Abstract
CD8+ T cells play a critical role in the immune response to viral pathogens. Persistent human cytomegalovirus (HCMV) infection results in a strong increase in the number of virus-specific, quiescent effector-type CD8+ T cells with constitutive cytolytic activity, but the molecular pathways involved in the induction and maintenance of these cells are unknown. We show here that HCMV infection induced acute and lasting changes in the transcriptomes of virus-reactive T cells collected from HCMV-seropositive patients at distinct stages of infection. Enhanced cell cycle and metabolic activity was restricted to the acute phase of the response, but at all stages, HCMV-specific CD8+ T cells expressed the Th1-associated transcription factors T-bet (TBX21) and eomesodermin (EOMES), in parallel with continuous expression of IFNG mRNA and IFN-γ–regulated genes. The cytolytic proteins granzyme B and perforin as well as the fractalkine-binding chemokine receptor CX3CR1 were found in virus-reactive cells throughout the response. During HCMV latency, virus-specific CD8+ T cells lacked the typical features of exhausted cells found in other chronic infections. Persistent effector cell traits together with the permanent changes in chemokine receptor usage of virus-specific, nonexhausted, long-lived CD8+ T cells may be crucial to maintain lifelong protection from HCMV reactivation.
Introduction
Depending on the clearance or persistence of a virus, virus-specific CD8+ T cells may have diverse fates after a primary immune response. In the case of cleared viruses, a stable fraction of memory cells remains after the initial effector T cell response. Upon reexposure to the same pathogen, these lymphocytes rapidly expand and acquire effector functions, providing protective immunity to the host (1). Memory cells persist in the absence of antigen through the actions of IL-7 and IL-15 (2), 2 members of the common γ (cγ) chain cytokine family. Effector and memory cells can derive from a single naive precursor (3), but the way in which these differentiated cells develop is still unknown. Although some studies suggest that memory cells derive from the original virus-specific effector T cell pool (4), others have recently suggested that segregation may already occur early in the response based on asymmetrical cell division between effector and memory cells (5).
Some persisting pathogens, such as particular strains of lymphocytic choriomeningitis virus (LCMV), induce an initial response that is highly similar to cleared viruses. However, cells that remain after the initial expansion appear to be addicted to antigen, since they are not maintained in antigen-naive mice (6). Moreover, these CD8+ T cells seem exhausted, as they are characteristically nonresponsive to cognate antigen stimulation. Importantly, blocking the function of inhibitory receptors such as programmed death 1 (PD1) and lymphocyte-activation gene 3 (LAG3) (7, 8) may relieve the nonresponsiveness of these T cells in vivo, demonstrating that exhaustion is not a fixed state, but rather is dynamically regulated by environmental cues. An exceptional type of response is induced by persistent murine CMV (MCMV), which induces a very robust T cell response that is maintained throughout life. As opposed to chronic LCMV-specific CD8+ T cells, MCMV-reactive cells obtained during latent infection are not functionally impaired, but rather highly active effector-type cells (9). Moreover, they lack the characteristic exhausted cell surface phenotype and can be typified by low levels of CD27 and CD28 and high expression of the NK cell–inhibitory killer cell lectin-like receptors KLRC1 (also known as NKG2A) and KLRG1 (9, 10). The diversity in the phenotype and function of virus-specific CD8+ T cells suggests that variations in (chronic) peptide presentation, together with the induction of specific cellular factors induced by infection with a specific virus (including costimulatory pathways), leads to a diverse spectrum of outcomes.
Dynamic changes induced by viruses in the CD8+ T cell compartment of healthy humans are more difficult to study because the exact time point of the primary infection is usually unknown. A limited number of observations during acute HIV, EBV, and human CMV (HCMV) infection revealed that, analogous to experimental infections in mice, the first phase of the response is characterized by robust expansion of virus-specific effector cells expressing activation markers (11–14). HCMV has infected the majority of the global population. In immunocompetent people, primary infection often remains unnoticed, but latent HCMV infection has been associated with vascular pathology and premature immunosenescence (15). These conditions seem to be related to the strong and chronic antiviral response rather than to direct cytopathic effects of the virus (15). We have previously documented virus-specific CD8+ T cell responses in HCMV-negative transplant recipients experiencing primary HCMV infection after receiving a HCMV-positive kidney (16–18). By meticulously monitoring the viral load by PCR, we could detect the precise onset of infection. Early HCMV-specific cells are strongly activated and cycling cells, as reflected by the expression of CD38, HLA-DR, and Ki-67. They express granzyme B (encoded by GZMB) and perforin and display peptide-specific cytotoxicity in vitro. In the months following the primary response, the majority of the HCMV-specific cells become resting cytolytic T cells that convert from a CD8+CD45R0+CD28+/–CD27+CD127– to a CD8+CD45R0–CD28–CD27–CD127– phenotype, which is prototypic for HCMV-specific T cells in healthy individuals (11, 19).
Recent studies in healthy people vaccinated with either live yellow fever or smallpox revealed that primary CD8+ T cell responses were largely similar to the HCMV response in the transplant recipients, yet these cells underwent a memory differentiation program, characterized, for example, by loss of GZMB but gain of CD127 expression, reminiscent of murine memory cells (20). Thus, although similar during the primary response, human CD8+ T cell differentiation pathways may also be highly divergent between T cells specific for either cleared or chronic viruses. Individual persistent viruses tend to elicit particular developmental programs (11). Specifically, whereas most CD8+ T cells reactive toward latent EBV epitopes resemble memory-type cells, lytic epitopes may drive CD8+ T cells toward a more differentiated phenotype, characterized by the lowering of CD28 expression (21). Interestingly, HCMV-specific cells have functional properties of acute effector cells, except for their quiescent state (11, 19). In concordance with the observations in mice, HCMV-specific CD8+ T cell responses are among the strongest immune responses that can be measured in humans (22), and the size of the virus-reactive pool markedly increases with age (23). The molecular pathways involved in the activation, differentiation, and maintenance of HCMV-specific CD8+ T cells are currently unknown. We therefore performed a comprehensive longitudinal molecular analysis of HCMV-specific CD8+ T cell populations from patients after primary HCMV infection. Starting with transcriptome analysis, differential regulation of selected molecules was confirmed at the protein level on CD8+ T cells specific for distinct CMV epitopes. Our data showed that differentiation into the prototypical HCMV-specific CD8+ T cell phenotype is a dynamic process involving both acute and chronic changes in gene expression patterns. Importantly, the key functional features and migratory properties of HCMV-specific CD8+ T cells were already installed during primary activation and stably persisted throughout the latency stage.
Results
Overall changes in gene expression patterns.
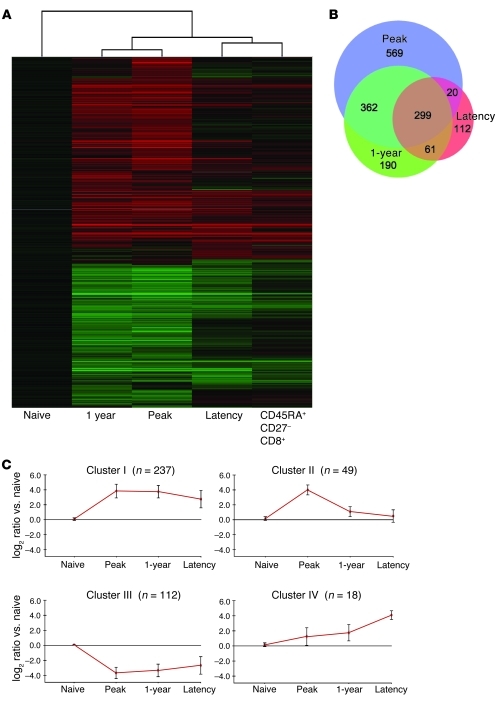
To measure changes in gene expression patterns in virus-specific CD8+ T cells in humans, HMCV-reactive cells were isolated at distinct stages of infection. Because of the inability to obtain large volumes of blood during the acute stage, activated effector cells from peripheral blood of kidney transplant recipients with primary HCMV infection were obtained by FACS sorting on CD8+HLA-DR+CD38+ T cells. Cells were sorted when the frequency of the antigen-specific CD8+ T cells reached its highest level (referred to herein as peak, n = 3; for sort windows, see Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI42758DS1). From the same patients, HCMV-specific cells were isolated between 40 and 60 weeks after the primary response (referred to herein as 1 year; for sort windows, see Supplemental Figure 1B) using CMV-specific pp65 tetramers. At these later stages, virus had become undetectable by PCR. HCMV-specific cells were also obtained from healthy CMV carriers via tetramer isolation (referred to herein as latency, n = 3). To relate the observed profiles in HCMV-specific cells to those of differentiated antigen-experienced CD8+ T cell subsets, CD45RA–CD27+ (memory-type) and CD45RA+CD27– (effector-type) CD8+ T cells from 6 healthy individuals were used (Supplemental Figure 1C). Gene expression profiles from all populations were compared with those of a pool of naive (CD45RA+CD27bright) CD8+ T cells on Agilent 44K arrays.
Initial analysis after Benjamini-Hochberg correction for multiple testing (corrected P ≤ 0.05) revealed that the expression of 8,542 genes was altered more than 2-fold at the peak of the anti-HCMV response. Unsupervised agglomerative 2D cluster analysis revealed that transcriptomes of CD8+ T cells obtained at peak and 1 year were more similar than those of CD8+ T cells from healthy individuals carrying latent HCMV (Figure 1, A and B). The latter clustered closely with total CD8+CD45RA+CD27– T cells, the prototypical phenotype of HCMV-specific cells (11, 24). Consensus clustering (25, 26) showed that clustering stability was high (Supplemental Figure 2). It seems remarkable that the 1 year sample did not cluster more closely to the latency sample, since at this point, the kidney transplant recipients do not have signs of active HCMV replication. Still, as we previously noted, HCMV-induced CD8+ T cell differentiation in transplant recipients appears to be a slow process that is possibly influenced by the immunosuppressive medication (16).
Figure 1. HCMV induces major changes in gene expression after priming of naive CD8+ T cells that are largely maintained during latency.
(A) Graphical representation of hierarchical clustering by Rosetta Resolver. Shown are mean changes in transcript levels of CMV-specific CD8+ cells induced at the peak and 1 year after primary infection as well as in latency, compared with naive samples. The gene set included all genes that were significant after the Benjamini-Hochberg correction for multiple testing (P ≤ 0.05) and were more than 2-fold up- or downregulated compared with RNA of naive CD8+ cells at 1 of the 4 time points. Samples were clustered together with mean expression value for total effector (CD8+CD45RA+CD27–) cells of HCMV+ healthy donors. (B) BioVenn (82) diagram showing the distribution between peak, 1 year (transplant recipient), and latency stage of infection (healthy donor) of all 1,613 genes that were significantly altered (2-fold change and corrected P ≤ 1 × 10–10). The relative size of the circle represents the size of the respective group. (C) Average gene expression patterns (± 1 SD) of the 4 clusters found using the CLICK algorithm. The gene set included all genes greater than 10-fold up- or downregulated compared with RNA of naive CD8+ T cells at 1 of the 4 time points with corrected P ≤ 1 × 10–10. n for each cluster is shown.
To obtain a manageable dataset, a cutoff was set at 10-fold change with corrected P ≤ 1 × 10–10. Averaging over all the probes mapped to the same gene revealed a dataset of 416 unique genes for us to further analyze. We clustered the expression patterns of these genes using the CLICK algorithm from the software package Expander 5.0 (27). Clustering detected 4 major trends describing the kinetics of the genes during HCMV infection. It appeared that the majority of changes (Figure 1C) occurred quickly and were maintained for different periods in the antigen-specific cells, whereas few genes were specifically regulated either in the long term (i.e., 1 year) or in latent HCMV-specific CD8+ T cells. The 4 clusters were analyzed for overrepresentation of Gene Ontology (GO) categories using the TANGO algorithm in Expander. Most changes were detected in GO categories related to immune response (profiles I and IV), cell cycle (profile II), or metabolism (profile III; Figure 1C and Supplemental Table 1). Genes that showed the most significant alterations in expression are discussed herein, categorized on the basis of their presumed role in cellular function. Additionally, we summarize expression characteristics of genes that, based on current knowledge, are important for CD8+ T cell function. The complete microarray dataset is available from the Gene Expression Omnibus (GEO; accession nos. GSE12589 and GSE24151).
Cell division and metabolism.
Viral infection induces naive antigen-specific CD8+ T cells to undergo vigorous proliferation (28). In line with this, concomitant with the emergence of the first tetramer+ cells, strong changes were observed in the expression of genes involved in cell cycle regulation (Figure 2A and Supplemental Table 2). The most pronounced changes in these genes were found at the peak of the response corresponding to profile II (Figure 1C).
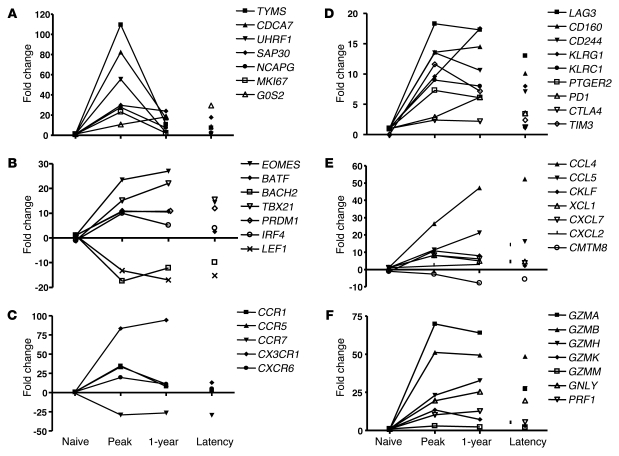
Figure 2. Alterations in gene expression of CD8+ T cells during primary HCMV infection.
Shown are kinetics of RNA transcripts in naive CD8+ T cells, HCMV-specific cells from a transplant recipient at peak of and 1 year after infection as well as HCMV-specific cells from healthy donors. The genes are subdivided into the following categories: (A) cell cycle; (B) transcription factors; (C) migration and adhesion markers; (D) exhaustion markers; (E) chemokines; and (F) CTL effector molecules. Expression is measured as fold change compared with a pool of naive CD8+ T cells.
Changes in thymidylate synthase (TYMS), an enzyme that maintains the thymidine monophosphate pool critical for DNA replication and repair, were most profound. The expression and activity of TYMS is related to the cell doubling time: the faster the cell proliferation, the greater the expression and activity of TYMS (29). Furthermore, we confirmed transient upregulation of the cell cycle protein Ki67 (encoded by MKI67), which we previously documented by intracellular FACS analysis (17). Moreover, BIRC5, which encodes survivin, was highly expressed at the peak of the response, consistent with survivin’s essential role in mitosis (30).
Other cell cycle genes that were highly regulated at the peak of the response were cell division cycle (CDC) genes CDC2, CDC6, CDC7, CDC8, and CDC50B as well as CDC-associated genes, notably CDCA5 and CDCA7. Proteins that work in concert with CDC genes to initiate eukaryotic genome replication, such as MCM4 and MCM10, were also elevated initially and declined thereafter (Supplemental Table 2). Histone deacetylation contributes to cell cycle regulation, and expression of UHRF1 and SAP30, 2 molecules involved in this process, was upregulation at the peak of the effector cell expansion. Apart from SAP30 and G0-G1 switch gene 2 (G0S2, which encodes a mitochondrial protein), other cell cycle–related genes were barely enhanced after the primary response in CMV-specific and total effector-type T cells (Supplemental Table 2), which indicates that extensive cycling occurs only at the peak of the response.
In parallel with the cell cycle regulators, major changes in genes involved in metabolism were only found during the early phases of the response (Supplemental Table 3). Based on the expression data, it appears that the HCMV-specific effectors cells do not permanently change their metabolic pathways and that they revert to a situation reminiscent of that for naive cells.
Apoptosis.
Apoptosis is essential for cellular homeostasis and removal of effector cells during the termination of an immune response. The balance between pro- and antiapoptotic genes of the Bcl-2 family is considered to be important to set the level of cell death sensitivity via the intrinsic pathway of apoptosis and to regulate the size of the antigen-specific T cell pool after the primary response (31). However, only the Bcl-2 members BAK and BIM showed relatively small, although significant, increases in mRNA levels compared with naive CD8+ T cells (3.1- and 2.7-fold, respectively). Other genes of this family, such as BCLXL, BAX, BAD, BIK, and BOK, did not show induction above the threshold of 2-fold (Supplemental Table 4).
Concerning extrinsic pathways of apoptosis, FAS (also known as CD95), Fas ligand (FASLG, also known as CD95L or TNFSF6), and TNFSF10 (also known as TRAIL) were all markedly increased at the peak of the primary response (Supplemental Table 5). These membrane molecules remained highly upregulated in the virus-specific CD8+ T cells, as well as in the cells obtained during viral latency. This could imply that the extrinsic pathways contribute to pool size regulation. Comparable but low expression of molecules involved in the extrinsic apoptosis pathways was found in total memory- and effector-type cells.
We found that the proapoptotic gene death-inducing protein (DIP) and caspase 1 were strongly induced early in the response (Supplemental Table 5). Another gene sharply upregulated in the acute effector cells was the proapoptotic protein BIRC4BP (also known as XAF1), which, by direct binding, negatively regulates the antiapoptotic activity of XIAP (32). Recent findings by Wang et al. showed that, by interaction with cell cycle kinases CHK1 and CDC25C, XAF1 induces G2/M arrest (33). Thus, at the peak of the antiviral response, XAF1 may possibly exert its function by controlling the size of the expanded effector pool, both through direct apoptosis and by modulating cell division.
Members of the annexin family of calcium-dependent phospholipid-binding proteins were regulated in HCMV-specific cells. At the peak of the response, ANXA2, ANXA4, and ANXA5 were markedly upregulated and remained elevated (except for ANXA4, which declined in latency). In effector LCMV-specific CD8+ T cells in mice, Wherry et al. observed a similar increase in Anxa1 and Anxa2 (34), which suggests these genes exert a conserved function in signal transduction in virus-primed CD8+ T cells.
Although a number of changes were observed in the mRNA levels of genes linked to apoptosis, no sharp picture of the pool size regulation of the CMV-specific CD8+ T cell populations at different stages of infection could be drawn from the dataset. This may be attributable, at least in part, to the fact that apoptosis is an intricately regulated process which makes it difficult to predict how changes in the expression of individual genes will specifically modify the sensitivity of cells to death-inducing signals. This complexity is enhanced by the observation that for some apoptosis modifiers, such as MCL1, regulation at the protein level rather than the mRNA level is crucial (35, 36). Moreover, as cells with a proapoptotic profile are likely to die and are rapidly removed from the T cell pool, they will not contribute in a representative way to microarray analysis performed at the population level.
Transcription factors.
By binding to promoter sites in the genome, transcription factors transform resting naive T cells that solely undergo homeostatic proliferation into cells that rapidly proliferate, exhibit cytolytic activity and cytokine secretion, and alter their migratory behavior. The transcription factor profile will determine the phenotype and function of CD8+ T cells that remain after the primary response as either memory, exhausted, or effector cells. Indeed, recent studies have indicated that expression of Blimp-1 (encoded by PRDM1) is instrumental in establishing a number of key features of the exhausted phenotype of chronic LCMV-specific cells (37–39). More than 40 transcription factors showed a greater than 10-fold change in expression when antigen-specific T cells arose after HCMV infection (P ≤ 1 × 10–10). As shown in Figure 2B and Supplemental Table 6, the most prominent changes took place at the peak of the immune response. However, many alterations in the expression of transcription factors installed during the initial priming of the cells were maintained into latency, which suggests that HCMV infection initiates stable, long-lasting changes in the virus-specific CD8+ T cell population (Figure 1C, profile I).
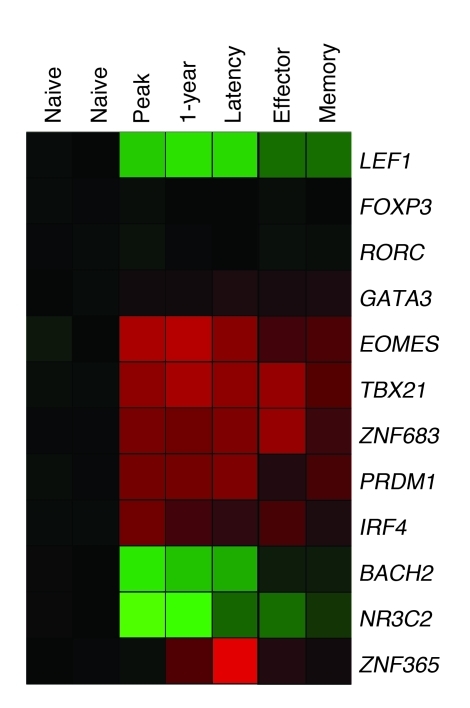
A selection of the proteins regarded as key regulators of T cell differentiation is depicted in Figure 2B and Figure 3. Lineage-determining transcription factors such as RORC (which encodes RORγt), FOXP3, and GATA3, which guide developing cells toward CD4+ Th17, Treg, or Th2 phenotypes, respectively, were not induced in CD8+ T cells. Interestingly, genes essential for murine CD8+ T cell differentiation — TBX21 (which encodes T-bet), EOMES (which encodes eomesodermin), and PRDM1 (40–43) — were significantly induced in HCMV-specific human CD8+ T cells and remained elevated during latency.
Figure 3. HCMV infection induces strong changes in transcription factor expression levels.
Shades of red and green represent up- and downregulation of genes, respectively, in naive cells compared with HCMV-specific CD8+ T cells at peak and 1 year of infection. Tetramer+ cells showed a similar profile at the 1-year time point in patient samples and in latency in a healthy control. Total effector and memory cells in latency showed the same trend.
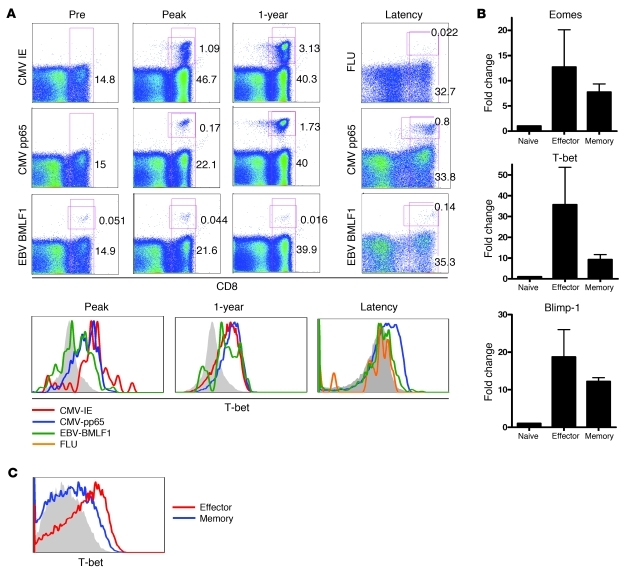
TBX21 and EOMES are transcription factors of the T-box family and involved in the regulation of IFN-γ, granzyme B, and perforin production by CD8+ T cells (42, 44). Mice harboring deletions for both Eomes and Tbx21 have a near-complete absence of protective CD8+ T cell responses (45). The availability of specific mAb (46) opened the possibility to analyze T-bet protein expression on a broader range of virus-specific cells. Analysis of HCMV-specific cells by tetramers directed against 2 different epitopes detected T-bet in most HCMV pp65- and immediate early–reactive (IE-reactive) T cells, regardless of whether they were retrieved early in the response or during latency (Figure 4A).
Figure 4. Transcription factors involved in effector molecule production are highly upregulated in HCMV-specific cells.
(A) HCMV-specific cells during a primary response were stained by tetramers for 2 epitopes of HCMV, IE and pp65, and EBV BMLF1 as a control. In latency, an influenza (FLU) tetramer and the pp65 tetramer, respectively, were used to visualize memory cells that remain after the infection is cleared. EBV-specific cells were stained by the BMLF1 tetramer. Numbers depict the percentage of CD8+ cells or tetramer+ cells within the CD8+ population. Histograms visualize T-bet measured in the tetramer+ samples and (C) in total effector-type (CD8+CD45RA+CD27–) and memory-type (CD8+CD45RA–CD27+) cells in latency. Shown is 1 representative patient of 3 and 1 representative healthy donor of 4 analyzed. (B) During latency, total effector and memory cells were sorted, and quantitative PCR was performed to measure transcription factors T-bet, Eomes, and Blimp-1. Graphs represent mean and SD of 3 donors analyzed. Filled gray histograms denote staining of naive CD8+ T cells.
Additionally, EBV- and influenza-specific T cells were analyzed to measure T-bet expression in CD8+ T cells specific for a different latent and a cleared virus, respectively. EBV BMLF1 epitope–specific T cells also expressed discernible amounts of T-bet protein, although levels appeared somewhat lower than in CMV-reactive cells. In agreement with previous observations on EBV-specific T cells (11), the BMLF1-reactive cells analyzed here were memory-type cells (Supplemental Figure 3 and data not shown), which suggests that separate stages of human T cell differentiation are associated with distinct T-bet levels. Indeed, T-bet expression in influenza-specific CD8+ T cells, which have an early memory-type differentiation status (47), was also low. Finally, when T-bet levels of total memory-type and effector-type cells were compared, the latter cells had significantly higher expression at both the mRNA and the protein level (Figure 4, B and C). This differential expression of T-bet in human subsets accords well with recent observations in mice showing relatively high expression of T-bet in short-lived effector cells (SLECs), in contrast to lower levels in memory precursor effector cells (MPECs) (48).
There was less distinction between memory-type and effector-type cells in Eomes expression (Figure 4B). Eomes and T-bet play similar and partially overlapping functions, yet respond differently to environmental cues. For example, the inflammatory cytokine IL-12 induces T-bet but represses Eomes (45, 49). Whereas T-bet expression leads to elevated SLEC formation, it has been suggested that Eomes participates by directing the differentiation of activated CD8+ T cells more toward the memory phenotype (50). Detailed protein expression analysis needs to be done to determine whether Eomes protein is differentially expressed between functionally distinct subsets of human CD8+ T cells.
Blimp-1 was originally discovered as a regulator of terminal B cell differentiation, but its major influence on CD8+ T cell development is now well established (reviewed in ref. 51). As Blimp-1–deficient CD8+ T cells are not able to maintain effector cell capacities, but acquire a memory cell phenotype, Blimp-1 seems to repress genes that lead to the formation of memory cells (37–39). As mentioned earlier, Blimp-1 may be involved in the establishment of the exhausted phenotype of virus-specific T cells in chronic LCMV infection (34). In HCMV-specific T cells, expression of Blimp-1 was found at the peak of the antiviral response and remained elevated after 1 year and in latency. Since no strong differences in expression were found between effector-type and memory-type cells, our data provided no clues as to the specific role of PRDM1 in human CD8+ T cell differentiation in latent virus infection. Interestingly, however, ZNF683 did show specific expression in latent HCMV-specific T cells and in total effector type cells (Figure 3A and Supplemental Table 6). Since ZNF683 is a close homolog of PRDM1 (with proteins 78.1% identical and 91.4% similar in the zinc finger region; K.M.L. Hertoghs, unpublished observations), we suggest that ZNF683 performs at least part of the functions of PRDM1 in long-lived human effector-type T cells. Finally, PRDM1 is thought to repress other transcriptional regulators, such as BACH2. Repression of BACH2 is necessary for generation of plasma cells during of B cell differentiation (52). Interestingly, BACH2 levels were downregulated in primed cells (Supplemental Table 6); thus, an inverse correlation also exists in human T cells between BACH2 and PRDM1 expression.
A number of transcription factors showed strong downregulation in primed CD8+ T cells (Figure 1C, profile III). Examples of these were the nuclear hormone receptor NR3C2, SCML1, and ZNF395 (also known as papillomavirus-binding factor; PBF). Although the functions of these genes in the immune system are currently unknown, the extensive decrease in their expression at the peak of the response suggests that their gene products have a function in naive cells that is incompatible with the functional changes that take place in antigen-stimulated CD8+ T cells. For example, SCML1, a gene that showed a greater than 100-fold reduction (Supplemental Table 6), is a member of the SCM group required to maintain the transcriptionally repressive state of homeotic genes by blocking chromatin (53). ZNF395 is a nuclear-cytoplasmic shuttling factor that has the ability to inhibit cell growth, and its expression must therefore be restricted to ensure the generation of proper cell numbers. One way to achieve this is by alleviating ZNF395-mediated repression of SAP30, a component of the mSIN3A–HDAC1 deacetylation complex. Indeed, we found vast upregulation of SAP30 (Supplemental Table 2), which could ensure cell cycle progression (54, 55).
The lymphoid enhancer factor 1 (LEF1, also known as TCF7) belongs to the LEF/TCF transcription factors that mediate canonical Wnt signaling (56). LEF1 can function as a transcriptional activator or repressor in cooperation with β-catenin, dependent on the context. Naive CD8+ T cells highly expressed LEF1 mRNA, and transcription was suppressed in antigen-primed cells. This was confirmed at the protein level by Western blot (Figure 3 and Supplemental Figure 4). LEF1 suppression was somewhat alleviated in memory cells (Supplemental Table 6), in agreement with Rutishauser and colleagues’ observed increase in Lef1 expression at 150 days after LCMV infection in mice (38).
Migration and adhesion.
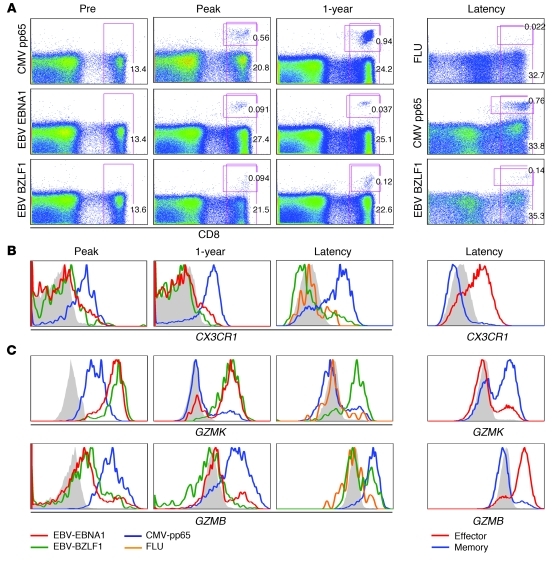
After priming, effector CD8+ T cells migrate from the lymphoid compartment toward sites of inflammation, where adherence to the endothelium is required for extravasation into the inflamed tissue. A specific set of chemokine receptors and cell adhesion molecules is induced to guide these homing processes. Figure 2C shows the predominant changes in chemokine receptor expression. As expected, expression of the chemokine receptor for lymph node homing, CCR7, was strongly downregulated and remained low on the HCMV-specific T cells. In contrast, CCR1, CCR5, CXCR6, and CX3CR1 transcripts were abundantly present at the peak response, but only CX3CR1 remained detectable in the latency stage and was elevated in resting effector-type cells (Supplemental Table 7). The induction of CX3CR1 expression on HCMV-specific cells could be important, since this receptor binds fractalkine, a cell-bound chemokine expressed by stressed endothelial cells, one of the target cells for HCMV.
We monitored CX3CR1 surface expression and observed that HCMV-specific T cells for either pp65- or IE-derived peptide expressed similar CX3CR1 levels during all phases of the response (Figure 5, A and B, and data not shown). In contrast, longitudinal analysis of a primary EBV infection revealed that CX3CR1 was not induced on EBV-specific CD8+ T cells (Figure 5, A and B). Likewise, influenza virus–reactive T cells obtained from a healthy donor also lacked expression of this chemokine receptor. Thus, CX3CR1 appears to be a discriminative marker for HCMV-specific effector cells in both acute and latent infection. This notion was corroborated by the observation that total effector-type cells, in marked contrast to memory-type cells, abundantly expressed CX3CR1 (Figure 5B).
Figure 5. Differential expression of CX3CR1, GZMB, and GZMK in virus-specific cells.
(A) Flow cytometric analysis of tetramers against HCMV epitope pp65 and EBV epitopes EBNA and BZLF1 during a primary response for both HCMV and EBV in the same donor and in a healthy donor during viral latency. Numbers depict the percentage of CD8+ or tetramer+ cells within the CD8+ population. Longitudinal stainings are shown for 1 representative patient of 2 and for 1 representative healthy virus carrier of 4 analyzed. (B and C) Kinetics of (B) CX3CR1 expression and (C) GZMK and GZMB expression on virus-specific T cells during primary HCMV and EBV infection and in latency. Total effector and memory cells in latency are shown at right. Filled gray histograms denote staining of naive CD8+ T cells.
Integrins are heterodimeric proteins made up of α and β subunits that together determine the structure and function of the complex and their roles in cell adhesion. In the acute phase, ITGAD (also known as CD11D) was extensively upregulated, but expression returned to that of naive cells 1 year after infection. At that time point, ITGB1, ITGB2, and ITGAL showed the strongest increase compared with naive CD8+ T cells. The latter 2 can form the adhesion protein LFA-1, which plays a central role in leukocyte adhesion and signaling through interactions with its ligands, ICAM1–ICAM3. Of the integrins, ITGA6 (also known as CD49F) was the only significantly downregulated gene, which suggests that ITGA6 exerts its main function in naive CD8+ T cells (Supplemental Table 7).
ADAM8, a member of the a disintegrin and metalloprotease domain (ADAM) family that is structurally related to snake venom disintegrins, remained elevated in all primed cells (Supplemental Table 7). ADAM family members have been implicated in a variety of biological processes, involving cell-cell and cell-matrix interactions, and the high expression of ADAM8 in effector-type cells may be crucial for their movement in tissues.
Transcript levels of CD62L (also known as SELL) were not reduced in the acute phase of the infection, although acutely activated CD8+ T cells have low CD62L surface expression (13). This may be explained by the rapid cleavage of CD62L from the cell surface after lymphocyte stimulation (57). In HCMV latency, a drop in CD62L mRNA levels was observed, but there were no apparent differences between resting effector- and memory-type cells (58).
Cytokine receptors.
In order to respond to environmental stimuli, naive cells that differentiate into effector cells altered the surface expression of a number of cytokine receptors (Supplemental Table 8). Transcript levels of IL-2 receptor α (IL2RA, also known as CD25) were not significantly changed (data not shown), but primed cells showed an upregulation of both the cγ chain (IL2RG, also known as CD132) and IL2RB (also known as CD122), which would allow increased responsiveness to IL-2 and IL-15. Furthermore, the amount of IL18R1 and IL28RA transcripts increased after activation, but no distinction between effector-type and memory-type cells was found.
Cell surface expression of IL7RA (also known as CD127) is downregulated after activation of naive T cells (59, 60), which was reflected by the massive drop of IL7RA message at the peak of the response (Supplemental Table 8). Expression remained low when the antiviral response progressed but, as also described at the protein level, memory-type cells expressed substantially more IL-7Rα than did resting effector-type cells (60).
Differentiation and exhaustion markers.
Naive and memory CD8+ T cells express the costimulatory receptors CD27 and CD28, but resting effector-type T cells typically lack surface expression of these molecules. In the acute stage of infection, CD27 expression was maintained on HCMV-specific CD8+ T cells; over the course of months, it was lost from the cell membrane (ref. 61 and Figure 6A). This change in phenotype does not occur during other persistent infections, such as EBV and HIV (11). Interestingly, CD27 mRNA levels changed only minimally in the course of HCMV infection, which suggests that CD27 surface expression is specifically regulated in resting effector type cells at the (post)translational level instead of the transcriptional level. The interaction between CD27 and its ligand, CD70, reduces cell surface CD27 expression presumably by proteolytic cleavage (62). In line with this possible regulation at the protein level, we found CD70 was strongly induced during the primary response and remained elevated thereafter (Supplemental Table 9). In contrast to CD27, CD28 showed gradual loss of mRNA transcripts during HCMV infection that correlated with protein expression (Figure 6B). In healthy donors, memory and naive cells had comparable levels of CD28 mRNA, whereas effector-type cells showed greater than 13-fold downmodulation (Supplemental Table 9).
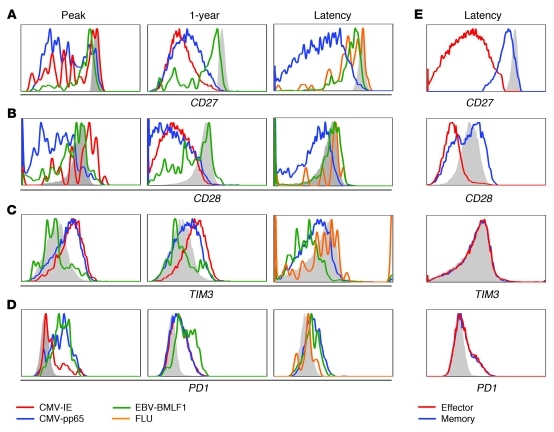
Figure 6. HCMV-specific cells show extensive differentiation, but do not become exhausted.
CD8+ T cells from a patient experiencing primary HCMV response and from a healthy latent virus carrier were stained with tetramers specific for HCMV and EBV epitopes. Shown is 1 representative patient of 3 and 1 representative healthy virus carrier of 4 analyzed. (A) CD27. (B) CD28. (C) TIM3. (D) PD1. (E) Expression on total memory and effector CD8+ T cells. Filled gray histograms denote staining of naive CD8+ T cells.
Recent studies in mice have defined a number of cell surface molecules associated with the CD8+ T cell exhaustion during persistent virus infection (reviewed in ref. 8). As HCMV also persists throughout life, we specifically scrutinized the dataset for these surface receptors (Supplemental Table 9). Kinetic analysis of the expression of inhibitory receptors revealed that during the peak of the antigen-dependent response against HCMV, the majority of the messages of PD1, LAG3, CD160, CD244 (also known as 2B4), TIM3, KLRG1, KLRC1, PTGER2, and CTLA4 (Figure 2D) were strongly increased. The expression of these genes remained elevated until 1 year after viral infection, and then declined during latency. Although transcript levels of the majority of genes were still slightly elevated above the level of naive CD8+ T cells, no major differences were found between resting effector-type and memory-type cells (Supplemental Table 9).
These findings were confirmed at the protein level for 2 of the molecules, PD1 and TIM3. TIM3 cell surface expression was clearly demonstrable on HCMV-specific CD8+ T cells during the primary response but declined thereafter (Figure 6C). Notably, TIM3 expression on latent EBV-specific T cells exceeded that of the HCMV-reactive ones (63). Moreover, although TIM3 expression was lowest on naive cells, no distinction was found between resting effector-type and memory-type cells. Likewise, PD1 was upregulated at the peak of the infection, but HCMV-reactive CD8+ T cells during latency had relatively low expression (Figure 6D).
Next, we compared our data on latent HCMV-specific CD8+ T cells with the microarray data on exhausted LCMV-reactive cells obtained by Wherry et al. (34), available from the Gene Expression Omnibus (GEO; see Methods). To work with a manageable dataset, we compared the genes that were upregulated 10-fold or greater in the latent HCMV-specific CD8+ T cells (corrected significance P ≤ 1 × 10–10) and that could be mapped to Affymetrix mouse probesets (n = 73) with the exhausted LCMV-reactive cells. Of these 73 genes, 52 (Supplemental Figure 5) did not show a marked change (fold increase >2), and only KLRD1 was discordantly regulated. Whereas 6 genes were downregulated in both HCMV- and LCMV-specific cells, 14 genes were upregulated in both populations (Supplemental Table 12). With the exception of LAG3, none of these genes has been implicated to our knowledge in the establishment of the exhausted phenotype.
In summary, neither the microarray data nor the FACS analyses provided evidence that latent HCMV-reactive cells are exhausted. This conclusion indeed fits well with previous studies showing that HCMV-specific CD8+ T cells can rapidly expand when stimulated with cognate peptide and cγ chain cytokines (64).
Chemokines and cytokines.
The differentiation of lymphocytes as well as their effect on other cells is orchestrated, at least in part, by cytokines. Surprisingly, not many cytokines were significantly upregulated in the CD8+ T cells themselves (Supplemental Table 10). The most enhanced expression was that of IL32, a cytokine originally described in activated NK cells. Its expression seemed to correlate with the emergence of acute effector T cells. IL32 can induce macrophages to produce TNF-α, MIP-2, and IL-8, which suggests that the cytokine plays a role in inflammatory responses to attract other leukocytes to the site of infection (65).
The chemokines CCL4 (also known as MIP1B) and CCL5 (also known as RANTES) were induced during the primary response and remained elevated in primed cells (Figure 2E). No clear differences between total memory- and effector-type cells were observed. The microarray data are in agreement with observations showing that virus-specific CD8+ T cells directed toward different viruses have the ability to secrete these chemokines (66).
Effector molecules.
The hallmark that distinguishes effector CD8+ T cells from naive T cells is the production of cytotoxic and other molecules that aid in killing of the target cells. Of the granzyme family of proteases, 5 members were induced in response to CMV infection (Figure 2F), albeit at different levels. Although both EBV- and CMV-specific cells upregulated GZMB during the primary response, during viral latency, its expression was specific for effector-type cells, as it was expressed in neither total memory-type cells nor in influenza- or EBV-specific CD8+ T cells (Figure 5C). This expression contrasted with that of GZMA, which remained high on primed CD8+ T cells regardless of differentiation status (ref. 67 and data not shown).
Like GZMB, GZMK was induced in both EBV- and CMV-specific CD8+ T cells during the primary response. However, in latent virus carriers, GZMK expression was only found in a subset of memory-type cells and EBV-specific cells (Figure 5C), but hardly in HCMV-specific cells or total effector-type CD8+ T cells. The absence of GZMK expression in influenza-specific T cells (Figure 5C) infers that its expression is limited to a very defined human CD8+ T cell differentiation stage. Thus, the regulation of GZMB and GZMK showed opposite patterns after the primary response (Figure 5C).
Of all effector molecules listed in Supplemental Table 10, only PROK2 (also known as BV8) was considerably increased as the infection progressed into latency (Figure 1C, profile II). Elevated PROK2 levels were only detectable in effector-type cells and not in the memory-type CD8+ T cells. PROK2 is a chemoattractant that already at very low concentrations is able to direct macrophages and monocytes to sites of inflammation, where it subsequently drives them into a more differentiated phenotype (68, 69). The addition of PROK2 to macrophages boosts the production of the proinflammatory cytokines IL-1β and IL-12 induced by LPS (70). The presence of very low levels of PROK2 RNA has been reported in CD8+ T cells (68). Our present data suggest that this may be due to the expression of PROK2 by effector-type cells. Thus, the secretion of this protein can be a tool for a subset of CD8+ cells to alter the (pro)inflammatory milieu.
Another gene with predominant expression in effector-type cells is the highly conserved extracellular matrix protein SPON2 (also known as Mindin). As proposed for PROK2, SPON2 is also involved in recruiting leukocytes like neutrophils and macrophages to sites of inflammation (71). Excreted SPON2 can bind to integrins on the cell surface of DCs. Real time imaging has shown that SPON2-deficient DCs are unable to efficiently engage with CD4+ T cells, thereby impeding the priming of T lymphocytes in vitro (72). SPON2 transcripts are elevated directly at the peak of the response and almost certainly remain elevated throughout life in vigilant effector cells. No SPON2 expression was found in the pool of memory cells in healthy individuals, which underscores the specificity of SPON2 production in HCMV-specific CD8+ T cells. Since more cell types can produce SPON2 (e.g., B cells, CD4+ T cells; ref. 72), further investigation will be needed to reveal the contribution of CD8+ T cells during the immune response and determine whether SPON2 expression is found in other chronic or acute settings.
We found continuous expression of IFNG mRNA in HCMV-specific CD8+ T cells isolated during the different stages of the response (Supplemental Table 11). This seems in line with the observation that the Th1 prototypic transcription factor T-bet was highly induced during the primary response and was maintained in these cells throughout the latency stage. The continuous T-bet expression did not lead to the expression of IFN-γ protein in unstimulated cells (data not shown). To determine whether high T-bet expression is associated with high inducible IFN-γ production, we performed term in vitro stimulations with PMA and ionomycin. As expected (see Figure 4), effector-type CD8+ T cells were enriched in the T-betbright fraction (Figure 7, left column). Moreover, the T-betbright fraction indeed contained more IFN-γ–producing cells than did the T-betdim population, both in the total CD8+ population and, importantly, in the memory- and effector-type fractions. The difference in TNF-α production between T-betbright and T-betdim cells was less pronounced. Thus, the continuous presence of IFNG mRNA in HCMV-specific and total effector-type cells may lead to rapid IFN-γ production in vivo. In support of this, HCMV-specific T cells showed upregulation of a number of IFN-γ–regulated genes (Supplemental Table 11).
Figure 7. CD8+ T cells expressing high levels of T-bet produce vast amounts of IFN-γ.
PBMCs from a healthy HCMV-positive donor were activated with PMA and ionomycin and were costained with antibodies specific for CD3, CD8, CD45RA, CD27, T-bet, IFN-γ, and TNF-α. T-betbright cells were defined as those that stained stronger than naive T cells (Figure 4C, filled gray histogram). Effector-type cells were gated as CD3+CD8+CD45RA+CD27–, memory-type cells as CD3+CD8+CD45RA–CD27+. Shown is 1 representative donor of 3.
Discussion
The remarkable feature of human CMV infection is that it generates a vast population of virus-specific CD8+ T cells with mainly an effector phenotype that increases with age. Our data showed that a number of key features of these cells, specifically cytolytic potential, IFN-γ production, and migratory potential, are already installed during the primary response in this population, a process likely governed by stable changes in transcription factor expression. From our findings, it was evident that these changes are specific for HCMV-reactive cells, as CD8+ T cells reactive toward various epitopes of EBV and influenza were markedly different with respect to function, chemokine receptor expression, and transcription factor expression, most notably in regard to TBX21 and ZNF683. Although high TBX21 expression and the cytolytic potential of HCMV-specific CD8+ T cells are reminiscent of SLECs, several important factors suggest that these cells display a unique phenotype. Human effector-type cells showed long-term survival, did not extensively divide, and had low metabolic activity (ref. 73 and the present study). Importantly, the expression of Blimp-1, which has recently been shown to be important for the establishment of an exhausted phenotype in mice (39), was not different between memory-type and effector-type cells. Collectively, these expression data agree with in vitro studies showing that HCMV-specific effector-type cells expand strongly when a combination of specific peptide and cγ chain cytokines is provided (18).
As we isolated memory-type cells by sorting on CD45RA and CD27, no distinction was made between central (i.e., CCR7+) and effector/memory cells (CCR7–). Likely, comparison of these populations will yield additional data on the functional diversity of primed human CD8+ T cells. An indication for this was obtained in the present study by our finding that GZMK expression was restricted to EBV-reactive T cells during latent infection. Since these cells, in contrast to influenza-specific CD8+ T cells, were predominantly of the effector/memory type, GZMK expression appears to be strongly linked to a discrete stage of T cell differentiation. The high expression of GZMK in EBV-reactive T cells may point to a specific role of this tryptase in maintaining EBV latency. Other than GZMB, the tryptase-like granzymes are not believed to be instrumental in inducing cell death in a physiological setting (74). We found no evidence for a putative extracellular role of GZMK, for example by modifying cell surface receptors directly involved in transmission of EBV (e.g., CD21), but it has been suggested that this class of proteases regulates immune reactions through cytokine modifications (75).
Part of the microarray and protein analyses was performed using virus-specific CD8+ T cells obtained from kidney transplant recipients that were on standard immunosuppressive therapy, a combination of prednisolone and cyclosporin. Although these patients developed protective anti-HCMV responses, their immune reactions may be different from those of immunocompetent individuals. Indeed, when comparing our kinetic data with rare observations that can be made in healthy individuals (76), the response in the transplant recipient appeared to be somewhat slower. Still, as changes taking place in key molecules are shared between the 1 year time point of the patients and the latency phase of healthy CMV carriers, the analysis of the first phase of this antiviral response may provide a proper perspective on the alterations that take place in immune CD8+ T cells occurring early after infection.
Coevolution of the human immune system and immune-evasive HCMV has led to the emergence of a prototypic HCMV-associated CD8+ T cell differentiation state, characterized not only by typical phenotypic and functional traits, but also by specific alterations of transcription factor expression. The changes take place at the population level, and establishment of the effector-type differentiation state that is characteristic for HCMV is found both in pp65- and in IE-specific T cells. Additionally, analysis of clonal diversity in phenotypically homogeneous pp65-specific CD8+ T cells revealed that not only were these populations not monoclonal but oligoclonal, but the relative presence of particular clones changed over time (E.B.M. Remmerswaal, unpublished observations). These data indicate that regardless of (fine) specificity of responsive T cells, HCMV has a dominant effect on the establishment of the prototypic phenotype in the majority of independent CD8+ T cell clones.
The predominant cytokine detected in HCMV-specific CD8+ T cells was IFN-γ, and both early and late virus-specific cells showed upregulation of a number of IFN-γ–regulated genes. The constitutive expression of IFN-γ message, which resembles stable GZMB expression in murine NK cells (77), may endow CD8+ effector-type T cells with a near-instantaneous capacity to secrete this cytokine upon TCR ligation, a mechanism that may be crucial in rapidly antagonizing virus replication upon reactivation of latent virus. On the downside, the high expression of IFN-γ may subtly increase the overall proinflammatory status in HCMV infection which may be causally related to pathogenic effects of HCMV on the vasculature (15). It is conceivable that the stable expression of IFN-γ in HCMV-reactive CD8+ T cells is controlled by a combination of transcription factors, such as T-bet, that are induced during the primary response and maintained throughout the latent state. Identification of the factors that induce the establishment of the robust cytotoxic, IFN-γ–secreting phenotype may be important for rational improvement of therapeutic vaccines to persisting viruses and tumors.
In summary, the longitudinal analyses of HCMV-specific CD8+ T cells that develop in humans after primary infection depicts a differentiation program that is distinct from those described for cleared and persistent viruses in mice. Importantly, we suggest that CD8+ T cell differentiation may not be linear, but divergent, depending on the evolutionary selected protective response that is essential to cope with individual viruses. Some aspects of this program may in fact be characteristic for humans, since the human immune system must be able to provide protection against HCMV that lasts for more than 7 decades, in the case of childhood infection. The factors that initiate and maintain this long-lasting, resting effector-type response are unknown, but it is conceivable that in addition to viral peptides, other cues provided either directly or indirectly by the virus to the immune system operate to shape this response.
Methods
Subjects.
Microarray analyses were performed using peripheral blood of 3 HLA-B7+ HCMV-seronegative renal transplant recipients of CMV-seropositive donor kidneys. For additional flow cytometry studies, 1 HLA-B35+ CMV- and EBV-seronegative renal transplant recipient of a CMV- and EBV-seropositive kidney and 1 HLA-A2+ CMV-seronegative renal transplant recipient of a CMV-seropositive kidney were studied longitudinally. Blood was also drawn from CMV- and EBV-seropositive HLA-A2+, HLA-B7+, and/or HLA-B8+ healthy volunteers. All renal transplant recipients were treated with basic immunosuppressive therapy (cyclosporine A, prednisolone, and mycophenolate mofetil), except for 1 of the patients in the array analysis, who received CD25mAb induction therapy followed by a basic immunosuppressive therapy of prednisolone A, mycophenolate mofetil, and tacrolimus. All subjects gave written informed consent, and the medical ethics committee of the Academic Medical Center approved the study.
Tetrameric complexes.
The following allophycocyanin-conjugated tetrameric complexes were obtained from Sanquin: HLA-A0201 tetramer loaded with the HCMV pp65-derived NLVPTMVATV peptide (HCMV A2 pp65), HLA-A0201 tetramer loaded with the influenza matrix protein–derived GILGFVFTL peptide (FLU A2 MP), HLA-A0201 tetramer loaded with the EBV BMLF1–derived GLCTLVAML peptide (EBV A2 BMLF1), HLA-B3501 tetramer loaded with the HCMV pp65–derived IPSINVHHY peptide (CMV B35 pp65), HLA-B3501 tetramer loaded with the EBV BZLF1–derived EPLPQGQLTAY peptide (EBV B35 BZLF1), HLA-B3501 tetramer loaded with the EBV EBNA1–derived HPVGEADYFEY peptide (EBV B35 EBNA1), HLA-B0801 tetramer loaded with the EBV BZLF1–derived RAKFKQLL peptide (EBV B8 BZLF1), HLA-B0702 tetramer loaded with the HCMV pp65–derived TPRVTGGGAM peptide (HCMV B7 pp65), and HLA-A0201 tetramer loaded with the HCMV IE–derived VLEETSVML peptide (HCMV A2 IE).
Sorting.
PBMCs were isolated from heparinized blood using standard density gradient centrifugation and subsequently cryopreserved in liquid nitrogen until the day of analysis. To isolate naive (CD8+CD45RAhiCD27hi), effector-type (CD8+CD45RAhiCD27lo), and memory-type (CD8+CD45RAloCD27+) cells from latently chronic-infected healthy donors, cells were isolated from buffy coats by a 2-step procedure. After Ficoll, CD8+ T cells were isolated by CD8+ microbeads (Miltenyi Biotec) and then labeled with CD27-FITC (7C9) (78), CD45RA-RD1 (Beckman Coulter), and CD8-allophycocyanin (BD Biosciences — Pharmingen) and subsequently sorted using a FACS Aria (BD Biosciences). For RNA isolation and microarray analysis, 3 independent donors and a pool of 3 additional healthy individuals were used.
To isolate HCMV-specific CD8+ effector cells at the peak of the HCMV response, PBMC were stained with HLA DR–FITC, CD38-PE (BD Biosciences), and CD8-allophycocyanin (BD Biosciences — Pharmingen). HLA-DRhiCD38hiCD8+ T cells were sorted using a FACS Aria. To obtain HCMV-specific CD8+ T cells in the latency phase, PBMCs obtained 40–60 weeks after transplantation (1 year after infection) were stained with allophycocyanin-conjugated tetramers; subsequently, allophycocyanin microbeads (Miltenyi Biotec) were used to isolate the cells. Upon reanalysis, the purified populations contained between 95% and 97% tetramer-binding cells. Due to the limited amount of blood that can be drawn from patients, virus-reactive CD8+ T cells from 3 patients with acute CMV infection were pooled before RNA isolation (see below). For comparison, RNA from HCMV-reactive CD8+ T cells from 3 latent CMV carriers was prepared in a similar fashion.
RNA isolation, amplification, labeling, and hybridization.
RNA was isolated using the nucleospin RNA isolation kit (Machery-Nagel) according to the manufacturer’s instructions. mRNA was amplified using the MessageAmp II Kit (Ambion). Labeling, hybridization, and data extraction were performed at ServiceXS, as described previously (79). Hybridization was performed with 600 ng of each labeled target along with fragmentation and hybridization buffer at 60°C for 17 hours on to Whole Human Genome (WHG) 44K Oligo Microarrays (Agilent Technologies) following the manufacturer’s protocol.
Microarray imaging and data analysis.
The microarray slides were washed according to the manufacturer’s instructions and scanned using an Agilent dual-laser DNA microarray scanner. Default settings of Agilent Feature Extraction preprocessing protocols were used to obtain normalized expression values from the raw scans. Exact protocol and parameter settings are described in the Agilent Feature Extraction Software User Manual (version 7.5). The default Agilent normalization procedure called Linear & Lowess was applied. Resulting data were imported in Rosetta Resolver (Rosetta Biosoftware), and replicates were combined using an error-weighted average. Hierarchical clustering of the resulting microarray data was performed using Pearson correlation as a distance measure and with complete linkage. An error-weighted 1-way ANOVA with the Benjamini-Hochberg false-discovery rate multiple testing correction was used to find differentially expressed genes across the virus-specific CD8+ T cell populations. Average expression ratios and corresponding P values for differential expression detection were exported from Rosetta Resolver. The resulting dataset presents the virus-specific CD8+ T cell subset’s mean fold change and P values for the combined dye swap experiments from the pooled RNA; for the purified non–antigen-specific subsets, the mean fold change and P values across the 4 microarrays per condition (3 individuals and a pool of 3 subjects) are shown.
Consensus clustering (25, 26) was used to assess the stability of hierarchical clustering. Consensus clustering evaluates the robustness of a clustering solution of the samples by repeatedly sampling with replacement (i.e., bootstrapping) from the probes and then reclustering the sampled data. For each pair of samples in the original data, a consensus matrix was used to keep track of the number of times they belonged to the same cluster in a bootstrap sample. Samples that remain consistently clustered together across different bootstrap samples have a high consensus and thus are stable clusters. Parameter settings for consensus clustering were as follows: 200 bootstrap samples, Pearson correlation distance for hierarchical clustering with complete linkage.
We clustered the expression patterns of a selected set of genes using the CLICK algorithm from the software package Expander 5.0 using Pearson correlation as a distance measure (27). Log ratio data was imported in Expander, and probes were annotated using the gene mapping for Agilent WHG arrays available in Expander. Log ratios for probes that mapped to the same gene were averaged. The CLICK algorithm was applied using default settings. CLICK does not require the user to choose the number of clusters a priori, but rather finds the optimal number of clusters by maximizing both the within-cluster homogeneity and the between-cluster separation, using a graph-based statistical algorithm. Resulting clusters were analyzed for enrichment of GO biological processes and molecular functions using the TANGO algorithm in Expander, applied using the default settings. Reported P values are the result of testing for the significance of overlap between gene groups and GO categories using Fisher’s exact test for independence. P values were corrected for multiple testing by sampling for each cluster 1,000 random gene sets of the same size as the original cluster and estimating the empirical P value distribution for the evaluated GO categories. A GO category was considered significantly enriched in a cluster if its corrected P value was 0.05 or less.
Cross-species analysis.
For the comparison of our CMV-specific CD8+ T cell data with published microarray data from exhausted murine CD8+ T cells, we downloaded the normalized data from Wherry et al. (34) available at GEO ( http://www.ncbi.nlm.nih.gov/projects/geo/; accession no. GSE9650) and selected all 15 Affymetrix U74A microarrays. All technical replicates per condition (naive, effector, memory, and exhausted) were averaged (34). Subsequently, we mapped the 41,058 Agilent WHG probes to the 12,488 Affymetrix MG-U74Av2 mouse probesets. First, we annotated probes on the Agilent array with their Entrez Gene IDs using the annotation available within Bioconductor (package hgug4112a.db, version 2.4.1). Next, we used the human gene IDs to mine the NCBI HomoloGene database (build64) and find orthologous gene IDs in the target species (Mus musculus). Finally, we used the Affymetrix NetAffx annotation for the MG-U74Av2 arrays to retrieve the corresponding (orthologous) probe sets. Thus, 13,082 of the Agilent probes could be mapped to 1 or more of 10,383 Affymetrix probesets. In case multiple mouse probesets were found for a given human probe, we selected the probeset with the largest absolute fold change versus naive for the condition of interest.
Immunofluorescent staining and flow cytometry.
PBMCs were washed in PBS containing 0.01% (w/v) NaN3 (Merck) and 0.5% (w/v) bovine serum albumin (Roche). PBMCs (2 × 106) were incubated with an appropriate concentration of tetrameric complexes in a small volume for 30 minutes at 4°C, protected from light. Fluorescence-labeled mAbs were then added and incubated for 30 minutes at 4°C, protected from light, at concentrations according to the manufacturer’s instructions. For intracellular stainings, cells were fixed with 2% PFA (Merck) after tetramer and surface staining and subsequently permeabilized with PBS containing 0.01% (w/v) NaN3, 0.5% (w/v) bovine serum albumin, 50 mM d-glucose (Merck), and 0.1% (w/v) saponin (Sigma-Aldrich). The following mAbs were used for surface marker expression analysis: TIM3 (R&D Systems); CD27 allophycocyanin-eFluor 780, PD1 PE, and CD27 PerCP-Cy5.5 (eBioscience Inc.); CX3CR1 PE (MBL International); CD45RA FITC, CCR7 PE-Cy7, CD3 PE-Cy7, and CD45RA PE-Cy7 (BD Biosciences); CD28 Alexa Fluor 700 (BioLegend); and CD8 PE-Alexa Fluor 610 (Invitrogen). The following mAbs were used for intracellular stainings: T-bet PerCP-Cy5.5 (eBioscience Inc.); GZMB PE (Sanquin); perforin FITC, TNF-α FITC, and IFN-γ PE (BD Biosciences); Ki67 FITC (BD Biosciences — Pharmingen); and GZMK FITC (Immunotools). Cells were washed and measured on a FACsCalibur or a FACsCanto flow cytometer (BD Biosciences) and analyzed with FlowJo software (FlowJo).
Intracellular cytokine staining.
For cytokine production and simultaneous T-bet staining, PBMCs were stimulated either with medium or with PMA (1 μg/ml) and ionomycin (1 ng/ml) (both from Sigma-Aldrich). Brefeldin A (Invitrogen) was added immediately, and the cultures were left to incubate for 4 hours. All cultures were performed in RPMI 1640 with l-glutamine and HEPES (Invitrogen) containing 10% heat-inactivated FCS (PAA), penicillin, and streptomycin. After cessation of the incubation, cells were washed and incubated with CD8 PE–Alexa Fluor 610 and CD3 PE-Cy7 followed by subsequent fixation and intracellular staining as described above with IFN-γ.
Quantitative PCR.
Total RNA was isolated using the Invisorb RNA isolation kit (Invitek). RNA was reverse transcribed with — and cDNA was synthesized by means of — oligodeoxythymidine and Superscript II RNase H-reverse transcriptase (Invitrogen). Amplification of specific targets by quantitative RT-PCR was performed with Lightcycler FastStart DNA Master SYBR Green I (Roche) using the following primers: EOMES forward, 5′-ACTGGTTCCCACTGGATGAG-3′; EOMES reverse, 5′-CCACGCCATCCTCTGTAACT-3′; ZNF683 forward, 5′-CATATGTGGCAAGAGCTTTGG-3′; ZNF683 reverse, 5′-AGAGCTTCACTCAACTTGCC-3′; LEF1 forward, 5′-AAGCAGAGTCCAAGGCAGAGAA-3′; LEF1 reverse, 5′-ACCTTGGCTCTCATCTCCTTCA-3′; PRDM1 forward, 5′-CAACAACTTTGGCCTCTTCC-3′; PRDM1 reverse, 5′-GCATTCATGTGGCTTTTCTC-3′; TBX21 forward, 5′-GGGAAACTAAAGCTCACAAAC-3′; TBX21 reverse, 5′-CCCCAAGGAATTGACAGTTG-3′.
After a 10-minute denaturation step at 95°C, 40 PCR cycles of 15 seconds at 94°C, 30 seconds at 60°C, 45 seconds at 72°C, and 5 seconds at 79°C were performed. For ZNF683, the melting temperature (second step) required 30 seconds at 66°C. Data were analyzed using LightCycler Software (version 3.5; Roche) and LinRegPCR (version 7.5; ref. 80). To confirm the purity and specificity of the reaction, a melting curve analysis was performed at the end of the PCR by slowly increasing (0.1°C/s) the temperature of the reaction from 65 to 95°C. S18 was used as an internal reference.
Western blot analysis.
Cell lysates for Western blotting were separated by 12% SDS-PAGE followed by Western blotting as described previously (81). Blots were probed with monoclonal anti-LEF1 antiserum (Abcam).
Statistics.
Genes differentially expressed between the different cell populations were determined using Rosetta Resolver, with a fold change cutoff of 1 of 4 parameters ≥10 and an error-weighted 1-way ANOVA with a P value cutoff of ≤1 × 10–10 with the Benjamini-Hochberg false-discovery rate correction.
Supplementary Material
Acknowledgments
The authors thank Martijn Nolte, Kris Reedquist, and Ester van Leeuwen for helpful discussions and critically reading the manuscript. K.M.L. Hertoghs and R.A.W. van Lier were supported by VICI grant 918.46.606 from the Netherlands Organization of Scientific Research; P.D. Moerland acknowledges the BioRange programme of NBIC, supported by a BSIK grant through the Netherlands Genomics Initiative (NGI); and A. van Stijn was supported by grant C03.2034, and S.L. Yong and P.J.E.J. van de Berg by grant C05.2141, from the Dutch Kidney Foundation.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(11):4077–4090. doi:10.1172/JCI42758.
References
- 1.Murali-Krishna K, et al. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity. 1998;8(2):177–187. doi: 10.1016/S1074-7613(00)80470-7. [DOI] [PubMed] [Google Scholar]
- 2.Schluns KS, Lefrancois L. Cytokine control of memory T-cell development and survival. Nat Rev Immunol. 2003;3(4):269–279. doi: 10.1038/nri1052. [DOI] [PubMed] [Google Scholar]
- 3.Stemberger C, et al. A single naive CD8+ T cell precursor can develop into diverse effector and memory subsets. Immunity. 2007;27(6):985–997. doi: 10.1016/j.immuni.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 4.Kaech SM, Hemby S, Kersh E, Ahmed R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell. 2002;111(6):837–851. doi: 10.1016/S0092-8674(02)01139-X. [DOI] [PubMed] [Google Scholar]
- 5.Chang JT, et al. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science. 2007;315(5819):1687–1691. doi: 10.1126/science.1139393. [DOI] [PubMed] [Google Scholar]
- 6.Wherry EJ, Ahmed R. Memory CD8 T-cell differentiation during viral infection. J Virol. 2004;78(11):5535–5545. doi: 10.1128/JVI.78.11.5535-5545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barber DL, et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439(7077):682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 8.Blackburn SD, et al. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol. 2009;10(1):29–37. doi: 10.1038/ni.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snyder CM, Cho KS, Bonnett EL, van Dommelen S, Shellam GR, Hill AB. Memory inflation during chronic viral infection is maintained by continuous production of short-lived, functional T cells. Immunity. 2008;29(4):650–659. doi: 10.1016/j.immuni.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karrer U, et al. Memory inflation: continuous accumulation of antiviral CD8+ T cells over time. J Immunol. 2003;170(4):2022–2029. doi: 10.4049/jimmunol.170.4.2022. [DOI] [PubMed] [Google Scholar]
- 11.Appay V, et al. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat Med. 2002;8(4):379–385. doi: 10.1038/nm0402-379. [DOI] [PubMed] [Google Scholar]
- 12.Callan MF, et al. Direct visualization of antigen-specific CD8+ T cells during the primary immune response to Epstein-Barr virus In vivo. J Exp Med. 1998;187(9):1395–1402. doi: 10.1084/jem.187.9.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roos MT, et al. Changes in the composition of circulating CD8+ T cell subsets during acute epstein-barr and human immunodeficiency virus infections in humans. J Infect Dis. 2000;182(2):451–458. doi: 10.1086/315737. [DOI] [PubMed] [Google Scholar]
- 14.Wills MR, Okecha G, Weekes MP, Gandhi MK, Sissons PJ, Carmichael AJ. Identification of naive or antigen-experienced human CD8(+) T cells by expression of costimulation and chemokine receptors: analysis of the human cytomegalovirus-specific CD8(+) T cell response. J Immunol. 2002;168(11):5455–5464. doi: 10.4049/jimmunol.168.11.5455. [DOI] [PubMed] [Google Scholar]
- 15.van de Berg PJ, van Stijn A, ten Berge IJ, van Lier RA. A fingerprint left by cytomegalovirus infection in the human T cell compartment. J Clin Virol. 2008;41(3):213–217. doi: 10.1016/j.jcv.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 16.Gamadia LE, Remmerswaal EB, Weel JF, Bemelman F, van Lier RA, ten Berge I. Primary immune responses to human CMV: a critical role for IFN-gamma-producing CD4+ T cells in protection against CMV disease. Blood. 2003;101(7):2686–2692. doi: 10.1182/blood-2002-08-2502. [DOI] [PubMed] [Google Scholar]
- 17.Rentenaar RJ, et al. Development of virus-specific CD4(+) T cells during primary cytomegalovirus infection. J Clin Invest. 2000;105(4):541–548. doi: 10.1172/JCI8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Leeuwen EM, ten Berge I, van Lier RA. Induction and maintenance of CD8+ T cells specific for persistent viruses. Adv Exp Med Biol. 2007;590:121–137. doi: 10.1007/978-0-387-34814-8_9. [DOI] [PubMed] [Google Scholar]
- 19.van Lier RA, ten Berge I, Gamadia LE. Human CD8(+) T-cell differentiation in response to viruses. Nat Rev Immunol. 2003;3(12):931–939. doi: 10.1038/nri1254. [DOI] [PubMed] [Google Scholar]
- 20.Miller JD, et al. Human effector and memory CD8+ T cell responses to smallpox and yellow fever vaccines. Immunity. 2008;28(5):710–722. doi: 10.1016/j.immuni.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 21.Hislop AD, et al. EBV-specific CD8+ T cell memory: relationships between epitope specificity, cell phenotype, and immediate effector function. J Immunol. 2001;167(4):2019–2029. doi: 10.4049/jimmunol.167.4.2019. [DOI] [PubMed] [Google Scholar]
- 22.Sylwester AW, et al. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J Exp Med. 2005;202(5):673–685. doi: 10.1084/jem.20050882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khan N, et al. Cytomegalovirus seropositivity drives the CD8 T cell repertoire toward greater clonality in healthy elderly individuals. J Immunol. 2002;169(4):1984–1992. doi: 10.4049/jimmunol.169.4.1984. [DOI] [PubMed] [Google Scholar]
- 24.Kuijpers TW, et al. Frequencies of circulating cytolytic, CD45RA+CD27-, CD8+ T lymphocytes depend on infection with CMV. J Immunol. 2003;170(8):4342–4348. doi: 10.4049/jimmunol.170.8.4342. [DOI] [PubMed] [Google Scholar]
- 25.Muller FJ, et al. Regulatory networks define phenotypic classes of human stem cell lines. Nature. 2008;455(7211):401–405. doi: 10.1038/nature07213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monti S, Tamayo P, Mesirov J, Golub T.Consensus Clustering: A resampling-based method for class discovery and visualization of gene expression microarray data. Machine Learning. 2003521–291–118. . 10.1023/A:1023949509487 [DOI] [Google Scholar]
- 27.Ulitsky I, et al. Expander: from expression microarrays to networks and functions. Nat Protoc. 2010;5(2):303–322. doi: 10.1038/nprot.2009.230. [DOI] [PubMed] [Google Scholar]
- 28.Latner DR, Kaech SM, Ahmed R. Enhanced expression of cell cycle regulatory genes in virus-specific memory CD8+ T cells. J Virol. 2004;78(20):10953–10959. doi: 10.1128/JVI.78.20.10953-10959.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Derenzini M, et al. Thymidylate synthase protein expression and activity are related to the cell proliferation rate in human cancer cell lines. Mol Pathol. 2002;55(5):310–314. doi: 10.1136/mp.55.5.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lens SM, Vader G, Medema RH. The case for Survivin as mitotic regulator. Curr Opin Cell Biol. 2006;18(6):616–622. doi: 10.1016/j.ceb.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 31.Strasser A, Pellegrini M. T-lymphocyte death during shutdown of an immune response. Trends Immunol. 2004;25(11):610–615. doi: 10.1016/j.it.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 32.Liston P, et al. Identification of XAF1 as an antagonist of XIAP anti-Caspase activity. Nat Cell Biol. 2001;3(2):128–133. doi: 10.1038/35055027. [DOI] [PubMed] [Google Scholar]
- 33.Wang J, et al. Identification of XAF1 as a novel cell cycle regulator through modulating G2/M checkpoint and interaction with checkpoint kinase 1 in gastrointestinal cancer. Carcinogenesis. 2009;30(9):1507–1516. doi: 10.1093/carcin/bgp155. [DOI] [PubMed] [Google Scholar]
- 34.Wherry EJ, et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27(4):670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 35.Akgul C. Mcl-1 is a potential therapeutic target in multiple types of cancer. Cell Mol Life Sci. 2009;66(8):1326–1336. doi: 10.1007/s00018-008-8637-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hallaert DY, et al. Crosstalk among Bcl-2 family members in B-CLL: seliciclib acts via the Mcl-1/Noxa axis and gradual exhaustion of Bcl-2 protection. Cell Death Differ. 2007;14(11):1958–1967. doi: 10.1038/sj.cdd.4402211. [DOI] [PubMed] [Google Scholar]
- 37.Kallies A, Xin A, Belz GT, Nutt SL. Blimp-1 transcription factor is required for the differentiation of effector CD8(+) T cells and memory responses. Immunity. 2009;31(2):283–295. doi: 10.1016/j.immuni.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 38.Rutishauser RL, et al. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 2009;31(2):296–308. doi: 10.1016/j.immuni.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shin H, et al. A role for the transcriptional repressor Blimp-1 in CD8(+) T cell exhaustion during chronic viral infection. Immunity. 2009;31(2):309–320. doi: 10.1016/j.immuni.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kallies A, et al. Transcriptional repressor Blimp-1 is essential for T cell homeostasis and self-tolerance. Nat Immunol. 2006;7(5):466–474. doi: 10.1038/ni1321. [DOI] [PubMed] [Google Scholar]
- 41.Martins GA, et al. Transcriptional repressor Blimp-1 regulates T cell homeostasis and function. Nat Immunol. 2006;7(5):457–465. doi: 10.1038/ni1320. [DOI] [PubMed] [Google Scholar]
- 42.Pearce EL, et al. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302(5647):1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 43.Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, Glimcher LH. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science. 2002;295(5553):338–342. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- 44.Glimcher LH, Townsend MJ, Sullivan BM, Lord GM. Recent developments in the transcriptional regulation of cytolytic effector cells. Nat Rev Immunol. 2004;4(11):900–911. doi: 10.1038/nri1490. [DOI] [PubMed] [Google Scholar]
- 45.Intlekofer AM, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol. 2005;6(12):1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 46.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100(6):655–669. doi: 10.1016/S0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 47.de Bree GJ, van Leeuwen EM, Out TA, Jansen HM, Jonkers RE, van Lier RA. Selective accumulation of differentiated CD8+ T cells specific for respiratory viruses in the human lung. J Exp Med. 2005;202(10):1433–1442. doi: 10.1084/jem.20051365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Joshi NS, et al. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27(2):281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takemoto N, Intlekofer AM, Northrup JT, Wherry EJ, Reiner SL. Cutting Edge: IL-12 inversely regulates T-bet and eomesodermin expression during pathogen-induced CD8+ T cell differentiation. J Immunol. 2006;177(11):7515–7519. doi: 10.4049/jimmunol.177.11.7515. [DOI] [PubMed] [Google Scholar]
- 50.Hamilton SE, Jameson SC. CD8(+) T cell differentiation: choosing a path through T-bet. Immunity. 2007;27(2):180–182. doi: 10.1016/j.immuni.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 51.Martins G, Calame K. Regulation and functions of Blimp-1 in T and B lymphocytes. Annu Rev Immunol. 2008;26:133–169. doi: 10.1146/annurev.immunol.26.021607.090241. [DOI] [PubMed] [Google Scholar]
- 52.Muto A, et al. The transcriptional programme of antibody class switching involves the repressor Bach2. Nature. 2004;429(6991):566–571. doi: 10.1038/nature02596. [DOI] [PubMed] [Google Scholar]
- 53.Shao Z, et al. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98(1):37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- 54.Pile LA, Schlag EM, Wassarman DA. The SIN3/RPD3 deacetylase complex is essential for G(2) phase cell cycle progression and regulation of SMRTER corepressor levels. Mol Cell Biol. 2002;22(14):4965–4976. doi: 10.1128/MCB.22.14.4965-4976.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sichtig N, Korfer N, Steger G. Papillomavirus binding factor binds to SAP30 and represses transcription via recruitment of the HDAC1 co-repressor complex. Arch Biochem Biophys. 2007;467(1):67–75. doi: 10.1016/j.abb.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 56.Hurlstone A, Clevers H. T-cell factors: turn-ons and turn-offs. EMBO J. 2002;21(10):2303–2311. doi: 10.1093/emboj/21.10.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Picker LJ, Treer JR, Ferguson-Darnell B, Collins PA, Buck D, Terstappen LW. Control of lymphocyte recirculation in man. I. Differential regulation of the peripheral lymph node homing receptor L-selectin on T cells during the virgin to memory cell transition. J Immunol. 1993;150(3):1105–1121. [PubMed] [Google Scholar]
- 58.Chao CC, Jensen R, Dailey MO. Mechanisms of L-selectin regulation by activated T cells. J Immunol. 1997;159(4):1686–1694. [PubMed] [Google Scholar]
- 59.Park JH, et al. Suppression of IL7Ralpha transcription by IL-7 and other prosurvival cytokines: a novel mechanism for maximizing IL-7-dependent T cell survival. Immunity. 2004;21(2):289–302. doi: 10.1016/j.immuni.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 60.van Leeuwen EM, et al. IL-7 receptor alpha chain expression distinguishes functional subsets of virus-specific human CD8+ T cells. Blood. 2005;106(6):2091–2098. doi: 10.1182/blood-2005-02-0449. [DOI] [PubMed] [Google Scholar]
- 61.Gamadia LE, et al. The size and phenotype of virus-specific T cell populations is determined by repetitive antigenic stimulation and environmental cytokines. J Immunol. 2004;172(10):6107–6114. doi: 10.4049/jimmunol.172.10.6107. [DOI] [PubMed] [Google Scholar]
- 62.Loenen WA, de Vries E, Gravestein LA, Hintzen RQ, van Lier RA, Borst J. The CD27 membrane receptor, a lymphocyte-specific member of the nerve growth factor receptor family, gives rise to a soluble form by protein processing that does not involve receptor endocytosis. Eur J Immunol. 1992;22(2):447–455. doi: 10.1002/eji.1830220224. [DOI] [PubMed] [Google Scholar]
- 63.Petrovas C, et al. PD-1 is a regulator of virus-specific CD8+ T cell survival in HIV infection. J Exp Med. 2006;203(10):2281–2292. doi: 10.1084/jem.20061496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Leeuwen EM, Gamadia LE, Baars PA, Remmerswaal EB, ten Berge I, van Lier RA. Proliferation requirements of cytomegalovirus-specific, effector-type human CD8+ T cells. J Immunol. 2002;169(10):5838–5843. doi: 10.4049/jimmunol.169.10.5838. [DOI] [PubMed] [Google Scholar]
- 65.Kim SH, Han SY, Azam T, Yoon DY, Dinarello CA. Interleukin-32: a cytokine and inducer of TNFalpha. Immunity. 2005;22(1):131–142. doi: 10.1016/j.immuni.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 66.DeVico AL, Gallo RC. Control of HIV-1 infection by soluble factors of the immune response. Nat Rev Microbiol. 2004;2(5):401–413. doi: 10.1038/nrmicro878. [DOI] [PubMed] [Google Scholar]
- 67.Hamann D, et al. Phenotypic and functional separation of memory and effector human CD8+ T cells. J Exp Med. 1997;186(9):1407–1418. doi: 10.1084/jem.186.9.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.LeCouter J, Zlot C, Tejada M, Peale F, Ferrara N. Bv8 and endocrine gland-derived vascular endothelial growth factor stimulate hematopoiesis and hematopoietic cell mobilization. Proc Natl Acad Sci U S A. 2004;101(48):16813–16818. doi: 10.1073/pnas.0407697101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Negri L, Lattanzi R, Giannini E, Melchiorri P. Bv8/Prokineticin proteins and their receptors. Life Sci. 2007;81(14):1103–1116. doi: 10.1016/j.lfs.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 70.Martucci C, et al. Bv8, the amphibian homologue of the mammalian prokineticins, induces a proinflammatory phenotype of mouse macrophages. Br J Pharmacol. 2006;147(2):225–234. doi: 10.1038/sj.bjp.0706467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jia W, Li H, He YW. The extracellular matrix protein mindin serves as an integrin ligand and is critical for inflammatory cell recruitment. Blood. 2005;106(12):3854–3859. doi: 10.1182/blood-2005-04-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li H, Oliver T, Jia W, He YW. Efficient dendritic cell priming of T lymphocytes depends on the extracellular matrix protein mindin. EMBO J. 2006;25(17):4097–4107. doi: 10.1038/sj.emboj.7601289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wallace DL, et al. Direct measurement of T cell subset kinetics in vivo in elderly men and women. J Immunol. 2004;173(3):1787–1794. doi: 10.4049/jimmunol.173.3.1787. [DOI] [PubMed] [Google Scholar]
- 74.Chowdhury D, Lieberman J. Death by a thousand cuts: granzyme pathways of programmed cell death. Annu Rev Immunol. 2008;26:389–420. doi: 10.1146/annurev.immunol.26.021607.090404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sower LE, Klimpel GR, Hanna W, Froelich CJ. Extracellular activities of human granzymes. I. Granzyme A induces IL6 and IL8 production in fibroblast and epithelial cell lines. Cell Immunol. 1996;171(1):159–163. doi: 10.1006/cimm.1996.0187. [DOI] [PubMed] [Google Scholar]
- 76.Wills MR, et al. Human virus-specific CD8+ CTL clones revert from CD45ROhigh to CD45RAhigh in vivo: CD45RAhighCD8+ T cells comprise both naive and memory cells. J Immunol. 1999;162(12):7080–7087. [PubMed] [Google Scholar]
- 77.Fehniger TA, et al. Acquisition of murine NK cell cytotoxicity requires the translation of a pre-existing pool of granzyme B and perforin mRNAs. Immunity. 2007;26(6):798–811. doi: 10.1016/j.immuni.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 78.van Lier RA, et al. Tissue distribution and biochemical and functional properties of Tp55 (CD27), a novel T cell differentiation antigen. J Immunol. 1987;139(5):1589–1596. [PubMed] [Google Scholar]
- 79.van Stijn A, et al. Human cytomegalovirus infection induces a rapid and sustained change in the expression of NK cell receptors on CD8+ T cells. J Immunol. 2008;180(7):4550–4560. doi: 10.4049/jimmunol.180.7.4550. [DOI] [PubMed] [Google Scholar]
- 80.Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003;339(1):62–66. doi: 10.1016/S0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 81.van der Kolk LE, et al. CD20-induced B cell death can bypass mitochondria and caspase activation. Leukemia. 2002;16(9):1735–1744. doi: 10.1038/sj.leu.2402559. [DOI] [PubMed] [Google Scholar]
- 82.Hulsen T, de Vlieg J, Alkema W. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.