Abstract
Caspase-9 is involved in the intrinsic apoptotic pathway and suggested to play a role as a tumor suppressor. Little is known about the mechanisms governing caspase-9 expression, but post-transcriptional pre-mRNA processing generates 2 splice variants from the caspase-9 gene, pro-apoptotic caspase-9a and anti-apoptotic caspase-9b. Here we demonstrate that the ratio of caspase-9 splice variants is dysregulated in non–small cell lung cancer (NSCLC) tumors. Mechanistic analysis revealed that an exonic splicing silencer (ESS) regulated caspase-9 pre-mRNA processing in NSCLC cells. Heterogeneous nuclear ribonucleoprotein L (hnRNP L) interacted with this ESS, and downregulation of hnRNP L expression induced an increase in the caspase-9a/9b ratio. Although expression of hnRNP L lowered the caspase-9a/9b ratio in NSCLC cells, expression of hnRNP L produced the opposite effect in non-transformed cells, suggesting a post-translational modification specific for NSCLC cells. Indeed, Ser52 was identified as a critical modification regulating the caspase-9a/9b ratio. Importantly, in a mouse xenograft model, downregulation of hnRNP L in NSCLC cells induced a complete loss of tumorigenic capacity that was due to the changes in caspase-9 pre-mRNA processing. This study therefore identifies a cancer-specific mechanism of hnRNP L phosphorylation and subsequent lowering of the caspase-9a/9b ratio, which is required for the tumorigenic capacity of NSCLC cells.
Introduction
Caspase-9 is a critical factor in the apoptotic pathway induced by various chemotherapies, stress agents, and radiation (1–9) and is implicated as a tumor suppressor (10). Although caspase-9 has roles in apoptosis and cellular transformation, little is known about the mechanisms regulating caspase-9 expression. Reports in the literature demonstrate that one aspect regulating the expression of caspase-9 is the post-transcriptional mechanism of alternative splicing (5, 6). In this regard, the caspase-9 gene produces 2 splice variants, caspase-9a and caspase-9b, via inclusion or exclusion of a 4-exon cassette (exons 3, 4, 5, and 6). Inclusion of this cassette into the mature transcript produces the pro-apoptotic caspase-9a, while the exclusion produces the anti-apoptotic caspase-9b (5, 6). The caspase-9b protein lacks the catalytic domain while retaining all other amino acid sequences such as the APAF-1 association region (5, 6). Caspase-9b inhibits activation of caspase-9a by competing with full-length caspase-9a for binding to the apoptosome, a large multiprotein structure formed in the process of apoptosis and responsible for activating caspase-9a. Caspase-9b has also been surmised to heterodimerize with caspase-9a, thus rendering the protein structurally incapable of auto-proteolysis (5). In essence, the regulation of the inclusion of this 4-exon cassette may be a critical factor in determining whether a cell is susceptible or resistant to apoptosis (5, 6, 11, 12).
The role of alternative RNA splicing in cancer phenotypes is largely understudied, but several recent reports have brought to light the possibility that RNA splicing factors can act as oncogenic factors. For example, Cheng and Sharp recently demonstrated that the RNA trans-factor SRm160 played a prominent role in regulating the alternative splicing of CD44 stimulated by H-rasV12 with a biological role in the anchorage-independent growth (AIG) ability of cancer cells (13). In another report, Krainer and colleagues demonstrated that overexpression of SRp30a acted as an oncogene cooperating to enhance the tumorigenic capacity of NIH 3T3 cells in nude mice (14). More recently, Carstens and colleagues found that the RNA trans-factors ESRP1 and ESRP2 are regulators of FGFR2 splicing. Importantly, when these regulators are downregulated in cells that express FGFR2-IIIb, a switch from an epithelial to a mesenchymal phenotype occurs (15). These recent reports show that alternative splicing plays a prominent role in the oncogenic potential of cancer cells.
In this study, our laboratory identified a purine-rich sequence within exon 3 of the caspase-9 pre-mRNA that acts as an exonic splicing silencer (ESS) (C9/E3-ESS). Heterogeneous nuclear ribonucleoprotein L (hnRNP L) was identified as the RNA trans-factor associated with this RNA cis-element. Further studies demonstrated that the phosphorylation state of hnRNP L is higher in non–small cell lung cancer (NSCLC) cells than in non-transformed lung epithelial cells, and phosphorylated hnRNP L modulates the alternative splicing of caspase-9. Our studies also show that hnRNP L regulated the tumorigenic capacity of NSCLC cells via post-transcriptional processing of caspase-9 pre-mRNA. Therefore, this study identifies the regulation of the phosphorylation state of hnRNP L and subsequent post-transcriptional processing of caspase-9 as key cellular mechanisms for the growth and maintenance of NSCLC tumors.
Results
The alternative splicing of caspase-9 is dysregulated in NSCLC tumors and cell lines.
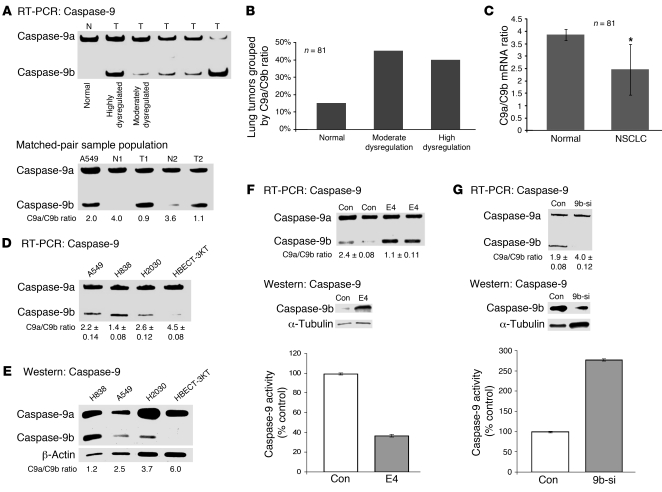
In this study, the ratio of caspase-9a/9b mRNA was compared between human lung tumor samples and normal human lung tissue (Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI43552DS1). Tumor samples were subdivided into 3 groups: normal, with a caspase-9a/9b mRNA ratio of at least 3.5; moderately dysregulated, with a 2.3–3.4 ratio of caspase-9a/9b mRNA; and highly dysregulated, with a caspase-9a/9b mRNA ratio of 2.2 or less (Figure 1, A and B). The normal group corresponded to the normal ratio of caspase-9a/9b mRNA observed in non-transformed cells; the moderately dysregulated group corresponded to a ratio of caspase-9a/9b reported to have a minor to moderate effect on caspase-9 activity (5, 6); and the highly dysregulated group corresponded to a ratio of caspase-9a/9b reported to significantly reduce caspase-9 activity and inhibit the association of caspase-9a with APAF-1 (5, 6). Quantitative/competitive RT-PCR analysis (16) of caspase-9 splice variants (17) showed that 40% of the NSCLC tumors examined (n = 81) presented with a highly dysregulated ratio of caspase-9a/9b mRNA compared with normal lung tissue. Only 15% of the lung tumor samples demonstrated a normal ratio of caspase-9a/9b mRNA, and 45% of the lung tumor samples demonstrated a moderately dysregulated ratio of caspase-9a/9b mRNA. The entirety of the human lung tumors were also compared directly with normal lung tissue, and demonstrated a reduction of 36% in the caspase-9a/9b mRNA ratio in human lung tumors (Figure 1C). Thus, the ratio of caspase-9a/9b mRNA was dramatically lower in a high percentage of clinically relevant NSCLC tumors, correlating with loss of activity of caspase-9a and favoring a pro-survival/apoptotic resistance phenotype.
Figure 1. Caspase-9a/9b ratio is dysregulated in lung adenocarcinoma tumors and transformed lung epithelial cells.
(A) Top: A sample population of cDNAs from pathologist-verified lung adenocarcinomas, which underwent quantitative/competitive PCR for caspase-9 splice variants. Bottom: The figure depicts a sample of the matched pair analysis. N, normal tissue; T, tumor tissue. (B) The percentage of lung tumors grouped by caspase-9a/9b mRNA ratio. (C) Human lung tumor samples directly compared with normal lung tissues. *P < 0.05, Student’s t test. (D and E) The caspase-9 splicing ratio observed in non-transformed HBEC-3KT cells versus NSCLC cell lines at the RNA (D) and protein level (E). Data are n = 3 on 2 separate occasions. (F and G) Top: Quantitative/competitive RT-PCR for caspase-9 splice variants from A549 cells treated with E4 ASRO (E4) or control (Con) ASRO (F) or control siRNA or siRNA against caspase-9b (9b-si) (G). Protein samples from simultaneous experiments were subjected to SDS-PAGE and Western immunoblotted (caspase-9b, α-tubulin). Bottom: Caspase-9 activity (LEHD) in the lysates of A549 cells treated with control ASRO and E4 ASRO. Caspase-9 activity (LEHD) was calculated as relative fluorescent units (RFU) per total protein (mg) and is represented as a percentage of control cells. The data presented in F and G are expressed as mean ± SEM and are n = 4 on 3 separate occasions.
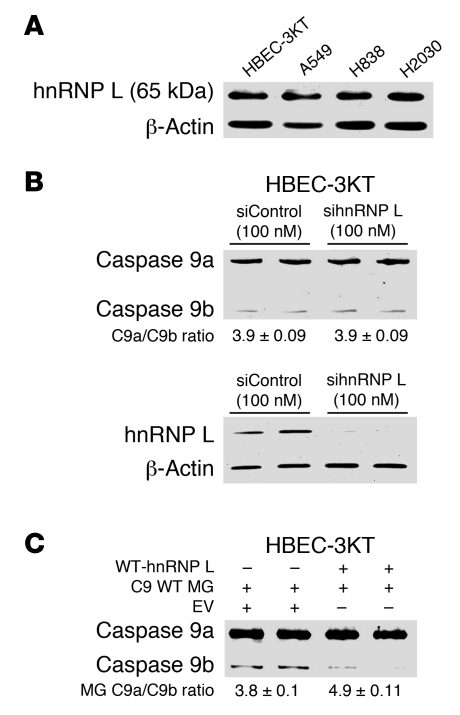
Although all of our samples were more than 50% NSCLC tumors, contamination of other cell types may have affected results. Therefore, we examined a pure population of non-transformed lung epithelial cells (specifically, HBEC-3KT cells) for their caspase-9a/9b ratio in comparison with the transformed lung adenocarcinoma cell lines A549, H838, and H2030 (see Supplemental Table 2 for cell line phenotypes). HBEC-3KT cells demonstrated a “normal” ratio of caspase-9a/9b mRNA of 4.5 ± 0.08 (Figure 1D). In comparison to the non-transformed cell line, all 3 adenocarcinoma cells lines (A549, H838, and H2030) demonstrated a dysregulated ratio of caspase-9a/9b mRNA: A549, 2.2 ± 0.14; H838, 1.4 ± 0.08; H2030, 2.6 ± 0.12 (Figure 1D). Importantly, the dysregulation in the ratio of caspase-9a/9b mRNA translated to the protein level (Figure 1E). These observations indicate that the alternative splicing of caspase-9 is dysregulated in lung adenocarcinoma cell lines as well as in NSCLC tumors.
To verify that the observed differences in caspase-9a/9b mRNA ratio translated to a significant effect on caspase-9 activity, siRNA and anti-sense RNA oligonucleotide (ASRO) technology was employed to directly manipulate the alternative splicing of caspase-9. Specifically, ASROs were designed to hybridize to the 5′ splice site (5′SS) of exon 3 (E3 ASRO), exon 4 (E4 ASRO), and exon 6 (E6 ASRO) in caspase-9 pre-mRNA to reduce the ratio of caspase-9a/9b mRNA as previously reported for other splicing events (18–20). The E4 ASRO effectively reduced the ratio of caspase-9a/9b mRNA in cells (Figure 1F), which translated to the protein level in comparison with control ASRO. ASROs targeting the 5′SS of exon 3 and exon 6 also significantly inhibited the inclusion of the exon 3,4,5,6 cassette into the mature mRNA, but the E4 ASRO was the most effective ASRO (data not shown). These results suggest that all exons within the exonic cassette require appropriate definition for caspase-9a mRNA to be produced, or the default exclusion of the exonic cassette, and subsequent production of caspase-9b, will occur.
To examine the functional translation of directly manipulating the alternative splicing of caspase-9, total caspase-9 activity was examined with E4 ASRO in comparison with the control ASRO. Figure 1F demonstrates that the total caspase-9 activity (LEHD activity) was lowered approximately 62% by the E4 ASRO, corresponding to a 46% reduction in the caspase-9a/9b mRNA ratio as compared with the control ASRO. Thus, the E4 ASRO directly manipulated the alternative splicing of caspase-9 to produce caspase-9b at the expense of caspase-9a, which significantly lowered caspase-9 activity in the cell. In order to specifically reduce caspase-9b levels, RNAi technology was employed to downregulate caspase-9b (siRNA against caspase-9b [9b-si]) versus caspase-9a. Treatment with 9b-si in comparison with control siRNA significantly increased the ratio of caspase-9a/9b mRNA in A549 cells producing a caspase-9a/9b mRNA ratio comparable to HBEC-3KT cells, which was conferred to the protein level (Figure 1G). Lastly, caspase-9 activity was assessed in A549 cells after 9b-si transfection in comparison with control siRNA, which demonstrated an approximate 2.7-fold increase in the caspase-9 activity (LEHD activity) (Figure 1G). The culmination of these data demonstrate that our observed changes in the caspase-9a/9b mRNA ratio in NSCLC cells and tumors correlate with a significant effect on the activity of caspase-9 and are thus physiologically relevant.
The alternative splicing of caspase-9 regulates the anchorage-independent growth and tumorigenic capacity of NSCLC cells.
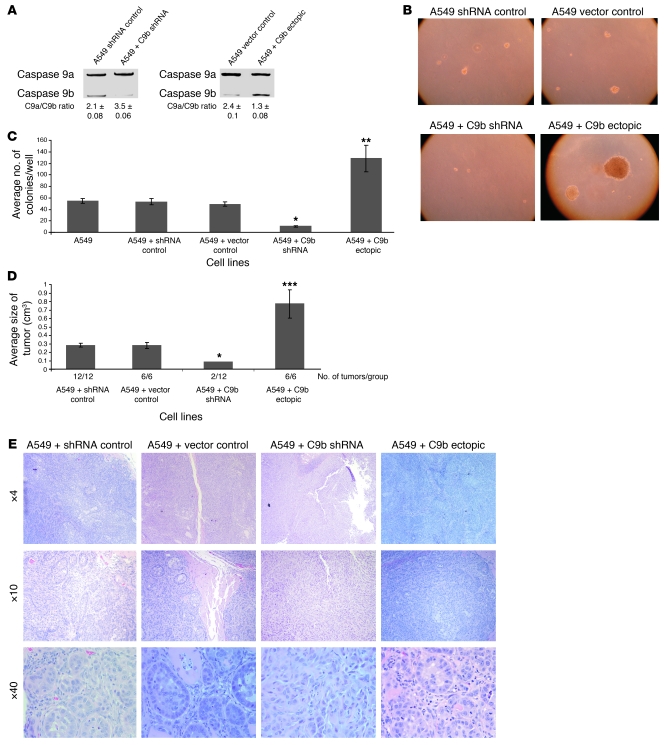
Based on the above findings and the knowledge that caspase-9a is a possible tumor suppressor (10), although controversial (21), we hypothesized that the alternative splicing of caspase-9 has a major role in maintaining the transformed phenotype of NSCLC cells. To test this hypothesis, multiple clones, and batch cultures of A549 cells stably expressing either caspase-9b cDNA or caspase-9b shRNA were produced (Figure 2A and Supplemental Table 3). Stable expression of caspase-9b shRNA essentially abolished the ability of A549 cells to grow in soft agar and reduced the mean diameter of cellular colonies (Figure 2, B and C, and Supplemental Figure 1). In contrast, low ectopic expression of caspase-9b significantly increased the ability of A549 cells to grow in soft agar (Figure 2, B and C). These effects did not require stable expression and did not result from integration artifacts, as short-term expression of caspase-9b shRNA and caspase-9b cDNA or use of stable batch cell lines produced comparable results (data not shown).
Figure 2. Caspase-9 regulates the tumorigenic capacity in A549 cells.
(A) Total RNA was isolated from the listed clonal cell lines and analyzed by competitive/quantitative RT-PCR for caspase-9 splice variants. (B) Cells (2 × 103) were plated into 6-well dishes in soft agar and cultured for 14 days before the colony count. (C) Quantization (mean) of the number of colonies for the indicated clonal cell lines. n = 6. Data are mean ± SEM. *P < 0.005, A549 + shRNA control versus A549 + C9b shRNA; **P < 0.001, A549 + vector control versus A549 + C9b ectopic; Student’s t test. (D) Cell lines in A were injected into SCID mice (5 × 106), and tumor volume was measured at the end of 28 days as represented as average size of tumor (cm3). Data are mean ± SEM. *P < 0.05, A549 + control shRNA versus A549 + C9b shRNA; ***P < 0.005, A549 + vector control versus A549 + C9b ectopic Student’s t test. (E) H&E stain of the tumors presented in D with magnification ranging from ×4 to ×40.
These observations translated to other NSCLC cell lines, as stable expression of caspase-9b shRNA in H2030 and H838 cell lines also induced a dramatic loss in the ability of these cells to grow in soft agar (Supplemental Figure 2). In contrast, low ectopic expression of caspase-9b demonstrated a marked increase in the ability of H838 and H2030 cells to grow in soft agar (Supplemental Figure 2). Furthermore, the clonogenic capacity in the formation of stable expression clones of caspase-9 shRNA was decreased by a factor of 10 compared with control shRNA expression using the same viral MOI (data not shown).
The tumorigenic capacity of these A549 cell lines was also characterized using human A549 xenograph tumor formation in SCID mice. Figure 2D shows that stable downregulation of caspase-9b by shRNA technology also induced a complete loss of tumor formation in 10 of 12 injections. The remaining tumors that formed in the caspase-9b shRNA–expressing cells demonstrated a dramatic loss in tumor volume (Figure 2D). Pathological analysis confirmed the complete loss of tumor formation for the caspase-9b shRNA experiments. In contrast, low ectopic expression of caspase-9b induced a large increase in tumor volume (Figure 2D). Interestingly, a different pathological phenotype was observed in the 2 minor tumors formed from cell lines expressing caspase-9b shRNA (Figure 2E). Specifically, caspase-9b shRNA xenographs demonstrated a greater quantity of matrix compared with the A549 vector control xenographs and absence of gland-like structures and invasion into the adjacent tissue. Furthermore, the caspase-9b shRNA tumors formed from the caspase-9b shRNA cells lost major architectural features of adenocarcinomas, and an inflammatory response was also observed. Large quantities of neutrophils were also present, along with visible apoptotic bodies, but there was a distinct absence of necrosis. Lastly, the effect of the downregulation of caspase-9b on AIG and tumorigenic capacity was not due to the enhancement of spontaneous apoptosis rates, as no significant effect on these rates in the A549 caspase-9b shRNA cells was observed compared with vector control cells (data not shown). Overall, these data demonstrate that the loss of caspase-9b dramatically alters the tumorigenic capacity of NSCLC cells.
Identification of a purine-rich exonic splicing silencer in exon 3 of caspase-9 pre-mRNA.
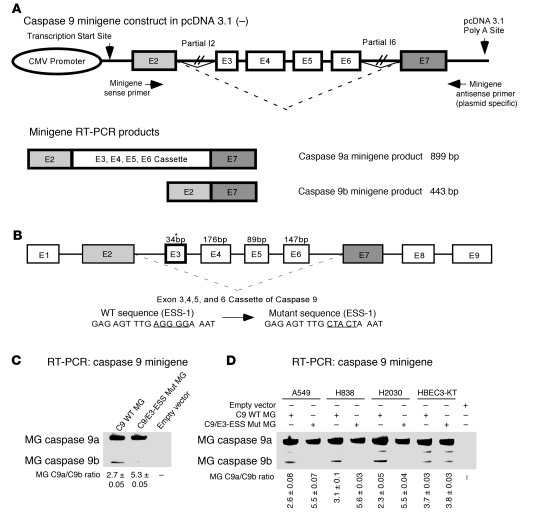
Given the biological findings, our laboratory examined the mechanistic regulation of the pre-mRNA processing of caspase-9. In an effort to identify exonic splicing enhancers (ESEs) and ESSs, our laboratory constructed a functional minigene to assay the inclusion or exclusion of the exon 3,4,5,6 cassette of caspase-9 in cells. This was accomplished by ligation of 3 DNA fragments of the caspase-9 gene into pcDNA 3.1–. The 3 fragments included a 603-bp fragment containing part of exon 2 and intron 2; a 3,962-bp fragment containing part of intron II, all of exons 3,4,5,6 and their intervening intronic sequences (all of the sequence for introns 3, 4, and 5), and part of intron 6; and a 431-bp DNA fragment containing part of intron 6 and part of exon 7 (Figure 3A).
Figure 3. Identification of an exonic splicing silencer in exon 3 of caspase-9 pre-mRNA.
(A) Schematic of the functional minigene construct for caspase-9 constructed in pcDNA 3.1– zeocin with arrows indicating the location of primers used in the RT-PCR assay. (B) Schematic representation of the exonic splicing silencer (C9/E3-ESS) located in exon 3 of the caspase-9 gene. Asterisk indicates the location of the C9/E3-ESS purine-rich sequence. This figure also depicts the wild-type and mutagenic sequence of the C9/E3-ESS utilized in C and D. (C and D) A549, H838, H2030, and HBEC-3KT cells were transfected with the pcDNA 3.1– zeocin plasmid containing the caspase-9 minigene (C9 WT MG) (1 μg), exon 3 mutant minigene (E3 Mut MG) (1 μg), or empty vector control (1 μg) for 24 hours. Total RNA was extracted and analyzed by competitive/quantitative RT-PCR for the ratio of minigene caspase-9a/9b mRNA. Data are n = 4 from 2 separate occasions.
Within the exon 3,4,5,6 cassette, 10 purine-rich sequences were identified as possible ESEs, intronic splicing enhancers, ESSs, or intronic splicing silencers by (a) screening the exon 3,4,5,6 cassette for known RNA cis-elements (e.g., purine-rich regions), (b) using ESE finder 2.0 (22), and (c) using Splicing Rainbow (23) (Supplemental Figure 3). Replacement mutagenesis of these sequences identified a particular purine-rich sequence in exon 3 (Figure 3B and Supplemental Figure 3) that acted as an ESS. Mutation of this purine-rich sequence in exon 3, AGGGGA, resulted in a significant increase of the caspase-9a/9b mRNA ratio in A549 cells from 2.7 ± 0.05 to 5.3 ± 0.05 (P < 0.0002) (Figure 3C). Of the 10 purine-rich sequences mutated in the caspase-9 minigene, only the mutagenesis of this particular sequence had the effect of increasing the ratio of caspase-9a/9b mRNA. Mutation of this ESS also induced an increase in the ratio of caspase-9a/9b minigene mRNA in H2030 and H838 cells, but interestingly no effect on the C9a/C9b ratio was observed in non-transformed HBEC3-KT cells (Figure 3D). Therefore, the purine-rich sequence, AGGGGA, in exon 3 of caspase-9 pre-mRNA acts as an ESS for the inclusion or exclusion of the exon 3,4,5,6 cassette in transformed NSCLC cells and is referred to throughout as caspase-9 exon 3 ESS (C9/E3-ESS).
hnRNP L binds specifically to the exonic splicing silencer in exon 3 of caspase-9 pre-mRNA.
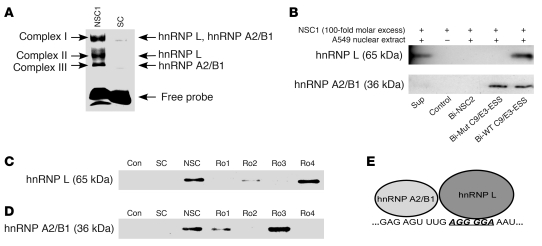
To study the RNA trans-factors interacting with C9/E3-ESS, a fluorescein-tagged RNA oligonucleotide (RO) was produced corresponding to the purine-rich regions of exon 3, FL-5′ GAGAGUUUGAGGGGAAAU. EMSAs coupled with competitor studies demonstrated 3 specific proteins, RNA complexes associated with this sequence designated complex I, II, and III (Figure 4A). To identify the RNA trans-factors bound to the C9/E3-ESS, nanospray LC-MS/MS analysis was employed on the 3 complexes. Besides the normal contaminants such as keratin and HSP70, 2 RNA trans-factors were identified in the 3 RNA/protein complexes, both with x-corr values over 20. Specifically, hnRNP A2/B1 (36 kDa) was identified in complexes I and III, while hnRNP L (65 kDa) was identified in complexes II and III (Figure 4A).
Figure 4. hnRNP L binds specifically to the exon splicing silencer in exon 3 of caspase-9.
(A) A 5′ FITC-tagged RO corresponding to the purine-rich sequences in exon 3 of caspase-9 was incubated in the presence of nuclear extract from A549 cells or IgG (control) and subjected to EMSAs. NSC1 or SC ROs were also added (100-fold molar excess) as indicated. Arrows indicate the 3 specific RNA-protein complexes. The RNA-protein complexes (complexes I, II, and III) were also subjected to nano-LC-MS/MS analysis. The RNA trans-factors depicted obtained x-corr values greater than 20. (B) A 5′ biotinylated wild-type C9/E3-ESS RO (Bio-C9/E3-ESS), 5′ biotinylated mutant ROs (Bio-Mut C9/E3-ESS), or a 5′ biotinylated nonspecific RO (Bio-NSC2) were incubated in the presence of nuclear extract from A549 cells or IgG (control), subjected to SDS-PAGE and Western immunoblotting analysis (anti-hnRNP L antibody; anti-hnRNP A2/B1 antibody). Unlabeled nonspecific ROs (e.g., NSC1) at a 100-fold molar excess were also added to the reactions as indicated. The corresponding supernatant from the Bio-WT C9/E3-ESS (Sup) shows the remaining RNA trans-factor after affinity purification. (C and D) The experiments in B were repeated, but with the addition (100-fold molar excess) of either unlabeled NSC1, unlabeled competitor ROs (RO1–RO4) or unlabeled SC, as indicated. Data in Figure 4 are n = 4 on 2 separate occasions. (E) A cartoon schematic indicating the locations of the hnRNP L and hnRNP A2/B1 interactions with exon 3 of caspase-9 pre-mRNA.
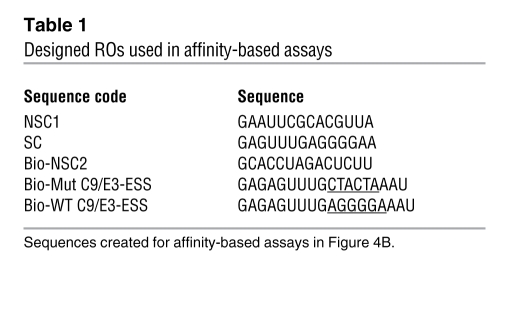
The specific interaction of these RNA trans-factors with exon 3 was verified by performing a biotin/streptavidin affinity assay using A549 nuclear extracts. Biotinylated ROs corresponding to the purine-rich regions of exon 3 were utilized for this affinity assay (Table 1): biotin-5′ GAGAGUUUGAGGGGAAAU (Bio-WT C9/E3-ESS), a mutant RO for the C9/E3-ESS; biotin-5′ GAGAGUUUGCTACTAAAU (Bio-C9/E3-ESS-mut); and a biotin-5′ non-specific control sequence (Bio-NSC2). Analysis verified that RNA trans-factors hnRNP L and A2/B1 specifically interacted with the purine-rich regions in exon 3 of caspase-9 (Figure 4B), as the non-specific RNA sequence (Bio-NSC2) did not show any interaction with these RNA trans-factors. More importantly, the Bio-C9/E3-ESS-mut, which possesses the same mutations utilized in the minigene assays in Figure 3, did not significantly interact with hnRNP L, in contrast to hnRNP A2/B1. Thus, both hnRNP L and hnRNP A2/B1 specifically interact with the purine-rich regions exon 3 of caspase-9 pre-mRNA, but only hnRNP L interacts specifically with the C9/E3-ESS sequence.
Table 1 .
Designed ROs used in affinity-based assays
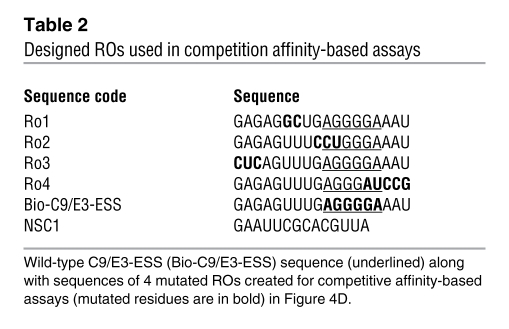
To determine where hnRNP A2/B1 specifically bound in exon 3 (e.g., AGGGGA) as well as identify other important nucleic acid residues for the hnRNP L interaction, ROs were designed that possessed mutations in various sites within the purine-rich regions of exon 3 (Table 2). In particular, RO4 and RO2 possessed similar and flanking mutations compared with the “core” mutations utilized to identify C9/E3-ESS using the caspase-9 minigene shown in Figure 3. The competition affinity-based assays used to obtain the results shown in Figure 4C were repeated using each mutant RO as an unlabeled “cold” competitor, in comparison with cold nonspecific competitor (NSC) and specific competitor (SC). RO4 was unable to compete for hnRNP L interaction with Bio-C9/E3-ESS (Figure 4C). Furthermore, RO2 was only a partial competitor for the hnRNP L/Bio-C9/E3-ESS interaction. In contrast, the hnRNP A2/B1 interaction with exon 3 was abolished to the same extent by RO4 and RO2 as an unmutated exon 3 SC. In further contrast, RO3 and to an extent RO1 were unable to compete for hnRNP A2/B1 binding in contrast to hnRNP L (Figure 4D). Therefore, hnRNP L specifically associates with C9/E3-ESS in exon 3 as well as flanking nucleotide sequences, whereas hnRNP A2/B1 associates with a purine-rich region 6–11 bp upstream of the previously identified C9/E3-ESS sequence (Figure 4E).
Table 2 .
Designed ROs used in competition affinity-based assays
hnRNP L regulates the pre-mRNA processing of caspase-9 in NSCLC cells.
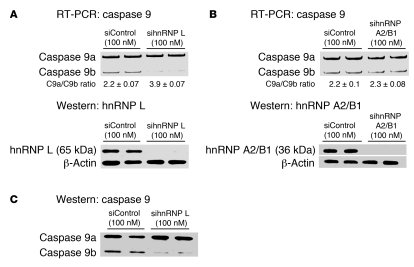
Although hnRNP A2/B1 did not bind specifically to the C9/E3-ESS, compared with hnRNP L, a role for this RNA trans-factor in repressing the inclusion of the exon 3,4,5,6 cassette was still a possibility. siRNA technology was used to examine the role of both hnRNP A2/B1 and hnRNP L in regulating the ratio of caspase-9 splice variants. Downregulation of hnRNP L using a multiplex siRNA (100 nM [25 nM for each sequence]) resulted in a significant increase in the ratio of caspase-9a/9b mRNA from 2.2 ± 0.07 to 3.9 ± 0.07 (P < 0.005; n = 4) (Figure 5A). On the other hand, multiplex siRNA treatment against hnRNP A2/B1 had no significant effect on the ratio of caspase-9a/9b mRNA (Figure 5B). Western immunoblot analysis confirmed a more than 80% downregulation of both hnRNP A2/B1 and hnRNP L compared with samples from cells treated with control siRNA (Figure 5, A and B). Importantly, the effect of hnRNP L siRNA on the caspase-9a/9b mRNA ratio translated to the protein level (Figure 5C).
Figure 5. Downregulation of hnRNP L, but not hnRNP A2/B1, increases the ratio of caspase-9a/9b mRNA.
A549 cells were transfected with control siRNA (100 nM), hnRNP L SMARTpool siRNA (100 nM), or hnRNP A2/B1 SMARTpool siRNA (100 nM) for 48 hours. Total RNA was isolated and analyzed by competitive/quantitative RT-PCR for caspase-9 splice variants. (A and B) hnRNP L siRNA (A) and hnRNP A2/B1 siRNA (B). Simultaneously, total protein lysates were also produced, subjected to SDS-PAGE analysis, and immunoblotted for hnRNP L, hnRNP A2/B1, caspase-9, and β-actin. (C) Results from the same Western blot membrane depicted in A. Data are n = 4 on 2 separate occasions.
The observed effect of hnRNP L downregulation on the caspase-9a/9b mRNA ratio could not be attributed to off-target effects, as multiple siRNA sequences targeted against hnRNP L produced the same result (Supplemental Figure 4), and a dose response curve for the most effective siRNA demonstrated an IC50 of less than 25 nM for hnRNP L siRNA with, again, no effect observed for hnRNP A2/B1 siRNA (Supplemental Figure 5, A and B). Lastly, several stable cell lines were produced from hnRNP L and hnRNP A2/B1 shRNA. All of the hnRNP L shRNA cell lines demonstrated a significant effect on the ratio of caspase-9a/9b mRNA (Supplemental Figure 5C). In contrast, none of the hnRNP A2/B1 shRNA cell lines presented with an increase in the ratio of caspase-9a/9b mRNA (Supplemental Figure 5D). As caspase-9a and 9b are transcribed from the same gene and possess the same 5′UTR and the same 3′UTR sequences, the effect of hnRNP L downregulation is likely attributable to the regulation of the alternative splicing of caspase-9 and not to effects on transcription, translation, or mRNA stability. Specifically, transcriptional activation/repression did not account for the changes in the C9a/9b mRNA ratio, as both were derived from the same promoter and actinomycin D treatment did not affect the caspase-9a/9b mRNA ratio (data not shown). The observed effect of hnRNP L siRNA also cannot be attributed to translation, as the observed changes in the ratio of caspase-9a/9b mRNA were mirrored at the protein level. There is the possibility that changes in mRNA stability played a role in our observations, as hnRNP L has been shown to regulate the stability of specific mRNAs (24, 25), but the half-lives of caspase-9a and -9b mRNA were previously shown by our laboratory to be unaffected by agonists (17). Furthermore, treatment of A549 control shRNA and A549 hnRNP L shRNA cell lines with actinomycin D demonstrated no significant difference in the decay rates (e.g., tν) of caspase-9a and caspase-9b mRNAs in the presence or absence of hnRNP L (data not shown). This finding suggests that hnRNP L does not regulate the mRNA stability of either splice variant, although known caveats in the use of actinomycin D make definitive conclusions difficult. Overall, the data support the conclusion that hnRNP L and not hnRNP A2/B1 acts as a repressor of the inclusion of the exon 3,4,5,6 cassette of caspase-9 via manipulation of the alternative splicing of caspase-9 pre-mRNA.
To determine whether this mechanism demonstrated translatability in NSCLC, hnRNP L siRNA was again used in H838 and H2030 cells. The role of hnRNP L was further corroborated in these NSCLC cell lines, as downregulation of hnRNP L by siRNA produced a concomitant increase in the ratio of caspase-9a/9b mRNA for H838 (2.0 ± 0.13 for siControl, 4.1 ± 0.09 for sihnRNP L) and H2030 (2.1 ± 0.13 for siControl, 3.7 ± 0.22 for sihnRNP L) (Supplemental Figure 6, A and B).
To demonstrate the specificity of hnRNP L in regulating the alternative splicing of caspase-9, other major factors of apoptosis were also examined (e.g., caspase-8, caspase-2, and Bcl-x) using quantitative/competitive RT-PCR. Downregulation of hnRNP L with siRNA demonstrated no effect on the alternative splicing of caspase-8, caspase-2, and Bcl-x (Supplemental Figure 7). Thus, the effect of downregulating hnRNP L is specific for regulating the alternative splicing of caspase-9, and not a generalized effect on pre-mRNA processing.
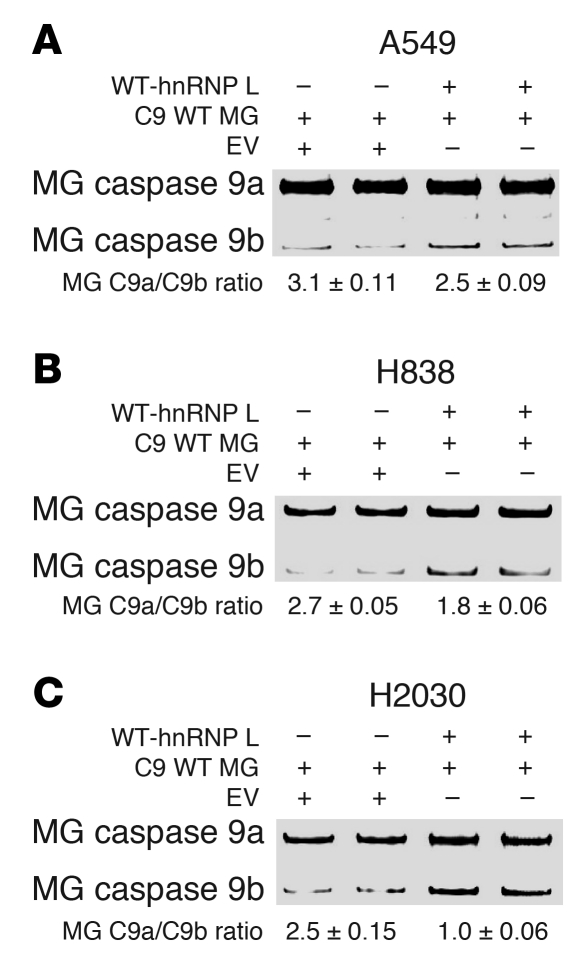
Since siRNA and shRNA can have numerous biological and sequence-specific effects, the reciprocal experiment was also undertaken in which low ectopic expression of hnRNP L in conjunction with the caspase-9 minigene was utilized. Transfection of A549 cells with hnRNP L (low ectopic expression) had the opposite effect of hnRNP L siRNA, which induced a moderate but significant reduction in the ratio of caspase-9a/9b minigene mRNA (Figure 6A). Note that the minigene caspase-9a/9b mRNA ratio was slightly higher in A549 cells (2.7–3.2) compared with the endogenous caspase-9a/9b ratio (1.9–2.2). To demonstrate translatability, H2030 and H838 cells were again utilized, and low ectopic expression of hnRNP L demonstrated the same effect on lowering the ratio of the caspase-9a/9b minigene mRNA (Figure 6, B and C).
Figure 6. Low ectopic expression of hnRNP L decreases the caspase-9a/9b splicing ratio.
(A) A549, (B) H838, and (C) H2030 cell lines were transfected with wild-type hnRNP L (WT-hnRNP L) (0.25 μg) in conjunction with caspase-9 minigene (C9 WT MG) (0.25 μg) or with caspase-9 minigene (C9 WT MG) (0.25 μg) in conjunction with empty vector (EV) (0.25 μg) for 24 hours. Total RNA was extracted and analyzed by competitive/quantitative RT-PCR for caspase-9 minigene splice variants. Data are n = 4 from 2 separate occasions. Note that the ratio of caspase-9a/9b minigene mRNA tended to present with a slightly higher ratio than endogenous caspase-9a/9b mRNA.
hnRNP L regulates the AIG and tumorigenic capacity of NSCLC cells via the pre-mRNA processing of caspase-9.
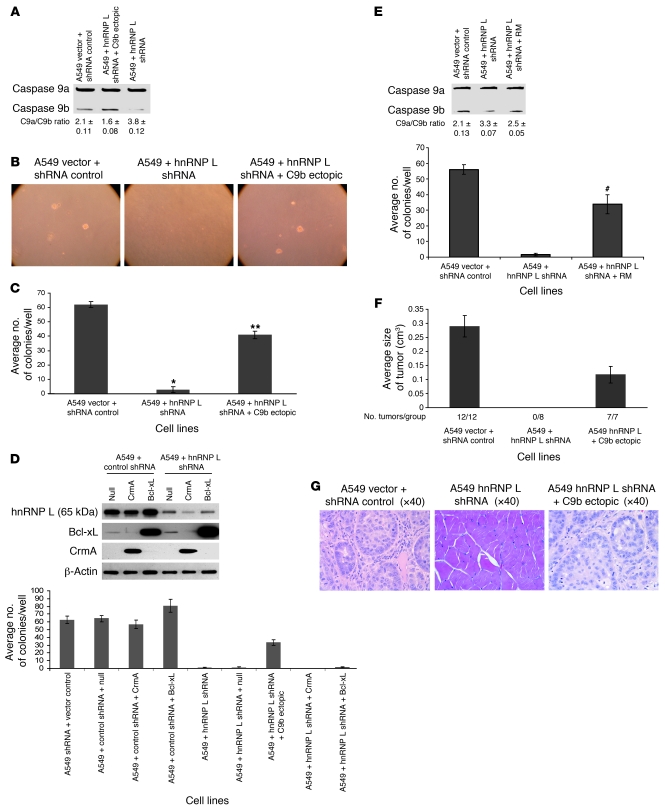
Modulation of the levels of splice variants of caspase-9 had dramatic effects on AIG and the tumorigenic capacity of NSCLC cells. Thus, we hypothesized that hnRNP L plays a major role in maintaining the transformed phenotype of NSCLC cells through modulation of the pre-mRNA processing of caspase-9. To test this hypothesis, multiple clones of A549 cells stably expressing hnRNP L shRNA were produced (Figure 7A and Supplemental Table 3). As with the studies utilizing caspase-9b shRNA cell lines, the first physiologic parameter examined was AIG. Stable expression of hnRNP L shRNA abolished the ability of A549 cells to grow in soft agar (Figure 7, B and C). More importantly, low ectopic expression of caspase-9b cDNA (e.g., the caspase-9a/9b mRNA ratio dropped from 3.8 to 1.6) in the hnRNP L shRNA cell lines rescued the loss of AIG in A549 cells (Figure 7, B and C). These effects did not require stable expression and were not due to integration artifacts, as short-term expression of hnRNP L shRNA or use of stable batch cell lines produced comparable results (data not shown). Furthermore, the effect on AIG induced by downregulation of hnRNP L was specific for the loss of caspase-9b, as ectopic of expression of apoptosis inhibitory factors CrmA and Bcl-x(L) did not rescue the phenotype (Figure 7D). Lastly, the stable re-expression of shRNA-resistant hnRNP L also rescued the loss of AIG induced by the loss of hnRNP L shRNA, demonstrating specificity for the loss of hnRNP L in both the RNA splicing and biological mechanisms (Figure 7E).
Figure 7. hnRNP L regulates the tumorigenic capacity in A549 cells via the alternative splicing of caspase-9.
(A) Total RNA from the designated cell lines was analyzed for caspase-9 splice variants. (B) Colony formation assays in soft agar for the designated cell lines. (C) Quantization of the number of colonies for the indicated clonal cell lines. n = 6; error bars represent SEM. *P < 0.005, **P < 0.001 versus A549 vector and shRNA control; Student’s t test. (D) Top: Total protein from the listed cell lines was subjected to SDS-PAGE analysis and immunoblotted for hnRNP L, Bcl-xL, CrmA, and β-actin. Bottom: Colony formation assays in soft agar for the cell lines shown at top. n = 9, error bars represent SD. A significant effect was observed compared with vector control cells (e.g., P < 0.005, Student’s t test). (E) Top: Total RNA from the listed clonal cell lines and analyzed for caspase-9 splice variants. Bottom: Quantization of the number of colonies for the indicated stable batch culture cell lines. n = 6; error bars represent SEM. #P < 0.05 compared with A549 + hnRNP L shRNA; Student’s t test. RM, resistant mutant. (F) The designated cell lines were injected subcutaneously into SCID mice. Tumor volume was measured at the end of 28 days. Error bars indicate SEM. (G) H&E stain of the tumors presented in D.
These observations translated to the NSCLC cell lines, H838 and H2030 cells, as no stable clones of either cell line were obtained for hnRNP L shRNA, in contrast to control shRNA and null lentivirus. Thus, both H838 and H2030 completely lost clonogenic capacity in the absence of hnRNP L, which correlated in the induction of apoptosis as assayed by PARP cleavage (e.g., caspase activation) (data not shown). Importantly, stable expression of caspase-9b cDNA rescued this phenotype, allowing for stable clones to be obtained for hnRNP L shRNA (data not shown). Thus, the loss of clonogenic capacity by stable expression of hnRNP L shRNA occurred at least in part via loss of caspase-9b expression. Therefore, the biological effects of hnRNP L downregulation and subsequent modulation of the pre-mRNA processing of caspase-9 translates to other NSCLC cell lines.
The tumorigenic capacity of these cell lines was also characterized using A549 xenograph tumor formation in severe combined immunodeficiency mice. Figure 7F shows that stable downregulation of hnRNP L in A549 cells led to a complete loss of tumor formation (0/8 cell injections). Pathological analysis confirmed the complete loss of tumor formation for hnRNP L shRNA cell lines. Comparable to the AIG effects, ectopic expression of caspase-9b completely rescued the loss of tumor formation induced by downregulation of hnRNP L (7/7 xenographs). Furthermore, pathological analysis of the hnRNP L shRNA/caspase-9b cell line demonstrated that ectopic expression of C9b also rescued the classification/grade of the hnRNP L shRNA cell line (Figure 7G). Overall, these data demonstrate that the loss of hnRNP L dramatically alters the tumorigenic capacity of NSCLC cells. Furthermore, these data demonstrate that hnRNP L shRNA predominantly affects the tumorigenic capacity via manipulation of the pre-mRNA processing of caspase-9.
The phosphorylation of hnRNP L on Ser52 regulates the alternative splicing of caspase-9 in NSCLC.
The expression of hnRNP L was not increased in NSCLC cells, and so hnRNP L expression did not correlate with a low ratio of caspase-9a/9b mRNA (Figure 8A). This disparity was further enhanced by the observation that neither downregulation of hnRNP L nor ectopic expression of hnRNP L in immortalized HBEC-3KT cells affected the inclusion/exclusion of the exon 3,4,5,6 cassette of caspase-9 in the same manner as NSCLC cells (Figure 8, B and C).
Figure 8. The role of hnRNP L in regulating the alternative splicing of caspase-9 in non-transformed human bronchial epithelial cells.
(A) Total protein lysates from A549, H838, H2030, and HBEC-3KT cell lines were subjected to SDS-PAGE analysis and immunoblotted for hnRNP L and β-actin. (B) HBEC-3KT cells were transfected with 100 nM control siRNA or 100 nM hnRNP L SMARTpool siRNA for 48 hours. Total RNA was isolated and analyzed by competitive/quantitative RT-PCR for caspase-9 splice variants. Simultaneously, protein lysates were also produced, subjected to SDS-PAGE, and immunoblotted for hnRNP L and β-actin. Data are n = 4 from 2 separate occasions. (C) HBEC-3KT cell lines were transfected with either wild-type hnRNP L (0.25 μg) in conjunction with caspase-9 minigene (0.25 μg) or caspase-9 minigene (0.25 μg) in conjunction with empty vector (0.25 μg) for 24 hours. Total RNA was extracted and analyzed by competitive/quantitative RT-PCR for caspase-9 minigene splice variants. Data are n = 3 on 2 separate occasions. Of note: the ratio of caspase-9a/9b minigene mRNA tended to present with a slightly higher ratio than endogenous caspase-9a/9b mRNA.
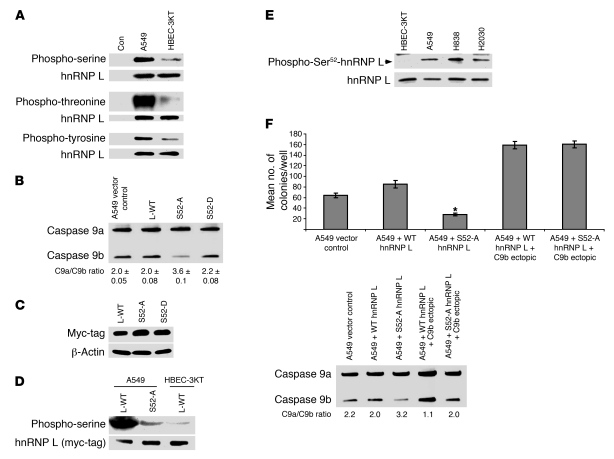
The above results suggested several possibilities for the contrasting findings in NSCLC cells versus non-transformed cells such as mislocalization (26), post-translational modifications of hnRNP L (27, 28), or a combination of mislocalization and post-translational modifications, which would modify the repressing capabilities of hnRNP L. To date, no confirmed phosphorylated site(s) have been established for hnRNP L functionality. Supplemental Figure 8 presents data that ruled out the mislocalization of hnRNP L; no difference was observed in cellular distribution between HBEC-3KT cells and several NSCLC cell lines. The phosphorylation state of hnRNP L between these cell lines was then examined using specific phosphorylated residue antibodies, and A549 cells demonstrated a dramatic enhancement of phosphorylated hnRNP L compared with HBEC-3KT cells (Figure 9A). Specifically, hnRNP L presented with a dramatic 10-fold increase in phospho-threonine, along with a marked increase in both serine (4-fold) and tyrosine (2-fold) phosphorylation in A549 cells compared with the non-transformed HBEC-3KT cells. To determine whether this observed phosphorylation demonstrated translatability in NSCLC, hnRNP L IP was used in H838 cells, and this NSCLC cell line also presented with a significant increase in serine and threonine phosphorylation (data not shown). Thus, an enhanced phosphorylation state of hnRNP L was observed in NSCLC cells versus non-transformed cells.
Figure 9. Ser52 of hnRNP L is hyperphosphorylated in NSCLC cells and regulates the alternative splicing of caspase-9.
(A) A549 and HBEC-3KT cell lines were seeded at the same confluency and in the same culture media 24 hours before IP. IP hnRNP L was resolved by SDS-PAGE and immunoblotted with phospho-specific and hnRNP L antibodies. (B) A549 cells were transfected with WT-hnRNP L (L-WT) (1 μg), Ser52Ala hnRNP L (S52-A) (1 μg), or Ser52Asp hnRNP L (S52-D) (1 μg). Total RNA was analyzed for caspase-9 splice variants. n = 4 from 3 occasions. (C) A549 cells were transfected with WT-hnRNP L, S52-A, or S52-D. Protein lysates were subjected to SDS-PAGE and immunoblotted for myc-tag and β-actin. Empty vector showed no expression of a myc-tagged protein. (D) A549 and HBEC-3KT cell lines seeded at the same confluency and in the same culture media for 24 hours before IP. Cell lines were transfected with WT-hnRNP L (1 μg) or S52-A (1 μg). Ectopically expressed hnRNP L was IP with c-myc tag Ab, resolved by SDS-PAGE, and immunoblotted with anti–phospho-serine and anti–c-myc tag antibodies. (E) Protein lysates were subjected to SDS-PAGE and immunoblot for phospho-Ser52 hnRNP L and hnRNP L. Phospho-Ser52 antibody for hnRNP L was validated by ELISA, hnRNP shRNA samples, and lack of identifying the Ser52Ala mutant of hnRNP L. (F) Colony formation assays in soft agar. n = 6; error bars represent SEM. *P < 0.005, A549 + S52-A hnRNP L + C9b ectopic versus A549 + S52-A hnRNP L.
Once hnRNP L was determined to be hyper-phosphorylated in a cancer-specific manner, we analyzed the amino acid sequence for hnRNP L for phosphorylation sites by mitogenic, cell survival, and oncogenic kinases. Several mass spectrometry databases were also consulted for references to identified/verified phosphorylated amino acids for hnRNP L. Supplemental Table 4 designates the possible phosphorylated sites for hnRNP L as predicted by PhosphositePlus, a phosphorylation site determination database (Cell Signaling Technology) (29), Human Protein Reference Database resource (30), Scansite (Massachusetts Institute of Technology) (31), and UniProtKB/Swiss-Prot (32). To determine which of these residues of hnRNP L regulated the exclusion of the exon 3,4,5,6 cassette of caspase-9, site-directed mutagenesis was used to convert relevant serines and threonines to alanine, and tyrosines to phenylalanine. A549 cells were transiently transfected (>50% transfection efficiency) with each mutant or wild-type hnRNP L, and the effect on the alternative splicing of endogenous caspase-9 was assayed by competitive/quantitative RT-PCR. Only ectopic expression of hnRNP L (Ser52Ala) induced a significant increase in the splicing ratio of caspase-9 in comparison with ectopic expression wild-type hnRNP L and empty vector controls (Figure 9B; Supplemental Table 4). To verify this result and rule out structural problems caused by mutation of this residue, a phosphomimic mutation, hnRNP L (Ser52Asp), was created. In contrast to the Ser52Ala mutation, transfection of hnRNP L (Ser52Asp) in A549 cells did not significantly affect the endogenous splicing ratio of caspase-9 compared with ectopic expression of wild-type hnRNP L, suggesting that either the levels of phosphorylated hnRNP L in NSCLC cells are sufficient to affect limiting levels of endogenous caspase-9 pre-mRNA, or that this mutation cannot appropriately mimic the phosphorylation of Ser52 (Figure 9B). Equivalent expression was verified by Western immunoblotting for the myc-tag associated with ectopically expressed hnRNP L (Figure 9C). The latter hypothesis is likely correct as ectopic expression of hnRNP L (Ser52Asp) in HBEC-3KT cells was not sufficient to reduce the endogenous caspase-9a/9b mRNA ratio (Supplemental Figure 10). Both phosphorylation mutants of hnRNP L also localized to only the nucleus (data not shown).
To demonstrate that phosphorylation on Ser52 was responsible for the serine hyper-phosphorylation of hnRNP L in NSCLC cells, wild-type hnRNP L and the Ser52Ala mutant of hnRNP L were transfected into A549 and HBEC-3KT cells, followed by specific IP of ectopically expressed/myc-tagged hnRNP L. Whereas ectopically expressed wild-type hnRNP L in A549 cells demonstrated a more than 3-fold increase in serine phosphorylation compared with ectopic expression in HBEC-3KT cells (in complete agreement with the endogenous findings for hnRNP L presented in Figure 9A), the Ser52Ala mutant of hnRNP L demonstrated a dramatic reduction in serine phosphorylation comparable to non-transformed HBEC-3KT cells (Figure 9D). To further demonstrate that hnRNP L is phosphorylated on Ser52 in a cancer-specific manner, we created a custom phospho-specific Ser52 hnRNP L antibody. Whole-cell lysates from A549, H838, H2030, and non-transformed HBEC-3KT cell lines were used to examine phospho-Ser52 state of hnRNP L by Western immunoblot analysis. All 3 cell lines demonstrated the higher expression of phospho-Ser52 hnRNP L compared with non-transformed HBEC-3KT cells, which demonstrated no discernible phospho-Ser52 hnRNP L (Figure 9E). Thus, hnRNP L is specifically phosphorylated on Ser52 in NSCLC cells, and the phosphorylation state of this residue of hnRNP L regulates the pre-mRNA processing of caspase-9.
To examine whether Ser52 of hnRNP L played a role in AIG of NSCLC cells, the hnRNP L (Ser52Ala) mutant and wild-type hnRNP L were stably expressed as batch cultures of A549 cells. Analogous to transient expression of the hnRNP L Ser52Ala mutant, stable expression of the hnRNP L Ser52Ala mutant induced an increase in the caspase-9a/9b mRNA ratio from 2.0 ± 0.13 to 3.2 ± 0.10. Stable transfection of the hnRNP L (Ser52Ala) mutant dramatically decreased the AIG ability of A549 cells in comparison with stable expression of wild-type hnRNP L (Figure 9F). Furthermore, re-expression of caspase-9b in the same cell lines completely rescued the effect of the hnRNP L (Ser52Ala) mutant on AIG (Figure 9F). Therefore, and in an analogous fashion to hnRNP shRNA, the phosphorylation of hnRNP L on Ser52 regulates the ability of NSCLC cells to grow in an anchorage-independent manner via the pre-mRNA processing of caspase-9.
Discussion
Lung cancer is the leading cause of cancer mortality, accounting for an estimated 28% of all cancer deaths in the United States, and the mortality rates for lung cancer have not significantly decreased over the years. Thus, there is an urgent need for further understanding of the molecular mechanisms of NSCLC and to find new molecular targets for the treatment of this disease. Indeed, the presented study identifies key molecular events specific to transformed/NSCLC cells (e.g., the pre-mRNA processing of caspase-9) that are required for the ability of these cells to form/maintain tumors. The mechanistic part of this study began with the simple observation that mutation of a purine-rich sequence in exon 3 of the exon 3,4,5,6 cassette of caspase-9 led to the loss of the caspase-9b splice variant. These original studies screened for possible ESEs interacting with SRp30a, a required RNA trans-factor for the inclusion of the exon 3,4,5,6 cassette of caspase-9 (33). The surprising finding of an ESS instead of an ESE led to the identification of hnRNP L, associated with exon 3 at the ESS. The observation that hnRNP L interacted with the purine-rich RNA sequence AGGGGA was also surprising, as several groups have shown that hnRNP L prefers CA repeats. Our findings raise the intriguing possibility that the phosphorylation status of hnRNP L may affect the RNA sequence specificity for this RNA trans-factor, which would explain the disparity of our findings. Alternatively, hnRNP L may be associated with a protein complex at C9/E3-ESS when phosphorylated and, therefore, does not interact directly with the RNA sequence. The data presented in this study support both possibilities, as hnRNP L was shown to interact with the C9/E3-ESS when phosphorylated and dephosphorylation of hnRNP L in A549 cells induced a significant reduction in the ability of hnRNP L to associate with the C9/E3-ESS (Supplemental Figure 9). Irrespective of the exact mechanism, and because of the requirement for phosphorylation in this interaction, the presented study adds a new dimension to our current knowledge of the role of RNA trans-factors in the development and maintenance of cancer cells/tumors. Specifically, the phosphorylation of hnRNP L, at Ser52 was required for this RNA trans-factor to act as a repressor of the exon 3,4,5,6 cassette of caspase-9 pre-mRNA. The requirement of a specific phosphorylation event in an RNA trans-factor regulating biological mechanisms related to cancer (e.g., AIG and tumorigenic capacity) was unexpected due to recent reports. In particular, several laboratories have published that expression/overexpression of a specific RNA trans-factor (e.g., SRp30a, ESRPs, and SRm160) was the critical mechanism for conferring AIG, epithelial-mesenchymal transition, or tumor formation (13–15). This study now provides an additional level of regulation of RNA trans-factors in relation to tumorigenesis, that is, specific phosphorylation of an RNA trans-factor.
Our findings on the phosphorylation state of hnRNP L regulating the pre-mRNA processing of caspase-9 agree with a recent report by Lynch and colleagues (27). That laboratory reported a correlation of an observed pH shift toward acidity in hnRNP L in T cells activated by phorbol esters, with effects on the inclusion of exon 4 into mature CD45 mRNA (27). Whereas that laboratory group did not determine whether the pH change was due to acetylation or phosphorylation, our data suggest the latter. Furthermore, Xie and colleagues recently demonstrated that hnRNP L mediates the alternative splicing of the Ca21/voltage-sensitive Slo potassium channel pre-mRNA via CAMKIV, which correlated with a 4-fold change in the expression of hnRNP L and an 8-fold increase phosphorylation status of the RNA trans-factor (28). The observed increase in the phosphorylation status of hnRNP L may have been due to basal stoichiometric phosphorylation, as this RNA trans-factor is phosphorylated on serine and tyrosine in non-transformed/unstimulated cells (28). Regardless, Xie and coworkers further showed that in vitro dephosphorylation of nuclear extracts reduced the affinity of hnRNP L to CA-repeats (28). Whereas both reports and our present study all support a role for phosphorylation in regulating the function of hnRNP L, our data do not support a role for Ser52 phosphorylation mediating the RNA affinity of hnRNP L. Whereas the phosphorylation of hnRNP L and the association of this RNA trans-factor with the C9/E3-ESS complex are required for exclusion of the exon 3,4,5,6 cassette, this event is unlikely to be sufficient to block the inclusion, as hnRNP L (Ser52Asp) mutant did not decrease the endogenous ratio of caspase-9a/9b mRNA in A549 cells. This may also simply mean that the endogenous pre-mRNA of caspase-9 is limiting in cells, and thus additional RNA trans-factor has no effect in further lowering the endogenous caspase-9a/9b ratio. This possibility is logical, as ectopic expression of wild-type hnRNP L reduces the ratio of caspase-9a/9b minigene mRNA but has a limited effect on the endogenous caspase-9a/9b mRNA ratio. Because of this conundrum, we undertook the critical experiment of expressing the phosphomimic of hnRNP L (Ser52Asp) in non-transformed HBEC3-KT cells and examined the ratio of caspase-9a/9b mRNA (Supplemental Figure 10). These data demonstrate that this single phosphorylation event is required for exclusion of the exon 3,4,5,6 cassette, but that it is not sufficient to induce the exclusion of the exon 3,4,5,6 cassette, suggesting a mechanism whereby hnRNP L phosphorylation on Ser52 is just one required step in a complex process that may also require the phosphorylation of hnRNP L on currently unidentified threonine or tyrosine residues. Importantly, our data show that expression of the hnRNP L (Ser52Ala) mutant competes with the phosphorylated form of hnRNP L in A549 cells to induce an increase in the caspase-9a/9b ratio. Thus, this particular phosphorylation site likely affects protein-protein interactions rather than the RNA affinity of hnRNP L. If phosphorylation of Ser52 was an event that enhanced the affinity of hnRNP L to the C9/E3-ESS or even CA-repeats, the hnRNP L (Ser52Ala) mutant would not have been able to compete with the high-affinity phosphorylated form of hnRNP L in A549 cells, and no effect would have been observed. Thus, the phosphorylation event reported by Xie and coworkers (28) that regulated the association of hnRNP L with CA repeats was not likely Ser52. This inference may also explain the lack of effect for other mutants examined in this study and cannot completely dismiss tyrosines and threonines as regulatory sites. Other serine residues in hnRNP L are still unlikely candidates, as Ser52 phosphorylation accounts for all of the observed increase in serine phosphorylation in NSCLC cells. Overall, our data suggest that phosphorylation of hnRNP L on Ser52 may regulate the association of hnRNP L with accessory RNA trans-factors, thus modifying the function of a protein complex; alternatively, this phosphorylation may inhibit the association of a required enhancing RNA trans-factor with the caspase-9 pre-mRNA.
In regards to a possible role for the phosphorylation of hnRNP L in cellular transformation, there are several candidate survival/oncogenic kinases (e.g., GSK3β, AKT, and casein kinase II) that are predicted to phosphorylate Ser52 of hnRNP L. Whereas these kinases have many targets and can affect cells via modulation of many signaling cascades, this study identifies the phosphorylation of hnRNP L by a currently unknown protein kinase as a key downstream target for oncogenic signaling in NSCLC cells. Based on these inferences, one can rationalize that a specific oncogenic pathway may require the phosphorylation of hnRNP L on Ser52, which would essentially block intrinsic apoptotic pathways, by lowering the ratio of caspase-9a/9b, and enhance cellular transformation. This is not surprising because previous studies have demonstrated that caspase-9a is a possible tumor suppressor (10). The presented study supports the findings of Lowe and coworkers, who showed enhanced cellular transformation of murine embryonic fibroblasts from caspase9–/– mice by H-RasV12/c-Myc (10), as well as studies utilizing a dominant-negative caspase-9 that completely alleviated pressure to inactivate p53 during lymphomagenesis (34). Our study applies these concepts to NSCLC, demonstrating the importance of the endogenous mechanism of blocking caspase-9 activity (e.g., alternative splicing) in the maintenance of NSCLC cells and tumors. Unfortunately, disparity and controversy exist over the studies of Lowe and coworkers in regards to caspase-9 as a tumor suppressor. For example, Ekert and coworkers showed caspase-9 is not necessary for apoptosis of factor-dependent cells, but merely affects its rate (35). Another contrasting report from Scott and colleagues demonstrated that caspase-9 did not play a role in lymphomagenesis (21). Furthermore, lymphocytes from caspase9–/– animals underwent apoptosis comparable to wild-type lymphocytes (21). On the other hand, Chipuk and Green pointed out in 2005 that no evidence was provided in these contrasting studies that caspase inhibitors blocked cell death (36). Thus, caspase-independent cell death was a plausible mechanism in the cell death observed in the Ekert and Scott studies (21, 35, 36). Although our data support the findings of Lowe and colleagues, the role of caspase-9b in regulating AIG and tumorigenic capacity may be outside the realm of direct inhibition of caspase-9a. In this regard, a role in cell signaling for caspase-9b has been reported by Latchman and coworkers (7). Specifically, the authors demonstrated that ectopic expression of caspase-9b induced the activation of NF-κB irrespective of caspase activity (7). Activation of NF-κB is known to cooperate with k-ras mutations in regard to cellular transformation, which thus may explain the function of caspase-9b (7). In essence, caspase-9b may elicit downstream signaling events outside the role of simple inactivation of caspase-9a. In support of this hypothesis, caspase-8 is suggested to play roles in the recruitment cell survival factors such as the p85 subunit of PI3K (37). Although these initiator caspases have roles in extrinsic pathways of programmed cell death, caspase-9b may play an analogous role in survival signaling to block the intrinsic pathway of programmed cell death.
Although this study produces a possible new paradigm in the regulation of alternative splicing with regards to oncogenic signaling, the mechanisms discussed above are likely an oversimplification of a complex and highly regulated RNA processing event. Whereas the phosphorylation of hnRNP L and the association of this RNA trans-factor with the C9/E3-ESS complex are required for exclusion of the exon 3,4,5,6 cassette, this exon 3 mechanism may be just one part of a large regulating complex that entails interplay with a number of RNA trans-factors to regulate the inclusion/exclusion of the exon 3,4,5,6 cassette. One such candidate RNA trans-factor that may interplay with hnRNP L is SRp30a (ASF/SF2). This RNA trans-factor has been identified by our laboratory as a required enhancing RNA trans-factor for the inclusion of the exon 3,4,5,6 cassette (33). Our laboratory has also shown correlation between the phosphorylation state of SR proteins and the inclusion of the exon 3,4,5,6 cassette with regard to ceramide treatment of NSCLC cells (33). As several past studies have shown that the phosphorylation state of SRp30a regulates its ability to modulate particular RNA splicing events, this is a logical hypothesis to test. Whether exclusion of the exon 3,4,5,6 cassette requires phosphorylation of hnRNP L on Ser52 as well as hyperphosphorylation of SRp30a is not currently known nor addressed in this study, but coordinated phosphorylation of repressing and enhancing RNA trans-factors interacting with multiple RNA cis-elements throughout the caspase-9 pre-mRNA may be required to repress the inclusion of the exon 3,4,5,6 cassette. Indeed, several high-affinity sites for SRp30a are predicted using Rainbow Splicer software (23), and in particular, one RNA cis-element for SRp30a is located adjacent to the C9/E3-ESS sequence. A recent study by Lynch and coworkers observed an interplay between SF2/ASF and hnRNP L for CD45 alternative splicing (38). Specifically, cell-based assays utilizing minigenes demonstrated that a balance between hnRNP L repression and SF2/ASF-enhancing activities ultimately determines the extent of exon 5 inclusion into CD45 (38). As more regulating factors for caspase-9 RNA splicing are identified, future studies will be geared toward investigating this complex interplay with regard to regulating the inclusion/exclusion of this exonic cassette. Unfortunately, these studies may require the identification of specific regulatory phosphorylation sites within SRp30a, and the number of constitutive phosphorylation sites in SRp30a alone, as well as the promiscuity of protein kinases for multiple sites in this RNA trans-factor, make this a rather daunting task.
It is important from a therapeutic stand point that only a 2-bp mutation was required to eliminate the association of hnRNP L with the C9/E3-ESS. Thus, the association of the C9/E3-ESS and the hnRNP L complex is highly specific. Since technologies are being developed to efficiently deliver small RNA sequences effectively to cells and animals, these data suggest that a scavenger RNA can be developed to the C9/E3-ESS. Another possibility is RNA aptamer technology, which is currently being employed toward nucleolin in phase 2 clinical trials (39). From a philosophical standpoint as to the development of new therapeutics for NSCLC patients, the targeting of cancer-specific as well as distal signaling mechanisms (e.g., the interaction of hnRNP L with the C9/E3-ESS, or the direct manipulation of the alternative splicing of caspase-9) is of key importance for 2 reasons. First, therapeutics based on a cancer-specific mechanism would limit the adverse effects of anticancer therapies by essentially only affecting the tumor cells. Second, the targeting of a distal mechanism limits the adaptability of a cancer cell to circumvent the anticancer therapy via a secondary pathway. The fewer opportunities that exist for a cancer cell to adapt, the more likely a specific therapy will show prolonged durability. Both hnRNP L and the alternative splicing of caspase-9 can be categorized as cancer-specific and distal points for therapeutic development in the treatment of NSCLC. The effect of using therapies targeted to either hnRNP L or caspase-9b would also be very limited on normal lung epithelial cells, as caspase-9b is not expressed in non-transformed epithelial cells and hnRNP L is dephosphorylated. In support of this hypothesis, unpublished data from our laboratory demonstrate that hnRNP L siRNA did not induce apoptosis in HBEC-3KT cells as assayed by PARP cleavage, in contrast to H838 and H2030 cells.
In conclusion, this report is a comprehensive study that began with the observation that the ratio of caspase-9a/9b mRNA was dysregulated in a large percentage of human lung adenocarcinoma tumors. Our studies demonstrated that the ratio of caspase-9a/9b mRNA played a significant and physiologically relevant role in the activity of caspase-9 and the tumorigenic capacity of NSCLC cells. Mechanistic studies then defined the C9/E3-ESS for the inclusion of the exon 3,4,5,6 cassette of caspase-9. This observation led to the identification of hnRNP L association with the C9/E3-ESS and regulation of the alternative splicing of caspase-9. Furthermore, this study demonstrated that an increase in the phosphorylation state of hnRNP L on Ser52 regulates the alternative splicing of caspase-9 in NSCLC. Lastly, these studies demonstrate that hnRNP L is required for the tumorigenic capacity of NSCLC cells via the regulation of this distal splicing event. Therefore, the presented study identifies the interaction of hnRNP L with the C9/E3-ESS in the exon 3 of caspase-9 pre-mRNA as well as the phosphorylation of hnRNP L on Ser52 as possible therapeutic targets for NSCLC.
Methods
Cell culture
Cell lines including A549, H2030, and H838s (ATCC) were cultured in RPMI 1640 supplemented with 10% (vol/vol) FBS (Sigma-Aldrich), 100 U/ml penicillin G sodium (Invitrogen), and 100 μg/ml streptomycin sulfate (Invitrogen). HBEC-3KT cells (40) were cultured with keratinocyte serum-free medium containing bovine pituitary extract and recombinant epidermal growth factor (Life Technologies). All cells were maintained at less than 80% confluency under standard incubator conditions. For all comparison studies between cell lines, 6 × 105 cells were plated in tissue culture plates (10 cm), which gave 80% confluency. The following day, medium was removed and cells were washed with 1× PBS and plated in DMEM without serum overnight. Total RNA and/or protein was then isolated for analysis.
EMSA
The following sequence was used for the FITC-tagged ROs (FL-C9/E3-ESS): exon 3 of caspase-9: (5′Fl-GAGAGUUUGAGGGGAAAU). The following competitor sequences were used: caspase-9 exon 3 NSC (5′-GAAUUCGCACGUUA) and caspase-9 exon 3 SC (5′-GAGUUUGAGGGGAA). RNA-binding reactions were performed in a final volume of 25 μl containing 100 ng of fluorescein-oligonucleotide, 50 μg A549 nuclear protein extract, 40 U RNASIN, and 11.3 μg tRNAs in buffer composed of 10 mM HEPES, 5 mM DTT, 120 mM KCl, 3 mM MgCl2, and 5% glycerol. The reactions were incubated on ice with 100-fold molar excess of either NSC or SC oligonucleotides for 5 minutes, followed by 20 minutes of incubation on ice with FL-C9/E3-ESS. Samples were loaded on a 5% TBE-acrylamide gel for electrophoretic separation of RNA-protein complexes. The gel was then scanned using Molecular Imager FX (Bio-Rad) with a 488-nm EX (530 nm BYPASS) laser.
Mass spectrometry analysis
Identification of RNA-protein complexes was performed at the Emory University Mass Spectrometry Center (Atlanta, Georgia, USA). Briefly, RNA-protein complexes were excised from the gel and in-gel trypsin digestion was performed. The tryptic mixtures were injected and analyzed by nano–LC-MS/MS. Proteins identified by nano–LC-MS/MS were based on independent sequencing of peptides. Nano–LC-MS/MS results were obtained by searching the Mascot database (Matrix Science).
Quantitative/competitive RT-PCR
Total RNA from cell lines listed above were isolated using the RNeasy Mini Kit X according to the manufacturer’s protocol. Total RNA RO was reverse transcribed using Superscript III reverse transcriptase and oligo (dT) as the priming agent. After 50 minutes of incubation at 42°C, the reactions were stopped by heating at 70°C for 15 minutes. Template RNA was then removed using RNase H. To quantitatively evaluate the expression of endogenous caspase-9 splice variants, an upstream 5′ primer to caspase-9 (5′-GCTCTTCCTTTGTTCATCTCC-3′) and a 3′ primer (5′-CATCTGGCTCGGGGTTACTGC-3′) were used. Using these primers, 20% of the reverse transcriptase reaction was amplified for 25 cycles (94°C, 30 seconds melt; 58°C, 30 seconds anneal; 72°C, 1 minute extension) using Platinum Taq DNA polymerase (Invitrogen Life Technologies). To evaluate the expression of the splice variant products of the caspase-9 minigene (Figure 1), a 5′ primer to caspase-9 minigene (5′-CATGCTGGCTTCGTTTCTG-3′) and a 3′ primer (5′-AGGGGCAAACAACAGATGG-3′) were used. Gene products produced from caspase-9 minigene PCR resulted in an 899-bp caspase-9a splice variant and a 443-bp caspase-9b splice variant. Using these primers, 20% of the reverse transcriptase reaction was amplified for 25 cycles (94°C, 30 seconds denaturation; 57°C, 30 seconds anneal; 72°C, 1 minute extension) using Platinum Taq DNA polymerase. The final PCR products were resolved on a 5% TBE acrylamide gel electrophoresis and visualized by staining with SYBR Gold (Invitrogen) and scanned using a Molecular Imager FX (Bio-Rad) with a 488-nm EX (530-nm BYPASS) laser.
Caspase-9 activity assay
The assay was performed by using Caspase-9 activity assay kit (Calbiochem; EMD Chemicals) according to the manufacturer’s instructions for examining LEHD activity. Caspase-9 activity was calculated as relative florescent units (RFU) per total protein (mg/ml). The LEHD activity is represented in the figures in percentage with respect to control or untreated cells.
Plasmid transfection
Cell lines were transfected with caspase-9 minigene constructs and additional plasmid vectors using Effectene Transfection Reagent (Qiagen Inc.) following the manufacturer’s protocol. Briefly, cell lines were plated at approximately 70% confluency in 6-well tissue culture dishes. Cells were transfected with plasmid constructs (0.25 to 1 μg), followed by 24 hours of standard incubation. After 24 hours of standard incubation, cells were examined for minigene expression by quantitative/competitive RT-PCR. This procedure routinely gave 50% transfection efficiency for A549 cells.
Streptavidin-agarose affinity purification
The following sequences were used for biotinylated-tagged ROs: caspase-9, exon 3 (Bio-C9/E3-ESS), (5′Bio-GAGAGUUUGAGGGGAAAUG 3′); caspase-9, exon 3 core mutant (Bio-C9/E3-ESS MUT), (5′Bio-GAGAGUUUGCTACTAAAUG 3′); NSC2, (5′Bio-ACUGCCAGUUACGAC); unlabeled SC, (5′-GAGUUUGAGGGGAA); and unlabeled NSC1, (5′-GAAUUCGCACGUUA) (Dharmacon Inc.). Also, see Supplemental Table 1 for additional cold unlabeled RO competitors used in these studies. Reaction mixtures were first pre-cleared for 2 hours at 4°C with streptavidin-agarose beads (20 μl) (50 μg A549 nuclear extract, 40 U RNASIN, 11.3 μg tRNAs, 10 mM HEPES, 5 mM DTT, 120 mM KCl, 3 mM MgCl2, and 5% glycerol). Reaction mixtures were then incubated with 100-fold molar excess of either NSC or SC oligo for 5 minutes following the addition of 400 ng Bio-C9/E3-ESS or Bio-NSC2 incubated on ice for 30 minutes. New streptavidin-agarose beads were then added to RNA-binding reactions and incubated at 4°C for 2 hours with gentle agitation. RNA-protein–binding reactions were washed with buffer containing 100 mM KCl, 20 mM Tris-HCl at pH 7.5, and 0.2 mM EDTA. After washing, the complex was pelleted by centrifugation. The pellet was resuspended in Laemmli buffer, dry-boiled for 10 minutes, and subjected to SDS-PAGE/Western immunoblot analysis.
siRNA transfection
A549, H838, H2030, and HBEC-3KT cell lines were transfected with 100 nM of hnRNP L SMARTpool multiplex, hnRNP A2/B1 SMARTpool multiplex, individual siRNAs to these RNA trans-factors, caspase-9b siRNA (5′-GATTTGGTGATGTCGAGCATT-3′), or control non-targeting siRNA using Dharmafect 1 transfection reagent (Dharmacon). Briefly, cell lines were plated in regular growth medium at 40%–50% confluence in a 6-well tissue culture dish 24 hours before transfection. Cells were placed in Opti-Mem I medium without antibiotics/FBS and transfected with 100 nM (dilution in 1× siRNA buffer) of the siRNA of choice (Dharmafect 1/Opti-MEM I reduced serum medium and incubated for 4 hours at standard incubator conditions). After incubation, 0.5 ml Opti-MEM I–reduced serum medium containing 3-fold the normal concentration of antibiotics/FBS was added to the transfected cells lines without removing the transfection mixture. After 48 hours, total RNA or total protein lysates were collected (33).
Oligonucleotide treatment
2′-O-(2-methoxy) ethyl 23-mer ASROs were designed to target the 5′SS of exons 3, 4, and 6 of caspase-9. The ASROs were designed following the methodology published in a collaborative report that was based on the report by Taylor and coworkers for manipulating Bcl-x pre-mRNA processing (20). E4 was essentially the best ASRO for lowering the caspase-9a/9b mRNA ratio, and thus, this study used the E4 ASRO (2-O-methoxy derivative) (5′-GAGUGUACCUUGGCAGUCAGGUC-3′) targeted to the 5′SS of caspase-9 exon 4. A549 cells were transfected with ASRO (400 nM) using Dharmafect reagent and following the same protocol as the siRNA transfection. Exon 3 and 6 sequences for the data not shown were 5′-CAUGGAGACUCACCAAAUCUGCA-3′ and 5′-GGAGCUGCUUACCCCCACCACAG-3′.
Statistics
Statistical differences between treatment groups were determined by a 2-tailed, unpaired Student’s t test when appropriate. P values less than or equal to 0.05 were considered significant.
Western immunoblotting
Cells were washed extensively with PBS, scraped, centrifuged to pellet, resuspended in cell lysis reagent/protease inhibitor cocktail buffer (Sigma-Aldrich), and sonicated. Total protein lysates were then diluted in SDS sample buffer and dry boiled for 10 minutes. Protein samples (10 μg) were subjected to 10% SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and blocked in 5% milk/1× PBS/0.1% Tween (M-PBS-T) for 2 hours. The membrane was incubated with anti-hnRNP L (1:1,000; Santa Cruz Biotechnologies Inc.), anti-hnRNP A2/B1 (1:1,000; Santa Cruz Biotechnologies Inc.), anti-PARP (1:1,000; Cell Signaling Technology), anti-caspase-9 (1:1,000; Assay Designs), anti–phospho-serine (1:1,000; Abcam), anti–phospho-threonine (1:1,000; Cell Signaling Technology), anti-phospho-tyrosine (1:1,000; Cell Signaling Technology), anti-Ser52 hnRNP L (1:5,000; custom produced via Genscript Corporation), or anti–β-actin (1:5,000; Santa Cruz Technology) for 2 hours in M-PBS-T, followed by 3 washes with PBS-T. The membrane was then incubated with a secondary antibody of horseradish peroxidase-conjugated goat anti-rabbit IgG antibody (1:5,000; Pierce Biotechnology) or goat anti-mouse IgM (1:5,000; Calbiochem) for 45 minutes, followed by 3 washes with PBS-T. Immunoblots were developed using Pierce enhanced chemiluminescence reagents and Bio-Max film.
Immunoprecipitation
Cells were washed with 1× PBS and scraped in 1× IP buffer containing: 1% Triton X-100, 150 mM NaCl, 10 mM Tris pH 7.4, 1 mM Na3VO4, 1 mM EDTA pH 8.0, 0.5% NP-40, 0.2 mM PMSF, 0.5 mM 4-DOP, and 1× complete protease inhibitor. The lysates were then sonicated and centrifuged, and the resulting supernatant was collected for analysis. Each sample lysate (200 μg) was precleared with prewashed protein G sepharose beads for 1 hour at 4°C with gentle agitation. After preclearing, samples were centrifuged and sample lysates were collected. For phosphorylation comparison among different cell lines, hnRNP L monoclonal antibody (3 μg) was placed in each sample and in 4°C with gentle agitation overnight. For Ser52 phosphorylation studies, 3 μg of myc-tag was placed into the samples. The following day, prewashed protein G Sepharose beads were added to each sample and placed in 4°C with gentle agitation for 1 hour. After 1 hour of incubation, bead complexes were centrifuged, washed extensively with 1× IP buffer, pelleted by centrifugation, resuspended in Laemmli buffer, dry-boiled for 10 minutes, and subjected to SDS-PAGE/Western immunoblotting analysis.
Stable transfections
See Supplemental Table 3 for specific plasmids and inserts utilized.
Lentivirus transfection/selection.
A549 cells (15 × 104) were transduced with lentivirus (20 MOI) for 24 hours as described by the manufacturer’s protocol. The following day, lentivirus-containing media was switched with prewarmed media for 24 hours, followed by puromycin (50 μg/ml) selection for 2 weeks. For batch culture cell lines, cells were consistently treated with puromycin selection marker for 2 weeks. For clonal cell lines, clonal rings were placed over individual colonies, trypsinized, and placed in 12-well plates.
Plasmid transfection and selection.
A549 cells (15 × 104) were transfected with the appropriate plasmid (1 μg) using Effectene following the manufacturer’s protocol (Qiagen). For selection purposes, cells were passaged into zeocin selection medium 24 hours after transfection, to produce batch and clonal cell lines.
Confocal microscopy
For confocal microscopy, the cells were seeded onto 22 mm2 coverslips in 35-mm plates in their appropriate medium and incubated at 37°C under 5% CO2 overnight. The next day, cells were washed twice with 1× PBS, then fixed with 100% cold methanol for 10 minutes at –20°C. The slides were then washed with 1× PBS containing 10 mM glycine and 0.2% sodium azide, followed by incubation (40 minutes) with the primary antibody, anti-hnRNP L (1:100) (Sigma-Aldrich). After the primary antibody incubation, cells were washed with 1× PBS, followed by incubation with anti-mouse Alexa Fluor 488 secondary antibody (1:500) for 30 minutes at room temperature. Coverslips were mounted in 10 mM n-propagalate in glycerol and viewed using a Leica confocal microscope (TCS-SP2 [AOBS]).
Imaging/quantification
All data presented in this manuscript were generated using multiple laser scans or film exposures and were not manipulated by adjusting brightness or contrast. Multiple exposures and various scans at multiple laser intensities were used to present the data for easy discernment of the appropriate effects by readers.
Animals and tumor models
Six-week-old male CB17-Prkdcscid/NCrCrl ORIGIN SCID mice were purchased from Charles River Laboratories. Animal experiments were conducted at Virginia Commonwealth University. The research protocol was approved by the VCU IACUC involving animal care in accordance with the US Department of Agriculture (USDA) Animal Welfare Regulations, the Public Health Service Policy on the Humane Care and Use of Laboratory Animals (41), and the US Government Principles for the Utilization and Care of Vertebrate Animals Used in Testing, Research, and Training (42). VCU is in compliance with all provisions of the Animal Welfare Act and other federal statues and regulations relating to animals. VCU is registered under the Animal Welfare Act as a class R research facility with the USDA Animal and Plant Health Inspection Service (APHIS) Animal Welfare (registration number 52-R-0007). The Office of Laboratory Animal Welfare (OLAW) approved the VCU Animal Welfare Assurance in compliance with the Public Health Service Policy (assurance number A3281-01). The SCID mice were acclimatized to the VCU Medical School Animal Facility for 1 week before they received injections of cancer cells; 5 × 106 A549 human NSCLC cells were resuspended in 100 μl Matrigel (BD Biosciences) and injected subcutaneously into the right hind flank. Tumors were allowed to develop for up to 4 weeks, at which point mice were sacrificed and tumors were extracted for further analysis.
Supplementary Material
Acknowledgments
This work was supported by grants from the Veteran’s Administration (VA Merit Review I and Research Career Scientist Award to C.E. Chalfant), the NIH (HL072925 and CA117950 to C.E. Chalfant, NH1C06-RR17393 to VCU for renovation), the National Cancer Institute Lung Cancer SPORE P50CA70907 (to J.D. Minna), the National Aeronautics and Space Agency NNJO5HD36G (to J.D. Minna), and the International Association for the Study of Lung Cancer as a Young Investigator Award (to D.R. Camidge). We thank Kristen Lynch (University of Texas Southwestern Medical Center) for the gift of the hnRNP L plasmids used for ectopic expression in this manuscript and for advice in examining hnRNP L/RNA interactions. We also thank Andrew Thorburn, Karen Knudsen, Sarah Spiegel, Yusuf Hannun, and Paul Dent for critical review of the data contained in this manuscript.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(11):3923–3939. doi:10.1172/JCI43552.
See the related Commentary beginning on page 3798.
References
- 1.Hajra KM, Liu JR. Apoptosome dysfunction in human cancer. Apoptosis. 2004;9(6):691–704. doi: 10.1023/B:APPT.0000045786.98031.1d. [DOI] [PubMed] [Google Scholar]
- 2.Kominsky DJ, Bickel RJ, Tyler KL. Reovirus-induced apoptosis requires both death receptor- and mitochondrial-mediated caspase-dependent pathways of cell death. Cell Death Differ. 2002;9(9):926–933. doi: 10.1038/sj.cdd.4401045. [DOI] [PubMed] [Google Scholar]
- 3.Liang Q, et al. 6–Hydroxydopamine induces dopaminergic cell degeneration via a caspase-9–mediated apoptotic pathway that is attenuated by caspase–9dn expression. J Neurosci Res. 2004;77(5):747–761. doi: 10.1002/jnr.20198. [DOI] [PubMed] [Google Scholar]
- 4.Philchenkov A, Zavelevich M, Kroczak TJ, Los M. Caspases and cancer: mechanisms of inactivation and new treatment modalities. Exp Oncol. 2004;26(2):82–97. [PubMed] [Google Scholar]
- 5.Seol DW, Billiar TR. A caspase-9 variant missing the catalytic site is an endogenous inhibitor of apoptosis. J Biol Chem. 1999;274(4):2072–2076. doi: 10.1074/jbc.274.4.2072. [DOI] [PubMed] [Google Scholar]
- 6.Srinivasula S M, et al. Identification of an endogenous dominant-negative short isoform of caspase-9 that can regulate apoptosis. Cancer Res. 1999;59(5):999–1002. [PubMed] [Google Scholar]
- 7.Stephanou A, Scarabelli TM, Knight RA, Latchman DS. Antiapoptotic activity of the free caspase recruitment domain of procaspase-9: a novel endogenous rescue pathway in cell death. J Biol Chem. 2002;277(16):13693–13699. doi: 10.1074/jbc.M108530200. [DOI] [PubMed] [Google Scholar]
- 8.Wu GS, Ding Z. Caspase 9 is required for p53-dependent apoptosis and chemosensitivity in a human ovarian cancer cell line. Oncogene. 2002;21(1):1–8. doi: 10.1038/sj.onc.1205020. [DOI] [PubMed] [Google Scholar]
- 9.Yacoub A, et al. MDA-7 regulates cell growth and radiosensitivity in vitro of primary (non-established) human glioma cells. Cancer Biol Ther. 2004;3(8):739–751. doi: 10.4161/cbt.3.8.968. [DOI] [PubMed] [Google Scholar]
- 10.Soengas MS, et al. Apaf-1 and caspase-9 in p53-dependent apoptosis and tumor inhibition. Science. 1999;284(5411):156–159. doi: 10.1126/science.284.5411.156. [DOI] [PubMed] [Google Scholar]
- 11.Ebert AD, Chen F, He X, Cryns VL, Bohn MC. A tetracycline-regulated adenovirus encoding dominant-negative caspase-9 is regulated in rat brain and protects against neurotoxin-induced cell death in vitro, but not in vivo. Exp Neurol. 2005;191 suppl 1:S80–S94. doi: 10.1016/j.expneurol.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 12.Hakem R, et al. Differential requirement for caspase 9 in apoptotic pathways in vivo. Cell. 1998;94(3):339–352. doi: 10.1016/s0092-8674(00)81477-4. [DOI] [PubMed] [Google Scholar]
- 13.Cheng C, Sharp PA. Regulation of CD44 alternative splicing by SRm160 and its potential role in tumor cell invasion. Mol Cell Biol. 2006;26(1):362–370. doi: 10.1128/MCB.26.1.362-370.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. 2007;14(3):185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type–specific regulators of FGFR2 splicing. Mol Cell. 2009;33(5):591–601. doi: 10.1016/j.molcel.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siebert PD, Larrick JW. Competitive PCR. Nature. 1992;359(6395):557–558. doi: 10.1038/359557a0. [DOI] [PubMed] [Google Scholar]
- 17.Chalfant CE, et al. De novo ceramide regulates the alternative splicing of caspase 9 and Bcl-x in A549 lung adenocarcinoma cells. Dependence on protein phosphatase-1. J Biol Chem. 2002;277(15):12587–12595. doi: 10.1074/jbc.M112010200. [DOI] [PubMed] [Google Scholar]
- 18.Kole R. Modification of pre-mRNA splicing by antisense oligonucleotides. Acta Biochim Pol. 1997;44(2):231–237. [PubMed] [Google Scholar]
- 19.Patel NA, et al. The protein kinase C beta II exon confers mRNA instability in the presence of high glucose concentrations. J Biol Chem. 2003;278(2):1149–1157. doi: 10.1074/jbc.M206797200. [DOI] [PubMed] [Google Scholar]
- 20.Taylor JK, Zhang QQ, Wyatt JR, Dean NM. Induction of endogenous Bcl-xS through the control of Bcl-x pre-mRNA splicing by antisense oligonucleotides. Nat Biotechnol. 1999;17(11):1097–1100. doi: 10.1038/15079. [DOI] [PubMed] [Google Scholar]
- 21.Scott CL, et al. Apaf-1 and caspase-9 do not act as tumor suppressors in myc-induced lymphomagenesis or mouse embryo fibroblast transformation. . J Cell Biol. 2004;164(1):89–96. doi: 10.1083/jcb.200310041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31(13):3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koscielny G, et al. ASTD: The alternative splicing and transcript diversity database. Genomics. 2009;93(3):213–220. doi: 10.1016/j.ygeno.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 24.Hui J, Reither G, Bindereif A. Novel functional role of CA repeats and hnRNP L in RNA stability. RNA. 2003;9(8):931–936. doi: 10.1261/rna.5660803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shih SC, Claffey KP. Regulation of human vascular endothelial growth factor mRNA stability in hypoxia by heterogeneous nuclear ribonucleoprotein L. J Biol Chem. 1999;274(3):1359–1365. doi: 10.1074/jbc.274.3.1359. [DOI] [PubMed] [Google Scholar]
- 26.Taguchi F, et al. hnRNP L enhances sensitivity of the cells to KW-2189. Int J Cancer. 2004;108(5):679–685. doi: 10.1002/ijc.11616. [DOI] [PubMed] [Google Scholar]
- 27.Melton AA, Jackson J, Wang J, Lynch KW. Combinatorial control of signal-induced exon repression by hnRNP L and PSF. Mol Cell Biol. 2007;27(19):6972–6984. doi: 10.1128/MCB.00419-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu J, Hai Y, Liu G, Fang T, Kung S K, Xie J. The heterogeneous nuclear ribonucleoprotein L is an essential component in the Ca2+/calmodulin-dependent protein kinase IV-regulated alternative splicing through cytidine-adenosine repeats. J Biol Chem. 2009;284(3):1505–1513. doi: 10.1074/jbc.M805113200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sui S, et al. Phosphoproteome analysis of the human Chang liver cells using SCX and a complementary mass spectrometric strategy. Proteomics. 2008;8(10):2024–2034. doi: 10.1002/pmic.200700896. [DOI] [PubMed] [Google Scholar]
- 30.Keshava Prasad TS, et al. Human Protein Reference Database-- 2009 update. Nucleic Acids Res. 2009;37(Database issue):D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31(13):3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.UniProt Consortium. The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 2009;37(Database issue):D169–D174. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Massiello A, Chalfant CE. SRp30a (ASF/SF2) regulates the alternative splicing of caspase-9 pre-mRNA and is required for ceramide-responsiveness. J Lipid Res. 2006;47(5):892–897. doi: 10.1194/jlr.C600003-JLR200. [DOI] [PubMed] [Google Scholar]
- 34.Schmitt CA, Fridman JS, Yang M, Baranov E, Hoffman RM, Lowe SW. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell. 2002;1(3):289–298. doi: 10.1016/S1535-6108(02)00047-8. [DOI] [PubMed] [Google Scholar]
- 35.Ekert PG, et al. Apaf-1 and caspase-9 accelerate apoptosis, but do not determine whether factor-deprived or drug-treated cells die. J Cell Biol. 2004;165(6):835–842. doi: 10.1083/jcb.200312031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chipuk JE, Green DR. Do inducers of apoptosis trigger caspase-independent cell death? Nat Rev Mol Cell Biol. 2005;6(3):268–275. doi: 10.1038/nrm1573. [DOI] [PubMed] [Google Scholar]
- 37.Senft J, Helfer B, Frisch SM. Caspase-8 interacts with the p85 subunit of phosphatidylinositol 3-kinase to regulate cell adhesion and motility. Cancer Res. 2007;67(24):11505–11509. doi: 10.1158/0008-5472.CAN-07-5755. [DOI] [PubMed] [Google Scholar]
- 38.Motta–Mena LB, Heyd F, Lynch KW. Context-dependent regulatory mechanism of the splicing factor hnRNP L. Mol Cell. 2010;37(2):223–234. doi: 10.1016/j.molcel.2009.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soundararajan S, Chen W, Spicer EK, Courtenay-Luck N, Fernandes DJ. The nucleolin targeting aptamer AS1411 destabilizes Bcl-2 messenger RNA in human breast cancer cells. Cancer Res. 2008;68(7):2358–2365. doi: 10.1158/0008-5472.CAN-07-5723. [DOI] [PubMed] [Google Scholar]
- 40.Sato M, et al. Multiple oncogenic changes (K-RAS(V12), p53 knockdown, mutant EGFRs, p16 bypass, telomerase) are not sufficient to confer a full malignant phenotype on human bronchial epithelial cells. Cancer Res. 2006;66(4):2116–2128. doi: 10.1158/0008-5472.CAN-05-2521. [DOI] [PubMed] [Google Scholar]
- 41. Office of Laboratory Animal Welfare. Public health service policy on humane care and use of laboratory animals [policy]. 2002. http://grants.nih.gov/grants/olaw/references/phspol.htm . [PubMed]
- 42.Office of Science and Technology Policy. U.S. Government principles for the utilization and care of vertebrate animals used in testing, research and training. Federal Register. 19855097 ) [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.