize functional regions in vertebrate genomes by the detection of evolutionary constrained sequences (purifying selection on functional sequences) [13], or for revealing directional selection operating on different parts of the genome. The available sequenced fish genomes span large evolutionary distances. The present work together with work on the European seabass, Dicentrarchus labrax, adds genomic information to much shorter distances. In addition, due to their interest for aquaculture these species have already piled up an important body of biological knowledge: they are dwelling in about the same geographic range with a relatively stable gradient of physicochemical parameters, but have distinct ecology, behaviour, sex determination systems, contrasting reproductive biology, differential stress tolerance to a set of stressors, differential tolerance to pathogens, physiology, and so forth. Therefore they offer a great case for comparative and population genomics approaches. These are applicable to closely related species for identifying adaptive evolution and functional variation by fine scale comparisons of genes and genomic regions and by simultaneous investigation of sequence polymorphism and divergence [14]. Whereas the assignment of homologues groups between closely related species can be achieved already by low density maps [15–17], chromosomes do not remain colinear over evolutionary time. Rearrangements take place by processes such as translocation, inversions, duplications, and deletions. Therefore, in addition to synteny mapping (on the same chromosome), reconstructing homologous colinearity by comparative BAC maps will enhance significantly the detection of new regions in the genome that are important for function as well as it will contribute to a better differentiation of lineage-specific constraint.
To date the comparative BAC mapping method is particularly suitable to map fish genomes, because several “good quality draft” reference fish genomes are already available in public databases for single or multispecies comparison. Five teleost fish genomes have been nearly completely sequenced (http://www.ensembl.org): the zebrafish (Danio rerio (Hamilton)) [18], the medaka (Oryzias latipes (Temminck and Schlegel)) [19] and the three-spined stickleback (Gasterosteus aculeatus L.) belonging to the order of Cypriniformes, Beloniformes, and Gasterosteiformes, respectively, as well as the two pufferfishes (Takifugu rubripes (Temminck and Schlegel) [20] and Tetraodon nigroviridis (Marion de Procé)) [21] belonging to the order of Tetraodontiformes. Sequencing of fish genomes is of particular interest as among vertebrates the ray-finned fishes (actinopterygians) are the most diverse groups comprising approximately 23,700 extant species [22]. Genome projects in teleosts were first focused on model species either due to their compact genome size (pufferfishes), their importance in developmental research (zebrafish), or as ecological model (three-spined stickleback). The genome analysis of the three main model fish species, the zebrafish (Danio rerio), pufferfish (Tetraodon nigroviridis) and medaka (Oryzias latipes), have shown that two rounds of genome duplications took place early in vertebrate evolution as well as a third round of genome duplication occurred in the ray-finned fish before the teleost radiation [23–25]. The interest to study nonmodel fish species first was based either on their importance in evolution or on their high commercial impact. The latter enhanced the research in the two main species of commercial interest in the Mediterranean, the gilthead sea bream Sparus aurata L. and the European seabass Dicentrarchus labrax L.
The gilthead sea bream belongs to the order Perciformes and Sparidae family which comprises about 100 species of which 24 are described from the North/Eastern Atlantic and Mediterranean coasts but also in the Red Sea and Australia-New Zealand. The family of Sparidae also includes several species such as Dentex dentex, Pagrus pagrus, Diplodus puntazzo which are important to Mediterranean aquaculture or Pagrus major which is routinely produced in Japan. Sparus aurata is one of the main target species for aquaculture and fisheries in Europe with 133,000 tons aquaculture production in 2008 (http://www.fao.org/fishery/culturedspecies/Sparus_aurata/en). Sea bream is a mass spawner having a 2-3 months sequential spawning of batches of eggs and a particular sex determination system that is characteristic also for other candidates in aquaculture. These species are sequential hermaphrodite, either protandrous or protogynous. Sea bream belongs to the protandrous hermaphrodite turning into females after the 2nd year [26–28]. Another characteristic behaviour of sea bream is cannibalism between juveniles of different size. In general sea bream is carnivorous. It feeds on molluscs, crustacean and small fish. We have recently published a comparative BAC map of the European seabass [29], which is closely related to the gilthead sea bream (both order of Perciformes). The three-spined stickleback Gasterosteus aculeatus genome was successfully used as reference genome in this regard. Stickleback does not belong to the order Perciformes, but is the most closely related genome to seabass among the currently available five sequenced teleost genomes. The comparative BAC mapping has already shown that some stickleback chromosomes have a nearly complete synteny with those of seabass. Here we present the results of comparative mapping of BAC end sequences of the gilthead sea bream to the three-spine stickleback as well as to contigs of the European seabass genome project (Kuhl et al. unpublished). The establishment of a dataset containing information about similarities, synteny relationships as well as colinearity will enhance significantly future approaches to identify and characterize new and novel functional genome regions. It also represents an important first step for gilthead sea bream whole-genome sequencing by second generation techniques as well as high-throughput SNP detection analysis, precise mapping of ESTs of specific interest, the identification of members of gene families and the differentiation of orthologous from paralogous genes.
2. Materials and Methods
2.1. BAC End Sequencing
The Sparus aurata BAC-library was constructed by Amplicon Express and was provided by the Institute of Marine Biology and Genetics of the Hellenic Centre for Marine Research (IMBG-HCMR). The library comprises BAC-clones arrayed in 144 × 384 well microtiter plates. The total genome coverage of the library is 7-8-fold with an average insert size of 120 kbp per BAC-clone. For template preparation, BAC-clones were inoculated in 2 × 384 deep well plates containing 190 μL of 2YT media and 12.5 mg/L chloramphenicol and cultivated for 18 hours at 37°C with rigorous shaking at 1100 rpm in Titramax 1000 incubators (Heidolph Instruments). E. coli cells were centrifuged at 2750 × g for 5 minutes (RT) and the supernatant was removed. The bacterial pellets were resolved in 20 μL Buffer 1 (50 mM TRIS HCl, 10 mM EDTA, pH 8 supplemented with 25 mg/L RNAse). Cell lysis was performed with 20 μL of Buffer 2 (0.2 M NaOH, 1% SDS) for 5 minutes and for neutralization 20 μL of Buffer 3 (0.933 M KAcetat, pH 4.8) was added. Cell debris was separated from DNA containing supernatant by centrifugation at 2750 × g for 30 minutes (RT). Supernatants from both plates (40 μL each) were combined in a new 384 deep well plate for size selective DNA precipitation with 38 μL of a polyethylene-glycol 6000/2-propanol mixture (75% 2-propanol(v/v)/100 g/L PEG-6000) and centrifuged at 2750 × g for 30 minutes at 4°C. Supernatants were depleted by centrifugation of the inverted plates at 172 × g for 1 minute (RT). The remaining DNA pellets were washed with 40 μL of 70% ethanol (v/v) to remove residual salts and PEG. Most of the BAC-DNA template preparations were done by an automated process that was developed at the MPI for molecular genetics.
BAC-DNA templates were end sequenced using ABI BigDyeV3.1 Terminator chemistry and T7 or SP6 primers. After postsequencing cleanup by ethanol/natrium acetate (NaOAc) precipitation, sequence analysis was performed on ABI3730xl capillary sequencers with 36 cm capillary arrays. Processing of raw sequencing data was done by the PHRED basecaller [30], quality clipping, and vector-clipping by LUCY [31].
2.2. Comparative Mapping Improved by Multiple Genome Alignment
Initially, BAC end sequences (BAC-ES) were aligned to the stickleback genome using BLASTN [32] with low stringency parameters (-W 7 -q -1 -e 0.01). The output tables were further screened for the best hits of BACs aligned with both ends that matched the same chromosome in the reference. A further screening step involved orientation (→←) and distance between paired BAC ends (insert size smaller than 300 kb).
To improve the number of mapped BACs we did a multiple genome alignment approach. We aligned all assembled contigs from the advanced seabass genome project (unpublished results, for a brief description see [33]) to the stickleback genome and rebuild the stickleback genome with the seabass contigs. We then performed the same alignment steps as above for the sea bream BACs on the “virtual” seabass chromosomes and transformed the hit coordinates back to the stickleback sequence.
Again read pairs were checked for consistency and alignments of both approaches were combined to result in an increased number of mapped BACs. For those BACs that mapped in both approaches we calculated the distance between the coordinates found by direct mapping and by mapping via seabass sequence, to check the placement error distribution induced by the coordinate transformation. If there were discrepancies between the same BACs in the two alignment approaches we considered those results that had better matches according to our consistency criteria.
2.3. Mapping Visualization
The start and end coordinates of the BAC clones were converted to GFF Format and included in the Ensembl genome browser for the stickleback genome (http://www.ensembl.org/Gasterosteus_aculeatus/Info/Index). BACs that form a minimal tiling path were chosen as described in [29] and may be browsed independently of the other placed BACs.
3. Results
3.1. BAC End Sequencing
After quality clipping and vector removal BAC end sequencing resulted in 92,468 reads. The number of BACs sequenced from both ends was 41,509 (physical coverage ~5.5x-6.5x). BACs sequenced from one direction accounted for 5,485 (only T7 dir./~0.7x-0.8x) and for 3,965 (only SP6 dir./~0.5x-0.6x). The average, quality clipped read length was 655 bp. The success rate of the sequencing reactions was 85.5%. About 1.9% of the clones were lacking an insert. The sequences were submitted to EMBL nucleotide database [EMBL: FR502695-EMBL: FR595162].
3.2. Comparative Mapping
Mapping all sequences directly to the stickleback genome resulted in 70,425 hits (76.2%/just best hit was counted). Further screening for paired BAC ends matching to a single reference chromosome resulted in 19,776 hits (23.8% of paired end sequenced BACs). This indicates that many hits in the raw blast are false positives as expected due to the low stringency parameters used. After checking for read orientation and distance 17,184 hits (8,592 BACs/20.7% of paired BAC ES) were consistently mapped. These BACs (1.16x–1.3x physical genome coverage) were already useful for building contigs of overlapping BAC-clones, but these covered only 58.1% of the stickleback chromosomes.
To further improve the mapping results we tried a multiple genome alignment approach. Contigs of the European seabass genome project (Kuhl et al. unpublished results) were concatenated according to the order of their ortholog sequences in the stickleback genome. As seabass belongs to the order of Perciformes, it is less diverged from sea bream than stickleback. We therefore expected more consistent hits using the seabass assembly. Additionally, synteny of genes is more conserved than nucleotide sequence in distantly related teleosts, which legitimizes the use of the nucleotide sequence of a closer relative of sea bream ordered by a species that is not as close in the phylogeny.
After aligning sea bream BAC ES to this sequence the hit coordinates were transformed back to stickleback coordinates. This way results from directly mapping to stickleback and indirectly mapping to stickleback (via orthologous seabass sequence) can be combined.
A total of 87,614 BAC ends (94.8%) had hits with low stringency blast in the multiple genome alignment approach. Paired BAC ends that mapped to the same reference chromosome accounted for 34,174 reads (41.2% of paired BAC ES). Consistency checks of read pairs resulted in 29,392 (14,696 BACs/35.4% of paired BAC ES). This was a 1.71-fold increase of consistently mapped BACs over the direct mapping approach.
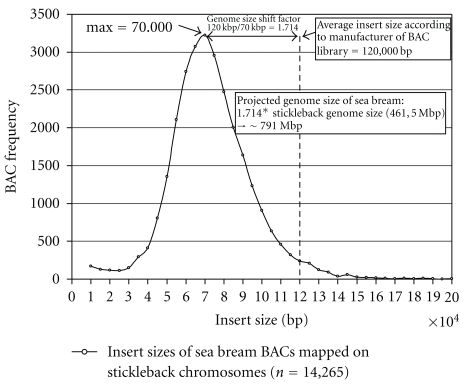
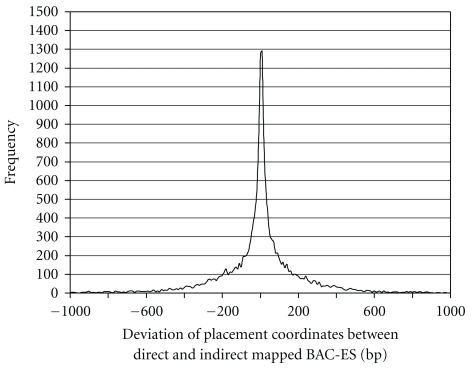
The combination of both mappings resulted in 28,530 BAC ES (14,265 BACs) with consistent mapping to the 21 stickleback chromosomes (1,488 BAC ES from direct and 27,042 from indirect mapping). The insert size distribution of mapped clones and its implications on genome size are shown in Figure 1. (See excel sheet with mapped BAC ES in Supplementary Material available online at doi: 10.1155∖2011∖329025].) As the transformation of the seabass sequence coordinates to coordinates on stickleback sequence introduces some placement error, we compared the coordinates of BAC clones (7,646) that were consistently mapped in both mapping approaches. Six BACs were mapped to different chromosomes by the different mapping strategies. All other BACs showed similar start/end coordinates on the same stickleback chromosomes. The median deviation of start and end between the two mapping strategies (see Figure 2) was 66 bp, the average deviation was 293 bp, and 96.7% of the mapped BACs had a deviation of less than 1,000 bp. As the placed BACs and their overlaps are much larger than these values, the fuzziness introduced by the seabass to stickleback coordinate transformation seems to be negligible.
Figure 1.
Insert size frequency distribution of consistently mapped sea bream BACs to chromosomes of three-spined stickleback (Gasterosteus aculeatus). Mapped BACs exhibit a shift to lower insert sizes compared to insert sizes of the BACs observed by pulse field gel-electrophoresis (according to manufacturer of the BAC library Amplicon Express, data not shown). This shift reflects the genome size difference between sea bream and stickleback.
Figure 2.
Placement error frequency distribution observed by BACs that were directly mapped to stickleback chromosomes as well as mapped indirectly by aligning them to seabass sequence that has been placed along the stickleback sequence before. Of the indirectly mapped BACs 96.7% had a placement error of less than 1000 bp.
The minimal tiling path calculated from consistently placed BACs covered 73.5% of stickleback chromosomes with about 13,415 gene loci predicted (70.2% of 19,121 genes predicted in stickleback chromosome sequences). It consists of 5,249 BACs, which constitute 1,485 BACtigs. The largest BACtig covers 2.15 Mbp and N50 BACtigs size is 337,253 bp. These sizes refer to the stickleback genome, as there is a large size difference between stickleback and sea bream the BACtig sizes should be multiplied by a factor of 1.7–2 to estimate their sizes in sea bream. The mapped clones may be browsed alongside the stickleback genome and the ordered seabass BAC clones at www.ensembl.org (Login: heiner7@hotmail.com/Passw: BREAMBACMAP2010). For a comparison of sea bream and seabass minimal tiling path clones mapped to stickleback chromosome VI and XXI, see Figure 3 (for all stickleback chromosomes see Supplementary Material. Clones with inconsistent matches of both ends in terms of distance or orientation were screened manually for potential intra-chromosomal rearrangements. If an inconsistency was indicated by two or more independent BAC clones it was considered a potential rearrangement. We identified 202 potential intra-chromosomal rearrangements that were covered by 804 BACs. The median distance of BAC ends assigned to rearrangements was 1.74 Mbp.
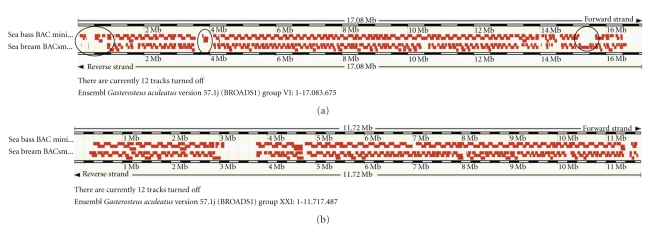
Figure 3.
Mapped minimal tiling path BAC clones of sea bream and seabass alongside the stickleback chromosomes VI (a) and XXI (b). While maps in (a) show some differences in regions covered or not covered (marked with black circles), maps in (b) are nearly identical.
4. Discussion
Studying the genomes of important commercial species is of direct value to the efficiency and exploitation of the species. Despite the fact that today access to other fish genomes, including the sister species of the gilthead sea bream the European seabass Dicentrarchus labrax, is available, additional genome information is of importance concerning identification of species specific characteristics like disease resistance, salinity, and temperature tolerance by comparative genomics. Additionally, it will offer the possibility to use population genomics approaches combining interspecific divergence with intraspecific diversity for identifying adaptive variation, or comparative genomics for identification of small noncoding regions of functional importance. The power of such approaches is in general decreasing with increasing evolutionary distance, either because of intervening confounding effects and increased complexity or because of alignment problems.
Comparative sequence analysis has become an important tool for studying genome function. The identification of ultraconserved elements (UCEs), similar genomic elements as well as conserved noncoding elements (CNEs) have pin pointed to genomic regulatory blocks, developmental regulatory target genes (s) and phylogenetically relevant and functionally genes [34].
Comparative mapping of BAC-ES is a fast and efficient method to deduce genome maps of nonmodel organisms from sequenced genomes of model organisms due to conserved synteny [12]. Thus the tremendous efforts that have been undertaken for model organisms in the past decade pay out now for the genomics of farmed animals. We have recently shown for the European seabass that the stickleback genome is a good choice for comparative studies in fish belonging to the order of Perciformes. In this study we have successfully mapped 14,265 sea bream BAC clones along the sequenced stickleback chromosomes. Nucleotide alignments in the present study indicate that sea bream and stickleback may be more diverged than seabass and stickleback. Thus we had to overcome a lower mapping efficiency by applying a multispecies alignment. The resulting mapping data represents a valuable tool for genomic research in this important aquaculture species. BAC clones covering regions of interest may be easily chosen for advanced analysis using the Ensembl genome browser tool. Besides these benefits some conclusions can be drawn on genome size of sea bream, when comparing the distribution of distances between two corresponding BAC ends mapped to the stickleback chromosomes and their real distance observed by pulse field gelelectrophoresis during BAC library construction.
4.1. Genome Sizes
To estimate an organism's genome size flow cytometry of cells with stained nuclei has been widely applied. According to www.genomesize.com the resulting C-value found by this method for G. aculeatus is 0.58 pg of haploid DNA content [35]. In D. labrax the C-value accounts for 0.78 pg and in S. aurata it is even higher with 0.95 pg [36]. If one relates the C-values to the lowest value observed in stickleback, the seabass genome should be 1.34-fold larger and the sea bream genome should be 1.64-fold larger than the stickleback genome. These values are in good concordance with values found by BAC insert size distributions of the comparative mappings: S. aurata/G. aculeatus = 1.71; D. labrax/G. aculeatus = 1.3. Nevertheless predicting the genome size by sequencing-based methods results in lower values for each genome compared to the size estimations of flow cytometry-based methods (G. aculeatus 462 Mbp versus 567 Mbp; D. labrax 601 Mbp versus 763 Mbp; S. aurata 791 Mbp versus 929 Mbp). One can speculate about the reasons for these findings. On the one hand, sequencing-based methods tend to underpredict genome sizes as some regions are hard to assemble and are later on missing in the published genome assemblies. On the other hand, flow cytometry might overestimate genome sizes due to including ongoing replication of DNA in the measurements or some staining background of RNA:RNA or RNA:DNA hybrids. Thus, we believe that the true genome size of the three teleosts lies in between the borders defined by the two approaches. The differences in genome-size among species may arise from insertion or deletion (indel) of large DNA fragments and/or insertion or deletion of small DNA sequences [37]. Given their frequency particularly the small deletions largely outnumber small insertions [37, 38], small indels have been recognised as major mechanisms responsible for genome-size evolution in eukaryotic organisms. Several studies have also shown that the wide range of variation in genome size among eukaryotic organisms is correlated with the amount of repetitive elements in the genome [39–41]. In fishes for example, the contribution of repetitive elements to genome-size difference between medaka and Takifugu is estimated about 54% [42]. Although the above interpretations are coherent and in agreement with results previously reported in fishes on the forces driving the evolution of genome size, they are still speculative since they are not based on comparisons at nucleotide sequence level. Further analysis of indels and repetitive elements are required to clarify the respective contribution to the differences in genome size between these species. This could be done when the genome of seabass and sea bream will be completely sequenced or in near future via a targeted analysis of large conserved sequence regions between the three species.
4.2. Comparison Seabass Map to Sea Bream Map
We have recently published a comparative BAC map for the European seabass [29]. The mapping of the BAC inserts was done in a similar way as described here for the sea bream. When comparing both maps, the seabass map outperforms the sea bream map in most parameters like reference coverage (87.0% versus 73.5%), covered stickleback gene predictions (85.4% versus 70.2%), N50 BACtig size (1.2 Mbp versus 0.34 Mbp), or total number of BACtigs (588 versus 1,485). Differences in map quality may be based on the lower physical genome coverage and average insert size of the gilthead sea bream BAC library than the European seabass BAC library. The lower coverage of mapped BACs for sea bream complicates the detection of chromosomal rearrangements. Nevertheless, we could identify a number of 202 potential intrachromosomal rearrangements between stickleback and sea bream that were spanned by two or more clones. This number is similar to what was found, when comparing stickleback to seabass (214 potential rearrangements, [29]). Seabass shares 114 of the 202 rearrangements with sea bream, thus the phylogenetic relationship between the three species seems to be reflected by the number of rearrangements.
Although the alignment coordinates of additional 1,402 sea bream BACs imply other large, scale intra-chromosomal rearrangements, their evidence is weak as they are covered by a single BAC clone only. These are likely to be chimeric clones or wrong alignments. Nevertheless, inconsistently mapped BACs (see Supplementary Material) may be subject to further investigations (e.g., checking overlaps by PCR) to proof further potential rearrangements.
5. Conclusions
The comparative sea bream BAC map described here is a valuable tool for researchers in the field. It allows the fast selection of BAC clones that cover genes or genomic loci of interest by simple BLAST searches in the stickleback genome and picking one of the BACs covering the hit location.
According to the coverage of the stickleback genome we estimate that about 75% of the sea bream genome is covered by the mapped clones. Additional genes, which are not represented by mapped BACs, may be found by BLAST searches against the BAC end sequences that comprise about 60.6 Mbp of the sea bream genome, the largest set of genomic sequences published for sea bream so far.
These data will be also of great value in future projects like a sea bream genome project and will allow scaffolding of WGS contigs or a sequencing strategy based on mapped clones, which will be especially useful, if short read sequencing technologies will be applied.
Thus, taking also into account the comparative seabass BAC map, ordered large insert clones for the two main aquaculture species in the Mediterranean are now available and may contribute to answer basic biological questions like what makes up the difference between hermaphrodite (sea bream) and temperature-driven sex differentiation (seabass) as well as aquaculture-related questions that aim at improved breeding stocks in both species.
Supplementary Material
Supplementary File 1: Excel table of mapped BAC end sequences to stickleback chromosomes.
Supplementary File 2: Minimal tiling path of comparative sea bream and sea bass BAC maps alongside the 21 stickleback chromosomes as displayed by the Ensembl genmome browser.
References
- 1.Cheema J, Dicks J. Computational approaches and software tools for genetic linkage map estimation in plants. Briefings in Bioinformatics. 2009;10(6):595–608. doi: 10.1093/bib/bbp045. [DOI] [PubMed] [Google Scholar]
- 2.Riera-Lizarazu O, Vales MI, Kianian SF. Radiation hybrid (RH) and HAPPY mapping in plants. Cytogenetic and Genome Research. 2008;120(3-4):233–240. doi: 10.1159/000121072. [DOI] [PubMed] [Google Scholar]
- 3.Shizuya H, Birren B, Kim U-J, et al. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(18):8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wild J, Hradecna Z, Szygbalski W. Conditionally amplifiable BACs: switching from single-copy to high-copy vectors and genomic clones. Genome Research. 2002;12(9):1434–1444. doi: 10.1101/gr.130502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katagiri T, Kidd C, Tomasino E, et al. A BAC-based physical map of the Nile tilapia genome. BMC Genomics. 2005;6, article 89 doi: 10.1186/1471-2164-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo M-C, Thomas C, You FM, et al. High-throughput fingerprinting of bacterial artificial chromosomes using the SNaPshot labeling kit and sizing of restriction fragments by capillary electrophoresis. Genomics. 2003;82(3):378–389. doi: 10.1016/s0888-7543(03)00128-9. [DOI] [PubMed] [Google Scholar]
- 7.Tao Q, Chang Y-L, Wang J, et al. Bacterial artificial chromosome-based physical map of the rice genome constructed by restriction fingerprint analysis. Genetics. 2001;158(4):1711–1724. doi: 10.1093/genetics/158.4.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khorasani MZ, Hennig S, Imre G, et al. A first generation physical map of the medaka genome in BACs essential for positional cloning and clone-by-clone based genomic sequencing. Mechanisms of Development. 2004;121(7-8):903–913. doi: 10.1016/j.mod.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 9.Bouzidi MF, Franchel J, Tao Q, et al. A sunflower BAC library suitable for PCR screening and physical mapping of targeted genomic regions. Theoretical and Applied Genetics. 2006;113(1):81–89. doi: 10.1007/s00122-006-0274-6. [DOI] [PubMed] [Google Scholar]
- 10.Yim Y-S, Moak P, Sanchez-Villeda H, et al. A BAC pooling strategy combined with PCR-based screenings in a large, highly repetitive genome enables integration of the maize genetic and physical maps. BMC Genomics. 2007;8 doi: 10.1186/1471-2164-8-47. Article ID 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujiyama A, Watanabe H, Toyoda A, et al. Construction and analysis of a human-chimpanzee comparative clone map. Science. 2002;295(5552):131–134. doi: 10.1126/science.1065199. [DOI] [PubMed] [Google Scholar]
- 12.Larkin DM, Everts-van der Wind A, Rebeiz M, et al. A cattle-human comparative map built with cattle BAC-ends and human genome sequence. Genome Research. 2003;13(8):1966–1972. doi: 10.1101/gr.1560203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Margulies EH, Birney E. Approaches to comparative sequence analysis: towards a functional view of vertebrate genomes. Nature Reviews Genetics. 2008;9(4):303–313. doi: 10.1038/nrg2185. [DOI] [PubMed] [Google Scholar]
- 14.Begun DJ, Holloway AK, Stevens K, et al. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biology. 2007;5(11, article e310) doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franch R, Louro B, Tsalavouta M, et al. A genetic linkage map of the hermaphrodite teleost fish Sparus aurata L. Genetics. 2006;174(2):851–861. doi: 10.1534/genetics.106.059014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarropoulou E, Kotoulas G, Power DM, Geisler R. Gene expression profiling of gilthead sea bream during early development and detection of stress-related genes by the application of cDNA microarray technology. Physiological Genomics. 2005;23(2):182–191. doi: 10.1152/physiolgenomics.00139.2005. [DOI] [PubMed] [Google Scholar]
- 17.Sarropoulou E, Nousdili D, Magoulas A, Kotoulas G. Linking the genomes of nonmodel teleosts through comparative genomics. Marine Biotechnology. 2008;10(3):227–233. doi: 10.1007/s10126-007-9066-5. [DOI] [PubMed] [Google Scholar]
- 18.Jekosch K. The Zebrafish genome project: sequence analysis and annotation. Methods in Cell Biology. 2004;2004(77):225–239. doi: 10.1016/s0091-679x(04)77012-0. [DOI] [PubMed] [Google Scholar]
- 19.Kasahara M, Naruse K, Sasaki S, et al. The medaka draft genome and insights into vertebrate genome evolution. Nature. 2007;447(7145):714–719. doi: 10.1038/nature05846. [DOI] [PubMed] [Google Scholar]
- 20.Aparicio S, Chapman J, Stupka E, et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297(5585):1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- 21.Jatllon O, Aury J-M, Brunet F, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431(7011):946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- 22.Nelson JS. Fishes of the World. 3rd edition. New York, NY, USA: John Wiley and Sons; 1994. [Google Scholar]
- 23.Amores A, Force A, Yan Y-L, et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282(5394):1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- 24.Amores A, Suzuki T, Yan Y-L, et al. Developmental roles of pufferfish Hox clusters and genome evolution in ray-fin fish. Genome Research. 2004;14(1):1–10. doi: 10.1101/gr.1717804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holland PW, Garcia-Fernàndez J, Williams NA, Sidow A. Gene duplications and the origins of vertebrate development. Development, Supplement. 1994:125–133. [PubMed] [Google Scholar]
- 26.Meiri I, Gothilf Y, Zohar Y, Elizur A. Physiological changes in the spawning gilthead seabream, Sparus aurata, succeeding the removal of males. Journal of Experimental Zoology. 2002;292(6):555–564. doi: 10.1002/jez.10072. [DOI] [PubMed] [Google Scholar]
- 27.Zohar Y, Abraham M, Gordin H. The gonadal cycle of the captivity-reared hermaphroditic teleost Sparus aurata (L.) during the first two years of life. Annales de Biologie Animale, Biochimie, Biophysique. 1978;18(4):877–882. [Google Scholar]
- 28.Zohar Y, Elizur A, Sherwood NM, et al. Gonadotropin-releasing activities of the three native forms of gonadotropin-releasing hormone present in the brain of gilthead seabream, Sparus aurata. General and Comparative Endocrinology. 1995;97(3):289–299. doi: 10.1006/gcen.1995.1029. [DOI] [PubMed] [Google Scholar]
- 29.Kuhl H, Beck A, Wozniak G, Canario AVM, Volckaert FAM, Reinhardt R. The European sea bass Dicentrarchus labrax genome puzzle: comparative BAC-mapping and low coverage shotgun sequencing. BMC Genomics. 2010;11(1, article 68) doi: 10.1186/1471-2164-11-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Research. 1998;8(3):186–194. [PubMed] [Google Scholar]
- 31.Chou H-H, Holmes MH. DNA sequence quality trimming and vector removal. Bioinformatics. 2001;17(12):1093–1104. doi: 10.1093/bioinformatics/17.12.1093. [DOI] [PubMed] [Google Scholar]
- 32.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 33.Kuhl H, Tine M, Hecht J, Knaust F, Reinhardt R. Analysis of single nucleotide polymorphisms in three chromosomes of European sea bass Dicentrarchus labrax. doi: 10.1016/j.cbd.2010.04.003. Comparative Biochemistry and Physiology D. In press. [DOI] [PubMed] [Google Scholar]
- 34.Kikuta H, Laplante M, Navratilova P, et al. Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome Research. 2007;17(5):545–555. doi: 10.1101/gr.6086307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vinogradov AE. Genome size and GC-percent in vertebrates as determined by flow cytometry: the triangular relationship. Cytometry. 1998;31(2):100–109. doi: 10.1002/(sici)1097-0320(19980201)31:2<100::aid-cyto5>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 36.Peruzzi S, Chatain B, Menu B. Flow cytometric determination of genome size in European seabass (Dicentrarchus labrax), gilthead seabream (Sparus aurata), thinlip mullet (Liza ramada), and European eel (Anguilla anguilla) Aquatic Living Resources. 2005;18(1):77–81. [Google Scholar]
- 37.Gregory TR. Insertion-deletion biases and the evolution of genome size. Gene. 2004;324(1-2):15–34. doi: 10.1016/j.gene.2003.09.030. [DOI] [PubMed] [Google Scholar]
- 38.Petrov DA. Mutational equilibrium model of genome size evolution. Theoretical Population Biology. 2002;61(4):533–546. doi: 10.1006/tpbi.2002.1605. [DOI] [PubMed] [Google Scholar]
- 39.Petrov DA. Evolution of genome size: new approaches to an old problem. Trends in Genetics. 2001;17(1):23–28. doi: 10.1016/s0168-9525(00)02157-0. [DOI] [PubMed] [Google Scholar]
- 40.Kidwell MG. Transposable elements and the evolution of genome size in eukaryotes. Genetica. 2002;115(1):49–63. doi: 10.1023/a:1016072014259. [DOI] [PubMed] [Google Scholar]
- 41.Gregory TR, Hebert PDN. The modulation of DNA content: proximate causes and ultimate consequences. Genome Research. 1999;9(4):317–324. [PubMed] [Google Scholar]
- 42.Imai S, Sasaki T, Shimizu A, Asakawa S, Hori H, Shimizu N. The genome size evolution of medaka (Oryzias latipes) and fugu (Takifugu rubripes) Genes and Genetic Systems. 2007;82(2):135–144. doi: 10.1266/ggs.82.135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File 1: Excel table of mapped BAC end sequences to stickleback chromosomes.
Supplementary File 2: Minimal tiling path of comparative sea bream and sea bass BAC maps alongside the 21 stickleback chromosomes as displayed by the Ensembl genmome browser.