Abstract
Many common disorders of pregnancy are attributed to insufficient invasion of the uterine lining by trophoblast, fetal cells that are the major cell type of the placenta. Interactions between fetal trophoblast and maternal uterine NK (uNK) cells — specifically interactions between HLA-C molecules expressed by the fetal trophoblast cells and killer Ig-like receptors (KIRs) on the maternal uNK cells — influence placentation in human pregnancy. Consistent with this, pregnancies are at increased risk of preeclampsia in mothers homozygous for KIR haplotype A (KIR AA). In this study, we have demonstrated that trophoblast expresses both paternally and maternally inherited HLA-C surface proteins and that maternal KIR AA frequencies are increased in affected pregnancies only when the fetus has more group 2 HLA-C genes (C2) than the mother. These data raise the possibility that there is a deleterious allogeneic effect stemming from paternal C2. We found that this effect also occurred in other pregnancy disorders (fetal growth restriction and recurrent miscarriage), indicating a role early in gestation for these receptor/ligand pairs in the pathogenesis of reproductive failure. Notably, pregnancy disorders were less frequent in mothers that possessed the telomeric end of the KIR B haplotype, which contains activating KIR2DS1. In addition, uNK cells expressed KIR2DS1, which bound specifically to C2+ trophoblast cells. These findings highlight the complexity and central importance of specific combinations of activating KIR and HLA-C in maternal-fetal immune interactions that determine reproductive success.
Introduction
The main tissue location where maternal allo-recognition of the fetus occurs is in the uterus at the site of placentation, where fetal extravillous trophoblast cells (EVTs) invade and intermingle with maternal leukocytes (1). Uterine NK (uNK) cells account for approximately 70% of decidual leukocytes and are likely to be involved in placentation and thus fetal growth and development. We proposed that placentation is regulated as a result of interactions between maternal killer immunoglobulin-like receptors (KIRs) expressed by uNK cells and their cognate ligands, HLA-C molecules, displayed by invading fetal trophoblast cells (2, 3). The importance of NK cell KIR/HLA-C interactions in mediating allo-recognition in the artificial context of BM transplantation (BMT) is well known (4). The only physiological situation in which NK allo-recognition occurs is during pregnancy.
The function of EVT is to access the maternal blood supply during placentation, when trophoblast invades the walls of the spiral arteries, converting them to high-conductance vessels (1). When trophoblast invasion into the uterus is defective, the blood flow is compromised, resulting in abnormal placental development and “starving” of the fetus. The clinical presentation of this failure of placentation is variable, and disorders such as preeclampsia, fetal growth restriction (FGR), unexplained stillbirth, or recurrent miscarriage (RM) can all occur (5, 6). These are the major disorders of human pregnancy, and an understanding of the mechanisms regulating trophoblast invasion will give insight into their primary pathogenesis.
Several preliminary pieces of evidence indicate that maternal KIR/trophoblast HLA-C interactions are important in placentation. Trophoblast cells that invade the uterus have a unique, tissue-specific pattern of HLA class I expression that includes invariant HLA-G and HLA-E and polymorphic HLA-C, but not highly polymorphic HLA-A and HLA-B (7). Therefore, of the class I molecules expressed by trophoblast, only HLA-C shows the variability necessary for a fetal allo-antigen. HLA-C molecules are mainly present in a stable conformation on the trophoblast, whereas on somatic cells they are also present as unstable, unfolded conformers (8). Significantly, despite the co-dominant expression of HLA class I molecules on somatic cells, it has never been established whether extra-embryonic EVTs express paternal HLA-C allotypes.
uNK cells are phenotypically distinct from their peripheral blood NK (PBNK) cell counterparts (9, 10). Of particular relevance to allo-recognition of the fetal trophoblast is that their KIR expression is skewed toward recognition of HLA-C (11, 12). This is reflected functionally by increased binding of HLA-C tetramers by uNK cells compared with PBNK cells. Reciprocally, KIR tetramers do bind specifically to the fully conformed HLA-C molecules on normal trophoblast. Thus, KIRs on uNK cells have a significantly enhanced ability to bind trophoblast HLA-C compared with KIRs on PBNK cells.
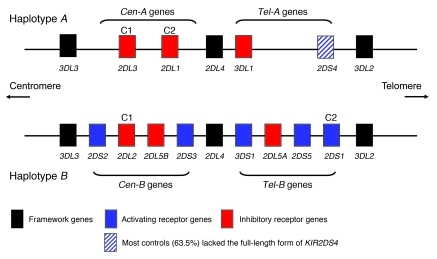
The regulation of NK responses is known to depend, in part, on 2 polymorphic gene systems, KIR and HLA, which are encoded on separate chromosomes, thus segregating independently (13, 14). Over 350 different KIR genotypes have been described, which highlights the great variability in the KIR gene family (15). There are 2 basic KIR haplotypes, however, A and B, which differ principally in that the B haplotype has additional activating receptors. In any pregnancy, the maternal KIR genotype could be AA (no activating KIRs) or AB/BB (1–10 activating KIRs). These 2 haplotypes can be further usefully subdivided into centromeric and telomeric regions, Cen-A, Tel-A, Cen-B, and Tel-B (16, 17) (Figure 1). KIR genes within each of these regions are in strong linkage disequilibrium (18, 19). The HLA-C ligands for KIRs are divided into 2 groups, defined by a dimorphism at position 80 of the α1 domain. Group 1 HLA-C (C1) allotypes bind inhibitory KIR2DL2/3, while group 2 HLA-C (C2) allotypes bind both inhibitory KIR2DL1 and activating KIR2DS1. While virtually all individuals have inhibitory KIRs for C1 and C2, less than 50% of white British individuals have the activating KIR2DS1 that is found on the B haplotype. This great diversity of maternal KIRs and fetal HLA-C ligands means that certain KIR/HLA-C combinations might be more favorable to reproductive success than others, due to the overall signals that the NK cell receives.
Figure 1. Representative KIR A and B haplotypes illustrate centromeric (Cen-A and Cen-B) and telomeric (Tel-A and Tel-B) gene regions, with the HLA-C ligands (C1 and C2) depicted above their cognate receptors.
In addition, KIR2DL2 also binds subsets of C2 allotypes (54), and KIR2DS4 binds some C1 and C2 allotypes (63). A KIR2DS5 locus may also be found in the Cen-B region, but this is seen infrequently in the white British population. Similarly, KIR2DS3 is most often associated with the KIR2DL5B locus in the Cen-B region, as shown here, but may occasionally occur in the Tel-B region (18).
Here we show that invading EVTs are the principal site of HLA-C expression in decidua basalis and that both maternal and paternal HLA-C allotypes are presented to KIRs on uNK cells. We demonstrate specific binding of both inhibitory and activating KIRs to trophoblast HLA-C. Thus, NK allo-recognition mediated by maternal KIR/fetal HLA-C interactions can occur at the maternal-fetal interface. Building on our previous work, we took a genetic approach to unravel how these KIR/ HLA-C interactions affect the process of placentation. Our genetic studies on 3 common disorders of pregnancy with similar underlying defects in placentation — preeclampsia, FGR, and RM — show that the inherent variability of maternal KIRs and fetal HLA-C acts in a predictable fashion to influence successful pregnancy.
Results
HLA-C is selectively expressed by EVT at the site of placentation.
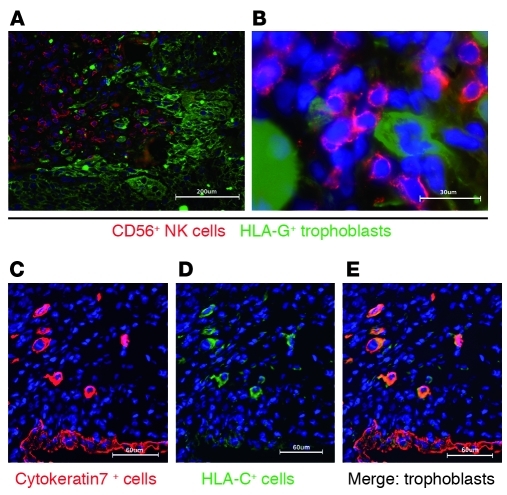
Since HLA-C is the predominant ligand for uNK KIRs, we examined expression of HLA-C in the decidua basalis. Previously, localization of HLA-C expression in tissues has been hampered by a lack of suitable antibodies. We used a newly characterized mAb (DT9) specific for HLA-C with minimal cross-reactivity with other class I antigens including HLA-G (20). In the decidua, infiltrating EVTs intermingled with and were closely apposed to maternal CD56+ NK cells that expressed KIRs (12) (Figure 2, A and B). HLA-C was strongly expressed selectively by EVTs (identified by cytokeratin or HLA-G expression) (Figure 2, C–E). We conclude that the HLA-C molecules expressed by the fetal trophoblast will be recognized preferentially by the maternal KIR on uNK cells at the site of placentation.
Figure 2. HLA-C is expressed predominately on the EVT in the decidua basalis.
(A and B) Immunofluorescence staining of frozen sections of human first-trimester implantation site showed infiltrating EVTs in sections of decidua basalis identified by staining for HLA-G. EVTs were observed mingling and in close apposition with CD56+ NK cells. (C) Cytokeratin staining identified EVTs and glandular epithelial cells. (D and E) Only EVTs were strongly labelled with the mAb DT9, specific for HLA-C allotypes. There was weak staining of other maternal cells in the decidua identified as CD45+ leukocytes (not shown). Scale bars: 200 μm (A); 30 μm (B); 60 μm (C–E).
EVTs express both maternal and paternal HLA-C allotypes.
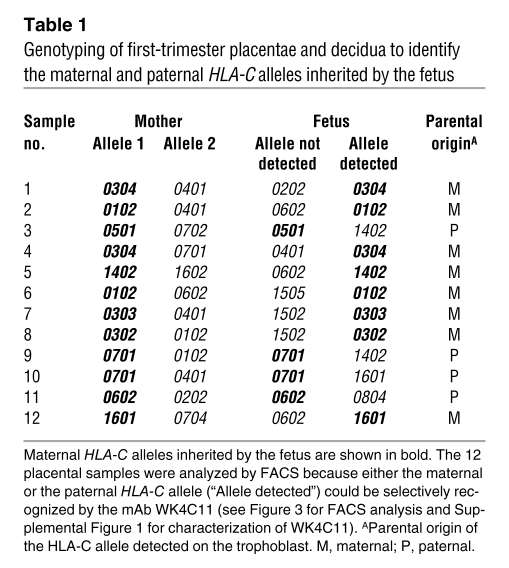
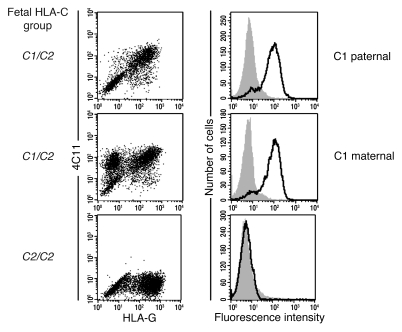
We next ascertained whether HLA-C on the surface of the extra-embryonic EVT is strictly maternal or paternal in origin, since many genes are imprinted in the placenta, or is co-dominantly expressed, as on all somatic cells. To do this we characterized the mAb WK4C11 (21) and found that it bound all C1 (apart from Cw7), but not C2, allotypes on normal cells including EVTs (Supplemental Table 1 and Supplemental Figure 1, A and B; supplemental material available online with this article; doi: 10.1172/JCI43998DS1). Consistent with this reactivity, WK4C11 blocked binding of KIR2DL3-Fc proteins to a C1 transfectant, but not of 2DL1-Fc to a C2 transfectant (Supplemental Figure 1C). Thus, we determined that WK4C11 can be used as a reliable marker of most C1 allotypes. Using WK4C11, we stained primary trophoblast cells in which parental origin of the HLA-C alleles had previously been determined by genotyping (Table 1). The results clearly showed that both maternal syngeneic and paternal allogeneic HLA-C allotypes were expressed by the trophoblast at equivalent levels (Figure 3). Thus, maternal KIR allo-recognition of paternal HLA-C, as well as recognition of self HLA-C, can occur at the site of placentation.
Table 1 .
Genotyping of first-trimester placentae and decidua to identify the maternal and paternal HLA-C alleles inherited by the fetus
Figure 3. Fetal EVTs express both maternally and paternally inherited HLA-C allotypes.
EVTs isolated from the genotyped placental samples shown in Table 1 were identified by flow cytometric staining for HLA-G. Double staining with the mAb WK4C11 showed that HLA-G+ EVTs expressed both paternal and maternal C1 allotypes. Examples are shown in which the C1 allele detected by mAb WK4C11 was inherited maternally or paternally (samples 8 and 9, respectively, in Table 1). No staining was seen on EVTs with WK4C11 when the trophoblast had the C2/C2 genotype.
Both inhibitory and activating KIRs for C2 are expressed by uNK cells and bind to trophoblast HLA-C molecules.
The only known activating KIR for HLA-C is 2DS1, which binds to C2 allotypes (22). Furthermore, a fundamental distinction between the 2 KIR haplotypes with respect to HLA-C is the presence in the B haplotype of the 2DS1 gene (Figure 1). It was therefore important to determine whether uNK cells are the same as PBNK cells in expressing 2DS1 when the gene is present (23). By using a combination of mAbs, we were able to show that uNK subsets express both 2DL1 and 2DS1 proteins (Supplemental Figure 2). We then asked whether these KIRs could bind trophoblast, given that, compared with PBLs, an unusually high proportion of HLA-C molecules are located in the KIR-reactive β2m-associated conformation on trophoblast (8). KIR-Fc proteins (inhibitory 2DL1 and activating 2DS1) both bound normal primary trophoblast and blood CD3+ T cells from C1/C2 but not C1/C1 donors. Binding was blocked by the anti-KIR mAb HP-3E4 (Figure 4). We conclude that KIR2DL1 and 2DS1 do indeed bind trophoblast C2 molecules.
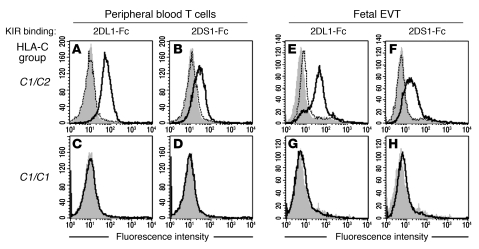
Figure 4. KIR2DS1 and KIR2DL1 bind specifically to C2 molecules on EVTs.
(A–H) Binding of KIR-Fc fusion proteins was measured by flow cytometry to peripheral blood CD3+ T cells (A–D) or HLA-G+ EVTs isolated from placentae of healthy first-trimester pregnancies (E–H). KIR2DL1 and 2DS1 bind cells from C2-positive (A, B, E, and F) but not C2-negative (C, D, G, and H) donors. The indicated KIR-Fc fusion protein histograms are outlined in black and secondary antibody histograms in gray. Pre-incubating with an anti-KIR mAb (HP-3E4) blocked KIR2DS1-Fc and KIR2DL1-Fc binding to both PBLs and trophoblasts (histograms with dotted line).
Paternal and maternal C2 groups both influence reproductive success.
Do the KIR/HLA-C interactions we have observed in the placental bed have any functional significance? Placentation is defective in several important clinical pregnancy disorders including preeclampsia, FGR, and RM. We confirmed and extended our previous observations with additional patient cohorts to show that the frequency of the KIR AA genotype is significantly increased compared with normal controls in preeclampsia, and here we found it was also increased in FGR and particularly in women with RM (Supplemental Figure 3). In addition, in all affected cohorts, both mother and fetus had increased C2 carrier frequencies (Supplemental Figure 4). These results suggest that in all these conditions associated with defective placentation, women with a KIR AA genotype seem to be more at risk when this genotype is combined with a fetal C2 group (Table 2). As the 2 additional cohorts of pre-eclamptic patients showed the same differences in maternal KIR genotype, individual KIR gene, and C2 frequencies compared with the controls, as observed with our initial cohort (2), we combined all 3 cohorts for further, more detailed analyses.
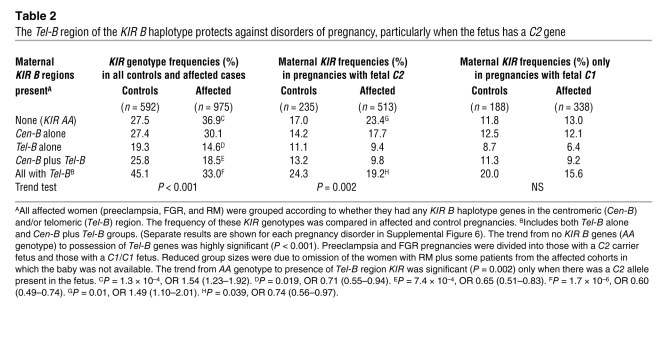
Table 2 .
The Tel-B region of the KIR B haplotype protects against disorders of pregnancy, particularly when the fetus has a C2 gene
uNK cells are likely to be educated during their development so that they are calibrated to the levels of maternal C2 present (24). The fetal trophoblast cells might have fewer, as many, or more C2 genes compared with the mother, and we predicted this could affect maternal uNK responses. We therefore analyzed the KIR AA frequencies in normal and affected pregnancies grouped according to the relative number of C2 genes in the mother and fetus (Figure 5A). In pregnancies where the fetus has more C2 genes than the mother (i.e., mother C1/C1 with fetal C1/C2 and mother C1/C2 with fetal C2/C2), the KIR AA genotype frequency was significantly increased in affected pregnancies compared with controls (P = 0.007, odds ratio [OR] 2.09) (Table 3). Thus, an extra copy of fetal C2 genes relative to the number of maternal C2 genes confers a significant risk. Furthermore, there was a statistically significant interaction detected between the KIR AA genotype and dosage of C2 genes (Table 3). This was shown by comparing the KIR AA frequencies between pregnancies in which the dose of C2 in the fetus was less or more than in the mother (P = 0.04, Mantel-Haenszel test). We conclude that there is a significant risk when the mother has a KIR AA genotype and the fetus has more C2 copies than the mother.
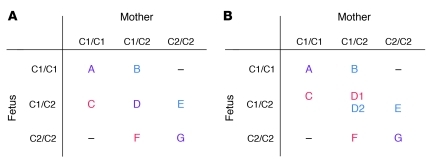
Figure 5. Categorization of pregnancy types according to the relative number of C2 genes or by parental origin of the fetal C2.
(A) Pregnancies were separated into 3 groups: those in which the fetus had fewer C2 genes than the mother (i.e., B and E), those in which the fetus and mother had the same number of C2 genes (i.e., A, D, and G), and those in which the fetus had more C2 genes than the mother (i.e., C and F). (B) Alternatively, pregnancies were selected in which the origin of the single fetal C2 gene could be definitively identified as being derived from either the father (i.e., C and D1) or the mother (i.e., D2 and E). HLA-C allele typing to 4 places was used to identify the parental origin of the fetal C2, distinguishing D1 from D2. See Table 3 for comparison of the groups.
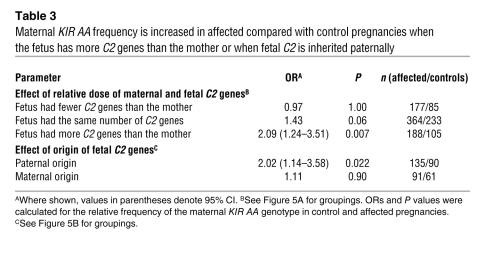
Table 3 .
Maternal KIR AA frequency is increased in affected compared with control pregnancies when the fetus has more C2 genes than the mother or when fetal C2 is inherited paternally
Where there is one more C2 allele in the fetus than in the mother, this allele must be derived from the father. To determine whether paternal C2 presents a risk distinguishable from a simple gene dosage effect, we performed another analysis, in which we selected only pregnancies with a single fetal C2 allele that we could definitively type as maternally or paternally inherited (Figure 5B). Even with these considerably reduced numbers, we could detect that only allogeneic paternal C2 ligands showed a significant risk (P = 0.022; OR 2.02) (Table 3). In both of these analyses, the fetus had inherited a paternal C2 allele. Based on these data, we propose that women with the KIR AA genotype are at an increased risk when confronted with additional fetal C2 alleles representing a paternal allogeneic C2 ligand on invading fetal trophoblast cells.
Genes in the telomeric region of KIR B haplotypes protect against disorders of pregnancy.
The corollary of our findings that KIR AA genotypes are detrimental is that KIR B haplotypes will be beneficial. To see which B-haplotype KIRs are protective, we grouped the women according to whether they had centromeric (Cen-B) and/or telomeric (Tel-B) genes (Figure 1). Although there was some protective effect when only the Cen-B region was present, the most striking observation in all pregnancy disorders (particularly in women with RM) was that the presence of the Tel-B region conferred highly significant protection (Table 2 and Supplemental Figure 5), which reached significance when the fetus carried a C2 group (Table 2). After comparing all controls (n = 592) with all affected pregnancies (n = 975), we found that the Tel-B region was present in 45% of controls but only in 33% of affected pregnancies (P = 1.7 × 10–6, OR 0.60 [95% CI 0.49–0.74]). A trend test analysis, comparing all cases with all controls, ordered from KIR AA to Tel-B (with and without Cen-B) gave a P value of 1 × 10–6 (Table 2). In spite of the consistently stronger deleterious effects of KIR AA in the mother when the fetus was C2, as opposed to C1, a Mantel-Haenszel test for an interaction of KIR AA and fetal C2 was not significant. This indicates that the deleterious effect of KIR AA is not dependent on fetal C2 alone. These observations, along with the significant difference between fetuses with fewer as opposed to more copies of C2 than the mother, emphasize the importance of maternal HLA-C status as well as the maternal/paternal derivation of fetal C2 (Table 3).
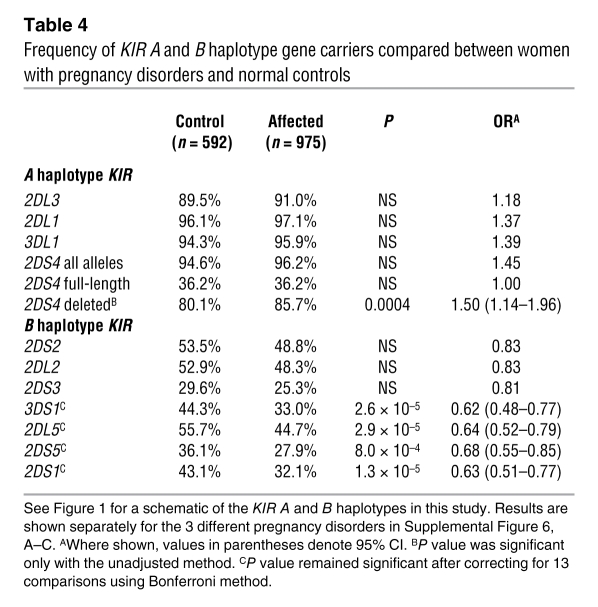
We also analyzed the frequency of individual KIR B haplotype genes to determine whether any particular KIR is responsible for the protective effect. We found a highly significant reduction in frequency for all KIR B genes telomeric to 2DL4 in all affected cohorts (Table 4 and Supplemental Figure 6, A–C). In particular, 2DS1, which encodes the activating KIR that binds to trophoblast C2 molecules, was present in 32% of affected pregnancies compared with 43% of controls (P = 1.3 × 10–5, OR 0.63 [95% CI 0.51–0.77]). Due to the strong linkage disequilibrium within the Tel and Cen regions, however, we cannot discard an additive effect of the other KIR genes within the Tel-B region.
Table 4 .
Frequency of KIR A and B haplotype gene carriers compared between women with pregnancy disorders and normal controls
Discussion
For successful establishment of the placenta, fetal trophoblast cells need to infiltrate the decidua and transform spiral arteries in the first few weeks of pregnancy. As a result, the fetus receives sufficient oxygen and nutrients for normal growth and development. This invasion must be balanced, so that excessive trophoblast penetration of the uterus does not occur (which would endanger the mother), or so that arterial transformation is not defective (which would starve the feto-placental unit). Our findings suggest that the immune system plays a part in defining this maternal-fetal boundary. The different experimental approaches we used all indicate that interactions of maternal KIRs with trophoblast HLA-C molecules influence placentation. Using the mAb WK4C11, we show that trophoblast cells strongly express both parental HLA-C allotypes. At the site of placentation, these fetal cells mingle with maternal NK cells expressing activating (2DS1) and inhibitory (2DL1) KIRs. Fresh uNK cells do not behave like PBNK cells in functional assays, and unless vigorously activated, they do not readily kill target cells or produce IFN-γ (9, 25–27). We therefore took a genetic approach to determine how polymorphism of KIR and HLA-C might affect maternal/fetal interactions. Genetic studies linking KIR and HLA variants with outcome in infectious diseases are now supported by functional studies (28–31). In preeclampsia and FGR, both of which are characterized by poor placentation, the pregnancies most at risk are those with a maternal KIR AA genotype in association with an extra C2 gene in the fetus relative to the mother. Furthermore, there is a clear protective effect when the mother has Tel-B KIR, the region containing KIR2DS1. Our findings indicate that maternal KIRs can mediate allo-recognition of paternal HLA-C on trophoblast, in a situation analogous to BMT (4). Indeed, recent studies reveal interesting parallels in transplantation for acute myelogenous leukemia, in that a reduced risk of relapse was found in transplantations involving donors with KIR B haplotypes (4, 32). However, the protective effect was strongest with Cen-B homozygosity, which is in contrast to our findings that Tel-B is protective. This probably reflects the dominant role of KIR2DS1 interaction with HLA-C in pregnancy compared with additional possible KIR interactions with a range of HLA and other ligands on leukemic cells.
The common genetic thread between these pregnancy disorders with diverse clinical presentations indicates that maternal KIR/fetal HLA-C interactions occur in the early weeks of gestation as the trophoblast moves into the decidua to effect the physiological conversion of uterine spiral arteries that is essential to successful pregnancy (33, 34). Interstitial trophoblasts invade decidua to surround and destroy the media of the spiral arteries, transforming them into high-conductance vessels. In addition, endovascular trophoblasts move down the spiral arteries during the first 8–10 weeks of gestation, forming a loose plug that prevents blood from flowing at high pressure into the delicate gestational sac. RM, defined as the loss of 3 or more consecutive pregnancies, affects 1%–3% of couples in the United Kingdom (35). The single most important predictive factor in identifying a woman’s risk of miscarriage is a previous miscarriage (36). The risk increases cumulatively as the number of previous miscarriages rises, suggesting that there are underlying predisposing causes. In addition, RM is linked to a poor reproductive outcome (preeclampsia or FGR) in subsequent pregnancies (37, 38). Defective placentation is known to occur in a substantial proportion of cases of early pregnancy loss, with reduced trophoblast invasion into both the decidua and spiral arteries (39–41). The underlying primary defect in preeclampsia, FGR, and stillbirth is also abnormal placentation, and these disorders share the resultant reduction in uteroplacental blood flow (5, 6, 42). Failure of placentation therefore underlies a spectrum of common pregnancy disorders.
The exact clinical presentation will depend on the degree of vascular transformation, the resultant disturbances in uterine artery blood flow, and the subsequent extent of stress responses in the developing placenta (5, 43). In miscarriage, trophoblast invasion is most severely deficient and the placenta is subjected to overwhelming oxidative stress, with fetal loss late in the first trimester. The decrease in frequency of KIR Tel-B genes and concomitant increase in KIR AA genotypes is most significant in women who experience this. By contrast, in pregnancies with FGR alone, trophoblast invades much further into the decidua, which only results in ER stress in the placenta (42, 43). Women who experienced only FGR had the least significant alteration in KIR gene frequencies. In preeclampsia, arterial vascular transformation is altered to an intermediate degree, with placental oxidative stress superimposed on the ER stress. Thus, in preeclampsia but not FGR, these severe stress responses trigger the release of inflammatory and anti-angiogenic factors into the maternal circulation. The severity of the resulting symptoms of preeclampsia depends on additional genes acting systemically in the mother. FGR and preeclampsia therefore exhibit different degrees of trophoblast transformation of spiral arteries, and this is reflected in the more significant findings in KIR/HLA-C genotypic variation in preeclampsia. These results may have clinical significance in that sperm from C1/C1 donors is predicted to be “safer”; indeed, C2/C2 males may be the “dangerous” males identified by epidemiological studies, since their sperm is guaranteed to lead to pregnancies in which the trophoblast cells express C2 (44, 45).
PBNK cells are calibrated during development by the strength of the input received by NK receptors from self-HLA, so they adjust to the specific HLA class I environment in an individual. This determines the responsiveness of mature NK cells, a process called NK education (24, 46). It is not yet known whether uNK cells are subjected to the same educational constraints as PBNK cells, and indeed there is now evidence that uNK cells may be educated in utero (47). Our findings do show an additional effect of maternal self–HLA-C on the risk conferred by maternal KIR AA in pregnancy. Women with the KIR AA genotype are particularly at risk when they have fewer C2 genes than the fetus, or when fetal C2 is paternally inherited. The size of our cohort did not provide the statistical power to determine whether it is the higher dose of fetal C2 or an allogeneic fetal C2 that results in harmful uNK cell responses in KIR AA women. However, our findings show that it is not just the number or identity of fetal C2 genes, but also their relation to maternal C2 that is important. This does implicate a role for maternal HLA-C in education during uNK cell development in determining the NK cell response to trophoblast.
We now show that in pregnancies in which trophoblast cells expressed C2, the most important protective maternal KIR genes for successful reproduction are those in the Tel-B region of the B haplotype. The only KIR on the Tel-B that is known to bind to HLA-C is KIR2DS1, the activating receptor for C2 groups (22). We showed specific binding of KIR2DS1 to normal trophoblast, providing confirmation that trophoblast C2 acts as a ligand for both KIR2DS1 and 2DL1. Although HLA-C molecules on trophoblast are all in a stable β2m-associated form with none of the unfolded conformers found on PBLs (8), the binding pattern was similar for both cell types with lower affinity of 2DS1 compared with 2DL1. One interpretation of these findings is that when the fetus has a C2 allele, uNK cell responses that result from KIR2DS1 binding to trophoblast C2 are beneficial for arterial transformation compared with those from a woman lacking this KIR. Several studies have shown that KIR2DS1+ PBNK cells can mediate allo-responses and produce IFN-γ after contact with C2+ target cells, but these 2DS1+ cells were only functional in a donor lacking C2 (48–50). Using PBNK cells, the presence of C2 in these 2DS1+ donors reduces the responsiveness of their NK cells, indicating that this activating KIR has an effect on PBNK cell education (51). The presence of maternal C1 or C2 groups might therefore affect uNK cell responsiveness not only in women with KIR AA genotypes, but also in those who possess KIR2DS1. In vitro assays such as CD107 or IFN-γ production are of limited use in assessing functions of freshly isolated uNK cells. Thus, despite some promising leads (52), the precise nature of uNK cell responses to C1+ or C2+ target cells is essentially unknown and will be a major challenge given the ethical and technical difficulties of studying these cells.
There are indications that other B haplotype KIR genes are important. Pregnancies with Cen-B KIR genes alone, without Tel-B region KIR, are at a reduced risk compared with KIR AA pregnancies (Table 4). Of these Cen-B KIR genes, KIR2DS3 appears non-functional (53) and the ligands for KIR2DS2 are unknown. However, KIR2DL2 does bind both C1 and C2 allotypes (54). In addition, the KIR2DL1*004 allele that is generally present on the KIR Cen-B haplotype is a poor NK cell educator and responses of uNK cells expressing this 2DL1 allele are likely to be weak (55). Thus, 2DL2 and 2DL1*004 Cen-B–associated KIRs might also influence successful placentation. Although C2 groups are clearly important, they appear not to be the only KIR ligands on the trophoblast, as there is a slight protective effect of Tel-B KIR in pregnancies even with a C1/C1 fetus. The ligands for KIR2DS5 and 2DL5 are unknown, but KIR3DS1 is unlikely to be important, as HLA-B antigens are never expressed by the trophoblast (7).
Using clinical, molecular, and genetic approaches, we now show that maternal KIR/fetal HLA-C interactions are determinants of successful placentation. These NK/trophoblast interactions occur in the first few weeks of pregnancy in the decidua basalis. Functional outcomes depend on inherent variability of maternal KIR and HLA-C as well as fetal HLA-C genes. Critical factors include the presence of an extra paternally derived C2 gene in the fetus and the advantageous effects of Tel-B KIR haplotype genes. The C2 group of MHC-C genes arose late and became fixed in primate evolution, but as yet there is little evidence that C2 is beneficial in infection (13, 28). This raises the question of what selective pressure keeps C2 in the human population. One possible explanation is that, although C2 is deleterious in the fetus, it appears to be beneficial in the mother, and thus reproductive performance might account for the persistence of C2 alleles in human populations. For KIR it is clear that KIR A haplotypes are beneficial in NK responses to infections (56). The fact that KIR A and B haplotypes have complementary functions in immunity and reproduction may serve to explain why they are found in all human populations (57).
Methods
Human tissue for cell biology.
Placental tissue was obtained from elective terminations of normal pregnancies between 6–12 weeks gestation. Ethical approval was obtained from the Cambridgeshire 2 Research Ethics Committee (reference no. 04/Q0108/23; Cambridge, United Kingdom), and informed written consent was obtained from each patient. Trophoblast cells and leukocytes were isolated as previously described (58).
Immunofluorescence staining of human implantation site.
Freshly cut 5-μm cryostat sections of first-trimester human implantation site were fixed for 10 minutes in cold acetone, rehydrated with PBS, and blocked for 1 hour with PBS/2% normal goat serum/1% BSA. Primary antibodies to HLA-C (clone DT9, mIgG2b) (20, 59) and cytokeratin-7 (clone OV-TL 12/30, mIgG1, Dako A/S) were applied together to sections overnight at 4°C. Isotype-specific goat anti-mouse IgG2b–Alexa Fluor 488 and goat anti-mouse IgG1–Alexa Fluor 568 (Invitrogen) were pre-absorbed with 10% human Ab serum and 2% normal goat serum prior to application to sections for 60 minutes at room temperature. Coverslips were mounted with Vectashield-DAPI and slides analyzed using a confocal microscope. Primary antibody to CD56 (clone My13, mIgG1; Zymed Laboratories Inc.) was applied as described above. Free anti-mouse sites were blocked with 200 μg/ml mouse IgG prior to the addition of mouse anti–HLA-G–FITC (clone MEM-G/9-FITC; AbD Serotec).
Fusion proteins.
Expression plasmids (provided by W. Carr, Massachusetts General Hospital, Charleston, Massachusetts, USA) containing extracellular regions of KIR2DL1 or KIR2DS1, matching the Ensemble sequences ENST00000336077 and ENST00000400867, respectively, fused to an IgG-Fc with the 18 C-terminal amino acids from IgA (60). A KIR2DL3 IgG-Fc expression plasmid was donated by E. Long (National Institute of Allergy and Infectious Diseases, Rockville, Maryland, USA) (61). Fusion proteins were expressed by transient transfection in 293T cells using Lipofectamine 2000 (Invitrogen) and purified from supernatant with protein G-Sepharose beads (Amersham Pharmacia Biotech). Fusion proteins resolved as single bands at approximately 75 kDa in reducing SDS-PAGE analysis. KIR2DL3-Fc bound C1- but not C2-transfected 721.221 cells. KIR2DL1-Fc and KIR2DS1-Fc bound C2- but not C1-transfected 721.221 cells.
Flow cytometry.
Freshly isolated or cultured trophoblasts were analyzed by flow cytometry as previously described (7). Briefly, cells were incubated with mAb or fusion protein, then with PE-conjugated polyclonal secondaries (Sigma-Aldrich and Dako), before staining with directly conjugated mAb to identify leukocyte or trophoblast cell populations using MEM-G/9-FITC to HLA-G (62) (AbD Serotec) and CD3-FITC (clone SK7) (Becton Dickinson). Fetal Hofbauer cells were gated out with CD45-PE-Cy5 (clone HI30) (BioLegend). Fusion protein binding was blocked by prior incubation with the mAb HP-3E4 (for KIR2DL1 and S1), a control isotype clone G155-228 (both Becton Dickinson) or the mAb WK4C11, generated from a hybridoma of B cells isolated from peripheral blood of multiparous women (21). WK4C11 reactivity was characterized against 96 common classical HLA-I allotypes using LABScreen Single Antigen beads (One Lambda) as previously described (7).
Expression of KIR2DS1 in CD56+CD14– uNK cells was examined by multicolor FACS analysis using the mAbs EB6 (to KIR2DL1 and KIR2DS1) (Beckman Coulter) and 8C11 (to most KIR2DL1 alleles but not KIR2DS1) (a gift from C. Retiere, University of Nantes, Nantes, France) (23). uNK cells were initially stained with 8C11, detected with goat anti-mouse IgG–Alexa Fluor 488 (Invitrogen), followed by EB6-allophycocyanin and CD56–peridinin chlorophyll protein (BioLegend).
Genomic DNA isolation and genotyping.
Genomic DNA was isolated from chorionic villi or decidual tissue and digested with proteinase K and RNase A (Roche) in combination with tissue lysis and protein precipitation buffers (Qiagen), prior to precipitation of DNA with isopropanol. HLA-C allele typing to 4 places was carried out at the Anthony Nolan Trust or in the Laboratory of Experimental Immunology.
Genomic DNA was obtained from maternal blood from 3 cohorts of women with preeclampsia, defined as new hypertension (140/90 mmHg) after week 20 of pregnancy, together with new proteinuria 300 mg/24 h (n = 742); from 2 groups with FGR (birth weight at the 5th percentile or less; n = 118); and from 1 cohort of couples in which the woman had suffered at least 3 unexplained miscarriages with no live births (RM women n = 115, RM men n = 81). Controls consisted of 3 cohorts of primiparous women with normal pregnancies and, where available, their male partners (total n = 592). Cord samples or mouth swabs were obtained from the babies for DNA isolation (preeclampsia babies n = 733, FGR babies n = 118, controls n = 423). All samples were from white British individuals. A study of our matched cohorts of preeclamptic and control pregnancies has been published previously (see Supplemental Figure 3 and ref. 2). The RM study group was an extension of our original study group (3). Genotyping data for the separate cohorts are provided in the supplemental figures. KIR and C1/C2 genotyping were performed as described previously (2, 3). KIR haplotype regions were defined by the presence of the following KIR genes: Cen-A/2DL3; Tel-A/3DL1 and 2DS4; Cen-B/2DL2 and 2DS2; Tel-B/2DS1 and 3DS1. Ethical approval for each study was obtained from the relevant committee: RM through the London Multiregional Ethical committee (reference no. 05/MRE02/20; London, United Kingdom), and preeclampsia and FGR through the Cambridge Research Ethics committee (reference nos. 01/197 and 05/Q0108/367; Cambridgeshire, United Kingdom). Informed written consent was obtained from all subjects.
Statistics.
The data were analyzed using the χ2 and 2-tailed Fisher’s exact test. A P value of 0.05 was considered to be statistically significant. When multiple testing was performed, the Bonferroni correction was applied. The magnitude of the effect was estimated by ORs and their 95% CIs (Windows 11.0.0.2001; SPSS Inc.). The trend tests were performed with SAS PROC FREQ procedure (SAS 9.1; SAS Institute). The Mantel-Haenszel test was used to test for interaction of KIR and HLA-C.
Supplementary Material
Acknowledgments
These studies have been supported by the Wellcome Trust, the British Heart Foundation, Wellbeing of Women, Centre for Trophoblast Research, and King’s College, University of Cambridge. We are grateful to Graham Burton and Charlie Loke for helpful discussions and Ying Qi for statistical advice. We would like to thank all the donors, the GOPEC Consortium, and research nurses for providing the samples, and the Anthony Nolan Trust for HLA-C typing and Luminex bead studies. This project was funded in whole or in part with federal funds from the National Cancer Institute, NIH, under contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported in part by the Intramural Research Program of the Center for Cancer Research (National Cancer Institute, NIH).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(11):4102–4110. doi:10.1172/JCI43998.
See the related Commentary beginning on page 3801.
References
- 1.Moffett A, Loke C. Immunology of placentation in eutherian mammals. Nat Rev Immunol. 2006;6(8):584–594. doi: 10.1038/nri1897. [DOI] [PubMed] [Google Scholar]
- 2.Hiby SE, et al. Combinations of maternal KIR and fetal HLA-C genes influence the risk of preeclampsia and reproductive success. J Exp Med. 2004;200(8):957–965. doi: 10.1084/jem.20041214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hiby SE, Regan L, Lo W, Farrell L, Carrington M, Moffett A. Association of maternal killer-cell immunoglobulin-like receptors and parental HLA-C genotypes with recurrent miscarriage. Hum Reprod. 2008;23(4):972–976. doi: 10.1093/humrep/den011. [DOI] [PubMed] [Google Scholar]
- 4.Cooley S, et al. Donors with group B KIR haplotypes improve relapse-free survival after unrelated hematopoietic cell transplantation for acute myelogenous leukemia. Blood. 2009;113(3):726–732. doi: 10.1182/blood-2008-07-171926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burton GJ, Jauniaux E. Placental oxidative stress: from miscarriage to preeclampsia. J Soc Gynecol Investig. 2004;11(6):342–352. doi: 10.1016/j.jsgi.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Burton GJ, Woods AW, Jauniaux E, Kingdom JC. Rheological and physiological consequences of conversion of the maternal spiral arteries for uteroplacental blood flow during human pregnancy. Placenta. 2009;30(6):473–482. doi: 10.1016/j.placenta.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Apps R, Murphy SP, Fernando R, Gardner L, Ahad T, Moffett A. Human leucocyte antigen (HLA) expression of primary trophoblast cells and placental cell lines, determined using single antigen beads to characterize allotype specificities of anti-HLA antibodies. Immunology. 2009;127(1):26–39. doi: 10.1111/j.1365-2567.2008.03019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Apps R, Gardner L, Hiby SE, Sharkey AM, Moffett A. Conformation of human leucocyte antigen-C molecules at the surface of human trophoblast cells. Immunology. 2008;124(3):322–328. doi: 10.1111/j.1365-2567.2007.02789.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moffett-King A. Natural killer cells and pregnancy. Nat Rev Immunol. 2002;2(9):656–63. doi: 10.1038/nri886. [DOI] [PubMed] [Google Scholar]
- 10.Koopman LA, et al. Human decidual natural killer cells are a unique NK cell subset with immunomodulatory potential. J Exp Med. 2003;198(8):1201–1212. doi: 10.1084/jem.20030305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verma S, King A, Loke YW. Expression of killer cell inhibitory receptors on human uterine natural killer cells. Eur J Immunol. 1997;27(4):979–983. doi: 10.1002/eji.1830270426. [DOI] [PubMed] [Google Scholar]
- 12.Sharkey AM, et al. Killer Ig-like receptor expression in uterine NK cells is biased toward recognition of HLA-C and alters with gestational age. J Immunol. 2008;181(1):39–46. doi: 10.4049/jimmunol.181.1.39. [DOI] [PubMed] [Google Scholar]
- 13.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5(3):201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 14.Bashirova AA, Martin MP, McVicar DW, Carrington M. The killer immunoglobulin-like receptor gene cluster: tuning the genome for defense. Annu Rev Genomics Hum Genet. 2006;7:277–300. doi: 10.1146/annurev.genom.7.080505.115726. [DOI] [PubMed] [Google Scholar]
- 15.Middleton D, Menchaca L, Rood H, Komerofsky R. New allele frequency database: http://www.allelefrequencies.net. . Tissue Antigens. 2003;61(5):403–407. doi: 10.1034/j.1399-0039.2003.00062.x. [DOI] [PubMed] [Google Scholar]
- 16.Hsu KC, Liu XR, Selvakumar A, Mickelson E, O’Reilly RJ, Dupont B. Killer Ig-like receptor haplotype analysis by gene content: evidence for genomic diversity with a minimum of six basic framework haplotypes, each with multiple subsets. J Immunol. 2002;169(9):5118–5129. doi: 10.4049/jimmunol.169.9.5118. [DOI] [PubMed] [Google Scholar]
- 17.Yawata M, Yawata N, Abi–Rached L, Parham P. Variation within the human killer cell immunoglobulin–like receptor (KIR) gene family. Crit Rev Immunol. 2002;22(5–6):463–482. [PubMed] [Google Scholar]
- 18.Hsu KC, Chida S, Geraghty DE, Dupont B. The killer cell immunoglobulin-like receptor (KIR) genomic region: gene order, haplotypes and allelic polymorphism. Immunol Rev. 2002;190:40–52. doi: 10.1034/j.1600-065X.2002.19004.x. [DOI] [PubMed] [Google Scholar]
- 19.Martin MP, Single RM, Wilson MJ, Trowsdale J, Carrington M. KIR haplotypes defined by segregation analysis in 59 Centre d’Etude Polymorphisme Humain (CEPH) families. Immunogenetics. 2008;60(12):767–774. doi: 10.1007/s00251-008-0334-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas R, et al. HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nat Genet. 2009;41(12):1290–1294. doi: 10.1038/ng.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulder A, et al. Human monoclonal HLA antibodies reveal interspecies crossreactive swine MHC class I epitopes relevant for xenotransplantation. Mol Immunol. 2010;47(4):809–815. doi: 10.1016/j.molimm.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Stewart CA, et al. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc Natl Acad Sci U S A. 2005;102(37):13224–13229. doi: 10.1073/pnas.0503594102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.David G, et al. Discrimination between the main activating and inhibitory killer cell immunoglobulin-like receptor positive natural killer cell subsets using newly characterized monoclonal antibodies. Immunology. 2009;128(2):172–184. doi: 10.1111/j.1365-2567.2009.03085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brodin P, Kärre K, Höglund P. NK cell education: not an on-off switch but a tunable rheostat. Trends Immunol. 2009;30(4):143–149. doi: 10.1016/j.it.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 25.Kopcow HD, et al. Human decidual NK cells form immature activating synapses and are not cytotoxic. Proc Natl Acad Sci U S A. 2005;102(43):15563–15568. doi: 10.1073/pnas.0507835102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eriksson M, Meadows SK, Basu S, Mselle TF, Wira CR, Sentman CL. TLRs mediate IFN-gamma production by human uterine NK cells in endometrium. J Immunol. 2006;176(10):6219–6224. doi: 10.4049/jimmunol.176.10.6219. [DOI] [PubMed] [Google Scholar]
- 27.Vacca P, et al. Regulatory role of NKp44, NKp46, DNAM-1 and NKG2D receptors in the interaction between NK cells and trophoblast cells. Evidence for divergent functional profiles of decidual versus peripheral NK cells. Int Immunol. 2008;20(11):1395–1405. doi: 10.1093/intimm/dxn105. [DOI] [PubMed] [Google Scholar]
- 28.Khakoo SI, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305(5685):872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 29.Carrington M, Martin MP. The impact of variation at the KIR gene cluster on human disease. Curr Top Microbiol Immunol. 2006;298:225–257. doi: 10.1007/3-540-27743-9_12. [DOI] [PubMed] [Google Scholar]
- 30.Alter G, et al. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J Exp Med. 2007;204(12):3027–3036. doi: 10.1084/jem.20070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin MP, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39(6):733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cooley S, et al. Donor selection for natural killer cell receptor genes leads to superior survival after unrelated transplantation for acute myelogenous leukemia [published online ahead of print June 25, 2010]. Blood. doi: 10.1182/blood-2010-05-283051. doi: 10.1182/blood-2010-05-283051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Robertson WB. Pathology of the pregnant uterus. In: Fox H, ed.Obstetrical and Gynaecological Pathology . London, United Kingdom: Churchill Livingstone; 1987:1149–1176. [Google Scholar]
- 34.Pijnenborg R, Vercruysse L, Hanssens M. The uterine spiral arteries in human pregnancy: facts and controversies. Placenta. 2006;27(9–10):939–958. doi: 10.1016/j.placenta.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 35.Li TC, Makris M, Tomsu M, Tickerman E, Laird S. Recurrent miscarriage:aetiology, management and prognosis. Hum Reprod Update. 2002;8(5):463–481. doi: 10.1093/humupd/8.5.463. [DOI] [PubMed] [Google Scholar]
- 36.Regan L, Braude PR, Trembath PL. Influence of past reproductive performance on risk of spontaneous abortion. BMJ. 1989;299(6698):541–545. doi: 10.1136/bmj.299.6698.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trogstad L, Magnus P, Moffett A, Soltenberg C. The effect of recurrent miscarriage and infertility on the risk of pre-eclampsia. BJOG. 2009;116(1):108–113. doi: 10.1111/j.1471-0528.2008.01978.x. [DOI] [PubMed] [Google Scholar]
- 38.van Oppenraaij RH, et al. Predicting adverse obstetric outcome after early pregnancy events and complications: a review. Hum Reprod Update. 2009;15(4):409–421. doi: 10.1093/humupd/dmp009. [DOI] [PubMed] [Google Scholar]
- 39.Khong TY, Liddell HS, Robertson WB. Defective haemochorial placentation as a cause of miscarriage: a preliminary study. Br J Obstet Gynaecol. 1987;94(7):649–655. doi: 10.1111/j.1471-0528.1987.tb03169.x. [DOI] [PubMed] [Google Scholar]
- 40. Pijnenborg R, Vercruysse L, Hanssens M, Van Assche A. Incomplete trophoblast invasion:the evidence. In: Critchley H, Maclean A, Poston L, Walker J, eds.Pre–eclampsia . London, United Kingdom: RCOG Press; 2003:15–26. [Google Scholar]
- 41.Jauniaux E, Burton GJ. Pathophysiology of histological changes in early pregnancy loss. Placenta. 2005;26(2–3):114–123. doi: 10.1016/j.placenta.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 42.Ness RB, Sibai BM. Shared and disparate components of the pathophysiologies of fetal growth restriction and preeclampsia. Am J Obstet Gynecol. 2006;195(1):40–49. doi: 10.1016/j.ajog.2005.07.049. [DOI] [PubMed] [Google Scholar]
- 43.Burton GJ, Yung HW, Cindrova-Davies T, Charnock-Jones DS. Placental endoplasmic reticulum stress and oxidative stress in the pathophysiology of unexplained intrauterine growth restriction and early onset preeclampsia. Placenta. 2009;30(suppl A):S43–S48. doi: 10.1016/j.placenta.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Esplin MS, et al. Paternal and maternal components of the predisposition to preeclampsia. N Engl J Med. 2001;344(12):867–872. doi: 10.1056/NEJM200103223441201. [DOI] [PubMed] [Google Scholar]
- 45.Skjaerven R, Vatten LJ, Wilcox AJ, Rønning T, Irgens LM, Lie RT. Recurrence of pre-eclampsia across generations: exploring fetal and maternal genetic components in a population based cohort. BMJ. 2005;331(7521):877–881. doi: 10.1136/bmj.38555.462685.8F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joncker NT, Raulet DH. Regulation of NK cell responsiveness to achieve self-tolerance and maximal responses to diseased target cells. Immunol Rev. 2008;224:85–97. doi: 10.1111/j.1600-065X.2008.00658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Male V, Hughes T, McClory S, Colucci F, Caliguri MA, Moffett A. Immature NK cells, capable of producing IL-22, are present in human uterine mucosa [published online ahead of print August 27, 2010]. J Immunol. doi: 10.4049/jimmunol.1001637. doi: 10.4049/jimmunol.1001637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chewning JH, Gudme CN, Hsu KC, Selvakumar A, Dupont B. KIR2DS1-positive NK cells mediate alloresponse against the C2 KLA-KIR ligand group in vitro. J Immunol. 2007;179(2):854–868. doi: 10.4049/jimmunol.179.2.854. [DOI] [PubMed] [Google Scholar]
- 49.Foley B, De Santis D, Lathbury L, Christiansen F, Witt C. KIR2DS1-mediated activation overrides NKG2A-mediated inhibition in HLA-C C2-negative individuals. Int Immunol. 2008;20(4):555–563. doi: 10.1093/intimm/dxn013. [DOI] [PubMed] [Google Scholar]
- 50.Morvan M, et al. Autologous and allogeneic HLA KIR ligand environments and activating KIR control KIR NK-cell functions. Eur J Immunol. 2008;38(12):3474–3486. doi: 10.1002/eji.200838407. [DOI] [PubMed] [Google Scholar]
- 51.Fauriat C, Ivarsson MA, Ljunggren HG, Malmberg KJ, Michaëlsson J. Education of human natural killer cells by activating killer cell immunoglobulin-like receptors. Blood. 2010;115(6):1166–1174. doi: 10.1182/blood-2009-09-245746. [DOI] [PubMed] [Google Scholar]
- 52.Hanna J, et al. Decidual NK cells regulate key developmental processes at the human fetal-maternal interface. Nat Med. 2006;12(9):1065–1074. doi: 10.1038/nm1452. [DOI] [PubMed] [Google Scholar]
- 53.VandenBussche CJ, Mulrooney TJ, Frazier WR, Dakshanamurthy S, Hurley CK. Dramatically reduced surface expression of NK cell receptor KIR2DS3 is attributed to multiple residues throughout the molecule. Genes Immun. 2009;10(2):162–173. doi: 10.1038/gene.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J Immunol. 2008;180(6):3969–3979. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- 55.Yawata M, Yawata N, Draghi M, Partheniou F, Little AM, Parham P. MHC class I-specific inhibitory receptors and their ligands structure diverse human NK-cell repertoires toward a balance of missing self-response. Blood. 2008;112(6):2369–2380. doi: 10.1182/blood-2008-03-143727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parham P. The genetic and evolutionary balances in human NK cell receptor diversity. Semin Immunol. 2008;20(6):311–316. doi: 10.1016/j.smim.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gendzekhadze K, et al. Co-evolution of KIR2DL3 with HLA-C in a human population retaining minimal essential diversity of KIR and HLA class I ligands. Proc Natl Acad Sci U S A. 2009;106(44):18692–18697. doi: 10.1073/pnas.0906051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Male V, et al. Natural killer cells in human pregnancy. Methods Mol Biol. 2010;612:447–463. doi: 10.1007/978-1-60761-362-6_30. [DOI] [PubMed] [Google Scholar]
- 59.Braud VM, Allan DS, Wilson D, McMichael AJ. TAP- and tapasin-dependent HLA-E surface expression correlates with the binding of an MHC class I leader peptide. Curr Biol. 1998;8(1):1–10. doi: 10.1016/S0960-9822(98)70014-4. [DOI] [PubMed] [Google Scholar]
- 60.Sørensen V, Rasmussen IB, Norderhaug L, Natvig I, Michaelsen TE, Sandlie I. Effect of the IgM and IgA secretory tailpieces on polymerization and secretion of IgM and IgG. J Immunol. 1996;156(8):2858–2865. [PubMed] [Google Scholar]
- 61.Winter CC, Gumperz JE, Parham P, Long EO, Wagtmann N. Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA-C allotype recognition. J Immunol. 1998;161(2):571–577. [PubMed] [Google Scholar]
- 62.Menier C, et al. Characterization of monoclonal antibodies recognizing HLA-G or HLA-E: new tools to analyze the expression of nonclassical HLA class I molecules. Hum Immunol. 2003;64(3):315–326. doi: 10.1016/S0198-8859(02)00821-2. [DOI] [PubMed] [Google Scholar]
- 63.Graef T, et al. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C. . J Exp Med. 2009;206(11):2557–2572. doi: 10.1084/jem.20091010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.