Abstract
Inducible nitric oxide synthase (NOS2) is involved in wound healing, angiogenesis, and carcinogenesis. NOS2 upregulation and increased nitric oxide (NO) production affect the redox state of cells and can induce protein, lipid, and DNA modifications. To investigate whether NOS2 levels influence survival of breast cancer patients, we examined NOS2 expression and its association with tumor markers and survival in 248 breast tumors. In multivariable survival analysis, increased NOS2 predicted inferior survival in women with estrogen receptor α–negative (ER-negative) tumors. Microdissected tumor epithelium from ER-negative tumors with high NOS2 had increased IL-8 and a gene expression signature characteristic of basal-like breast cancer with poor prognosis. In cell culture, NO only induced selected signature genes in ER-negative breast cancer cells. ER transgene expression in ER-negative cells inhibited NO-induced upregulation of the stem cell marker CD44 and other proteins encoded by signature genes, but not of IL-8. Exposure to NO also enhanced cell motility and invasion of ER-negative cells. Last, pathway analysis linked the tumor NOS2 gene signature to c-Myc activation. Thus, NOS2 is associated with a basal-like transcription pattern and poor survival of ER-negative patients.
Introduction
Breast cancer is not composed of one single homogeneous disease, but rather is made up of a number of subtypes. Recent results from large-scale gene expression profiling studies showed that estrogen receptor α–negative (ER-negative) and ER-positive breast tumors exhibit large differences in their gene expression signatures (1). These studies revealed that ER-negative and ER-positive tumors should be further subdivided based on their gene expression profiles. Molecular signatures characterize basal-like, HER2-positive, and normal-like subtypes among the ER-negative tumors and luminal A, luminal B, and luminal C subtypes among the ER-positive tumors. Among them, basal-like breast cancer has been recognized as the most aggressive subtype and the one most difficult to treat. Unlike ER-positive tumors, ER-negative tumors are hormonally unresponsive and not treatable by endocrine-targeted therapy such as tamoxifen and aromatase inhibitors. Patients with ER-negative breast tumors also have a worse prognosis than individuals with ER-positive breast tumors independent of therapy, particularly in the first 5 years following diagnosis (2), with almost twice the risk of death when compared with those with ER-positive tumors (3). There is an urgent need to identify novel targets for the treatment of ER-negative breast cancer in general, and even more for the basal-like subtype. Recently, a causative role of chronic inflammation in the progression of breast cancer has been proposed (4, 5). Tumor-infiltrating macrophages and expression of proinflammatory factors such as cyclooxygenase-2 are markers of poor outcome for the disease, and targeting markers of inflammation in breast cancer may provide new opportunities for therapy in ER-negative breast cancer (6, 7).
Inducible NOS2 is an inflammation-responsive enzyme that is upregulated in acute and chronic inflammation as part of host defense and the wound-healing process (8, 9). Upregulation of this enzyme leads to increased NO production, by which it affects the redox state of cells (10, 11). Aberrant expression of NOS2 has been observed in many types of solid tumors, including breast and colon cancer, and also in melanoma (12–16). NO can both cause DNA damage and protect from cytotoxicity and either inhibit or stimulate cell proliferation and migration as well as apoptosis (17–21). The effect of NO will be dependent on the microenvironment, including tissue oxygen tension and local superoxide concentrations. Thus, NO’s effects in human solid tumors are difficult to predict and are best estimated from the expression analysis of nitric oxide synthase enzymes in these tumors and their association with tumor markers and survival. We and others have previously discovered that NOS2 expression correlates with increased Akt phosphorylation in breast tumors and that NO induces Akt phosphorylation and activation of the oncogenic Akt pathway in breast cancer cells in culture (21, 22). Here, we tested the hypothesis that NOS2 is a predictor of breast cancer survival and examined the mechanism by which NO may induce a poor outcome phenotype. Our approach led to the discovery of a relationship between NOS2 expression, increased NO production, and the induction of a basal-like transcription pattern and poor prognosis in ER-negative breast tumors.
Results
High NOS2 is associated with increased vascularization and prevalence of p53 mutations.
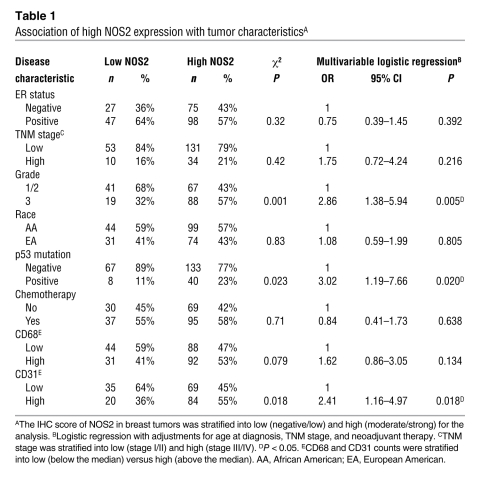
Expression of NOS2 was determined immunohistochemically in 248 surgically resected breast carcinoma specimens, as previously described by us and others (22, 23). The demographic and clinicopathological features of the patients and the immunohistochemistry (IHC) results are shown in Supplemental Table 1. NOS2 was moderately to strongly expressed in 173 of the 248 breast tumors (70%). The protein was present in both the tumor epithelium and tumor-infiltrating cells (Figure 1). However, only a few tumors showed a marked expression of NOS2 in infiltrating immune cells, e.g., tumor-infiltrating macrophages, monocytes, and lymphocytes (Figure 1D). We estimate that about 10%–20% of the NOS2 immunostaining in these tumors colocalizes with CD11b-positive cells. CD11b is a marker for macrophages, granulocytes, natural killer cells, and blood monocytes. Thus, we conclude that the major source of NO in breast tumors is most likely the tumor epithelium. Next, we examined the relationship of NOS2 with the tumor p53 mutational status, tumor microvessel density, and tumor grade, because others have previously observed relationships between NOS2 and these markers in human cancer (12, 13, 24–26). Consistent with previous reports, high NOS2 expression significantly correlated with an increased p53 mutation frequency — the first such report in breast cancer to the best of our knowledge — and with high tumor grade and increased density of CD31-positive microvessels (Table 1). No significant association was seen between NOS2 expression and ER status, race/ethnicity, tumor-node-metastasis (TNM) stage, phagocyte density (CD68 count), or receipt of neoadjuvant therapy. Multivariable logistic regression was performed to calculate odds ratios (ORs) for association of NOS2 with markers of tumor aggressiveness (Table 1). After adjustment for age at diagnosis, race/ethnicity, TNM stage, and neoadjuvant therapy, NOS2 expression remained independently associated with the tumor p53 mutational status (OR, 3.02; 95% CI, 1.19–7.66), tumor grade (OR, 2.86; 95% CI, 1.38–5.94), and microvessel density (OR, 2.41; 95% CI, 1.16–4.97).
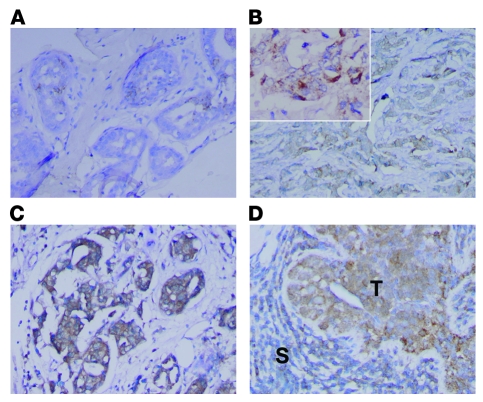
Figure 1. NOS2 expression in human breast tumors.
IHC analysis of invasive breast carcinomas for expression of inducible NOS2 (A–D). NOS2 was mainly expressed in the cytoplasm of tumor cells, with some cells showing a marked perinuclear localization of NOS2 protein. NOS2 expression ranged from (A) absent or occasional expression in the tumor epithelium, to (B) moderate expression in numerous tumor cells, to (C and D) a rather strong expression in the tumor epithelium that was either found localized within the tumor or present in most tumor cells. (D) Shown is a tumor with NOS2 expression in both the tumor epithelium and a lymphoid infiltrate (T, tumor epithelium; S, surrounding lymphoid infiltrate). Original magnification, B: ×100, ×200 (inset); A, C, and D: ×200. Counterstain: hematoxylin.
Table 1 .
Association of high NOS2 expression with tumor characteristicsA
NOS2 expression predicts poor survival in ER-negative breast cancer.
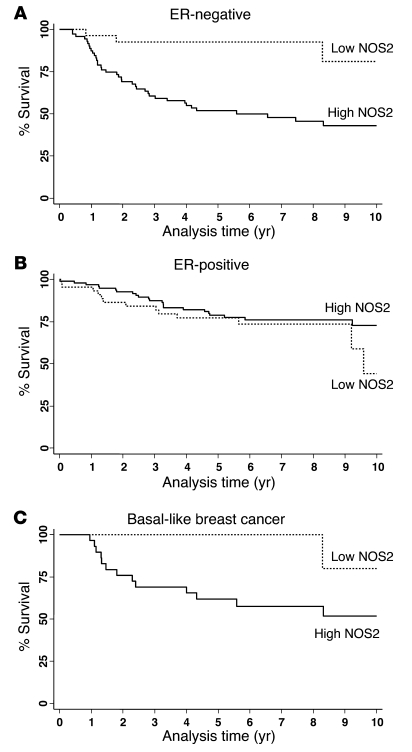
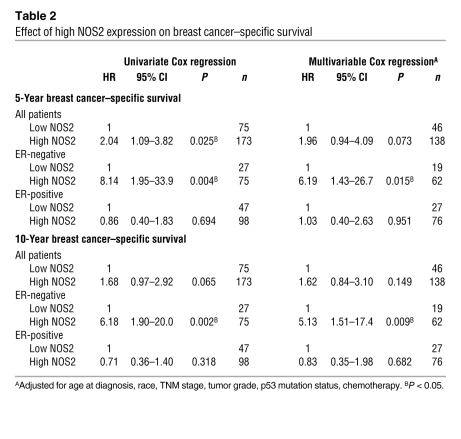
We next examined the effect of NOS2 expression on predicting patient survival using the Kaplan-Meier method (Figure 2) and multivariable Cox regression analysis (Table 2). The Kaplan-Meier analysis showed that NOS2 predicts breast cancer–specific survival in ER-negative patients. High NOS2 expression was significantly associated with decreased survival of these patients (Figure 2A), but not of ER-positive breast cancer patients (Figure 2B). This finding was further corroborated by a test for interaction showing that the tumor ER status modifies the effect of NOS2 on breast cancer survival (P = 0.002). The association of NOS2 with poor survival in ER-negative breast cancer patients remained significant in the multivariable analysis. Cox regression analysis showed that NOS2 is an independent predictor of 5- and 10-year survival after adjustment for age at diagnosis, tumor grade, TNM stage, receipt of chemotherapy, and tumor p53 mutation status (Table 2). Other factors, such as race/ethnicity or menopausal status, did not influence the association between NOS2 and breast cancer survival in ER-negative breast cancer.
Figure 2. Association between NOS2 expression and breast cancer survival by ER status.
(A) Kaplan-Meier cumulative breast cancer–specific survival curves of ER-negative breast cancer patients by NOS2 status (n = 98). The survival of patients with high NOS2 expression (n = 71) was significantly poorer than the survival of patients with low NOS2 expression (n = 27). P < 0.001, log-rank test. (B) ER-positive breast cancer patients by NOS2 status (n = 139). The survival of patients with high NOS2 expression (n = 95) was not significantly different from the survival of patients with low NOS2 expression (n = 44). P = 0.315. (C) Basal-like breast cancer (ER–/HER2–, and EGFR- or cytokeratin5/6-positive) by NOS2 status (n = 41). The survival of patients with high NOS2 expression (n = 29) was significantly poorer than the survival of patients with low NOS2 expression (n = 12). P = 0.038.
Table 2 .
Effect of high NOS2 expression on breast cancer–specific survival
High NOS2 expression is associated with a distinct gene signature in ER-negative tumors.
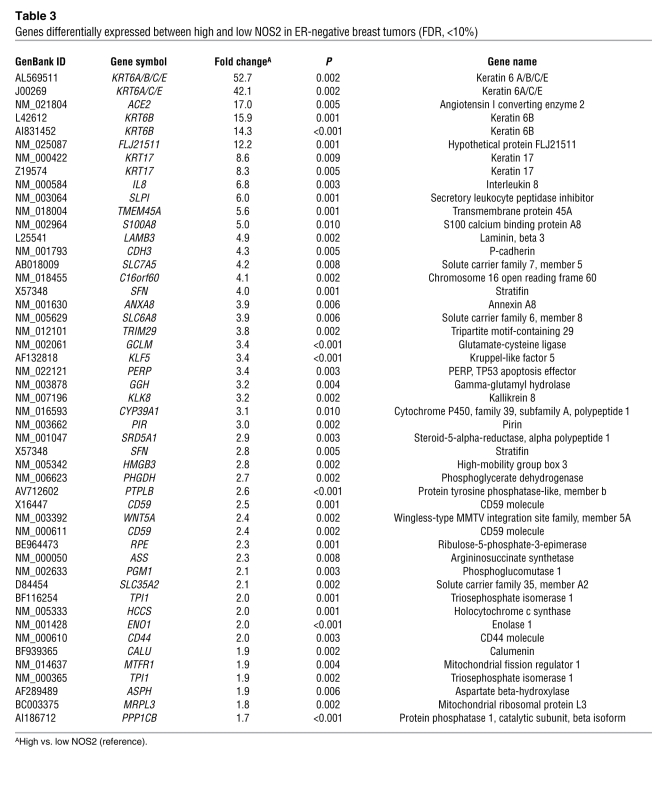
In order to determine why NOS2 is associated with poor survival in ER-negative but not ER-positive breast cancer, we analyzed the gene expression profiles of microdissected tumor epithelium from 32 breast tumors, comparing high NOS2 versus low NOS2 expression in ER-negative cases and ER-positive cases (9 NOS2 high/ER-negative and 8 NOS2 low/ER-negative; 6 NOS2 high/ER-positive and 9 NOS2 low/ER-positive). This analysis revealed a NOS2 gene signature in ER-negative tumors but not in ER-positive tumors. No gene was differentially expressed in the ER-positive tumors at any acceptable false discovery rate (FDR). In ER-negative tumors, 49 mRNA transcripts corresponding to 44 genes were consistently differentially expressed between NOS2 high and NOS2 low tumors. The genes are listed in Table 3. Strikingly, among the genes most differentially expressed were the genes encoding cytokeratins 6 and 17 and P-cadherin, which are markers of the basal-like breast cancer phenotype (27–29), and also IL-8. To determine whether this gene signature had more common features with basal-like breast cancer, we examined two previously published basal-like breast cancer gene signatures for similarities (1, 30). Cross-referencing the NOS2/ER-negative gene signature with these data sets (Supplemental Table 2) revealed that 11 of the 44 (25%) genes were also present in the Sorlie signature (1) and 15 of 44 (34%) in the Charafe-Jauffret signature (30), with 21 of 44 (48%) genes being previously identified as basal-like signature genes. Because of this finding, we further characterized the 248 breast tumors by IHC and identified those patients with basal-like breast cancer in our cohort (ER-negative, HER2-negative, and either EGFR-positive or cytokeratin 5/6-positive, as described in ref. 31). In the Kaplan-Meier survival analysis, high NOS2 was associated with poor survival in this patient subset (P = 0.038) (Figure 2C). The result suggests that in addition to inducing a basal-like signature in ER-negative tumors, the presence of NOS2 may further enhance disease aggressiveness in the presence of this signature.
Table 3 .
Genes differentially expressed between high and low NOS2 in ER-negative breast tumors (FDR, <10%)
The gene signature associated with NOS2 is a predictor of outcome in an independent cohort of ER-negative breast cancer patients.
We examined the ability of our NOS2 gene signature to predict both overall survival and relapse-free survival in ER-negative breast cancer patients (n = 77) in an independent and publicly available data set from the Karolinska Institute in Sweden ( http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE1456) (32), applying the survival prediction algorithm provided by the publicly available BRB-ArrayTools software. We conducted this analysis to assess whether the NOS2 gene signature from our study, which is similar but not identical to the reported basal-like gene signature in breast cancer, is also predictive of outcome in other breast cancer data sets. Indeed, the NOS2 signature predicted both relapse-free survival (P = 0.006) and overall survival (P = 0.019) in the ER-negative patients from the Karolinska data set (Supplemental Figure 1).
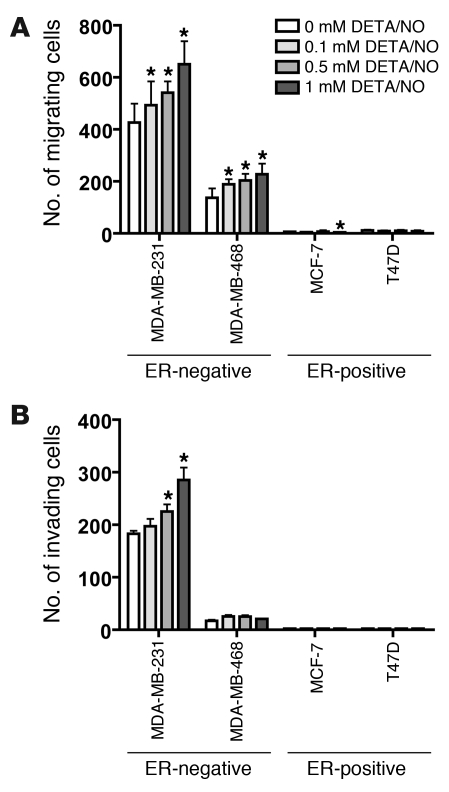
NO induces increased cell motility and invasion in ER-negative breast cancer cells.
To better understand the mechanism by which NOS2 induces an aggressive phenotype in ER-negative breast cancer, the NO donor diethylenetriamine/NO (DETA/NO) was used to examine the effects of NO, the product of NOS2, on breast cancer cell migration and cell invasion. To assess whether NO can induce or inhibit breast cancer cell migration and invasion, 4 breast cancer cell lines, 2 ER-negative (MDA-MB-231, MDA-MB-468) and 2 ER-positive (MCF-7, T47D), were exposed to escalating doses of 0, 0.1, 0.5, and 1 mM DETA/NO (Sigma-Aldrich), equivalent to about 0.2 and 0.4 μM NO concentrations at the highest doses of 0.5 and 1 mM DETA/NO over a 24-hour period (33). DETA/NO was found to induce cell motility in a dose-dependent manner in the two ER-negative cell lines, but not in the ER-positive cell lines as shown in Figure 3A. A regression analysis indicated a statistically significant trend toward increased levels of cell motility with increasing DETA/NO concentration in MDA-MB-231 (P < 0.001) and MDA-MB-468 cells (P = 0.009). DETA/NO also induced cell invasion in a dose-dependent manner in the ER-negative cell line MDA-MB-231 (P < 0.001) but not in MDA-MB-468 (P = 0.637) or in the ER-positive cell lines T47D and MCF-7, as shown in Figure 3B.
Figure 3. Induction of cell motility and cell invasion in ER-negative breast cancer cell lines after exposure to the NO donor DETA/NO.
(A) DETA/NO induced significantly increased cell migration in two ER-negative cell lines, MDA-MB-231 and MDA-MB-468, over a 24-hour exposure, but not in the ER-positive cell lines T47D and MCF-7. (B) DETA/NO significantly enhanced cell invasion of the invasive ER-negative cell line MDA-MB-231 over a 24-hour exposure, but not in the less-invasive ER-negative MDA-MB-468 line or the two ER-positive lines T47D and MCF-7. Shown are mean ± SD. *P < 0.05 versus no treatment, Student’s t test.
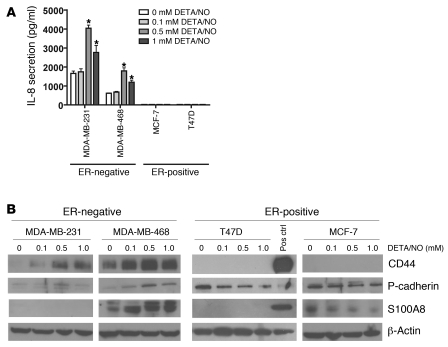
DETA/NO induces increased IL-8, CD44, S100A8, and P-cadherin protein expression in ER-negative breast cancer cell lines.
Next, we determined whether the gene expression profile associated with high NOS2 expression in ER-negative breast tumors was at least partly due to the effects of NO by treating cells with DETA/NO. For that, we focused on 4 gene products: IL-8, which has previously been found to be expressed only in ER-negative breast cancer (34, 35); S100 calcium-binding protein A8 (S100A8); and P-cadherin (CDH3), which are both poor outcome markers and markers of basal-like breast cancer (28, 29, 36); and the hyaluronate receptor (CD44), a marker of breast cancer stem cells and poor disease outcome (37). We examined the effects of NO on IL-8 secretion by ELISA (Figure 4A) and on S100A8, P-cadherin, and CD44 expression by Western blot analysis (Figure 4B) in 4 breast cancer cell lines, the ER-positive MCF-7 and T47D cells and the ER-negative MDA-MB-231 and MDA-MB-468 cells. Consistent with our gene expression data, exposure to NO increased the expression of IL-8, S100A8, P-cadherin, and CD44 expression in ER-negative breast cancer cells, but not the ER-positive cells. In the ER-positive cells, NO rather inhibited the expression of S100A8 (MCF-7) and P-cadherin (MCF-7 and T47D). To further corroborate the finding that the induction of these markers by NO in ER-negative human breast cancer cells is applicable across multiple cell lines with different genetic alterations, we performed the same experiments in two additional ER-negative human breast cancer cell lines, MDA-MB-157 and Hs578T, and also in ER-negative, non-tumorigenic MCF-10A human breast epithelial cells. As shown in Supplemental Figure 2, NO increased marker expression in these cell lines as well, although some differences were observed between these cell lines and the MDA-MB-231 and MDA-MB-468 cells.
Figure 4. Induction of IL-8, S100A8, CD44, and P-cadherin in ER-negative breast cancer cell lines after exposure to the NO donor DETA/NO.
(A) DETA/NO induced IL-8 secretion in the two ER-negative cell lines MDA-MB-231 and MDA-MB-468, over a 48-hour exposure, but not in the ER-positive cell lines T47D and MCF-7. Shown are mean ± SD. *P < 0.05 versus no treatment, Student’s t test. (B) DETA/NO induced increased protein expression of CD44, P-cadherin, and S100A8 in the ER-negative cell lines MDA-MB-231 and MDA-MB-468 over a 48-hour exposure, with no or opposite effects in the ER-positive cell lines T47D and MCF-7. ELISA was used for IL-8. Pos ctrl, positive control.
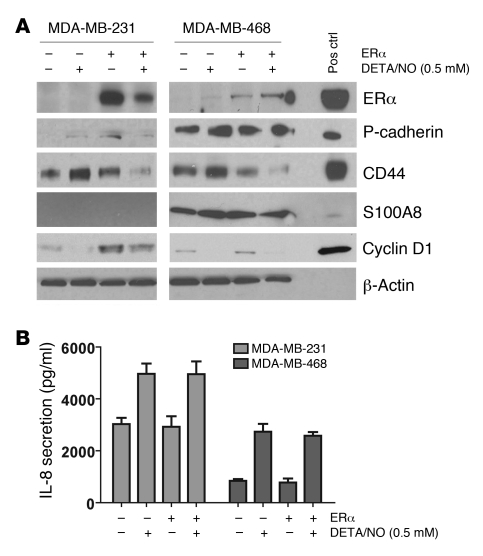
ER transgene expression in ER-negative breast cancer cells inhibits NO-induced upregulation of 3 markers but not IL-8.
The lack of an association between NOS2 and survival in ER-positive breast cancer and the absence of a noticeable gene expression signature related to high NOS2 expression in ER-positive tumors, coupled with the inability of NO to induce S100A8, P-cadherin, CD44, or IL-8 expression in ER-positive cells, suggested that the ER may modify the effects of NOS2 and its product NO on breast cancer cell phenotype. To determine whether the expression of the ER in the ER-negative cell lines MDA-MB-231 and MDA-MB-468 would modify NO-induced upregulation of P-cadherin, CD44, S100A8, or IL-8 in these cells, we transiently transfected them with a plasmid encoding the ER and exposed them to 0.5 mM DETA/NO for 48 hours. As shown in Figure 5A, introduction of the ER-encoding transgene (ESR1) into MDA-MB-231 and MDA-MB-468 cells at least partly reversed the effects of DETA/NO on P-cadherin, CD44, and S100A8 expression. However, expression of ESR1 in these cells had no effect on basal and NO-induced levels of IL-8, indicating that an intrinsic mechanism independent of ER status suppresses IL-8 expression in ER-negative cells (Figure 5B). To ensure that the ESR1-transfected cells had an intact ER pathway, we examined as indicators of ER pathway activation the expression of the ER-responsive gene cyclin D1 (38) and the activation of an ER-responsive luciferase construct in the MDA-MB-231 and MDA-MB-468 cells. As shown in Figure 5 and Supplemental Figure 3, ESR1 transgene expression increased expression of cyclin D1 (Figure 5A) and transcriptionally activated the reporter construct in a β-estradiol–dependent manner (Supplemental Figure 3), indicating that transgene expression led to intact ER signaling in our cell culture experiments.
Figure 5. Inhibition of DETA/NO effects in ER-negative breast cancer cells after ectopic expression of estrogen receptor α.
MDA-MB-231 and MDA-MB-468 cells were transfected with either an expression vector for estrogen receptor α (ERα) or a vector control, and protein expression was examined. (A) ERα expression partly reversed the induction of P-cadherin, CD44, and S100A8 by 0.5 mM DETA/NO (B) but had no effect on IL-8 induction by DETA/NO. Shown are mean ± SD for the IL-8 secretion data. (A) ERα expression led to a marked induction of the ERα-responsive gene cyclin D1 in MDA-MB-231 cells. This response was weaker in the MDA-MB-468 cells. In both cell lines, cyclin D1 was downregulated by DETA/NO. ELISA was used for IL-8.
MetaCore pathway analysis links NOS2 gene signature in ER-negative tumors to c-Myc.
Bioinformatics tools allow us to find transcription factors and mechanisms that are the underlying causes of a gene expression signature in human tissues. We used MetaCore software to identify candidate transcription factors that could be key regulators of the NOS2 gene signature in ER-negative tumors. This approach identified c-Myc as the top candidate transcription factor underlying the NOS2 gene signature, as shown in Supplemental Figure 4A. To experimentally follow up on this finding, we exposed MDA-MB-231 and MDA-MB-468 cells to 0.5 mM DETA/NO and isolated both total and nuclear fraction protein at 0, 20, 40, and 60 minutes of exposure. Then, we examined c-Myc protein expression by Western blot analysis and c-Myc activation using a transcription factor DNA binding ELISA. Incubation with DETA/NO resulted in an increase in both c-Myc protein expression (Supplemental Figure 4B) and c-Myc DNA binding activity (Supplemental Figure 4C) within 60 minutes in the MDA-MB-468 cells, which are known to have a basal-like phenotype. These early and preliminary mechanistic observations are consistent with the hypothesis that c-Myc activation by NO links NOS2 expression in ER-negative breast tumors to the observed NOS2 gene signature in these tumors.
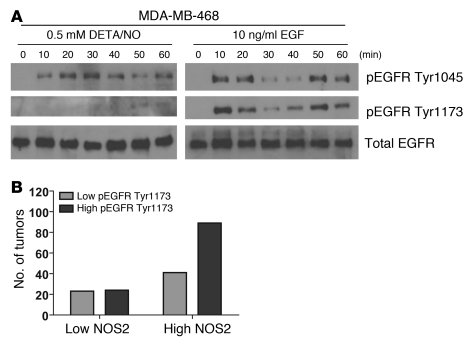
Phosphorylation of EGFR by NO in MDA-MB-468 cells.
Overexpression of EGFR is a marker of basal-like breast cancer (31). To further explore mechanisms by which NO may influence survival of breast cancer patients with a basal-like gene signature in their tumors, we studied the phosphorylation status of EGFR in response to NO exposure. Phosphorylation of this oncogenic receptor at residues Tyr1045 and Tyr1173 leads to receptor activation and downstream signaling. Exposure of MDA-MB-468 cells to 0.5 mM DETA/NO indeed induced rapid phosphorylation of both tyrosine residues in these cells (Figure 6A). Because of this finding, we performed an immunohistochemical analysis of EGFR Tyr1173 phosphorylation in a series of EGFR-positive breast tumors. The IHC for EGFR and pEGFR at Tyr1173 is shown in Supplemental Figures 5 and 6, respectively. This analysis revealed that EGFR-positive tumors with high NOS2 are significantly more likely to have a high EGFR phosphorylation score than EGFR-positive tumors with low NOS2 (Figure 6B). Together, these data indicate that NOS2 and NO may induce increased EGFR phosphorylation and cause EGFR pathway activation in breast cancer.
Figure 6. Induction of EGFR phosphorylation by the NO donor DETA/NO.
(A) DETA/NO (0.5 mM) induced increased phosphorylation of EGFR at tyrosine phosphorylation sites 1045 and 1173, within 60 minutes. (B) Increased NOS2 expression is associated with increased phosphorylation of EGFR at Tyr1173 in human breast tumors. High EGFR phosphorylation at Tyr1173 (pEGFR Tyr1173) was significantly more common in tumors with high NOS2 than in tumors with low NOS2 (IHC analysis; P = 0.033, χ2 test). In tumors with low NOS2: low pEGFR Tyr1173, n = 23 (48%), high pEGFR Tyr1173, n = 25 (52%). In tumors with high NOS2: low pEGFR Tyr1173, n = 41 (32%), high pEGFR Tyr1173, n = 89 (68%).
Discussion
In this study, we examined whether NOS2 influences breast cancer survival and investigated mechanisms by which NOS2 and NO may cause a poor outcome phenotype. The research led to the clinically significant observation that NOS2 expression is associated with a prognostic basal-like transcription pattern and is a predictor of inferior survival in women with ER-negative tumors. Moreover, the presence of NOS2 correlated with other poor outcome markers, such as an increased microvessel density and p53 mutation frequency as well as activated EGFR. Additional work found that NO induces basal-like genes and IL-8 only in ER-negative breast cancer cells. Some of these genes that were found to be induced by NO, such as CD44 and c-Myc, have been linked to an embryonic stem cell–like phenotype in breast cancer that is enriched in basal-like breast tumors (39, 40), raising the possibility that the expression of NOS2 and increased NO release into the tumor microenvironment lead to poorly differentiated and aggressive tumors with a distinct gene signature, enhanced angiogenesis, and the inactivation of the p53 tumor suppressor gene.
Previous reports indicated that elevated NOS2 expression may be linked to a high grade and poor prognosis in breast cancer (12, 41, 42), and also to poor outcome in other human epithelial cancers. In addition, mammary tumor latency is increased in NOS2-knockout mice (43). Aberrant expression of NOS2 has been observed in human breast (12, 13), colon (14, 15), stomach (44, 45), non–small cell lung (23), esophageal (46), and squamous cell head and neck cancer (47). In the two breast cancer studies, increased NOS2 correlated with tumor grade (12) and tumor angiogenesis and p53 protein accumulation (13). Our observations are consistent with these reports. NO has proangiogenic activities (48–50) and also promotes carcinogenesis through the inactivation of wild-type p53 function, by either causing loss of DNA-binding activity (51) and/or selecting for mutant p53 (26). Thus, we also evaluated in the current study to what extent tumor grade, tumor p53 status, and tumor microvessel density may influence the association of NOS2 with survival among the ER-negative breast cancer patients. This analysis showed that high NOS2 is the single most significant predictor among them and predicts poor outcome of ER-negative breast cancer patients largely independent of these 3 factors (Supplemental Figure 7). Two other studies of breast cancer analyzed NOS2 expression in tumors and examined the relationship between NOS2 and patient outcome (41, 42). In agreement with our findings and the study by Vakkala et al. (13), NOS2 was found to be commonly expressed by the cancer cells themselves. However, the two studies were relatively small-sized, with mainly ER-positive patients, which did not allow an independent assessment of NOS2 in ER-negative breast cancer. Both studies found a significant association of NOS2 with high-grade tumors, as we did in the present study, and a borderline association of high NOS2 with decreased survival in the univariate but not multivariable analysis. From their results, it appears that NOS2 expression may have some prognostic value in ER-positive breast tumors, although our study did not find this.
In an effort to determine why high NOS2 expression is associated with poor survival in ER-negative breast cancer, we performed a gene expression analysis of microdissected breast tumor epithelium and also assayed phenotypes of ER-negative breast cells that were exposed to NO. These approaches led to the discovery of a distinct and robust basal-like gene expression signature for high NOS2 in ER-negative tumor epithelium and NO-induced phenotypic alterations in ER-negative cells that indicated increased aggressiveness. Moreover, the NOS2 signature per se is prognostic, as found by us with the analysis of an independent sample set with gene expression data from 77 ER-negative breast tumors (32). An oncogenic effect of NO in the ER-negative MDA-MB-231 breast cancer cell line has been observed by others (21), consistent with a NO-induced poor outcome phenotype and our data. The global gene expression analysis did not find a major influence of NOS2 status on gene expression in ER-positive tumors, and furthermore, NO did not induce an aggressive phenotype in ER-positive breast cancer cells, as indicated by the cell migration and invasion results.
It is likely that both the difference in ER expression and intrinsic differences between ER-negative and ER-positive breast cancer cells independent of the tumor ER status contribute to this diminished responsiveness to NO in ER-positive cancer cells. We found that ER expression inhibited NO-induced upregulation of the stem cell marker CD44, and also S100A8 and P-cadherin, but not of IL-8. IL-8 is preferentially expressed by ER-negative breast tumors and breast cancer cell lines (34). Atypical methylation of the IL8 gene is a candidate mechanism for the differential expression of IL-8 in breast tumors (52). Regulation of IL-8 expression by NO has previously been observed in pancreatic cancer cells, human monocytes, and lung epithelial cells (53–55). IL-8 is of particular importance in breast tumor biology and is associated with increased breast cancer cell invasion, neutrophil infiltration, and microvessel density (35). IL-8 mediates metastasis in breast cancer and other cancers, and its secretion correlates with early dissemination and poor survival (56). Moreover, it has been shown that IL-8 supports breast cancer stem cell renewal and invasion (57). Our findings suggest that NOS2 may lead to poor survival among ER-negative patients partly because it induces IL-8 selectively in tumors of these patients and not in ER-positive tumors, leading to increased stem cell renewal, cell invasion, angiogenesis, and metastasis. This hypothesis is supported by our two findings that NO induces IL-8 only in ER-negative breast cancer cells and that IL-8 expression is increased in ER-negative tumors with high NOS2 but not in ER-positive tumors with high NOS2. Whether ER expression regulates IL-8 levels in breast cancer cells remains controversial. We did not find that IL-8 secretion is influenced by transient ER expression in ER-negative cells. In contrast, it was reported that an MDA-MB-231 clone that constitutively expressed an ER transgene had lower IL-8 production than the parent cell line (35). However, the same authors did not find that estrogen influenced IL-8 expression in the ER-expressing cells, arguing that perhaps clonal selection rather than ER expression may have contributed to this finding.
Alternatively, NOS2 may not produce NO as efficiently in ER-positive tumors as in ER-negative tumors, or NO signaling may occur differentially in ER-positive and ER-negative tumors because of differences in the tumor microenvironment. Effects of estrogen on NOS activity have been described; however, these appear to be mainly restricted to the endothelial isoform. Estrogen has been found to alter NOS2 expression in murine macrophages, splenocytes, and vascular smooth muscle cells by mechanisms involving ER (58–60), but because of the known differences in the promoter regulation of human and murine NOS2 (61), we cannot be sure that estrogen would have similar effects on human NOS2. Differences in the tumor microenvironment between ER-positive and ER-negative tumors have been observed that may affect NO signaling. ER-negative tumors tend to have more tumor-associated macrophages than ER-positive tumors (62), and many proinflammatory cytokines are expressed at a higher level in ER-negative tumors than ER-positive tumors (63). Macrophages and these cytokines will alter the tumor microenvironment and may lead to a proinflammatory state. Thus, the NO biochemistry and signaling, which are greatly influenced by reactive oxygen species, may be different in ER-negative and ER-positive breast cancer.
The gene signature induced by NOS2 in ER-negative breast tumors showed some striking similarities to two gene signatures associated with basal-like breast cancer (1, 30). Basal-like breast cancer is an ER-negative breast cancer subtype with an aggressive phenotype and limited therapy options (1, 31). Our evaluation of NOS2 in basal-like tumors indicated that its expression is associated with poor outcome. This suggests that NOS2 is a novel candidate prognostic marker for basal-like breast cancer and possibly a therapeutic target, whose activity could be selectively inhibited. Recently, increased expression of stem cell markers in basal-like tumors has been reported (40, 64). Among them was CD44, which is the major receptor for hyaluronan. Expression of CD44 is a poor outcome marker in breast cancer (65), and CD44-positive breast cancer cells exhibit increased invasive properties (66), increased resistance to radiotherapy (67), and increased resistance to chemotherapeutics (68). We observed that CD44 protein is induced by NO in ER-negative breast cancer cells and its gene expression was significantly elevated in ER-negative tumors with high NOS2. This observation links NOS2 and increased NO production to the development of a poorly differentiated breast cancer phenotype with stem cell–like characteristics. NO may induce this phenotype by activation of transcription factors, such as c-Myc, or by inducing the release of stem cell renewal factors like IL-8 (57), a property that NO has, as we have shown in this study. To gain an understanding of what may drive the NOS2 gene signature in ER-negative tumors, we used a bioinformatics tool to identify candidate factors that would induce these genes. c-Myc advanced as the top candidate from this analysis. Because of this finding, and because c-Myc is a part of an embryonic stem cell–like gene expression signature in breast cancer (39), we tested whether NO induces this transcription factor in ER-negative breast cancer cells. Indeed, NO upregulated c-Myc in MDA-MB-468 cells, which have basal-like characteristics. We consider this finding as preliminary evidence that NO may induce the basal-like signature in ER-negative breast tumors in part by upregulation of c-Myc.
EGFR is one of the signature genes in basal-like breast cancer (31). NO has previously been shown to activate EGFR in lung cancer cells (69). Furthermore, nuclear EGFR signaling has been shown to lead to upregulation of NOS2 in breast cancer cells (70, 71). Because of the association of the NOS2 signature with the basal-like gene expression signature and the role of the EGFR in basal-like breast cancer, we explored the hypothesis that NO may activate this receptor through phosphorylation in EGFR-positive MDA-MB-468 cells and breast tumors. This work led to the finding that NO induces EGFR phosphorylation at Tyr1045 and Tyr1173 in breast cancer cells and of a significant association between EGFR phosphorylation and NOS2 expression in breast tumors. Together, these early mechanistic data indicate that besides inducing a poor prognosis signature and enhancing angiogenesis, NOS2 may also cause a poor outcome phenotype by activating the oncogenic EGFR pathway.
In conclusion, our study provides evidence that NOS2 is predictor of survival and determinant of disease aggressiveness associated with ER-negative breast cancer, and as such is a candidate target for therapy. The underlying mechanisms that lead to an NO-induced poor outcome phenotype may include a combination of events, such as the induction of a basal-like phenotype, activation of the EGFR pathway, increased angiogenesis, and selection for mutant p53 cells. We propose that selective NOS2 inhibitors may be particularly efficacious in patients with basal-like breast cancer.
Methods
Tissue collection.
Fresh-frozen (n = 32) and paraffin-embedded (n = 248) tumor specimens were obtained from breast cancer patients that resided in the greater Baltimore area, as described previously (22, 72). Patients were recruited at the University of Maryland Medical Center (UMD), the Baltimore Veterans Affairs Medical Center, Union Memorial Hospital, Mercy Medical Center, and Sinai Hospital in Baltimore between 1993 and 2003. All patients signed a consent form. Clinical and pathological information (e.g., tumor receptor status) was obtained from medical records and pathology reports. Disease staging was performed according to the TNM system of the American Joint Committee on Cancer/Union Internationale Contre le Cancer (AJCC/UICC) (73). The Nottingham system was used to determine the tumor grade (74). The collection of tumor specimens and clinical and pathological information was reviewed and approved by the University of Maryland Institutional Review Board for the participating institutions (UMD protocol 0298229). IRB approval of this protocol was then obtained at all institutions (Veterans Affairs Medical Center, Union Memorial Hospital, Mercy Medical Center, and Sinai Hospital).
IHC.
IHC was performed as described previously (22). In brief, protein expression was evaluated using the following primary antibodies: 1:250 diluted monoclonal antibody (no. 610328; BD Biosciences) for NOS2; ready-to-use monoclonal antibody (Lab Vision Corp.) for CD31; ready-to-use monoclonal Ab-3 antibody (Lab Vision Corp.) for CD68; 1:250 diluted monoclonal antibody (no. 2729-1; Epitomics) for CD11b; ready-to-use mouse monoclonal antibody for EGFR (HER1) (Lab Vision Corp.); 1:250 diluted mouse monoclonal antibody for cytokeratin 5 (Lab Vision Corp.); 1:100 diluted monoclonal (no. 4407) anti–phospho-EGFR Try1173 (Cell Signaling Technology); and ready-to-use monoclonal (clone 6F11) antibody (Ventana Medical Systems) for ER. A combined score of intensity and distribution was used to categorize the immunohistochemical staining for NOS2 and other proteins, as previously described by us and others (13, 22), with the exception of the ER expression. Intensity received a score of 0–3 if the staining was negative, weak, moderate, or strong. The distribution received a score of 0–4 if the staining distribution was <10% positive cells, 10%–30%, >30%–50%, >50%–80%, and >80%. A sum score was then divided into 4 groups as follows: (a) negative, 0–1; (b) weak, 2–3; (c) moderate, 4–5; and (d) strong, 6–7. ER expression was determined according to the reference range set by ChromaVision ACIS assisted quantitative image analysis software (Clarient Diagnostic Services), and the ER status was then scored negative/positive according to clinical guidelines. The number of phagocytes in the tumor specimens was determined by counting the number of CD68-positive monocytes per ×250 field in 3 representative fields. The quantification of the tumor microvessel density was performed on CD31-positive microvessels, as opposed to single CD31-positive cells, according to the method described by Weidner et al. (75). The number of microvessels per ×200 field was determined in the most vascular regions of the tumor and is presented as the average of 3 representative fields.
TP53 mutational analysis.
Tumors were screened for p53 mutations as previously described (76).
Laser capture microdissection.
Enriched tumor epithelium from 32 fresh-frozen surgical breast tumors was obtained by laser capture microdissection (LCM) as previously described (72). Of those, 17 had low NOS2 expression and 15 had high NOS2 expression. In brief, frozen 8-μm serial sections from OCT-preserved frozen tissues were prepared and mounted on plain, uncharged microscope slides. One H&E-stained section of each specimen was reviewed by a pathologist who indicated which representative sections of the tumors should be microdissected. LCM was performed at the NIH Collaborative Research LCM Core Laboratory with the Pixcell II LCM system (Arcturus). At least 3,000–5,000 cells were obtained per specimen. Total RNA was isolated using the PicoPure protocol (Arcturus). The mRNA was amplified with two linear amplification steps by in vitro transcription using the MEGAscript T7 kit (Ambion) followed by the labeling step using the BioArray HighYield RNA Transcript Labeling Kit (T3) from Enzo Life Sciences. Labeled cRNA was hybridized onto Affymetrix HG-U133A GeneChips. Cel files with the normalized expression data were deposited in the GEO repository ( http://www.ncbi.nlm.nih.gov/projects/geo/; accession no. GSE5847).
Analysis of gene expression data.
All chips were normalized with the robust multichip analysis procedure (77). Gene lists comparing the effects of high versus low NOS2 expression on mRNA expression in the tumor epithelium of both ER-positive and ER-negative breast tumors were generated using moderated t scores to obtain significance testing and P values that are controlled for multicomparison analysis and false discovery (72). In addition, we calculated FDR for the differentially expressed genes, as described previously (78). BRB-ArrayTools software ( http://linus.nci.nih.gov/pilot/index.html) was used to evaluate the association of a NOS2 gene signature with prognosis in a publicly available data set. The pathway analysis and data mining tool MetaCore (GeneGo Inc.) was used to identify and visualize the relationship of a candidate transcription factor with the NOS2 gene signature in ER-negative breast tumors. GeneCo pathway maps and networks in MetaCore can be used to link a transcription factor to a gene expression pattern.
Treatment of human breast cancer cell lines with DETA/NO.
T47D, MCF-7, MDA-MB-231, MDA-MB-468, MDA-MB-157, Hs578T, and MCF-10A human breast cell lines were obtained from ATCC. With the exception of the MCF-10A cell line, cells were maintained at 37°C in RPMI 1640 medium (GIBCO, Invitrogen) supplemented with 10% fetal bovine serum and antibiotics. MCF-10A cells were grown in DMEM/F12 with 15 mM HEPES buffer, 5% horse serum, 10 μg/ml insulin, 20 ng/ml EGF, 100 ng/ml choleratoxin, 0.5 μg/ml hydrocortisone, and antibiotics. Cells were grown to 80% confluence and serum starved overnight, followed by replacement with serum-free medium containing DETA/NO (Sigma-Aldrich). After treatment, cell culture supernatants were collected for IL-8 quantification and spun at 180 g for 5 minutes to remove debris, then stored at –20°C. Cells were rinsed twice with cold PBS and lysed directly on the dish with cold RIPA buffer (no. 89900, Pierce) supplemented with protease inhibitors (no. 78410, Pierce), scraped, and spun at 14,000 g for 15 minutes at 4°C. Supernatant was collected and stored at –20°C for Western blot analysis of protein expression.
Western blot analysis.
Protein concentrations were determined with the Bio-Rad Protein Assay. Western blot analysis was performed according to standard procedures, and 50–100 μg of total protein was loaded per lane. The protein bands were visualized using the SuperSignal West Pico Chemiluminescent Substrate (Pierce). The following antibodies were used to detect proteins of interest: mouse monoclonal anti–P-cadherin (no. 610228), 1:500, from BD Biosciences; goat polyclonal anti-S100A8 (no. AF3059), 1:1,000, from Santa Cruz Biotechnology Inc.; mouse monoclonal anti-ER (no. PP-H4624-00), 1:1,000, and mouse monoclonal anti-CD44 (no. BBA10), 1:1,000, both from R&D Systems; mouse monoclonal anti–cyclin D1 (DSC6), 1:2,000, from Cell Signaling Technology; rabbit polyclonal anti–phospho-EGFR Tyr1045 (no. 2237), 1:1,000, and rabbit monoclonal anti–phospho-EGFR Tyr1173 (no. 4407), 1:1,000, and rabbit polyclonal anti-EGFR (no. 2232), 1:1,000, all from Cell Signaling Technology; rabbit polyclonal c-Myc (sc-764), 1:1,000, from Santa Cruz Biotechnology Inc.
Preparation of nuclear fraction.
Cells were plated on 100-mm tissue culture plates and grown to 80% confluence in complete medium. Cells were serum starved for 24 hours, after which they were exposed to 0.5 mM DETA/NO for 0, 20, 40, and 60 minutes. The nuclear and cytoplasmic protein fractions were isolated per the manufacturer’s instructions using the Nuclear Extract Kit (Active Motif). In brief, cells were washed with ice-cold PBS/phosphatase inhibitors, removed by gently scraping with cell lifter, and spun for 5 minutes at 45 g at 4°C, to pellet cells. Cells were resuspended in 500 μl hypotonic buffer and chilled for 15 minutes on ice, 25 μl detergent was added, and samples were vortexed for 10 seconds, followed by a 30-second spin at 14,000 g at 4°C. Supernatant (cytoplasmic fraction) was isolated and stored at –80°C. The pellet (nuclear fraction) was resuspended in 50 μl complete lysis buffer, vortexed for 10 seconds, and incubated/rocked for 30 minutes on ice. The pellet was vortex for a further 30 seconds, and spun at 14,000 g for 10 minutes at 4°C. Resulting supernatant was isolated and stored at –80°C.
c-Myc activation assay.
c-Myc activation was examined using the TransAM c-Myc ELISA (Active Motif). In brief, 40 μl complete binding buffer and 10 μl of diluted sample/positive control/protein standard were added to each well and incubated for 1 hour at room temperature (RT) with mild agitation on a rocking platform, to allow binding to a c-Myc consensus sequence. Wells were washed 3 times with 200 μl 1× wash buffer. Diluted c-Myc antibody (100 μl) was added to each well and incubated for 1 hour at RT. Wells were washed 3 times with 200 μl 1× wash buffer. Diluted HRP-conjugated antibody (100 μl) was added to each well and incubated for 1 hour at RT. Wells were washed 4 times with 200 μl 1× wash buffer. Developing solution (100 μl) was added to each well and incubated for 2–10 minutes at RT. Stop solution (100 μl) was added and absorbance read on a spectrophotometer within 5 minutes at 450 nm/655 nm.
IL-8 ELISA.
IL-8 concentrations in culture supernatants were determined using the human IL-8 Quantikine ELISA kit (R&D Systems), following the manufacturer’s protocol. Briefly, supernatants from ER-negative breast cancer cell lines were diluted 1:5, while ER-positive cell line supernatants were used undiluted. Sample (50 μl) and assay buffer (100 μl) were added in triplicate to an IL-8–precoated 96-well ELISA plate, incubated for 2 hours at RT. After washing, 200 μl HRP-conjugated IL-8 antibody was added and incubated for 1 hour at RT. After another washing step, 200 μl detection reagent was added and incubated for 30 minutes at RT. The plates were read at 450 nm in a microplate reader. Each independent experiment was carried out 3 times.
Invasion and motility assays.
Invasion and motility assays were performed as previously described (79). In brief, cells were harvested and suspended in RPMI 1640 containing 0.2% FCS at a concentration of 1 × 106 cells/ml. Cell suspension (100 μl) and 100 μl of DETA/NO at 2× concentration was added to either BD Matrigel Invasion Chambers (for invasion assays) or BD HTS multiwell insert systems (for motility assays), and 500 μl of appropriate medium containing 0.2% FCS was added to the well underneath the chamber. Cells were incubated at 37°C for 24 hours for motility assays and 48 hours for invasion assays. After this time, the inner side of the insert was wiped with a wet swab to remove the cells, while the outer side of the insert was gently rinsed with PBS and stained with 0.25% crystal violet for 5 minutes, rinsed again, and then allowed to dry. The inserts were then viewed under the microscope and the cells over a total of 5 random fields were counted at ×200 magnification, to determine the relative number of invading/migrating cells.
Transient transfection of ER-negative cell lines with ESR1.
MDA-MB-231 and MDA-MB-468 cells were plated at 5 × 105 cells per 100-mm dish in antibiotic-free medium. Twenty-four hours later, the cells were transfected using Lipofectamine LTX and PLUS reagents (Invitrogen) with either 12.5 μg of empty vector plasmid pCMV6-XL4 or pCMV6-XL4-ESR1 plasmid from Origene. Cells were transfected as follows: For each plate, 12.5 μg plasmid DNA and 12.5 μl PLUS reagent were diluted in 2.5 ml Opti-MEM-I–reduced serum medium, mixed gently, and incubated for 15 minutes at RT. Lipofectamine LTX (12.5 μl for MDA-MB-231 and 17.5 μl for MDA-MB-468) was added to the DNA/Plus mix and incubated for a further 30 minutes at RT. The mixture was then added to the plates dropwise and allowed to incubate at 37°C overnight. Twenty-four hours after transfection, medium was replaced with serum-free medium to serum starve overnight, followed by replacement with serum-free medium containing DETA/NO (Sigma-Aldrich). After treatment, cell culture supernatants were collected for IL-8 quantification and spun at 180 g for 5 minutes to remove debris, then stored at –20°C. Cells were rinsed twice with cold PBS and lysed directly on the dish with cold RIPA buffer (no. 89900, Pierce) supplemented with protease inhibitors (no. 78410, Pierce), scraped, and spun at 14,000 g for 15 minutes at 4°C. Supernatant was collected and stored at –20°C for Western blot analysis of protein expression.
Reporter construct–based activity assay for ER.
MDA-MB-468 and MDA-MB-231 cells were grown in phenol red–free RPMI containing 10% estrogen-depleted (charcoal-stripped) serum for 72 hours. The cells were transfected with the reporter construct (3X ERE-Luc containing 3 copies of the vitellogenin estrogen response element) and/or the ER expression plasmid (pCMV6-XL4-ESR1) using Lipofectamine LTX and Plus reagents (Invitrogen) following the Invitrogen protocol. Briefly, 30,000 cells were plated into 96-well plates along with DNA transfection reagent mixtures totaling 225 ng of total DNA (200 ng 3X ERE-Luc plasmid reporter construct [Addgene Inc.], 20 ng pCMV-ERα expression plasmid, 5 ng pRL-TK Renilla luciferase control vector, or control plasmid) and 0.5 μl LTX reagent per well. Transfections were performed in sextuplicate, and both pGL2-basic (Promega) and empty pCMV6-XL4 (Origene) were used as control plasmids to ensure that there were equal amounts of transfected DNA in each experiment. Twenty-four hours after transfection, the estrogen-depleted medium was changed to medium containing either 10 μM β-estradiol (Sigma-Aldrich) or an equal volume of ethanol (control). Luciferase activity was measured with the Dual Luciferase Reporter Assay System as described by the manufacturer (Promega), using a Luminoskan Ascent luminometer (Thermo Scientific). Normalized luciferase activity (or estrogen promoter activity) was expressed as firefly luciferase values normalized by pRL-TK Renilla luciferase values ± SD.
Statistics.
Data analysis was performed using Stata/SE 10.1 statistical software (Stata Corp). All statistical tests were 2-sided, and an association was considered statistically significant with P values less than 0.05. A t test was used to analyze differences between treatment groups in cell culture. χ2 and Fisher’s exact tests and multivariable logistic regression were used to analyze dichotomized data and to calculate ORs, respectively. Survival was determined for the period from the date of hospital admission to the date of the last completed search for death entries in the Social Security Death Index (date of search: February 3, 2006) for the 248 case patients with available tumor specimens. The mean and median follow-up times for breast cancer survival were 71 months and 68 months, respectively (range, 12–166 months). We obtained death certificates for the deceased case patients and censored all patients whose causes of death were not related to breast cancer. The Kaplan-Meier method and the log-rank test were used for univariate survival analysis. Cox regression was used for multivariable survival analysis to calculate an adjusted hazard ratio (HR). The following covariates were included in the analyses: age at diagnosis (as a continuous variable), estrogen receptor status (categorized as positive vs. negative), TNM stage (categorized as stage II vs. >stage II), tumor grade (categorized as grade 2 vs. >grade 3), and p53 mutation (categorized as positive vs. negative). Proportional hazards assumptions were verified by log-log plots and with the non-zero slope test of the scaled Schoenfeld residuals. A statistical test for interaction was performed in Stata to determine whether the association of NOS2 with breast cancer survival was modified by other factors.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, the National Cancer Institute, and the Center for Cancer Research. We thank Raymond Jones, Audrey Salabes, Leoni Leondaridis, Glennwood Trivers, Elise Bowman, and personnel at the University of Maryland and the Baltimore Veterans Administration and the Surgery and Pathology Departments at the University of Maryland Medical Center, Baltimore Veterans Affairs Medical Center, Union Memorial Hospital, Mercy Medical Center, and Sinai Hospital for their contributions. Sharon A. Glynn is the recipient of an All-Ireland Cancer Consortium National Cancer Institute Cancer Prevention Fellowship, sponsored in part by the Health Research Board of Ireland.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(11):3843–3854. doi:10.1172/JCI42059.
References
- 1.Sorlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bentzon N, During M, Rasmussen BB, Mouridsen H, Kroman N. Prognostic effect of estrogen receptor status across age in primary breast cancer. Int J Cancer. 2008;122(5):1089–1094. doi: 10.1002/ijc.22892. [DOI] [PubMed] [Google Scholar]
- 3.Dunnwald LK, Rossing MA, Li CI. Hormone receptor status, tumor characteristics, and prognosis: a prospective cohort of breast cancer patients. Breast Cancer Res. 2007;9(1):R6. doi: 10.1186/bcr1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leek RD, Lewis CE, Whitehouse R, Greenall M, Clarke J, Harris AL. Association of macrophage infiltration with angiogenesis and prognosis in invasive breast carcinoma. Cancer Res. 1996;56(20):4625–4629. [PubMed] [Google Scholar]
- 5.Denardo DG, Coussens LM. Inflammation and breast cancer. Balancing immune response: crosstalk between adaptive and innate immune cells during breast cancer progression. Breast Cancer Res. 2007;9(4):212. doi: 10.1186/bcr1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420(6917):860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ristimaki A, et al. Prognostic significance of elevated cyclooxygenase-2 expression in breast cancer. Cancer Res. 2002;62(3):632–635. [PubMed] [Google Scholar]
- 8.Nathan C, Xie QW. Nitric oxide synthases: roles, tolls, and controls. Cell. 1994;78(6):915–918. doi: 10.1016/0092-8674(94)90266-6. [DOI] [PubMed] [Google Scholar]
- 9.Yamasaki K, et al. Reversal of impaired wound repair in iNOS-deficient mice by topical adenoviral-mediated iNOS gene transfer. J Clin Invest. 1998;101(5):967–971. doi: 10.1172/JCI2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wink DA, Vodovotz Y, Laval J, Laval F, Dewhirst MW, Mitchell JB. The multifaceted roles of nitric oxide in cancer. Carcinogenesis. 1998;19(5):711–721. doi: 10.1093/carcin/19.5.711. [DOI] [PubMed] [Google Scholar]
- 11.Wink DA, Mitchell JB. Chemical biology of nitric oxide: insights into regulatory, cytotoxic, and cytoprotective mechanisms of nitric oxide. Free Radic Biol Med. 1998;25(4–5):434–456. doi: 10.1016/S0891-5849(98)00092-6. [DOI] [PubMed] [Google Scholar]
- 12.Thomsen LL, Miles DW, Happerfield L, Bobrow LG, Knowles RG, Moncada S. Nitric oxide synthase activity in human breast cancer. Br J Cancer. 1995;72(1):41–44. doi: 10.1038/bjc.1995.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vakkala M, Kahlos K, Lakari E, Paakko P, Kinnula V, Soini Y. Inducible nitric oxide synthase expression, apoptosis, and angiogenesis in in situ and invasive breast carcinomas. Clin Cancer Res. 2000;6(6):2408–2416. [PubMed] [Google Scholar]
- 14.Ambs S, et al. Frequent nitric oxide synthase-2 expression in human colon adenomas: implication for tumor angiogenesis and colon cancer progression. Cancer Res. 1998;58(2):334–341. [PubMed] [Google Scholar]
- 15.Yu JX, et al. Expression of NOS and HIF-1alpha in human colorectal carcinoma and implication in tumor angiogenesis. World J Gastroenterol. 2006;12(29):4660–4664. doi: 10.3748/wjg.v12.i29.4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ekmekcioglu S, Ellerhorst JA, Prieto VG, Johnson MM, Broemeling LD, Grimm EA. Tumor iNOS predicts poor survival for stage III melanoma patients. Int J Cancer. 2006;119(4):861–866. doi: 10.1002/ijc.21767. [DOI] [PubMed] [Google Scholar]
- 17.Gal A, Wogan GN. Mutagenesis associated with nitric oxide production in transgenic SJL mice. Proc Natl Acad Sci U S A. 1996;93(26):15102–15107. doi: 10.1073/pnas.93.26.15102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wink DA, Hanbauer I, Krishna MC, DeGraff W, Gamson J, Mitchell JB. Nitric oxide protects against cellular damage and cytotoxicity from reactive oxygen species. Proc Natl Acad Sci U S A. 1993;90(21):9813–9817. doi: 10.1073/pnas.90.21.9813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ambs S, Hussain SP, Harris CC. Interactive effects of nitric oxide and the p53 tumor suppressor gene in carcinogenesis and tumor progression. FASEB J. 1997;11(6):443–448. doi: 10.1096/fasebj.11.6.9194524. [DOI] [PubMed] [Google Scholar]
- 20.Kim PK, Zamora R, Petrosko P, Billiar TR. The regulatory role of nitric oxide in apoptosis. Int Immunopharmacol. 2001;1(8):1421–1441. doi: 10.1016/S1567-5769(01)00088-1. [DOI] [PubMed] [Google Scholar]
- 21.Pervin S, Singh R, Hernandez E, Wu G, Chaudhuri G. Nitric oxide in physiologic concentrations targets the translational machinery to increase the proliferation of human breast cancer cells: involvement of mammalian target of rapamycin/eIF4E pathway. Cancer Res. 2007;67(1):289–299. doi: 10.1158/0008-5472.CAN-05-4623. [DOI] [PubMed] [Google Scholar]
- 22.Prueitt RL, et al. Inflammation and IGF-I activate the Akt pathway in breast cancer. Int J Cancer. 2007;120(4):796–805. doi: 10.1002/ijc.22336. [DOI] [PubMed] [Google Scholar]
- 23.Marrogi AJ, et al. Nitric oxide synthase, cyclooxygenase 2, and vascular endothelial growth factor in the angiogenesis of non-small cell lung carcinoma. Clin Cancer Res. 2000;6(12):4739–4744. [PubMed] [Google Scholar]
- 24.Forrester K, et al. Nitric oxide-induced p53 accumulation and regulation of inducible nitric oxide synthase (NOS2) expression by wild-type p53. Proc Natl Acad Sci U S A. 1996;93(6):2442–2447. doi: 10.1073/pnas.93.6.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiarugi V, Magnelli L, Gallo O. Cox-2, iNOS and p53 as play-makers of tumor angiogenesis. Int J Mol Med. 1998;2(6):715–719. doi: 10.3892/ijmm.2.6.715. [DOI] [PubMed] [Google Scholar]
- 26.Ambs S, et al. Relationship between p53 mutations and inducible nitric oxide synthase expression in human colorectal cancer. J Natl Cancer Inst. 1999;91(1):86–88. doi: 10.1093/jnci/91.1.86. [DOI] [PubMed] [Google Scholar]
- 27.Livasy CA, et al. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol. 2006;19(2):264–271. doi: 10.1038/modpathol.3800528. [DOI] [PubMed] [Google Scholar]
- 28.Potemski P, et al. Relationship of P-cadherin expression to basal phenotype of breast carcinoma. Pol J Pathol. 2007;58(3):183–188. [PubMed] [Google Scholar]
- 29.Paredes J, Lopes N, Milanezi F, Schmitt FC. P-cadherin and cytokeratin 5: useful adjunct markers to distinguish basal-like ductal carcinomas in situ. Virchows Arch. 2007;450(1):73–80. doi: 10.1007/s00428-006-0334-y. [DOI] [PubMed] [Google Scholar]
- 30.Charafe-Jauffret E, et al. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene. 2006;25(15):2273–2284. doi: 10.1038/sj.onc.1209254. [DOI] [PubMed] [Google Scholar]
- 31.Nielsen TO, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10(16):5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 32.Pawitan Y, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7(6):R953–R964. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei L, Gravitt PE, Song H, Maldonado AM, Ozbun MA. Nitric oxide induces early viral transcription coincident with increased DNA damage and mutation rates in human papillomavirus-infected cells. Cancer Res. 2009;69(11):4878–4884. doi: 10.1158/0008-5472.CAN-08-4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Freund A, et al. IL-8 expression and its possible relationship with estrogen-receptor-negative status of breast cancer cells. Oncogene. 2003;22(2):256–265. doi: 10.1038/sj.onc.1206113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yao C, et al. Interleukin-8 modulates growth and invasiveness of estrogen receptor-negative breast cancer cells. Int J Cancer. 2007;121(9):1949–1957. doi: 10.1002/ijc.22930. [DOI] [PubMed] [Google Scholar]
- 36.Matos I, Dufloth R, Alvarenga M, Zeferino LC, Schmitt F. p63, cytokeratin 5, and P-cadherin: three molecular markers to distinguish basal phenotype in breast carcinomas. Virchows Arch. 2005;447(4):688–694. doi: 10.1007/s00428-005-0010-7. [DOI] [PubMed] [Google Scholar]
- 37.Ponti D, et al. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65(13):5506–5511. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- 38.Charpentier AH, et al. Effects of estrogen on global gene expression: identification of novel targets of estrogen action. Cancer Res. 2000;60(21):5977–5983. [PubMed] [Google Scholar]
- 39.Ben Porath, I, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40(5):499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Honeth G, et al. The CD44+/CD24- phenotype is enriched in basal-like breast tumors. Breast Cancer Res. 2008;10(3):R53. doi: 10.1186/bcr2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loibl S, et al. The role of early expression of inducible nitric oxide synthase in human breast cancer. Eur J Cancer. 2005;41(2):265–271. doi: 10.1016/j.ejca.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 42.Bulut AS, Erden E, Sak SD, Doruk H, Kursun N, Dincol D. Significance of inducible nitric oxide synthase expression in benign and malignant breast epithelium: an immunohistochemical study of 151 cases. Virchows Arch. 2005;447(1):24–30. doi: 10.1007/s00428-005-1250-2. [DOI] [PubMed] [Google Scholar]
- 43.Ellies LG, et al. Mammary tumor latency is increased in mice lacking the inducible nitric oxide synthase. Int J Cancer. 2003;106(1):1–7. doi: 10.1002/ijc.11178. [DOI] [PubMed] [Google Scholar]
- 44.Wang YZ, Cao YQ, Wu JN, Chen M, Cha XY. Expression of nitric oxide synthase in human gastric carcinoma and its relation to p53, PCNA. World J Gastroenterol. 2005;11(1):46–50. doi: 10.3748/wjg.v11.i1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen CN, Hsieh FJ, Cheng YM, Chang KJ, Lee PH. Expression of inducible nitric oxide synthase and cyclooxygenase-2 in angiogenesis and clinical outcome of human gastric cancer. J Surg Oncol. 2006;94(3):226–233. doi: 10.1002/jso.20372. [DOI] [PubMed] [Google Scholar]
- 46.Matsumoto M, Furihata M, Kurabayashi A, Araki K, Sasaguri S, Ohtsuki Y. Association between inducible nitric oxide synthase expression and p53 status in human esophageal squamous cell carcinoma. Oncology. 2003;64(1):90–96. doi: 10.1159/000066519. [DOI] [PubMed] [Google Scholar]
- 47.Brennan PA, Dennis S, Poller D, Quintero M, Puxeddu R, Thomas GJ. Inducible nitric oxide synthase: correlation with extracapsular spread and enhancement of tumor cell invasion in head and neck squamous cell carcinoma. Head Neck. 2008;30(2):208–214. doi: 10.1002/hed.20675. [DOI] [PubMed] [Google Scholar]
- 48.Jenkins DC, et al. Roles of nitric oxide in tumor growth. Proc Natl Acad Sci U S A. 1995;92(10):4392–4396. doi: 10.1073/pnas.92.10.4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ziche M, et al. Nitric oxide mediates angiogenesis in vivo and endothelial cell growth and migration in vitro promoted by substance P. J Clin Invest. 1994;94(5):2036–2044. doi: 10.1172/JCI117557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ambs S, et al. p53 and vascular endothelial growth factor regulate tumour growth of NOS2-expressing human carcinoma cells. Nature Med. 1998;4(12):1371–1376. doi: 10.1038/3957. [DOI] [PubMed] [Google Scholar]
- 51.Calmels S, Hainaut P, Ohshima H. Nitric oxide induces conformational and functional modifications of wild-type p53 tumor suppressor protein. Cancer Res. 1997;57(16):3365–3369. [PubMed] [Google Scholar]
- 52.de Larco JE, Wuertz BR, Yee D, Rickert BL, Furcht LT. Atypical methylation of the interleukin-8 gene correlates strongly with the metastatic potential of breast carcinoma cells. Proc Natl Acad Sci U S A. 2003;100(24):13988–13993. doi: 10.1073/pnas.2335921100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiong Q, Shi Q, Le X, Wang B, Xie K. Regulation of interleukin-8 expression by nitric oxide in human pancreatic adenocarcinoma. J Interferon Cytokine Res. 2001;21(7):529–537. doi: 10.1089/10799900152434411. [DOI] [PubMed] [Google Scholar]
- 54.Ma P, et al. Nitric oxide post-transcriptionally up-regulates LPS-induced IL-8 expression through p38 MAPK activation. J Leukoc Biol. 2004;76(1):278–287. doi: 10.1189/jlb.1203653. [DOI] [PubMed] [Google Scholar]
- 55.Sparkman L, Boggaram V. Nitric oxide increases IL-8 gene transcription and mRNA stability to enhance IL-8 gene expression in lung epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2004;287(4):L764–L773. doi: 10.1152/ajplung.00165.2004. [DOI] [PubMed] [Google Scholar]
- 56.Steele RJ, Eremin O, Brown M, Hawkins RA. Oestrogen receptor concentration and macrophage infiltration in human breast cancer. Eur J Surg Oncol. 1986;12(3):273–276. [PubMed] [Google Scholar]
- 57.Benoy IH, et al. Increased serum interleukin-8 in patients with early and metastatic breast cancer correlates with early dissemination and survival. Clin Cancer Res. 2004;10(21):7157–7162. doi: 10.1158/1078-0432.CCR-04-0812. [DOI] [PubMed] [Google Scholar]
- 58.Charafe-Jauffret E, et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69(4):1302–1313. doi: 10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.You HJ, Kim JY, Jeong HG. 17 beta-estradiol increases inducible nitric oxide synthase expression in macrophages. Biochem Biophys Res Commun. 2003;303(4):1129–1134. doi: 10.1016/s0006-291x(03)00477-7. [DOI] [PubMed] [Google Scholar]
- 60.Dai R, Phillips RA, Karpuzoglu E, Khan D, Ahmed SA. Estrogen regulates transcription factors STAT-1 and NF-kappaB to promote inducible nitric oxide synthase and inflammatory responses. J Immunol. 2009;183(11):6998–7005. doi: 10.4049/jimmunol.0901737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsutsumi S, et al. Differential regulation of the inducible nitric oxide synthase gene by estrogen receptors 1 and 2. J Endocrinol. 2008;199(2):267–273. doi: 10.1677/JOE-07-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Vera ME, et al. Transcriptional regulation of human inducible nitric oxide synthase (NOS2) gene by cytokines: initial analysis of the human NOS2 promoter. Proc Natl Acad Sci U S A. 1996;93(3):1054–1059. doi: 10.1073/pnas.93.3.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chavey C, et al. Oestrogen receptor negative breast cancers exhibit high cytokine content. Breast Cancer Res. 2007;9(1):R15. doi: 10.1186/bcr1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rennstam K, et al. Numb protein expression correlates with a basal-like phenotype and cancer stem cell markers in primary breast cancer. Breast Cancer Res. 2009;9(1):R15. doi: 10.1186/bcr1648. [DOI] [PubMed] [Google Scholar]
- 65.Zhou L, et al. The prognostic role of cancer stem cells in breast cancer: a meta-analysis of published literatures. Breast Cancer Res Treat. 2010;122(3):795–801. doi: 10.1007/s10549-010-0999-4. [DOI] [PubMed] [Google Scholar]
- 66.Sheridan C, et al. CD44+/CD24- breast cancer cells exhibit enhanced invasive properties: an early step necessary for metastasis. Breast Cancer Res. 2006;8(5):R59. doi: 10.1186/bcr1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Phillips TM, McBride WH, Pajonk F. The response of CD24(-/low)/CD44+ breast cancer-initiating cells to radiation. J Natl Cancer Inst. 2006;98(24):1777–1785. doi: 10.1093/jnci/djj495. [DOI] [PubMed] [Google Scholar]
- 68.Bourguignon LY, Peyrollier K, Xia W, Gilad E. Hyaluronan-CD44 interaction activates stem cell marker Nanog, Stat-3-mediated MDR1 gene expression, and ankyrin-regulated multidrug efflux in breast and ovarian tumor cells. J Biol Chem. 2008;283(25):17635–17651. doi: 10.1074/jbc.M800109200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee HC, et al. Activation of epidermal growth factor receptor and its downstream signaling pathway by nitric oxide in response to ionizing radiation. Mol Cancer Res. 2008;6(6):996–1002. doi: 10.1158/1541-7786.MCR-08-0113. [DOI] [PubMed] [Google Scholar]
- 70.Lo HW, et al. Nuclear interaction of EGFR and STAT3 in the activation of the iNOS/NO pathway. Cancer Cell. 2005;7(6):575–589. doi: 10.1016/j.ccr.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 71.Lo HW, Hung MC. Nuclear EGFR signalling network in cancers: linking EGFR pathway to cell cycle progression, nitric oxide pathway and patient survival. Br J Cancer. 2006;94(2):184–188. doi: 10.1038/sj.bjc.6602941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boersma BJ, et al. A stromal gene signature associated with inflammatory breast cancer. Int J Cancer. 2007;122(6):1324–1332. doi: 10.1002/ijc.23237. [DOI] [PubMed] [Google Scholar]
- 73.Singletary SE, Connolly JL. Breast cancer staging: working with the sixth edition of the AJCC Cancer Staging Manual. CA Cancer J Clin. 2006;56(1):37–47. doi: 10.3322/canjclin.56.1.37. [DOI] [PubMed] [Google Scholar]
- 74.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19(5):403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 75.Weidner N, Semple JP, Welch WR, Folkman J. Tumor angiogenesis and metastasis — correlation in invasive breast carcinoma. N Engl J Med. 1991;324(1):1–8. doi: 10.1056/NEJM199101033240101. [DOI] [PubMed] [Google Scholar]
- 76.Boersma BJ, et al. Association of breast cancer outcome with status of p53 and MDM2 SNP309. J Natl Cancer Inst. 2006;98(13):911–919. doi: 10.1093/jnci/djj245. [DOI] [PubMed] [Google Scholar]
- 77.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Glynn SA, O’Sullivan D, Eustace AJ, Clynes M, O’Donovan N. The 3-hydroxy-3-methylglutaryl-coenzyme A reductase inhibitors, simvastatin, lovastatin and mevastatin inhibit proliferation and invasion of melanoma cells. BMC Cancer. 2008;8:9. doi: 10.1186/1471-2407-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.