Abstract
Eukaryotic chromosomal DNA is packaged into nucleosomes to form a dynamic structure known as chromatin. The compaction of DNA within chromatin poses a unique hindrance with regards to the accessibility of the DNA to enzymes involved in replication, transcriptional regulation, and repair. The physical structure and physiological activity of chromatin are regulated through a diverse set of posttranslational modifications, histone exchange, and structural remodeling. Of the covalent chromatin modifications, the acetylation of lysine residues within histone proteins by acetyltransferase enzymes, such as GCN5, is one of the most prevalent and important steps in the regulation of chromatin function. Alteration of histone acetyltransferase activity can easily result in the dysregulation of gene transcription and ultimately the onset of a disease state. Many transcription factors contain polyglutamine regions within their primary sequence. Mutations resulting in the elongation of these polyglutamine tracts are associated with a disease family known as the polyglutamine expansion disorders. Spinocerebellar ataxia type 7 (SCA7) is one of the nine diseases that are grouped in this family and is caused by polyglutamine expansion of the ataxin-7 protein, which is a component of the GCN5-containing human SAGA histone acetyltransferase complex. Mutation of ataxin-7 in this manner has been shown to disrupt the structural integrity of the SAGA complex and result in aberrant chromatin acetylation patterns at the promoters of genes involved in the normal function of tissues that are affected by the disease. The specific aspects of molecular pathology are not currently understood; however, studies carried out in laboratory systems ranging from the budding yeast Saccharomyces cerevisiae to transgenic mouse models and cultured human cells are poised to allow for the elucidation of disease mechanisms and subsequent therapeutic approaches.
I. Packaging and Accessibility of DNA in the Context of Chromatin
Despite its molecular simplicity, DNA encodes information that serves as the blueprint for the structure and function of every living organism. The relative abundance of DNA precludes the possibility of a simple mechanism for its packaging and accessibility within the eukaryotic nucleus. Within the nucleus, DNA is wrapped around nucleosomes, which allow for its compaction into the structure called chromatin. The nucleosome core particle binds 146bp of DNA and is composed of two histone H2A/H2B dimers that interact with a heterotetramer containing two copies each of histones H3 and H4 (Luger et al., 1997). Each of the histone proteins within the nucleosome contains alpha helical histone fold domains, which facilitate their interaction with adjacent histones within individual nucleosomes and DNA. Histone proteins also possess unstructured amino-(N-) and carboxy-terminal (C-terminal) tails that extend outside the nucleosome particle and serve as a substrate for the enzymatic activities of histone-modifying enzymes. Histones are extensively modified by a myriad of reactions; examples include acetylation, methylation, ubiquitination, SUMOylation, phosphorylation, and ADP-ribosylation. These modifications alter chromatin dynamics and influence histone–DNA interactions, as well as the recruitment and binding of protein factors to chromatin (reviewed in Taverna et al., 2007). The tight correlation between histone modification patterns and their subsequent regulation of chromatin activity led to the hypothesis by Strahl and Allis (2000) that posttranslational modification of histones encodes a language that is read and followed by chromatin-interacting proteins. These so-called chromatin-binding proteins interact with chromatin through several distinct domains, each of which has the ability to recognize distinct histone modifications and can be responsible for the targeting of multisubunit protein complexes to specific chromatin regions.
Histone acetylation is a posttranslational modification that performs two known functions on chromatin. First, acetylation neutralizes the positive charge of lysine residues within the histone tails thereby reducing their ability to form charge-based interactions with the negatively charged phosphodiester backbone of associated DNA. The acetylation of histones in general is associated with chromatin decondensation, although it is not necessarily causative. One exception is the acetylation of histone H4 on lysine 16, which has been shown to directly promote a decondensed chromatin state (Shogren-Knaak et al., 2006). The decondensation of chromatin is associated with the formation of a euchromatin state, which increases the accessibility of chromatin to transcription factors. In contrast, chromatin condensation leads to the formation of heterochromatin, which is transcriptionally silent. The second known function of histone acetylation is to provide a platform for the recruitment of transcription factors through acetyl–lysine interacting motifs, known as bromodomains (Dhalluin et al., 1999; Jacobson et al., 2000).
Histone acetylation is accomplished by a family of enzymes known as histone acetyltransferases (HATs), which were originally divided into two groups based on their subcellular localization, the nuclear HATs (Type A) and cytoplasmic HATs (Type B). The activity and substrate recognition of many HATs is typically regulated by their association with other factors in the context of multiprotein complexes (reviewed in Grant and Berger, 1999; Roth et al., 2001; Torok and Grant, 2004). These protein complexes often contain smaller subcomplexes, termed “modules,” each of which contribute a discrete activity to the greater complex. The modular nature of these multiprotein complexes allows them to couple multiple histone modification activities to effectively regulate chromatin accessibility and function. While not always required, the substrate recognition and activity level of subcomplex modules can be dependent on their association with the parent complex. Overall, histone acetylation serves as a significant regulatory mechanism governing chromatin accessibility and providing a substrate for the association of factors with DNA in chromatin.
II. Gcn5 is a Histone Acetyltransferase that Functions in the Context of SAGA and its Related Complexes in Eukaryotes
The general control of amino acid biosynthesis 5 (Gcn5) protein in budding yeast was originally identified as a transcription factor involved in the cellular response to amino acid starvation and as a factor required for the optimal activity of other transcriptional activators (Georgakopoulos and Thireos, 1992). Sequence comparison of the p55 protein of Tetrahymena, the first confirmed Type-A HAT protein (Brownell and Allis, 1995), indicated significant similarity to the yeast Gcn5 protein and further analysis demonstrated that purified, recombinant yeast Gcn5 was also able to acetylate free core histones in vitro (Brownell et al., 1996). Subsequent characterization of recombinant Gcn5 demonstrated its ability to catalyze the acetylation of both histone H3 at lysine 9 and 14 and histone H4 at lysine 8 and 16. Despite its ability to acetylate lysine residues on either histone H3 or H4, Gcn5 exhibits a clear preference for histone H3 as an acetylation substrate. Within histone H3, lysine 14 is preferentially acetylated in vitro while lysine 9 appears to be the favored target in vivo (Kuo et al., 1996). Despite its documented ability to acetylate free core histones, recombinant Gcn5 did not appear to be capable of acetylating histones within the nucleosome core particle, the context in which histones exist within chromatin. The lack of nucleosomal acetyltransferase activity suggested that other factors were required to regulate either the recognition of histone substrates within the nucleosome or HAT activity of the enzyme in this altered context.
Early studies involving the yeast Gcn5 protein demonstrated its ability to functionally interact both in vitro and in vivo with two adaptor proteins, Ada2 and Ada3, to form a trimeric complex with the ability to promote transcriptional activation (Marcus et al., 1994, Horiuchi et al., 1995; Candau et al., 1996). While it was subsequently shown by Balasubramanian et al. (2002) that the heterotrimeric complex formed by Gcn5, Ada2, and Ada3 had the ability to acetylate nucleosomal histones, its native association with other polypeptides to form larger chromatin-modifying complexes had not yet been explored. The search for these complexes led to an unbiased interrogation of multiprotein complexes possessing HAT activity through a multiple step, native chromatographic purification from yeast extracts. This exploration ultimately resulted in the identification of four distinct complexes with the ability to acetylate nucleosomal histones (Grant et al., 1997). Two of these four complexes were shown to contain Gcn5 and the adaptor protein Ada2, suggesting that they possessed the necessary catalytic activity and targeting ability to function as multisubunit acetyltransferase complexes. Subsequent characterization of the highest molecular weight HAT complex, estimated at 1.8 megadaltons, identified several additional subunits, including several members of the suppressor of Ty (Spt) protein family, Spt3, Spt7, and Spt20. The discovery of these subunits in association with the high molecular weight HAT complex led its naming as the Spt-Ada-Gcn5-acetyltransferase (SAGA) complex. During initial characterization of the yeast SAGA complex it was shown that deletion of Gcn5, Ada2, or Ada3 resulted in loss of HAT activity, indicating that Gcn5 was the sole acetyltransferase responsible for SAGA-mediated histone acetylation. Additionally, these experiments demonstrated that the adaptor proteins Ada2 and Ada3 were required for mediating the activity of Gcn5 in a physiologically relevant context. While Gcn5, Ada2, and Ada3 were not required for the overall integrity of the complex, they were shown to be critical for its ability to recognize and acetylate nucleosomal substrates and defined the Gcn5-Ada2-Ada3 heterotrimer as the catalytic core of the SAGA complex (Grant et al., 1997). It was later determined that there was a second Gcn5-containing protein complex in yeast that functioned in stress-response pathways and shared a similar subunit composition and histone substrate specificity with SAGA (Pray-Grant et al., 2002; Sterner et al., 2002). Due to its similarity to SAGA, this second Gcn5-containing complex was named the SAGA-like (SLIK) complex. Both the SAGA and SLIK complexes serve as examples of how histone acetylation by Gcn5 is controlled through its interactions with accessory factors in the context of multiprotein chromatin-modifying complexes. The composition and functions of these yeast acetyltransferase complexes has served as the basis for the identification and characterization of similar complexes in higher eukaryotes.
Unlike yeast, metazoans possess two GCN5 isoforms that are believed to arise from the alternative splicing of a single gene product (Smith et al., 1998). The lower molecular weight isoform (GCN5-S) contains both a HAT domain and a single bromodomain and is similar in size and function to the single form of Gcn5 found in yeast. The second, higher molecular weight isoform (GCN5-L) has been shown to contain an additional N-terminal domain that shows high similarity to the N-terminal domain of the GCN5 homolog p300/CREB-binding protein-associated factor, known as PCAF (Xu et al., 1998). Additionally, the vertebrate-specific histone acetyltransferase PCAF, identified as a paralog of human GCN5 (Yang et al., 1996), is also a constituent of a multisubunit HAT complex.
GCN5 isoforms serve as the catalytic acetyltransferases in several multiprotein complexes in human cells, including the TATA-binding protein-free TAF complex (TFTC), ADA-two-a-containing complex (ATAC), and the SPT3-TAFII31-GCN5-L acetylase (STAGA) complex (reviewed in Nagy and Tora, 2007). The human STAGA complex was purified from cultured cells shortly after the identification and characterization of the homologous yeast SAGA complex (Martinez et al., 1998). The STAGA complex preferentially incorporates GCN5-L over both its homolog PCAF and the smaller GCN5 isoform and favors histone H3 as a substrate for acetylation compared to other histone proteins (Ogryzko et al., 1998; Martinez et al., 2001). Similar to yeast SAGA, the human STAGA complex contains human homologs of Ada2 and Ada3, the components of the GCN5 catalytic trimer, and has the ability to activate transcription from in vitro-assembled chromatin templates (Martinez et al., 2001). The subunit compositions of the yeast SAGA and human STAGA complexes are well conserved and demonstrate the ability to regulate gene expression through several different functions, which will be described below.
III. Substrate Targeting of Gcn5 Histone Acetyltransferase Activity is Regulated by Interactions with Adaptor Proteins
The control of substrate specificity and catalytic function by association with accessory proteins is a recurring theme in enzyme regulation throughout the cell. Proteins that exhibit HAT activity in the absence of accessory proteins often demonstrate the ability to target multiple lysine residues within individual, or multiple, histones. The degree of promiscuity demonstrated by acetyltransferases, in the absence of regulatory proteins, raises the question of how their specificities are controlled to ensure the appropriate spatiotemporal acetylation of histone targets to preserve the fidelity of transcriptional regulation. The exact mechanism of histone substrate targeting by these acetyltransferases is the subject of rigorous study as the answers to these and other related questions may facilitate the understanding of how gene expression in both normal and disease states is controlled by histone acetylation.
The monomeric Gcn5 protein in yeast acetylates both histone H3 and H4 when they are free in solution, but not when they are in the context of the nucleosome core particle in vitro (Xu et al., 1998). Microsequencing of histones acetylated by Gcn5 in vitro indicated the preferential acetylation of histone H3 at lysine 14 and histone H4 at lysine 8 and 16 (Kuo et al., 1996). These data provided the foundation for the characterization of HAT activity exerted by Gcn5; however, it was determined later that the substrate specificity of Gcn5 in the context of both the Gcn5-Ada2-Ada3 catalytic core and the greater SAGA complex varied from that observed for monomeric Gcn5 (Grant et al., 1999). The preferential targeting of lysine residues within the nucleosome core particle by Gcn5 was further studied and was shown to be dependent on its association with the adaptor proteins Ada2 and Ada3. In addition to the lysine specificity exhibited by monomeric Gcn5, the Gcn5-Ada2 heterodimer can acetylate lysine 18, although with a lower efficiency compared to lysine 14. Subsequent addition of Ada3, to complete the Gcn5-Ada2-Ada3 catalytic core, further alters target lysine specificity leading to acetylation of histone H3 on lysine 9, 14, 18, and 23 with variable efficiency. In the context of the entire SAGA complex, overall acetylation of lysine residues on histone H3 is decreased compared to the catalytic core; however, the substrate specificity and relative preference for specific lysine residues are similar between the two conditions (Balasubramanian et al., 2002). Similar to the SAGA complex, the lower molecular weight ADA complex efficiently acetylates histone H3; however, the ADA complex appears to demonstrate a more robust ability to acetylate histones H2B and H4 when compared to SAGA. The difference in substrate targeting of ADA coincides with the incorporation of a different complement of proteins into this functionally distinct, Gcn5-containing complex (Grant et al., 1997, 1999; Sterner et al., 1999). These observations suggest the potential for Gcn5 to regulate transcription, or other chromatin-related activities, through multiple mechanisms based on protein–protein interactions within multisubunit complexes.
In contrast to yeast Gcn5, the larger GCN5 isoform found in higher eukaryotes can acetylate histones H3 and H4 in both the free and nucleosome-incorporated states in vitro without the presence of accessory proteins (Xu et al., 1998). The ability of free GCN5-L protein to acetylate nucleosomal histones in more complex organisms is presumably dependent on the presence of the additional N-terminal domain, which is absent from yeast Gcn5. As mentioned above, Ada2 is involved in facilitating the recognition of nucleosomal histone substrates by Gcn5 in yeast. During the course of evolution, the single Ada2 gene in yeast developed into two functionally distinct ADA2 paralogs in metazoans, ADA2-alpha (ADA2a) and ADA2-beta (ADA2b). Interestingly, GCN5 functionally interacts with both of these ADA2 isoforms in Drosophila (Muratoglu et al., 2003), mice (Candau et al., 1996), and humans (Barlev et al., 2003), despite their occurrence in different multiprotein complexes that exhibit alternative lysine target specificity.
Biochemical purifications from both Drosophila and human cells have demonstrated the distinct nature of complexes containing either ADA2a or ADA2b despite their relatively high level of sequence similarity (Barlev et al., 2003; Kusch et al., 2003; Guelman et al., 2006; Gamper et al., 2009). The ADA2b protein is exclusively found in the STAGA complex while its paralog, ADA2a, is excluded from these complexes but associates instead with the ATAC and PCAF complexes. Interestingly, the adaptor protein ADA3, which is a well-characterized component of the acetyltransferase catalytic core in complexes containing GCN5, is present in both ADA2a- and ADA2b-containing complexes. Deletion studies in Drosophila have demonstrated that the different ADA2 paralogs play a regulatory role in the targeting of GCN5-containing complexes to individual histone substrates in the context of chromatin (Qi et al., 2004). Deletion of Ada2a does not affect the acetylation of histone H3; however, it does compromise the acetylation of histone H4 at specific lysine residues. Interestingly, loss of Ada2b results in a significant reduction in the acetylation of lysine residues on histone H3 targeted by GCN5 but does not appear to alter the acetylation of histone H4 (Qi et al., 2004). Further, deletion of GCN5 also causes a loss in histone H4 acetylation, which overlaps with that seen in the ADA2a deletion mutant (Ciurciu et al., 2006). These results imply a role for the ADA2a-regulated, GCN5-mediated acetylation of histone H4 and further that the substrate specificity of the GCN5 acetyltransferase can be specifically modulated by its interaction with either of the ADA2 paralogs.
IV. SAGA and Related Complexes Possess Histone Deubiquitination Activity
The activity of Gcn5 in the context of the catalytic heterotrimer that it forms with Ada2 and Ada3 represents a critical function within the greater SAGA complex; however, SAGA-related complexes exhibit several distinct chromatin-modifying activities and therefore play multiple roles in the regulation of transcription. Several functions in addition to acetyltransferase activity are afforded to the SAGA as a result of the presence of the aforementioned modules, which contribute to the overall composition of the complex. One of these SAGA modules exhibits ubiquitin hydrolase activity and is involved in the regulation of histone H2B monoubiquitination (Köhler et al., 2006, 2008; Lee et al., 2009). Posttranslational modification of the histone H2B C-terminal tail by monoubiquitination is catalyzed by Rad6, a ubiquitin ligase, in yeast or its homologs in other organisms (Robzyk et al., 2000; reviewed in Osley, 2006). Rad6-mediated monoubiquitination of histone H2B is dependent on the recruitment of both RNA polymerase II (RNAPII) and other transcription factors and has been shown to influence di- and trimethylation levels of histone H3 (reviewed in Weake and Workman, 2008). The SAGA subunit Ubp8 and its homologs Nonstop and USP22 in Drosophila and humans, respectively, are ubiquitin hydrolases that can specifically remove the monoubiquitin moiety from histone H2B (Henry et al., 2003; Daniel et al., 2004; Weake et al., 2008; Zhao et al., 2008). In yeast, Ubp8 physically interacts with accessory proteins, Sgf11 and Sus1, to form the functional deubiquitination module (hereafter referred to as the Ubp8 module) that is found in association with the SAGA complex (Köhler et al., 2006, 2008; Lee et al., 2009). Similarly, USP22, the human homolog of Ubp8, is bound to the STAGA complex through interactions with ATXN7L3 and ENY2, which are homologs of Sgf11 and Sus1, respectively (Zhao et al., 2008). These histone H2B deubiquitination modules, Ubp8/Sgf11/Sus1 in yeast and USP22/ATXN7L3/ENY2 in humans, are anchored to SAGA complexes through their interactions with homologs of the ataxin-7 protein from their respective organisms.
While neither the deubiquitination of histone H2B nor the presence of the deubiquitination module itself appear to overtly influence the HAT activity of their parent SAGA complexes, the monoubiquitination of histone H2B plays an important role in regulating transcription. In fact, the presence of a functional STAGA deubiquitination module has been directly implicated in nuclear receptor-mediated transcriptional activation (Zhao et al., 2008). Deletion analysis in yeast has demonstrated that Ubp8 is required for the recruitment of Ctk1 kinase and its subsequent phosphorylation of the RNAPII C-terminal domain (CTD) on serine 2 within the heptapeptide-repeat domain at SAGA-regulated genes (Wyce et al., 2007). Phosphorylation of the RNAPII CTD is required for the initiation and elongation of transcription by RNAPII, making the function of Ubp8 critical for proper regulation of SAGA-dependent gene expression. Additionally, the studies by Wyce et al. (2007) indicated that the persistence of monoubiquitinated histone H2B, due to the deletion of Ubp8, prevented the recruitment of the Set2 methyltransferase and as a result led to a reduction in levels of histone H3 methylation at lysine 36, a mark associated with transcriptionally active genes (Shilatifard, 2006). Given the regulatory effect that monoubiquitinated histone H2B exerts on transcriptional initiation and subsequent elongation of the nascent transcript by RNAPII, it seems clear that the SAGA complexes facilitate transcriptional activation and elongation not only through acetylation of chromatin, but also through the modulation of histone ubiquitination.
V. Gcn5 and PCAF Acetyltransferases Exhibit Distinct, Yet Partially Overlapping, Functions
As discussed above, both Gcn5 and PCAF are highly homologous proteins that serve as acetyltransferases in different multiprotein complexes. Somewhat predictably due to their high degree of sequence similarity, Gcn5 and PCAF exhibit partially overlapping, yet distinct, functions within the cellular context. While the PCAF complex contains a similar complement of proteins as the Gcn5-containing complexes, there are several differences that may explain both their overlapping and dissimilar functions. To better understand the roles of both Gcn5 and PCAF, the effects of their deletion on the development and adult phenotype of transgenic, knock-out mice were examined. Embryogenesis is a highly regulated process during which the up- and downregulation of many genes is required for the proper formation of organs and tissues within the developing animal. While the functions of Gcn5 had been well studied in both yeast and cultured mammalian cells, the role of this acetyltransferase, and its closely related homolog PCAF, in developmental processes was not as well characterized. To address questions relating to the roles of these enzymes in mouse development, several transgenic models were generated harboring deletions of the Gcn5 locus, the PCAF locus, or both. Despite the high similarity between the two proteins, their control over embryogenesis was shown to be quite disparate. Surprisingly, homozygous deletion of the PCAF gene resulted in a lack of physical impairment in adult mice (Yamauchi et al., 2000). Loss of PCAF expression, however, did lead to neurological dysfunction in young adult mice, including memory deficit and an exaggerated response to externally applied stress (Maurice et al., 2008). In contrast, despite a lack of phenotype in heterozygous Gcn5 knock-out mice, loss of both Gcn5 alleles led to embryonic lethality that was associated with an increase in apoptosis and a failure to complete neural tube development (Xu et al., 2000; Yamauchi et al., 2000). Homozygous deletion of both PCAF and Gcn5 alleles led to embryonic lethality occurring at earlier stages in development than that observed in Gcn5-null mice, further suggesting that the two proteins were required for functionally distinct processes. Not surprisingly, considering the distinct phenotypes of Gcn5and PCAF-null mice, Gcn5 mRNA was found to be highly expressed until later stages in embryonic development while PCAF mRNA was barely detectable until mice reached adulthood (Xu et al., 2000). Additional analysis of tissues from these mouse models indicated that expression of Gcn5 protein increased in response to loss of PCAF (Yamauchi et al., 2000). Coupled with the more severe developmental phenotype observed in mice lacking both PCAF and Gcn5 it can be concluded that these two acetyltransferases exhibit distinct but partially overlapping functions. These findings indicate a vital role for the Gcn5 protein, even at a reduced level of expression, in the completion of proper nervous system development and suggest two possible avenues for Gcn5 dependence during mouse embryonic development. The first possibility is that the requirement for specific acetyltransferases at individual gene loci may be flexible in the case of certain genes and more stringent for others. Some genes may specifically require the activity of Gcn5 to be properly regulated, while others may be appropriately regulated by either PCAF or Gcn5. This scenario may explain why Gcn5 is upregulated in PCAF-null mice and how these animals can complete development normally. PCAF may be a more “dispensable” acetyltransferase during embryonic development since it has arisen more recently, evolutionarily speaking, compared to its paralog Gcn5, which is found in more ancestral organisms. A second possibility is that the Gcn5 protein may be providing or facilitating a yet-to-be discovered activity that is essential for the normal regulation of genes required for developmental processes.
The idea that Gcn5 and PCAF have overlapping functions has been further supported by experiments performed in cultured chicken cells, which demonstrated a robust upregulation of PCAF expression in response to loss of Gcn5 (Kikuchi et al., 2005). The observed compensation for loss of Gcn5 was PCAF-specific since the expression of other acetyltransferases was not significantly altered. It is not difficult to believe that loss of either Gcn5 or PCAF results in the increased expression of the other since these two proteins share a very high degree of sequence similarity and function in closely related, multiprotein complexes. Interestingly, the altered steady-state levels of either Gcn5 or PCAF observed after loss of one or the other apparently compensates for the overall level of acetyltransferase protein but leads to differential patterns of histone H3 and H4 acetylation (Kikuchi et al., 2005). While not unexpected, this observation does raise the question of how compensation of Gcn5, or PCAF, function is truly achieved in the absence of a known pattern of overlapping histone acetylation. The answer could suggest that the generalized acetylation of chromatin by these proteins may play a more important role in regulating gene expression than Gcn5- or PCAF-specific acetylation.
VI. Polyglutamine Stretches are Common Attributes of Transcription Factors
The initiation of gene transcription is an intricate process involving the coordinated actions of both protein–DNA and protein–protein interactions. DNA-binding transcriptional activators have been shown to minimally contain one of four general domains, acidic, glutamine-rich, proline-rich, and DNA binding, that influence their ability to promote transcription (Courey and Tijan, 1988). Of the aforementioned four domains, the acidic, glutamine-rich, and proline-rich domains have been shown to impact the ability of a protein to facilitate transcription (Mitchell and Tijan, 1989). The obvious necessity for the DNA-binding domain aside deletion of any of the three remaining domains negatively impacts the ability of a transcription factor to promote transcription (Courey and Tijan, 1988; Gerber et al., 1994). The significance of glutamine-rich domains in the transactivation potential of transcription factors was explored on the global level in the budding yeast Saccharomyces cerevisiae by two high-throughput screens that yielded partially complementary results (Uetz et al., 2000; Ito et al., 2001). The amino acid sequences of transcriptional activators that were identified in these screens were analyzed to identify patterns that correlated with their transactivation potential. While there were several physiochemical properties that appeared to be common to many of these proteins, those that contained a glutamine-rich region were shown to be strongly associated with a greater potential to stimulate transcription (Titz et al., 2006). More detailed studies in mammalian cells have shown that the presence of glutamine-rich domains in transcriptional activators, such as the cAMP response element binding (CREB) protein and Sp1, are necessary for their association with chromatin in vivo (Mayr et al., 2005). Additionally, glutamine-rich domains are interchangeable between these transcriptional activators, indicating their overall importance in this context.
Interrogation of information within protein databases for the identities and functions of proteins that contain homopolymeric tracts of glutamine (containing at least 20) or proline (containing at least 10) residues revealed that the majority of hits returned, 82% and 78%, respectively, were factors associated with transcription (Gerber et al., 1994). Exploration of these findings indicated that an increase in the number of glutamine or proline residues in their respective homopolymer tracts correlated with an increased ability to stimulate transcription in vitro. Interestingly, the effect of increasing homopolymeric glutamine tract length on transcriptional activation was limited to peptides containing fewer than 40 glutamines (Gerber et al., 1994). A group of otherwise unrelated genes have been shown to be subject to a mutation that results in lengthening of polyglutamine tracts within their protein products. Polyglutamine expansion of these proteins results in altered function and polyglutamine tract-dependent aggregation of proteins to form nuclear inclusions (reviewed in Orr and Zoghbi, 2007). Understanding the normal role of glutamine-rich domains within transcriptional activators is of particular interest in the context of elucidating the pathological mechanisms associated with polyglutamine-expansion disorders. One of these diseases, spinocerebellar ataxia type 7 (SCA7), will be the discussed below.
VII. Polyglutamine-Expansion Disorders
The polyglutamine-expansion disease family encompasses at least nine heritable disorders (Table I), including Huntingtin’s disease (HD), dentatorubral pallidoluysian atrophy (DRPLA), spinal and bulbar muscular atrophy (SBMA), and the spinocerebellar ataxias SCA1, SCA2, SCA3, SCA6, SCA7, and SCA17 (reviewed in Orr and Zoghbi, 2007). Each of these disorders results from the expansion of a polyglutamine tract that is present in the wild-type protein. Polyglutamine expansion of disease-associated proteins is the direct effect of instability within the CAG trinucleotide coding region at the genomic locus of the affected gene. Proteins that are subject to polyglutamine expansion are thought to undergo conformational change (reviewed in Nagai and Popiel, 2008) and in some cases proteolytic cleavage that results in products exhibiting a cytotoxic function (Wellington et al., 1998; Ellerby et al., 1999). The autosomal dominant inheritance patterns observed in eight of the nine polyglutamine-expansion disorders suggest that the altered proteins associated with disease exert a functional dominance over their wild-type counterparts. Interestingly, polyglutamine-expanded proteins are widely expressed throughout the body; however, pathology is primarily restricted to neuronal tissue. The mechanisms of pathology are not precisely known for members of this disease family, although many of the genes affected encode proteins that are normally involved in transcriptional regulation in some capacity. The following will discuss the implications of findings from the literature on the elucidation of mechanisms associated with the pathology of polyglutamine-expansion disorders. An emphasis will be placed on SCA7 since the protein that becomes expanded in this condition, ataxin-7 (ATXN7), is an integral component of the SAGA family of acetyltransferase complexes that are conserved from yeast to humans.
Table I.
Basic Information on Genes that are Associated with Polyglutamine Expansion Disorders
| Disease | Gene | Normal CAG repeat length | Pathogenic CAG repeat length | Normal protein function |
|---|---|---|---|---|
| HD | Huntingtin | 6–34 | 36–121 | Multifunctional scaffold |
| SCA1 | Ataxin-1 | 8–44 | 39–83 | Transcriptional regulation |
| SCA2 | Ataxin-2 | 13–33 | 32–77 | RNA metabolism |
| SCA3 | Ataxin-3 | 12–40 | 54–89 | Ubiquitin protease |
| SCA6 | CACNA-1 | 4–18 | 19–33 | Voltage-gated calcium channel |
| SCA7 | Ataxin-7 | 4–35 | 37–306 | Structural component of GCN5 HAT complexes |
| SCA17 | TBP | 29–42 | 47–55 | General transcriptional factor |
| SBMA | Androgen receptor | 6–39 | 40–63 | Nuclear receptor |
| DRPLA | Atrophin-1 | 6–36 | 49–84 | Unknown |
VIII. Spinocerebellar Ataxia Type 7
Spinocerebellar ataxia type 7, formerly known as autosomal-dominant cerebellar ataxia with retinal degeneration (ADCA Type II), is in the family of neurodegenerative disorders known as trinucleotide repeat, or polyglutamine-expansion, disorders. The gene associated with SCA7 was mapped to chromosome 3p12-p13 (Benomar et al., 1995; David et al., 1996) and further examination of the genomic region indicated that the disease-associated allele was subject to expansion of a CAG trinucleotide repeat sequence (Lindblad et al., 1996). Cloning of the gene responsible for SCA7, hereafter referred to as ataxin-7 (ATXN7), followed and demonstrated a broad range of CAG repeats, between 38 and 130, associated with development of SCA7 within individuals that were surveyed in the study (David et al., 1997). Trends observed in families harboring the pathogenic ATXN7 allele demonstrated that the age of symptom onset decreased, which was accompanied by an increased rate of disease progression, in affected offspring compared to their parents, suggesting clinical anticipation. Additionally, paternal transmission of the mutant gene resulted in greater anticipation and therefore an earlier age of onset when compared to maternal transmission, which raises the possibility of paternal imprinting at the ATXN7 gene locus (Benomar et al., 1994).
Trinucleotide repeat expansion within the ATXN7 gene results in a protein product containing a polyglutamine-expanded region. Clinically, polyglutamine expansion of the ATXN7 protein is characterized by progressive degeneration of both the cerebellum and the retina. Cerebellar degeneration leads to ataxia, which is characterized by a generalized reduction in the ability to coordinate muscle movements. Less-penetrant phenotypes include muscle weakness, amyotrophy, difficulty speaking (dysarthria), difficulty swallowing (dysphagia), hearing loss, and intellectual impairment (Benomar et al., 1994). While ataxia is a symptom that is common to all of the trinucleotide repeat expansion disorders, SCA7 is unique in the additional development of retinal degeneration, which is characterized by cone-rod dystrophy and eventual blindness. The highly complex nature of cellular dysfunction in the context of SCA7 has led to the development and use of many experimental systems and the publication of studies that contribute to the understanding of disease mechanisms from multiple experimental approaches.
IX. Transgenic Mouse Models of SCA7
Transgenic, knock-in mouse models of SCA7 have been produced with expression of polyglutamine-expanded ATXN7 in Purkinje cells (Yvert et al., 2000), retina (Yvert et al., 2000; La Spada et al., 2001), whole brain (Yvert et al., 2001), central nervous system with the exception of Purkinje cells (La Spada et al., 2001), Bergmann glia of the cerebellum (Custer et al., 2006), or throughout the entire body (Yoo et al., 2003). The transgenic mouse models that have been designed to express mutant ATXN7 in different regions of the brain exhibit several commonalities that mimic clinical manifestations of SCA7 in humans. With the exception of those expressing polyglutamine-expanded ATXN7 under control of a retinal-specific promoter, these mice demonstrated symptoms of progressively worsening ataxia. Further, the age of onset and rate of degeneration varied depending on the specific tissue in which the polyglutamine-expanded ATXN7 protein was expressed. In addition to variability relating to the tissue in which the mutant ATXN7 protein was expressed, the observed disparity in disease progression in these transgenic mouse models was likely to have resulted from the different polyglutamine tract expansion lengths used in each model.
Purkinje cell-specific expression of mutant ATXN7 led to the observation of a detectable phenotype in transgenic mice at approximately 11 months of age (Yvert et al., 2000). Despite the observation of impaired motor function in these animals, histological examination of cerebellum tissue samples from 11-month-old individuals did not show obvious signs of degeneration in Purkinje cells, or any other cell type within the cerebellum. In this model, nuclear accumulation of polyglutamine-expanded ATXN7 protein was observed when mice were 1 month of age, followed by the formation of nuclear inclusions in rostral lobes of the cerebellum at 4 months. By the time mice were 16 months of age, most Purkinje cells contained at least one nuclear inclusion. Inclusions observed at later time points appeared to localize adjacent to the nucleolus when observed by either indirect immunofluorescence or electron microscopy. In contrast to observations made in their 11-month-old counterparts, Purkinje cells in cerebellar tissue sections obtained from 16-month-old mice exhibited a reduction in dendritic arborization and were present in a generally lower density compared to matched controls (Yvert et al., 2000). Expression of polyglutamine-expanded ATXN7 in Bergmann glial cells, which are responsible for clearing glutamate from Purkinje cell synapses, also led to the development of ataxia in transgenic mice (Custer et al., 2006). Bergmann glia expressing mutant ATXN7 exhibited a reduction in levels of the glutamate transporter GLAST and glutamate uptake. Purkinje cells associated with Bergmann glia expressing polyglutamine-expanded ATXN7 demonstrated signs of dark cell degeneration, which is associated with excitotoxicity linked to glutamate overstimulation (Leranth and Hamori, 1970; Strahlendorf et al., 1998; Strahlendorf and Strahlendorf, 1999). These two studies show that neurological dysfunction associated with expression of polyglutamine-expanded ATXN7 can result from either cell autonomous or non-cell-autonomous Purkinje cell degeneration.
Transgenic mice expressing mutant ATXN7 in the central nervous system with the exception of Purkinje cells exhibited symptoms of neurological dysfunction, including ataxia, beginning as early as 8 weeks of age (Garden et al., 2002). Sections of cerebellum prepared from 20-week-old individuals demonstrated a reduction in the size and dendritic arborization of Purkinje cells, despite lack of mutant ATXN7 protein expression. Further, apoptotic cells were not observed in these tissue sections, indicating the neurological dysfunction in these individuals occurred in the absence of significant neuronal cell loss. The onset of neurological symptoms in these transgenic mice correlates with the appearance of nuclear inclusions in the granule and molecular cell layers of the cerebellum, along with a reduction in the detection of the Purkinje cell marker calbindin by immunohistochemistry (Garden et al., 2002). The results of this study are of particular importance because they demonstrate that Purkinje cell degeneration in SCA7 can occur in a non-cell-autonomous manner and therefore may indicate a more significant role of mutant ATXN7 expressed in other cell types of the cerebellum in SCA7 pathology. Consistent with these findings, Bergmann glia-specific expression of mutant ATXN7 was sufficient to produce ataxia and degeneration (Custer et al., 2006), suggesting that glial cell dysfunction leads to Purkinje cell degeneration.
Transgenic mice expressing polyglutamine-expanded ATXN7 in all tissues of the brain demonstrated yet another timeline for disease onset and progression (Yvert et al., 2001). Individuals with whole-brain expression of mutant ATXN7 were observed to exhibit initial symptoms of neurological dysfunction between 3 and 8 months of age. The formation of nuclear inclusions in these animals often coincided, or preceded, the onset of disease symptoms and progressed in a manner similar to that observed in other transgenic mouse models. Additionally, the distribution of nuclear inclusions between affected and unaffected neuronal populations was indistinguishable, suggesting that the formation of nuclear inclusions was not directly linked to toxicity in all cell types (Yvert et al., 2001).
The pathology of SCA7 is set apart from the other polyglutamine-expansion disorders by the occurrence of progressive retinal degeneration in affected patients. To better understand the mechanisms of SCA7 retinopathy in the absence of other neurodegenerative phenotypes, a transgenic mouse model was developed in which either normal or polyglutamine-expanded ATNX7 was expressed under control of the retinal-specific rhodopsin promoter (Yvert et al., 2000). Following initial characterization of the retinal-specific ATXN7 transgenic model, subsequent studies using the same transgenic mouse line demonstrated that these mice exhibited loss of ATXN7 transgene expression during the first few months of their lives. The reduction and eventual loss of transgenic ATXN7 expression was shown to result from downregulation of the rhodopsin promoter by the mutant ATXN7 protein (Helmlinger et al., 2004a). These observations led to the identification of this model as a system for studying how loss of polyglutamine-expanded ATXN7 at an early age influenced the onset and progression of SCA7 retinopathy. Rhodopsin promoter activity, affecting both endogenous rhodopsin and ATXN7 transgene expression, was greatly reduced in 3-week-old mice and continued to decline with age until becoming negligible by the time mice reached 9 weeks of age. Despite the loss of detectable exogenous ATXN7 mRNA and soluble ATXN7 protein by week 9, nearly all of the cells in retinal tissue from 10-week-old mice contained nuclear inclusions that stained positively with antibodies specific for the N-terminal region of the ATXN7 protein. Nuclear inclusions were also observed in these mice up to 2 years of age, indicating their maintenance over time despite early loss of mutant ATXN7 expression. The persistence of nuclear inclusions in this SCA7 mouse model is of particular interest because the breakdown of nuclear inclusions in a mouse model of HD was shown to occur in as few as 3 weeks after loss of polyglutamine-expanded Huntingtin protein expression (Martin-Aparicio et al., 2001). The lack of agreement between the nuclear inclusion dynamics observed in these two distinct mouse models suggests that there is a fundamental difference in the way that cells handle these highly complex structures when they are initiated by different proteins. Interestingly, photoreceptor cell loss did not coincide with the downregulation of rhodopsin promoter activity in young mice; however, retinal dysfunction did begin several weeks later and progressed to a complete loss of photoreceptor function despite the eventual abolition of exogenous ATXN7 expression. These results indicate that the presence of ATXN7 nuclear inclusions in photoreceptors of the retina, but not continuous expression of the expanded ATXN7 gene, is required for both repression of the rhodopsin promoter and retinal dysfunction.
Cone-rod dystrophy and subsequent retinal degeneration are hallmarks of SCA7 disease that are observed in both human and transgenic mouse models. Outside the context of SCA7, cone-rod dystrophy has been clinically associated with the presence of mutations in the cone-rod homeobox (CRX) transcription factor, which is involved in the regulation of retinal-specific genes (reviewed in Hennig et al., 2008). La Spada et al. (2001) produced a transgenic, knock-in mouse model with the ATXN7 transgene under control of the neuron-specific murine prion protein promoter to study the phenotype that develops when polyglutamine-expanded ATXN7 expression is limited to neuronal tissue. Similar to other SCA7 mouse models, mice expressing polyglutamine-expanded ATXN7 in this model developed progressive retinal degeneration, which was accompanied by photoreceptor cell loss. While both normal and mutant ATXN7 were shown to interact with CRX, polyglutamine-expanded ATXN7 recruited CRX into nuclear inclusions. Sequestration of CRX by mutant ATXN7 was also coincident with reduced expression of several retinal-specific genes. Repression of these retinal-specific genes was attributed to a mutant ATXN7-dependent inhibition of the interaction between CRX and the rhodopsin promoter, which was observed in electrophoretic mobility shift assays. Subsequent studies further dissected the physical interaction between ATXN7 and CRX and its effect on the expression of retinal-specific genes.
Like many transcriptional activators, the CRX protein contains a polyglutamine-rich region; however, it has been shown to be dispensable for CRX-dependent activation of retinal-specific promoters (Chen et al., 2004). Expectedly, the interaction between CRX and ATXN7 is mediated through the glutamine-rich region of CRX and the polyglutamine domain in the N-terminus of ATXN7. Despite the requirement of the ATXN7 N-terminal region for the ATXN7–CRX interaction, the C-terminus of the ATXN7 protein is essential for maximal repression of CRX-dependent promoters by mutant ATXN7 (Chen et al., 2004). The necessity of the ATXN7 C-terminal region in the modulation of CRX-dependent promoters may reflect either its role in anchoring ATXN7 to the SAGA complex (Köhler et al., 2008) or the presence of nuclear localization signals within this region of the protein (Chen et al., 2004).
The ATXN7 protein was later shown to be required for the interaction of CRX with the STAGA complex and its subsequent recruitment to CRX-dependent promoters (Palhan et al., 2005). Additional experiments conducted on the retinas of transgenic mice by Palhan et al. (2005) demonstrated that the expression of polyglutamine-expanded ATXN7 led to reduction in the recruitment of CRX to some, but not all, retinal-specific promoters. These promoters were shown to be hypoacetylated on histone H3 on lysine 9 and 14; however, the reduction in histone H3 acetylation was often more dramatic than the corresponding change in CRX occupancy at the same promoter. To elucidate this apparent discrepancy, the presence of GCN5 at the promoters of interest was determined. In contrast to the reduced recruitment of CRX and acetylation of histone H3, the occupancy of GCN5 at the regulatory regions of these genes was not significantly different when either normal or mutant ATXN7 was being expressed. This finding indicated that the observed hypoacetylation was not the result of a defect in the ability of mutant ATXN7-containing STAGA complexes to effectively associate with target genes (Palhan et al., 2005). From these findings the authors concluded that polyglutamine-expanded ATXN7 was inhibiting the acetyltransferase activity of STAGA complexes without affecting their recruitment to chromatin and association with tissue-specific transcription factors. In addition to the loss of normal histone acetylation by STAGA, the presence of these nonfunctional complexes may prevent compensatory acetylation of these regions by other redundant or overlapping acetyltransferases implying a dominant-negative effect of mutant ATXN7. Hypoacetylation of histone H3 is associated with a lack of transcriptional activation and provides a mechanistic explanation of how polyglutamine-expanded ATXN7 expression leads to aberrant regulation of retinal-specific genes in both affected humans and mouse models. These results also highlight the importance of HAT complex recruitment and function in the activation of tissue-specific promoters and how their dysregulation can result in disease onset and progression.
X. Proteolytic Processing of ataxin-7
The formation and accumulation of ATXN7-containing nuclear inclusions over time was observed in all transgenic mouse models. Nuclear inclusions from several of these mouse models were shown to immunostain positive with antibodies recognizing either the N-terminal region of the protein or the polyglutamine tract, but not the ATXN7 C-terminal region (Yvert et al., 2000, 2001; Garden et al., 2002). These observations suggest that the N-terminal region of the ATXN7 protein, which contains the polyglutamine tract, is enriched in these protein aggregates. Later studies demonstrated that like several other polyglutamine-expanded proteins, ATXN7 is subject to cleavage by activated caspase proteases. These experiments established that both normal and polyglutamine-expanded ATXN7 are substrates of activated caspase-7 and cleavage can occur at either of two consensus sites within the ATXN7 protein. Other proteins that can undergo polyglutamine expansion, such as the androgen receptor (AR), atrophin-1, huntingtin, and ataxin-3, are also cleaved in a caspase-dependent manner, suggesting that this event may represent an important step in the cellular pathology associated with their respective diseases (Wellington et al., 1998). In some cases, such as that of the polyglutamine-expanded AR and ATXN7, caspase-mediated cleavage of polyglutamine proteins is required for their formation of nuclear inclusions (Ellerby et al., 1999; Young et al., 2007).
Caspase-7-mediated proteolysis of ATXN7 is required for polyglutamine-induced aggregation, transcriptional dysregulation, and cytotoxicity (Young et al., 2007). Cleavage of the ATXN7 protein at one of two known caspase-7 consensus motifs examined by Young et al. (2007) yields a product that corresponds to the N-terminal ATXN7 fragment that was observed by Garden et al. (2002) in extracts prepared from transgenic mice expressing mutant ATXN7. Interestingly, the preferred site of caspase-7-mediated cleavage of ATXN7 in vivo differs from the preferred site in vitro. The differential preference of caspase-7 for the consensus site that is preferred in vivo may reflect the influence of accessory factors that were absent during in vitro experiments. The requirement for proteolytic cleavage of ATXN7 prior to its inclusion into nuclear aggregates may indicate that the N- and C-terminal regions of the protein serve different functions in the cell or that protein turnover is regulated by proteolytic processing. Either way, the processing of the ATXN7 protein adds an additional level of complexity to the overall understanding of how it functions in both normal and disease states.
XI. Ataxin-7 is a Subunit in SAGA and SAGA-Related Histone Acetyltransferase Complexes
The function of the normal ATXN7 protein has yet to be clearly determined; however, it has been shown to reside in both yeast and human HAT complexes and has been implicated as a potential transcriptional activator by global proteomics studies performed in yeast (Titz et al., 2006). Human ataxin-7 (hATXN7) and its yeast homolog, Sgf73, hereafter referred to as yATXN7, are constituents of the SAGA/SLIK and STAGA/TFTC HAT complexes, respectively (Sanders et al., 2002; Helmlinger et al., 2004b; McMahon et al., 2005; Palhan et al., 2005). Normal fractionation of SAGA is disrupted by deletion of yATXN7 despite maintenance of HAT activity on free core histones by recoverable complexes. Interestingly, deletion of yATXN7 abolishes the ability of purified SAGA to acetylate histones in the context of the nucleosome octamer in vitro. Restoration of nucleosomal HAT activity is observed in chromatographically purified samples when deletion of yATXN7 is complemented by expression of normal human ATXN7 (hATXN7), but not by polyglutamine-expanded hATXN7 (McMahon et al., 2005). These findings were mirrored when transiently expressed normal and polyglutamine-expanded ATXN7 were purified and assayed for their relative abilities to acetylate oligonucleosomal substrates in vitro from cultured human cells (Palhan et al., 2005). Maintenance of free HAT activity accompanied by loss of the ability to acetylate nucleosomes suggests that polyglutamine expansion of the ATXN7 protein in either species disrupts the substrate targeting of SAGA complexes, but not acetyltransferase activity of GCN5. To lend additional support for the role of ATXN7 in targeting the SAGA complex to its nucleosome substrate, yATXN7 has been shown to be essential for the recruitment of SAGA to the upstream activation sequences of genes that it regulates. Further, yATXN7 is required for the recruitment and assembly of the transcriptional preinitiation complex (PIC) at the promoters of SAGA-dependent genes in a HAT-dependent or independent manner (Shukla et al., 2006).
Normal and mutant ATXN7 have been shown to copurify with the Gcn5 protein in yeast and both GCN5 isoforms in human cells (McMahon et al., 2005; Palhan et al., 2005). As discussed above, the longer GCN5 isoform is thought to represent a differentially spliced GCN5 transcript and has been shown to contain an additional N-terminal domain that is highly homologous to the N-terminal domain of its homolog PCAF. The additional N-terminal domain allows GCN5-L to recognize and acetylate histones in the context of nucleosomes, which is a capability that is not exhibited by its shorter homolog when assayed in vitro with recombinant protein (Xu et al., 1998). Given the copurification of hATXN7 and GCN5-L and the appearance of GCN5-L as the preferred GCN5 isoform in STAGA, it seems likely that mutant hATXN7 could disrupt the normal acetylation of chromatin by interfering with the nucleosomal targeting ability of GCN5-L or the STAGA complex as a whole. Alternatively, improper substrate recognition may result from the depletion of specific SAGA subunits, including Ada2/Ada2b, Ada3, TAF12, and Spt3 in both yeast and humans (McMahon et al., 2005; Palhan et al., 2005). Loss of these subunits may be of particular importance to the mutant yATXN7-dependent SAGA dysfunction in yeast due to the absence of the N-terminal domain and subsequent inability of yGcn5 to target nucleosomal substrates without the aid of other factors. Consistent with this observation, Ada3 and TAF12 are required for Gcn5 to efficiently acetylate histones within nucleosomes (Grant et al., 1998; Balasubramanian et al., 2002). Of additional importance, STAGA complexes that copurify with polyglutamine-expanded hATXN7 are deficient in the adaptor protein ADA2b (Palhan et al., 2005). Exclusion of ADA2b from the mutant STAGA complex would also be expected to significantly alter the targeting and substrate specificity of its GCN5-dependent acetyltransferase activity. This could be expected to result in either generalized loss of GCN5-dependent histone acetylation or hyperacetylation of aberrant histone substrates. Exclusion of ADA2b also has the potential to alter chromatin occupancy of mutant ATXN7-containing STAGA. The dominant-negative function of polyglutamine-expanded ATXN7, suggested by Palhan et al. (2005), may reflect a situation in which the mutant STAGA complex binds to chromatin, is unable to acetylate associated nucleosomes, and fails to dissociate, therefore impeding other transcription factors from accessing chromatin efficiently. Alternatively, loss of ADA2b may selectively prohibit mutant STAGA from interacting with its chromatin substrate at certain targets and, as a result, facilitate the aberrant acetylation of normal STAGA targets by alternative, promiscuously acting acetyltransferase complexes. Abolition of the normal enzyme–substrate interactions between STAGA and nucleosomal histones is likely to facilitate the establishment of atypical chromatin acetylation patterns from either similar, yet nonredundant, acetyltransferases, such as PCAF, or other more dissimilar acetyltransferases.
Several studies have demonstrated the loss of nucleosomal HAT activity in polyglutamine-expanded ATXN7-containing SAGA complexes (McMahon et al., 2005; Palhan et al., 2005). Interestingly, Helmlinger et al. (2006) observed severe decondensation of chromatin within the nuclei of retinal cells expressing polyglutamine-expanded ATXN7, containing 90 glutamine repeats, under the control of a retinal-specific promoter. This observation was counterintuitive to the previously established loss of acetyltransferase activity due to the incorporation of mutant ATXN7 since chromatin decondensation is characteristic of an acetylated, transcriptionally active state. Follow-up experiments demonstrated that both STAGA/TCTF complex recruitment, as measured by chromatin immunoprecipitation using antibodies that recognize the STAGA subunit SPT3, and acetylation of histone H3 on lysine 9 and 14 increased significantly at retinal-specific genes that were dysregulated. Taken together, these observations indicate that STAGA/TFTC complexes containing polyglutamine-expanded ATXN7 would exhibit prolonged chromatin occupancy and lead to hyperacetylation of associated target genes. The inference of STAGA/TFTC presence as a result of SPT3 localization was accurate given the known association of SPT3 with these complexes at the time (Martinez et al., 1998; Martinez et al., 2001); however, more recently published data suggest the incorporation of SPT3 into other multiprotein complexes (Wang et al., 2008). While the aforementioned observations do not directly evaluate the ability of other SPT3-containing complexes to acetylate histones, they do demonstrate its coimmunoprecipitation with PCAF, in addition to GCN5, and distribution over a broad molecular weight range during size exclusion chromatography of human nuclear extracts. Of related interest, when yeast harboring a deletion of the yATXN7 gene were analyzed for the transcriptional activity of SAGA-dependent genes it was shown that the expression of yATXN7 protein was required for both the recruitment of intact SAGA to the UAS and the assembly of the transcriptional PIC (Shukla et al., 2006). In the same study, it was shown that deletion of yATXN7 resulted in an increased presence of lysine 9 and 14 acetylated histone H3 at SAGA-dependent promoters; however, the increased acetylation level was determined to be an artifact of reduced histone H3 eviction from these regions in the yATXN7-deleted strain. Later studies also suggest that Spt3 functions in a SAGA-independent manner to stabilize global levels of histones H3 and H4 in yeast strains harboring temperature-sensitive alleles of Not1, a structural component of the Ccr4–Not complex (James et al., 2007). In this scenario, the existence of a SAGA-independent role for Spt3, proposed by James et al. (2007), is also supported by their observation of a synthetic phenotype between spt3Δ and spt7Δ yeast strains since deletion of Spt7 is sufficient to abolish SAGA complex activity (Grant et al., 1997).
Tissues derived from SCA7 transgenic mice have been shown to exhibit a progressively decreasing amount of full-length, soluble polyglutamine-expanded ATXN7 protein, which is likely to reflect the results of both proteolytic processing by caspase-7 and incorporation of the mutant protein into insoluble inclusions over time. Interestingly, despite its preferential incorporation into nuclear inclusions, the abundance of a soluble N-terminal ATXN7 fragment appears to be comparable to its counterpart that is derived from nonexpanded ATXN7 (Garden et al., 2002). The significance of the N-terminal region of ATXN7 has yet to be fully characterized; however, it has been implicated in several important aspects of normal ATXN7 function, including physical interaction with both transcription factors (Chen et al., 2004) and the deubiquitination module of SAGA complexes (Köhler et al., 2008; Zhao et al., 2008). The net outcome is an apparent reduction in the availability of the soluble ATXN7 C-terminal region, which is required for the interaction between ATXN7 and the SAGA complex. Therefore, decreased availability of the ATXN7 C-terminus may have a substantial effect on the regulation of SAGA-dependent genes since it is required not only for the structural integrity of the SAGA complex, but also for its ability to efficiently associate with its chromatin substrate. Further, recombinant, full-length polyglutamine-expanded ATXN7 was able to inhibit nucleosomal acetyltransferase activity of purified PCAF complex when added to in vitro acetylation reactions. Interestingly, addition of a polyglutamine-expanded N-terminal ATXN7 fragment had no effect under the same conditions (Palhan et al., 2005). These findings support the proposed interaction of ATXN7 with the SAGA complex through its C-terminal region and the importance of the C-terminal region in the ability of ATXN7 to control the activity of other acetyltransferase complexes. Regardless of the acetyltransferase, there is clear potential for disruption of the normal pattern of histone acetylation, in addition to other histone modifications that constitute the regulatory histone code by polyglutamine-expanded ATXN7. Despite its ability to interfere with nucleosomal acetylation, mutant ATXN7 does not directly abolish the ability of the GCN5 protein to catalyze the acetyltransferase reaction, since purified mutant yeast and human SAGA complexes retain the ability to acetylate free histones in vitro (McMahon et al., 2005; Palhan et al., 2005). Rather, incorporation of mutant ATXN7 into these complexes appears to disrupt their normal ability to acetylate histones within the context of nucleosomes, implying that polyglutamine expansion of ATXN7 disturbs the targeting or substrate presentation mechanisms of these complexes that allow them to interact with their physiologically relevant targets.
In yeast, the N-terminal region of yATXN7 is not required for its association with SAGA; however, it is required for its ability to link the Ubp8 deubiquitination module with the complex (Köhler et al., 2008; Lee et al., 2009). Additionally, purification of the N-terminal region of yATXN7 results in the purification of the Ubp8 deubiquitination module, but not the SAGA complex as a whole. Deletion studies in yeast demonstrated that the N-terminal region of yATXN7 is both necessary and sufficient for the association of the deubiquitination module, consisting of Sus1, Sgf11, and the deubiquitinating enzyme Ubp8, with other subunits of the SAGA complex. These findings in yeast are corroborated by similar studies implicating the necessity of ATXN7 for the association and activity of the deubiquitination module in STAGA (Zhao et al., 2008). As discussed above, these SAGA deubiquitination modules are involved in the removal of monoubiquitin from histone H2B, which is correlated with RNAPII elongation. It is also possible that the STAGA complex regulates transcription by modulating the ubiquitination states of other transcription factors in a manner similar to that of other polyglutamine-expansion proteins, such as ataxin-3 (ATXN3). The non-expanded ATXN3 protein functions to repress gene activation through the inhibition of CBP/p300- and PCAF-dependent histone acetylation and the recruitment and maintenance of transcriptional corepressors to chromatin (Li et al., 2002). Transcriptional repression mediated by ATXN3 is dependent on its ability to inhibit the proteosome-mediated degradation of corepressor complexes by both deubiquitylating polyubiquitylated constituent proteins and preventing recognition of these polyubiquitin chains by the ubiquitin-proteosome pathway (Berke et al., 2005; Burnett and Pittman, 2005). Polyglutamine expansion of ATXN3 does not significantly impact its interactions with the transcriptional repressors HDAC3 and NCoR in vivo or in vitro; however, it does inhibit the deacetylase activity of ATXN3-containing complexes, which is dependent on the ubiquitin protease activity of the ATXN3 protein (Evert et al., 2006).
While current evidence indicates that SAGA complexes are involved in the removal of monoubiquitin from histone H2B, the effect of polyglutamine-expanded ATXN7 on the integrity and association of the deubiquitination module with the SAGA complex has not yet been studied. The normal ATXN7 protein may also facilitate the targeting of other, nonhistone substrates for deubiquitination, or simply anchor the deubiquitination module to the greater complex. Either way, the potential for alteration of this SAGA module in molecular pathogenesis associated with polyglutamine expansion of ATXN7 is obvious and warrants additional exploration.
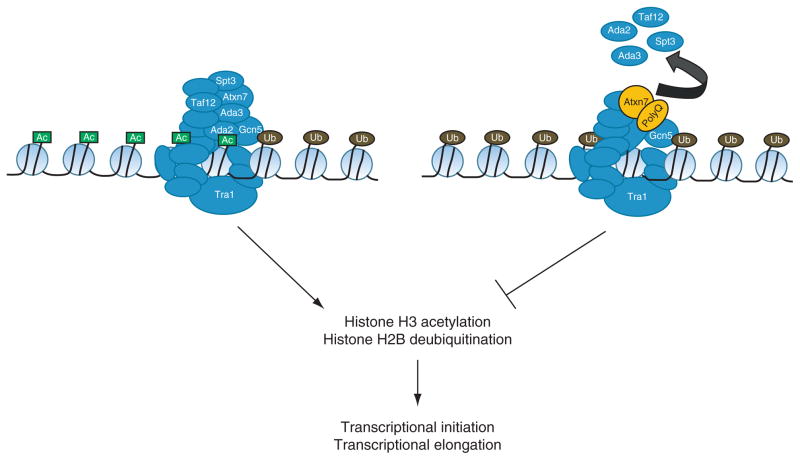
The multifunctional nature of the ATXN7 protein, and the SAGA complexes in which it resides, makes elucidation of its precise role in the molecular mechanisms of SCA7 pathology difficult. Clear associations have been made between the expression of polyglutamine-expanded ATXN7 protein and alteration of acetyltransferase activity in histone-modifying multiprotein complexes; however there appears to be other avenues by which this protein operates in the control of cellular functions. Interestingly, despite the structural alterations that accompany polyglutamine expansion of ATXN7, its association with transcription factors, such as CRX, does not appear to be significantly altered. In contrast, interaction with mutant ATXN7 prevents transcriptional activation by CRX and results in an altered gene expression profile that is associated with SCA7 retinal pathology. Although interactions with CRX are preserved, the inability of mutant ATXN7-containing protein complexes to effectively activate transcription is likely to be the result of lost associations with other proteins and therefore the inability to perform its normal functions. An example would be the disassociation of Ada2, Ada3, Spt3, and Taf12 from SAGA complexes containing mutant ATXN7 (Fig. 1). Loss of these subunits, particularly Ada2 and Ada3, would be likely to prevent the recognition and acetylation of nucleosomal substrates by Gcn5 without affecting the ability of the SAGA complex to associate with other factors. Interestingly, the scientific literature does not contain documented studies of the implications of ATXN7 polyglutamine expansion on the function of the SAGA complex deubiquitination module. It is easy to conceive that disruption of SAGA complex HAT activity would be sufficient to cause dysregulation of a large number of genes on its own; however, it is also difficult to ignore the potential for the additional alteration of deubiquitination activity given the data demonstrating the role of ATXN7 in anchoring the functional deubiquitination activity to the greater SAGA complex. Hopefully, future studies will elucidate whether polyglutamine expansion of ATXN7 alters the deubiquitination activity of the SAGA complex and its effects on the regulation of SAGA-dependent genes. The establishment of a better understanding of the molecular mechanisms underlying SCA7 pathology will facilitate the development of effective treatment methods for SCA7 and other polyglutamine-expansion disorders.
Fig. 1.
Incorporation of polyglutamine-expanded ATXN7 into SAGA complexes alters subunit composition and catalytic activities. SAGA complexes containing polyglutamine-expanded ATXN7 exhibit reduced levels of Ada2, Ada3, Spt3, and TAF12. Several of these subunits, such as Ada2 and Ada3, are necessary for the ability of Gcn5 to acetylate nucleosomal histones. While experiments have not yet addressed the issue, there is potential for polyglutamine expansion of ATXN7 to disrupt the histone deubiquitination activity of SAGA complexes since the N-terminal region of ATXN7 is involved in the attachment of the deubiquitination module to the greater complex. Both histone acetylation and deubiquitination are involved in the initiation of gene transcription and subsequent elongation by RNA polymerase II.
XII. Chemotherapeutic Options for the Treatment of Polyglutamine-Expansion Disorders
As highlighted above, substantial evidence exists to implicate the loss of HAT activity as a significant contributor to the pathology associated with polyglutamine-expansion disorders. As a result, the class of drugs known as histone deacetylase (HDAC) inhibitors has been tested as a therapeutic approach to the treatment of these disorders in laboratory models. HDAC inhibitors have been under investigation and development as chemotherapeutic approaches to the treatment of a variety of cancers with variable efficacy. Treatment with these drugs has been shown to reduce the expression of proteins involved in DNA synthesis, upregulate expression of proapoptotic factors, promote cell cycle arrest and differentiation, and more generally, modulate the expression of a broad range of genes (reviewed in Smith and Workman, 2009). While promising results have been observed, the specific mechanisms through which HDAC inhibitors influence cancer cell growth and overall disease progression are not yet fully clear. Fortunately, the development of these inhibitors as anticancer therapies has led to findings that support their use in the treatment of other diseases.
Sufficient evidence has been set forth in the scientific literature to support a link between altered histone acetylation and altered gene expression in several polyglutamine-expansion disorders. In this context, the application of HDAC inhibitors as potential chemotherapeutic agents for their treatment seems intuitive and is currently under exploration. Treatment of transgenic mice expressing different polyglutamine-expanded alleles, androgen receptor or atrophin-1, with the HDAC inhibitor sodium butyrate (SB) before the onset of symptoms demonstrated its ability to slow the rate of onset and overall disease severity (Minamiyama et al., 2004; Ying et al., 2006). In these experiments, brain tissue from transgenic mice was shown to exhibit an overall reduction in histone acetylation, especially of histone H3, compared to control mice that did not carry the polyglutamine-expanded allele. Treatment of these mice with SB resulted in an increase in histone H3 acetylation that was equivalent to or greater than matched wild-type controls, suggesting that SB treatment could offset the loss of histone acetylation that resulted from expression of these individual polyglutamine-expanded alleles. Further, the neuropathological events associated with onset of polyglutamine disease, such as neuronal loss and tissue degeneration, were either reduced or absent in drug-treated animals. While interpreting the results of HDAC inhibitor studies in mouse models of polyglutamine disease it is important to consider the effects of drug dosage on the outcome of treated animals. Interestingly, treatment of animals with higher, but still reasonable, doses of SB caused an earlier onset and more rapid progression of disease symptoms (Minamiyama et al., 2004). Treatment of matched control animals with the same dose yielded no observable phenotype in the study. These observations suggest potential for the use of HDAC inhibitors in the treatment of polyglutamine-expansion disorders; however, they also emphasize the care that must be taken to establish effective dosing guidelines to prevent exacerbation of disease symptoms.
In addition to studies in transgenic animals, the effect of HDAC inhibitors in preventing, or reversing, cytopathology is being examined in cultured cells expressing polyglutamine-expanded proteins. Expression of polyglutamine-expanded ATXN7 in cultured cells leads to an increase in the number of cells undergoing apoptosis (McCampbell et al., 2001). Treatment of these cells with micromolar concentrations of SB resulted in a nearly 50% reduction in the number of apoptotic cells indicating a protective effect of this HDAC inhibitor on cells expressing the toxic polyglutamine-expanded ATXN7 protein. Similar results were obtained from experiments in the same study that were designed to test the efficacy of several other HDAC inhibitors, trichostatin A (TSA) and suberoylanilide hydroxamic acid (SAHA), in a cell culture model of SMBA. Surprisingly, treatment of cells in these experiments with yet another HDAC inhibitor, phenylbutyric acid (PBA), failed to prevent toxicity for reasons that were not determined. While still promising, studies examining the application of HDAC inhibitors in the treatment of HD have been less forthcoming than other cell culture models of polyglutamine-expansion disorders. Expression of a polyglutamine-expanded form of the Htt protein in cultured cells led to a decreased association of acetylated histone H3 with genes whose mRNA is specifically downregulated in HD models. These findings indicate a correlation between the hypoacetylation of histones and the repression of certain HD-associated genes. Interestingly, treatment of these cells with SB resulted in a global increase in acetyl histone H3, an increase in the association of acetylated histone H3 with affected genes, and the restoration of their normal expression levels (Sadri-Vakili et al., 2007). Further, the polyglutamine-expanded Htt protein has been shown to sequester the HAT protein CBP into aggregates in transgenic mouse models of HD (Steffan et al., 2000; Nucifora et al., 2001). Interestingly, treatment of cells not expressing polyglutamine-expanded Htt with curcumin, a specific inhibitor of p300/CBP HAT activity (Balasubramanyam et al., 2004; Marcu et al., 2006), led to the global hypoacetylation of histone H3 and the nonspecific downregulation of gene expression that did not mimic an HD gene profile (Sadri-Vakili et al., 2007). The conclusions reached from these results implicate a factor, or factors, other than the inhibition of CBP-dependent histone acetylation by mutant Htt protein in conferring the specific pathology that is associated with HD.
The tissue-specific nature of polyglutamine-expansion disorders implies that their associated polyglutamine-expanded proteins affect a discreet set of regulatory factors and specific subsets of genes that are particularly vital for the normal function of neuronal cells and tissues. Interestingly, treatment of transgenic animals with HDAC inhibitors, which increase histone acetylation in a nonspecific manner, can alleviate symptoms and some of their underlying causes in these relatively specific diseases. While moving the field in a positive direction, this only highlights the need for a more thorough understanding of how polyglutamine expansion of proteins cause cellular dysfunction and, more generally, how histone acetylation influences cellular functions on a global level. These findings highlight two important observations involving the potential use of HDAC inhibitors in the treatment of polyglutamine-expansion disorders. First, clear evidence exists indicating that HDAC inhibitors can at least partially prevent cytotoxicity associated with the expression of polyglutamine-expanded proteins in both cell culture and transgenic animal models. Second, while several of these chemotherapeutic agents have shown promise in alleviating the toxic effects of polyglutamine-expanded proteins, no specific mechanism has been elucidated and not all HDAC inhibitors exert the same protective activity. While the ameliorative effects of HDAC inhibitors in the context of polyglutamine-expansion disorders is promising in the laboratory setting, the translation of these findings to accepted and effective clinical applications is still years away. Despite this realization, it appears that recent advances in the understanding of disease mechanisms and possible avenues for clinical treatment of polyglutamine-expansion disorders provide a strong foundation for future findings.
Acknowledgments
The authors would like to thank Paul J. Bonthuis and Madison E. Stellfox for their critical review of this chapter. This work is supported by NIH grant #R01NS049065.
References
- Balasubramanian R, Pray-Grant MG, Selleck W, Grant PA, Tan S. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J Biol Chem. 2002;277:7989–7995. doi: 10.1074/jbc.M110849200. [DOI] [PubMed] [Google Scholar]
- Balasubramanyam K, Varier RA, Altaf M, Swaminathan V, Siddappa NB, Ranga U, et al. Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J Biol Chem. 2004;279:51163–51171. doi: 10.1074/jbc.M409024200. [DOI] [PubMed] [Google Scholar]
- Barlev NA, Emelyanov AV, Castagnino P, Zegerman P, Bannister AJ, Sepulveda MA, et al. A novel human Ada2 homologue functions with Gcn5 or Brg1 to coactivate transcription. Mol Cell Biol. 2003;23:6944–6957. doi: 10.1128/MCB.23.19.6944-6957.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benomar A, Krols L, Stevanin G, Cancel G, LeGuern E, David G, et al. The gene for autosomal dominant cerebellar ataxia with pigmentary macular dystrophy maps to chromosome 3p12-p21.1. Nat Genet. 1995;10:84–88. doi: 10.1038/ng0595-84. [DOI] [PubMed] [Google Scholar]
- Benomar A, Le Guern E, Durr A, Ouhabi H, Stevanin G, Yahyaoui M, et al. Autosomal-dominant cerebellar ataxia with retinal degeneration (ADCA type II) is genetically different from ADCA type I. Ann Neurol. 1994;35:439–444. doi: 10.1002/ana.410350411. [DOI] [PubMed] [Google Scholar]
- Berke SJ, Chai Y, Marrs GL, Wen H, Paulson HL. Defining the role of ubiquitin-interacting motifs in the polyglutamine disease protein, ataxin-3. J Biol Chem. 2005;280:32026–32034. doi: 10.1074/jbc.M506084200. [DOI] [PubMed] [Google Scholar]
- Brownell JE, Allis CD. An activity gel assay detects a single, catalytically active histone acetyltransferase subunit in Tetrahymena macronuclei. Proc Natl Acad Sci USA. 1995;92:6364–6368. doi: 10.1073/pnas.92.14.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, et al. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- Burnett BG, Pittman RN. The polyglutamine neurodegenerative protein ataxin 3 regulates aggresome formation. Proc Natl Acad Sci USA. 2005;102:4330–4335. doi: 10.1073/pnas.0407252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candau R, Moore PA, Wang L, Barlev N, Ying CY, Rosen CA, et al. Identification of human proteins functionally conserved with the yeast putative adaptors ADA2 and GCN5. Mol Cell Biol. 1996;16:593–602. doi: 10.1128/mcb.16.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Peng GH, Wang X, Smith AC, Grote SK, Sopher BL, et al. Interference of Crx-dependent transcription by ataxin-7 involves interaction between the glutamine regions and requires the ataxin-7 carboxy-terminal region for nuclear localization. Hum Mol Genet. 2004;13:53–67. doi: 10.1093/hmg/ddh005. [DOI] [PubMed] [Google Scholar]
- Ciurciu A, Komonyi O, Pankotai T, Boros IM. The Drosophila histone acetyltransferase Gcn5 and transcriptional adaptor Ada2a are involved in nucleosomal histone H4 acetylation. Mol Cell Biol. 2006;26:9413–9423. doi: 10.1128/MCB.01401-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courey AJ, Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988;55:887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- Custer SK, Garden GA, Gill N, Rueb U, Libby RT, Schultz C, et al. Bergmann glia expression of polyglutamine-expanded ataxin-7 produces neurodegeneration by impairing glutamate transport. Nat Neurosci. 2006;9:1302–1311. doi: 10.1038/nn1750. [DOI] [PubMed] [Google Scholar]
- Daniel JA, Torok MS, Sun ZW, Schieltz D, Allis CD, Yates JR, 3rd, et al. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J Biol Chem. 2004;279:1867–1871. doi: 10.1074/jbc.C300494200. [DOI] [PubMed] [Google Scholar]
- David G, Abbas N, Stevanin G, Durr A, Yvert G, Cancel G, et al. Cloning of the SCA7 gene reveals a highly unstable CAG repeat expansion. Nat Genet. 1997;17:65–70. doi: 10.1038/ng0997-65. [DOI] [PubMed] [Google Scholar]
- David G, Giunti P, Abbas N, Coullin P, Stevanin G, Horta W, et al. The gene for autosomal dominant cerebellar ataxia type II is located in a 5-cM region in 3p12-p13: genetic and physical mapping of the SCA7 locus. Am J Hum Genet. 1996;59:1328–1336. [PMC free article] [PubMed] [Google Scholar]
- Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- Eberharter A, Sterner DE, Schieltz D, Hassan A, Yates JR, 3rd, Berger SL, et al. The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:6621–6631. doi: 10.1128/mcb.19.10.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellerby LM, Hackam AS, Propp SS, Ellerby HM, Rabizadeh S, Cashman NR, et al. Kennedy’s disease: caspase cleavage of the androgen receptor is a crucial event in cytotoxicity. J Neurochem. 1999;72:185–195. doi: 10.1046/j.1471-4159.1999.0720185.x. [DOI] [PubMed] [Google Scholar]
- Evert BO, Araujo J, Vieira-Saecker AM, de Vos RA, Harendza S, Klockgether T, et al. Ataxin-3 represses transcription via chromatin binding, interaction with histone deacetylase 3, and histone deacetylation. J Neurosci. 2006;26:11474–11486. doi: 10.1523/JNEUROSCI.2053-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamper AM, Kim J, Roeder RG. The STAGA subunit ADA2b is an important regulator of human GCN5 catalysis. Mol Cell Biol. 2009;29:266–280. doi: 10.1128/MCB.00315-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garden GA, Libby RT, Fu YH, Kinoshita Y, Huang J, Possin DE, et al. Polyglutamine-expanded ataxin-7 promotes non-cell-autonomous purkinje cell degeneration and displays proteolytic cleavage in ataxic transgenic mice. J Neurosci. 2002;22:4897–4905. doi: 10.1523/JNEUROSCI.22-12-04897.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgakopoulos T, Thireos G. Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J. 1992;11:4145–4152. doi: 10.1002/j.1460-2075.1992.tb05507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber HP, Seipel K, Georgiev O, Hofferer M, Hug M, Rusconi S, et al. Transcriptional activation modulated by homopolymeric glutamine and proline stretches. Science. 1994;263:808–811. doi: 10.1126/science.8303297. [DOI] [PubMed] [Google Scholar]
- Grant PA, Berger SL. Histone acetyltransferase complexes. Semin Cell Dev Biol. 1999;10:169–177. doi: 10.1006/scdb.1999.0298. [DOI] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- Grant PA, Eberharter A, John S, Cook RG, Turner BM, Workman JL. Expanded lysine acetylation specificity of Gcn5 in native complexes. J Biol Chem. 1999;274:5895–5900. doi: 10.1074/jbc.274.9.5895. [DOI] [PubMed] [Google Scholar]
- Grant PA, Schieltz D, Pray-Grant MG, Steger DJ, Reese JC, Yates JR, 3rd, et al. A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell. 1998;94:45–53. doi: 10.1016/s0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- Guelman S, Suganuma T, Florens L, Swanson SK, Kiesecker CL, Kusch T, et al. Host cell factor and an uncharacterized SANT domain protein are stable components of ATAC, a novel dAda2A/dGcn5-containing histone acetyltransferase complex in Drosophila. Mol Cell Biol. 2006;26:871–882. doi: 10.1128/MCB.26.3.871-882.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmlinger D, Abou-Sleymane G, Yvert G, Rousseau S, Weber C, Trottier Y, et al. Disease progression despite early loss of polyglutamine protein expression in SCA7 mouse model. J Neurosci. 2004;24:1881–1887. doi: 10.1523/JNEUROSCI.4407-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmlinger D, Hardy S, Abou-Sleymane G, Eberlin A, Bowman AB, Gansmuller A, et al. Glutamine-expanded ataxin-7 alters TFTC/STAGA recruitment and chromatin structure leading to photoreceptor dysfunction. PLoS Biol. 2006;4:e67. doi: 10.1371/journal.pbio.0040067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmlinger D, Hardy S, Sasorith S, Klein F, Robert F, Weber C, et al. Ataxin-7 is a subunit of GCN5 histone acetyltransferase-containing complexes. Hum Mol Genet. 2004;13:1257–1265. doi: 10.1093/hmg/ddh139. [DOI] [PubMed] [Google Scholar]
- Hennig AK, Peng GH, Chen S. Regulation of photoreceptor gene expression by Crx-associated transcription factor network. Brain Res. 2008;1192:114–133. doi: 10.1016/j.brainres.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, et al. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi J, Silverman N, Marcus GA, Guarente L. ADA3, a putative transcriptional adaptor, consists of two separable domains and interacts with ADA2 and GCN5 in a trimeric complex. Mol Cell Biol. 1995;15:1203–1209. doi: 10.1128/mcb.15.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- James N, Landrieux E, Collart MA. A SAGA-independent function of SPT3 mediates transcriptional deregulation in a mutant of the Ccr4-not complex in Saccharomyces cerevisiae. Genetics. 2007;177:123–135. doi: 10.1534/genetics.107.076299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi H, Takami Y, Nakayama T. GCN5: a supervisor in all-inclusive control of vertebrate cell cycle progression through transcription regulation of various cell cycle-related genes. Gene. 2005;347:83–97. doi: 10.1016/j.gene.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Kohler A, Pascual-Garcia P, Llopis A, Zapater M, Posas F, Hurt E, et al. The mRNA export factor Sus1 is involved in Spt/Ada/Gcn5 acetyltransferase-mediated H2B deubiquitinylation through its interaction with Ubp8 and Sgf11. Mol Biol Cell. 2006;17:4228–4236. doi: 10.1091/mbc.E06-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Schneider M, Cabal GG, Nehrbass U, Hurt E. Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat Cell Biol. 2008;10:707–715. doi: 10.1038/ncb1733. [DOI] [PubMed] [Google Scholar]
- Kuo MH, Brownell JE, Sobel RE, Ranalli TA, Cook RG, Edmondson DG, et al. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- Kusch T, Guelman S, Abmayr SM, Workman JL. Two Drosophila Ada2 homologues function in different multiprotein complexes. Mol Cell Biol. 2003;23:3305–3319. doi: 10.1128/MCB.23.9.3305-3319.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Spada AR, Fu YH, Sopher BL, Libby RT, Wang X, Li LY, et al. Polyglutamine-expanded ataxin-7 antagonizes CRX function and induces cone-rod dystrophy in a mouse model of SCA7. Neuron. 2001;31:913–927. doi: 10.1016/s0896-6273(01)00422-6. [DOI] [PubMed] [Google Scholar]
- Lee KK, Swanson SK, Florens L, Washburn MP, Workman JL. Yeast Sgf73/Ataxin-7 serves to anchor the deubiquitination module into both SAGA and Slik(SALSA) HAT complexes. Epigenetics Chromatin. 2009;2:2. doi: 10.1186/1756-8935-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leranth C, Hamori J. “Dark” Purkinje cells of the cerebellar cortex. Acta Biol Acad Sci Hung. 1970;21:405–419. [PubMed] [Google Scholar]
- Li F, Macfarlan T, Pittman RN, Chakravarti D. Ataxin-3 is a histone-binding protein with two independent transcriptional corepressor activities. J Biol Chem. 2002;277:45004–45012. doi: 10.1074/jbc.M205259200. [DOI] [PubMed] [Google Scholar]
- Lindblad K, Savontaus ML, Stevanin G, Holmberg M, Digre K, Zander C, et al. An expanded CAG repeat sequence in spinocerebellar ataxia type 7. Genome Res. 1996;6:965–971. doi: 10.1101/gr.6.10.965. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Marcu MG, Jung YJ, Lee S, Chung EJ, Lee MJ, Trepel J, et al. Curcumin is an inhibitor of p300 histone acetylatransferase. Med Chem. 2006;2:169–174. doi: 10.2174/157340606776056133. [DOI] [PubMed] [Google Scholar]
- Marcus GA, Silverman N, Berger SL, Horiuchi J, Guarente L. Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors. EMBO J. 1994;13:4807–4815. doi: 10.1002/j.1460-2075.1994.tb06806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Aparicio E, Yamamoto A, Hernandez F, Hen R, Avila J, Lucas JJ. Proteasomal-dependent aggregate reversal and absence of cell death in a conditional mouse model of Huntington’s disease. J Neurosci. 2001;21:8772–8781. doi: 10.1523/JNEUROSCI.21-22-08772.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E, Kundu TK, Fu J, Roeder RG. A human SPT3-TAFII31-GCN5-L acetylase complex distinct from transcription factor IID. J Biol Chem. 1998;273:23781–23785. doi: 10.1074/jbc.273.37.23781. [DOI] [PubMed] [Google Scholar]
- Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, et al. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol Cell Biol. 2001;21:6782–6795. doi: 10.1128/MCB.21.20.6782-6795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice T, Duclot F, Meunier J, Naert G, Givalois L, Meffre J, et al. Altered memory capacities and response to stress in p300/CBP-associated factor (PCAF) histone acetylase knockout mice. Neuropsychopharmacology. 2008;33:1584–1602. doi: 10.1038/sj.npp.1301551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr BM, Guzman E, Montminy M. Glutamine rich and basic region/leucine zipper (bZIP) domains stabilize cAMP-response element-binding protein (CREB) binding to chromatin. J Biol Chem. 2005;280:15103–15110. doi: 10.1074/jbc.M414144200. [DOI] [PubMed] [Google Scholar]
- McCampbell A, Taye AA, Whitty L, Penney E, Steffan JS, Fischbeck KH. Histone deacetylase inhibitors reduce polyglutamine toxicity. Proc Natl Acad Sci USA. 2001;98:15179–15184. doi: 10.1073/pnas.261400698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon SJ, Pray-Grant MG, Schieltz D, Yates JR, 3rd, Grant PA. Polyglutamine-expanded spinocerebellar ataxia-7 protein disrupts normal SAGA and SLIK histone acetyltransferase activity. Proc Natl Acad Sci USA. 2005;102:8478–8482. doi: 10.1073/pnas.0503493102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamiyama M, Katsuno M, Adachi H, Waza M, Sang C, Kobayashi Y, et al. Sodium butyrate ameliorates phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Hum Mol Genet. 2004;13:1183–1192. doi: 10.1093/hmg/ddh131. [DOI] [PubMed] [Google Scholar]
- Mitchell PJ, Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Muratoglu S, Georgieva S, Papai G, Scheer E, Enunlu I, Komonyi O, et al. Two different Drosophila ADA2 homologues are present in distinct GCN5 histone acetyltransferase-containing complexes. Mol Cell Biol. 2003;23:306–321. doi: 10.1128/MCB.23.1.306-321.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai Y, Popiel HA. Conformational changes and aggregation of expanded polyglutamine proteins as therapeutic targets of the polyglutamine diseases: exposed beta-sheet hypothesis. Curr Pharm Des. 2008;14:3267–3279. doi: 10.2174/138161208786404164. [DOI] [PubMed] [Google Scholar]
- Nagy Z, Tora L. Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene. 2007;26:5341–5357. doi: 10.1038/sj.onc.1210604. [DOI] [PubMed] [Google Scholar]
- Nucifora FC, Jr, Sasaki M, Peters MF, Huang H, Cooper JK, Yamada M, et al. Interference by huntingtin and atrophin-1 with cbp-mediated transcription leading to cellular toxicity. Science. 2001;291:2423–2428. doi: 10.1126/science.1056784. [DOI] [PubMed] [Google Scholar]
- Ogryzko VV, Kotani T, Zhang X, Schiltz RL, Howard T, Yang XJ, et al. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- Osley MA. Regulation of histone H2A and H2B ubiquitylation. Brief Funct Genomic Proteomic. 2006;5:179–189. doi: 10.1093/bfgp/ell022. [DOI] [PubMed] [Google Scholar]
- Palhan VB, Chen S, Peng GH, Tjernberg A, Gamper AM, Fan Y, et al. Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration. Proc Natl Acad Sci USA. 2005;102:8472–8477. doi: 10.1073/pnas.0503505102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pray-Grant MG, Schieltz D, McMahon SJ, Wood JM, Kennedy EL, Cook RG, et al. The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Mol Cell Biol. 2002;22:8774–8786. doi: 10.1128/MCB.22.24.8774-8786.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi D, Larsson J, Mannervik M. Drosophila Ada2b is required for viability and normal histone H3 acetylation. Mol Cell Biol. 2004;24:8080–8089. doi: 10.1128/MCB.24.18.8080-8089.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- Sadri-Vakili G, Bouzou B, Benn CL, Kim MO, Chawla P, Overland RP, et al. Histones associated with downregulated genes are hypo-acetylated in Huntington’s disease models. Hum Mol Genet. 2007;16:1293–1306. doi: 10.1093/hmg/ddm078. [DOI] [PubMed] [Google Scholar]
- Sanders SL, Garbett KA, Weil PA. Molecular characterization of Saccharomyces cerevisiae TFIID. Mol Cell Biol. 2002;22:6000–6013. doi: 10.1128/MCB.22.16.6000-6013.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- Shukla A, Bajwa P, Bhaumik SR. SAGA-associated Sgf73p facilitates formation of the preinitiation complex assembly at the promoters either in a HAT-dependent or independent manner in vivo. Nucleic Acids Res. 2006;34:6225–6232. doi: 10.1093/nar/gkl844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ER, Belote JM, Schiltz RL, Yang XJ, Moore PA, Berger SL, et al. Cloning of Drosophila GCN5: conserved features among metazoan GCN5 family members. Nucleic Acids Res. 1998;26:2948–2954. doi: 10.1093/nar/26.12.2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KT, Workman JL. Histone deacetylase inhibitors: anticancer compounds. Int J Biochem Cell Biol. 2009;41:21–25. doi: 10.1016/j.biocel.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Steffan JS, Kazantsev A, Spasic-Boskovic O, Greenwald M, Zhu YZ, Gohler H, et al. The Huntington’s disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc Natl Acad Sci USA. 2000;97:6763–6768. doi: 10.1073/pnas.100110097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Belotserkovskaya R, Berger SL. SALSA, a variant of yeast SAGA, contains truncated Spt7, which correlates with activated transcription. Proc Natl Acad Sci USA. 2002;99:11622–11627. doi: 10.1073/pnas.182021199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Strahlendorf JC, Brandon T, Miles R, Strahlendorf HK. AMPA receptor-mediated alterations of intracellular calcium homeostasis in rat cerebellar Purkinje cells in vitro: correlates to dark cell degeneration. Neurochem Res. 1998;23:1355–1362. doi: 10.1023/a:1020742404945. [DOI] [PubMed] [Google Scholar]
- Strahlendorf JC, Strahlendorf HK. Enduring changes in Purkinje cell electrophysiology following transient exposure to AMPA: correlates to dark cell degeneration. Neurosci Res. 1999;33:155–162. doi: 10.1016/s0168-0102(98)00126-6. [DOI] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titz B, Thomas S, Rajagopala SV, Chiba T, Ito T, Uetz P. Transcriptional activators in yeast. Nucleic Acids Res. 2006;34:955–967. doi: 10.1093/nar/gkj493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torok MS, Grant PA. Histone acetyltransferase proteins contribute to transcriptional processes at multiple levels. Adv Protein Chem. 2004;67:181–199. doi: 10.1016/S0065-3233(04)67007-0. [DOI] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Wang YL, Faiola F, Xu M, Pan S, Martinez E. Human ATAC Is a GCN5/PCAF-containing acetylase complex with a novel NC2-like histone fold module that interacts with the TATA-binding protein. J Biol Chem. 2008;283:33808–33815. doi: 10.1074/jbc.M806936200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Lee KK, Guelman S, Lin CH, Seidel C, Abmayr SM, et al. SAGA-mediated H2B deubiquitination controls the development of neuronal connectivity in the Drosophila visual system. EMBO J. 2008;27:394–405. doi: 10.1038/sj.emboj.7601966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Wellington CL, Ellerby LM, Hackam AS, Margolis RL, Trifiro MA, Singaraja R, et al. Caspase cleavage of gene products associated with triplet expansion disorders generates truncated fragments containing the polyglutamine tract. J Biol Chem. 1998;273:9158–9167. doi: 10.1074/jbc.273.15.9158. [DOI] [PubMed] [Google Scholar]
- Wyce A, Xiao T, Whelan KA, Kosman C, Walter W, Eick D, et al. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol Cell. 2007;27:275–288. doi: 10.1016/j.molcel.2007.01.035. [DOI] [PubMed] [Google Scholar]
- Xu W, Edmondson DG, Evrard YA, Wakamiya M, Behringer RR, Roth SY. Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat Genet. 2000;26:229–232. doi: 10.1038/79973. [DOI] [PubMed] [Google Scholar]
- Xu W, Edmondson DG, Roth SY. Mammalian GCN5 and P/CAF acetyltransferases have homologous amino-terminal domains important for recognition of nucleosomal substrates. Mol Cell Biol. 1998;18:5659–5669. doi: 10.1128/mcb.18.10.5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi T, Yamauchi J, Kuwata T, Tamura T, Yamashita T, Bae N, et al. Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc Natl Acad Sci USA. 2000;97:11303–11306. doi: 10.1073/pnas.97.21.11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Ogryzko VV, Nishikawa J, Howard BH, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- Ying M, Xu R, Wu X, Zhu H, Zhuang Y, Han M, et al. Sodium butyrate ameliorates histone hypoacetylation and neurodegenerative phenotypes in a mouse model for DRPLA. J Biol Chem. 2006;281:12580–12586. doi: 10.1074/jbc.M511677200. [DOI] [PubMed] [Google Scholar]
- Yoo SY, Pennesi ME, Weeber EJ, Xu B, Atkinson R, Chen S, et al. SCA7 knockin mice model human SCA7 and reveal gradual accumulation of mutant ataxin-7 in neurons and abnormalities in short-term plasticity. Neuron. 2003;37:383–401. doi: 10.1016/s0896-6273(02)01190-x. [DOI] [PubMed] [Google Scholar]
- Young JE, Gouw L, Propp S, Sopher BL, Taylor J, Lin A, et al. Proteolytic cleavage of ataxin-7 by caspase-7 modulates cellular toxicity and transcriptional dysregulation. J Biol Chem. 2007;282:30150–30160. doi: 10.1074/jbc.M705265200. [DOI] [PubMed] [Google Scholar]
- Yvert G, Lindenberg KS, Devys D, Helmlinger D, Landwehrmeyer GB, Mandel JL. SCA7 mouse models show selective stabilization of mutant ataxin-7 and similar cellular responses in different neuronal cell types. Hum Mol Genet. 2001;10:1679–1692. doi: 10.1093/hmg/10.16.1679. [DOI] [PubMed] [Google Scholar]
- Yvert G, Lindenberg KS, Picaud S, Landwehrmeyer GB, Sahel JA, Mandel JL. Expanded polyglutamines induce neurodegeneration and transneuronal alterations in cerebellum and retina of SCA7 transgenic mice. Hum Mol Genet. 2000;9:2491–2506. doi: 10.1093/hmg/9.17.2491. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, et al. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell. 2008;29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]