Abstract
Activating mutations in the Ras alleles are found frequently in tumors, making the proteins they encode highly attractive candidate therapeutic targets. However, Ras proteins have proven difficult to target directly. Recent approaches have therefore focused on identifying indirect targets to inhibit Ras-induced oncogenesis. For example, RNAi-based negative selection screens to identify genes that when silenced in concert with activating Ras mutations are incompatible with cellular proliferation, a concept known as synthetic lethality. In this issue of the JCI, Vicent et al. report on the identification of Wilms tumor 1 (Wt1) as a Kras synthetic-lethal gene in a mouse model of lung adenocarcinoma. Silencing of Wt1 in cells expressing an endogenous allele of activated Kras triggers senescence in vitro and has an impact on tumor progression in vivo. These findings are of significant interest given previous studies suggesting that the ability of oncogenic Kras to induce senescence versus proliferation depends on its levels of expression.
Activating mutations in the Ras genes (Kras, Hras, and Nras) are a common occurrence in a broad spectrum of tumors (1). It is therefore not surprising that extensive efforts have been made to develop therapies to directly target oncogenic Ras. However, the Ras proteins have proven to be formidable foes, and success with direct targeting approaches has been limited so far. One strategy to circumvent this issue has been to search for targets that could serve as a potential Achilles’ heel to be exploited to have an impact on Ras indirectly. This approach is based on the idea that expression of oncogenic Ras results in tumor cell alterations so that they become dependent on one or more pathways or particular molecular targets, which then represent ideal hits to strike the tumor cells while sparing normal cells (2). The concept of synthetic lethality perfectly fits this idea. Synthetic lethality between two genes occurs when loss of function of one gene results in cell death only in the presence of genetic alteration of the other, while mutation of either gene alone is compatible with viability. Several genes and pathways have been identified by this approach and, interestingly, many of these are not oncogenic themselves but become essential for cells in the tumorigenic state (see below for discussion of specific examples). It is thus clear why a gene that exhibits a synthetic-lethal interaction with activated Ras would represent a high-value target for the development of therapeutics. In this issue of the JCI, Vicent et al. describe the identification of Wilms tumor 1 (Wt1) as a novel synthetic-lethal gene in a mouse model of Kras-induced tumorigenesis in the lung (3).
Negative selection screens to identify Ras synthetic-lethal interactors
In the past, synthetic-lethal interactions have been largely studied in model organisms such as yeast, Drosophila melanogaster, and Caenorhabditis elegans. More recently, thanks to the development of RNAi technology (4–6), it has become feasible to extend the concept of synthetic lethality to mammalian cells to identify genes whose loss of function causes growth arrest or cell death (negative selection). Furthermore, the employment of systematic high-throughput platforms has allowed for screening of significant numbers of targets within a relatively short period of time and in an unbiased manner. In the case of Ras, after knocking down one or more specific targets, it is possible to evaluate the effects on viability through side-by-side comparison of cells with and without oncogenic Ras expression. However, the limitations of these approaches should also be noted. For example, the response to the inhibition of any given target will be highly dependent on multiple factors including cell type and screen conditions. Furthermore, to achieve a strong signal-to-background ratio and to control for off-target effects related to RNAi approaches, the experimental conditions of the screening require careful optimization (7). Nevertheless, the power of such approaches has been demonstrated recently by a number of studies (8–10).
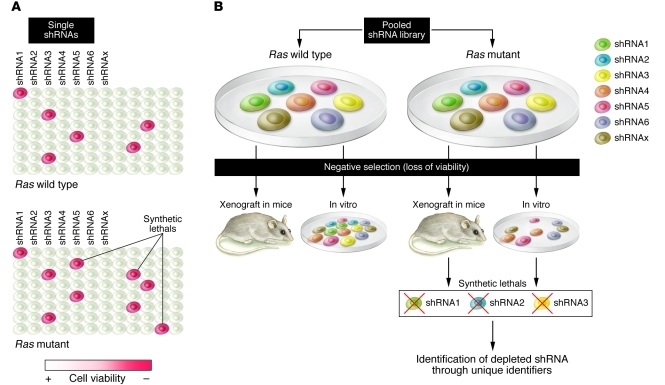
Negative selection screens can be conducted using a well-by-well array or a pooled approach (Figure 1). In the well-by-well array, the impact of each shRNA on cell growth and survival is scored individually, requiring a high-throughput platform to perform the screen (Figure 1A). A successful example of this approach is the work of Scholl et al. (10), which led to the identification of serine/threonine kinase 33 (STK33), a kinase not previously known to be associated with cancer, as a Kras synthetic lethal. An shRNA library targeting approximately 1,000 genes encoding the majority of known and predicted protein kinases, selected phosphatases, and other known cancer-related targets was screened across a panel of Kras wild-type and mutant human cancer cell lines. Interestingly, STK33 is thought to function in this context by regulating the activity of the cell death agonist BCL2-associated agonist of cell death (BAD) (10). This work is an elegant example of the power of synthetic lethality screens; STK33, indeed, was not known as a component of the Kras signaling pathway and does not behave as an oncogene in transformation assays; therefore, its role in Kras tumorigenesis would have been difficult to unveil by other approaches.
Figure 1. Negative selection shRNA screening approaches: single-well format versus pooled libraries.
(A) In single-well arrays, shRNAs are transduced at 1 per well. Negative effect on growth and survival is evaluated individually by direct comparison of corresponding wells in different plates containing Kras wild-type (reference) and Kras mutant cells. (B) Pooled screens are conducted by infecting Kras wild-type (reference) and Kras mutant cells with an shRNA library. The cell populations are then selected either in vitro or in vivo, resulting in shRNAs with unfavorable effects on viability being depleted from the pool of cells. The depleted shRNAs are then identified through bar code tags.
Another arrayed-format screen, which underscored the importance of the NF-κB pathway as a critical effector of Ras signaling, revealed the dependence on TANK-binding kinase 1 (TBK1) for the survival of cells harboring activating Kras mutations (8). TBK1 is a noncanonical IκB kinase and regulates innate immunity through activation of NF-κB, interferon regulatory factor 3 (IRF3), and IRF7 (11). According to the report by Barbie et al., in the context of oncogenic Kras-driven lung tumorigenesis, TBK1 preferentially activates the NF-κB pathway rather than the interferon response and loss of TKB1 ultimately results in inhibition of antiapoptotic signaling (8).
An alternative strategy for negative selection RNAi screening is the pooled approach, in which a bulk population of cells is infected with an entire pooled shRNA library at a low multiplicity of infection and selected for loss of viability either in vitro or upon implantation into mice (Figure 1B). shRNAs that are selectively eliminated from the pool can then be identified through “bar codes.” This approach allows for rapid evaluation of large, virtually genome-wide gene sets and for the identification of pathways and classes of genes rather than single candidates. An interesting example is the identification of mitotic genes such as polo-like kinase 1 (PLK1) and adenomatous polyposis coli (APC) as Kras synthetic lethals (9). Luo et al. propose that activated Kras subjects the cells to mitotic stress so that they require certain mitotic effectors to progress through mitosis (9). In the absence of these factors, mitosis is stalled or arrested, leading to cell death.
Identification of Wt1 as a regulator of senescence and proliferation downstream of Kras
In this issue of the JCI, Vicent et al. report on their performance of a pooled negative selection shRNA screen to interrogate a library focused on targets that had been previously implicated in non–small cell lung carcinoma (NSCLC) and/or downstream of Kras (3). The study was performed in cell lines derived from tumors that develop in an endogenous mouse model of Kras-driven lung adenocarcinoma. The list of top-scoring genes obtained from this primary screen was validated in a secondary screen in primary mouse embryo fibroblasts (MEFs) expressing either wild-type or oncogenic Kras. Of the targets validated, the authors focused on Wt1, a potential transcriptional regulator of the Kras signature.
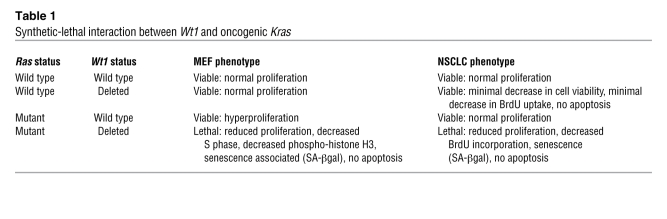
The authors went on to validate the synthetic-lethal interaction between Kras and Wt1 at multiple levels (3). First, they demonstrated that the effect of Wt1 loss was specific to oncogenic Kras-expressing cells by repeating the secondary screen in a mouse lung epithelial cell line expressing wild-type Kras. Moreover, they showed that loss of Wt1 reduced the tumorigenicity of a Kras-mutant lung tumor cell line in a xenograft model and provided strong genetic evidence of the functional interaction between Kras and Wt1 through the employment of a mouse model harboring a conditionally expressed oncogenic allele of Kras combined with a conditional knockout allele of Wt1. MEFs derived from these animals displayed a cell-cycle distribution compatible with an antiproliferative effect of Wt1 loss, specifically in oncogenic Kras-expressing cells (Table 1). Finally, the loss of Wt1 in an endogenous model of Kras-induced lung adenocaricnoma resulted in decreased tumor burden compared with mice expressing Wt1, confirming the requirement for Wt1 in Kras-driven lung tumorigenesis.
Table 1 .
Synthetic-lethal interaction between Wt1 and oncogenic Kras
WT1 was originally identified as a tumor suppressor in Wilms tumor (12). It is a zinc-finger transcription factor expressed as multiple splice forms and has been shown to have either tumor suppressive or oncogenic functions in a manner that appears to be cell-type and context dependent (13). Considering that Wt1 functions as a transcription factor, the transcriptional profiles of oncogenic Kras-expressing cells with or without Wt1 expression were analyzed (3). The authors reasoned that if WT1 targets were indeed important for RAS-induced tumorigenesis, it would be expected that patients with activated RAS alleles and decreased expression of WT1 target genes would have an improved prognosis. The transcriptional profiles obtained from the MEFs were used to develop “WT1 high” and “WT1 low” signatures. Using these signatures, the authors showed that the WT1-signature status allowed stratification of lung cancer patients into poor and good prognosis groups, but only in the presence of a contemporaneous KRAS gene signature. This suggests that WT1 modulates expression of genes that are specifically relevant to KRAS-driven lung cancers. While the data are highly circumstantial, they support the notion of a KRAS/WT1 synthetic-lethal interaction in patients. Clearly, additional studies are required to further explore this hypothesis.
Senescence and the anticancer barrier
How does the status of Wt1 so profoundly have an impact on cells harboring an activating Kras mutation? Vicent et al. show that the loss of Wt1 expression in the context of activated Kras in MEFs results in decreased cellular proliferation and increased senescence (3). Importantly, they corroborated these observations, made in primary MEFs and in mouse lung tumor cells, in human NSCLC cell lines (Table 1). Kras represents the paradigm of oncogene-induced senescence (OIS), being the first oncogene for which paradoxical growth arrest after ectopic expression was observed (14). Work over the past few years has suggested that the ability of oncogenic Kras to induce senescence versus proliferation depends on the levels of its expression above a certain threshold (15, 16). In addition, it has been recently proposed that induction of senescence and resistance to oncogenic Kras transformation is tissue specific and correlates with the strength of tumor suppression, as assessed by the transcriptional status of the p19Arf locus (17). In this study, Young and Jacks show that physiological levels of oncogenic Kras expression induce transformation rather than senescence in the lung due to a stable transcriptional silencing of the p19Arf locus, whereas in other tissues, where this locus is in a more permissive state, expression of oncogenic Kras strongly induces p19Arf expression and consequently the onset of the senescence response.
The question of how Wt1 controls senescence remains open (neither alterations in the levels of p16Ink4a, p19Arf, and trp53 expression nor increased activation of downstream MAPK signaling was found by Vicent et al. after knockdown of Wt1 expression) (3). Nevertheless, the data from Vicent et al. are of significant interest because they demonstrate that in mouse primary cells, under certain conditions such as Wt1 loss, senescence can occur in response to physiologic levels of oncogenic Kras. These observations also point to the involvement of what could potentially be a novel tumor suppressive mechanism that likely does not involve the usual suspects previously implicated in control of senescence.
Future directions
While a number of studies have recently pointed at OIS, originally identified as an in vitro phenomenon, as a barrier to tumorigenesis in vivo, the mechanistic details involved in the process remain incompletely understood (18–21). An interesting question stemming from the work of Vicent et al. (3) is whether their observations regarding senescence also occur in vivo. The authors show that deletion of Wt1 in the context of oncogenic Kras in the lung results in a significant reduction in tumor volume but not in the total number of lesions, indicating that Wt1 loss likely affects tumor progression rather than initiation. This fits with a role for Wt1 in repressing the senescence response to oncogenic Kras as observed in vitro. However, more studies are needed to conclusively prove that Wt1 controls Kras-induced senescence in vivo.
Acknowledgments
The work in the Kissil laboratory is supported in part by NIH grant R01CA124495 and the American Cancer Society.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(11):3804–3807. doi:10.1172/JCI44901.
See the related article beginning on page 3940.
References
- 1.Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9(7):517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaelin WG., Jr The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5(9):689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 3.Vicent S, et al. Wilms tumor 1 (WT1) regulates KRAS-driven oncogenesis and senescence in mouse and human models. J Clin Invest. 2010;120(11):3940–3952. doi: 10.1172/JCI44165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Westbrook TF, Stegmeier F, Elledge SJ. Dissecting cancer pathways and vulnerabilities with RNAi. Cold Spring Harb Symp Quant Biol. 2005;70:435–444. doi: 10.1101/sqb.2005.70.031. [DOI] [PubMed] [Google Scholar]
- 5.Downward J. Use of RNA interference libraries to investigate oncogenic signalling in mammalian cells. Oncogene. 2004;23(51):8376–8383. doi: 10.1038/sj.onc.1208073. [DOI] [PubMed] [Google Scholar]
- 6.Bernards R, Brummelkamp TR, Beijersbergen RL. shRNA libraries and their use in cancer genetics. Nat Methods. 2006;3(9):701–706. doi: 10.1038/nmeth921. [DOI] [PubMed] [Google Scholar]
- 7.Sharma S, Rao A. RNAi screening: tips and techniques. . Nat Immunol. 2009;10(8):799–804. doi: 10.1038/ni0809-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbie DA, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462(7269):108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo J, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137(5):835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scholl C, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137(5):821–834. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 11.Hacker H, Karin M. Regulation and function of IKK and IKK-related kinases. Sci STKE. 2006;2006(357):re13. doi: 10.1126/stke.3572006re13. [DOI] [PubMed] [Google Scholar]
- 12.Rivera MN, Haber DA. Wilms’ tumour: connecting tumorigenesis and organ development in the kidney. . Nat Rev Cancer. 2005;5(9):699–712. doi: 10.1038/nrc1696. [DOI] [PubMed] [Google Scholar]
- 13.Yang L, Han Y, Suarez Saiz F, Minden MD. A tumor suppressor and oncogene: the WT1 story. Leukemia. 2007;21(5):868–876. doi: 10.1038/sj.leu.2404624. [DOI] [PubMed] [Google Scholar]
- 14.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88(5):593–602. doi: 10.1016/S0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 15.Tuveson DA, et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5(4):375–387. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- 16.DeNicola GM, Tuveson DA. RAS in cellular transformation and senescence. Eur J Cancer. 2009;45(Suppl 1):211–216. doi: 10.1016/S0959-8049(09)70036-X. [DOI] [PubMed] [Google Scholar]
- 17.Young NP, Jacks T. Tissue-specific p19Arf regulation dictates the response to oncogenic K-ras. Proc Natl Acad Sci U S A. 2010;107(22):10184–10189. doi: 10.1073/pnas.1004796107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braig M, et al. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436(7051):660–665. doi: 10.1038/nature03841. [DOI] [PubMed] [Google Scholar]
- 19.Chen Z, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436(7051):725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collado M, et al. Tumour biology: senescence in premalignant tumours. Nature. 2005;436(7051):642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 21.Michaloglou C, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436(7051):720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]