Abstract
Protein expression in humans is controlled by numerous RNA processing steps that occur between transcription of a gene and translation of protein. However, the importance of such regulatory steps to human diseases, especially cancer, is just now coming to light. Changes in the alternative splicing or stability of mRNA transcribed from genes involved in cell-cycle control, cell proliferation, and apoptosis has been linked to tumor formation and progression. Nevertheless, in the majority of these cases, the identity of the regulators that control the expression of such cancer-related genes is poorly understood. In this issue of the JCI, Goehe et al. demonstrate that heterogeneous nuclear ribonuclear protein family member L (hnRNP L), a member of the hnRNP family of RNA processing factors, is specifically phosphorylated in non–small cell lung cancer (NSCLC). The phosphorylated hnRNP L, in turn, promotes expression of the antiapoptotic form of caspase-9, thereby contributing to tumorigenesis.
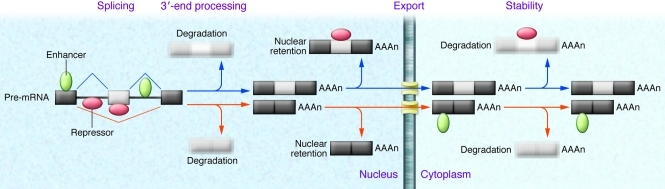
In humans, as in other higher eukaryotes, nascent pre-mRNA transcripts are subject to multiple processing steps prior to translation (1). Each one of these steps is a potential point of regulation and can control both on/off decisions in gene expression as well as allow for preferential expression of distinct mRNA and protein isoforms (Figure 1). Alternative splicing, which involves differential ligation of exons to generate distinct mRNAs from a common transcript, controls the expression of more than 90% of human genes (2, 3). Differential isoform expression can also be controlled at the level of selective mRNA export to the cytoplasm and/or mRNA stability (1). Alternative splicing as well as isoform-specific export and stability are typically controlled by the binding of regulatory proteins to cis-regulatory sequences (Figure 1). Notably, members of the heterogenous nuclear ribonuclear protein family (hnRNPs) have been shown to have activity at all of these checkpoints in mRNA processing (4).
Figure 1. The processing of a nascent transcript involves numerous potential points of regulation.
Pre-mRNA molecules synthesized in the nucleus are subjected to specific RNA processing steps in both the nucleus and cytosol of cells. The RNA processing events in the nucleus include capping, splicing, and 3′-end processing (and, in some cases, RNA editing), while regulation of poly(A) tail length typically occurs in the cytosol. Each of these RNA processing steps can be regulated so as to generate multiple mRNA isoforms of a given gene. The depicted pre-mRNA molecule (boxes represent exons, while the line denotes introns) can be differentially spliced to generate mRNA isoforms that encode functionally distinct proteins. Pre-mRNA alternative splicing can also dictate isoform expression by regulating mRNA stability via inducing/blocking NMD. Differential isoform expression can further be achieved by regulating the mRNA export of specific isoforms. In the cytosol, mRNAs containing degradation signals can be selectively degraded, in turn leading to differential protein isoform expression. In each of the above scenarios, regulation is brought about by recognition of cis-acting elements in the pre-mRNA/mRNA by cognate trans-acting factors that promote (green ovals) or inhibit (red ovals) particular events. Light gray boxes indicate mRNAs subject to degradation. "AAAn" refers to the addition of a poly(A) tail.
As each given mRNA isoform has the capacity to encode a protein of unique function, the generation of multiple distinct mRNA species from a common gene greatly expands the proteome capacity of the human genome. Moreover, a considerable proportion of alternative processing events are tissue- and developmental stage–specific, which helps to shape cell type–specific protein expression and function (5). It is therefore not surprising that aberrant regulation of splicing, export, stability, and other processing steps often perturbs normal cellular function and leads to disease pathology. Indeed, a substantial number of disease-causing mutations in humans alter mRNA processing events (6). However, the mechanisms by which such mutations function are typically poorly understood, thus limiting our ability to predict and treat human disease. In this issue of the JCI (7), Goehe and colleagues provide a comprehensive study demonstrating that hnRNP family member L (hnRNP L) is specifically phosphorylated in non–small cell lung cancer cells (NSCLC cells) and that this regulates its association with caspase-9 pre-mRNA. The interaction of hnRNP L with caspase-9 pre-mRNA in NSCLC cells promotes preferential expression of the 9b isoform of caspase-9, which is antiapoptotic, and promotes tumor growth, thereby providing a direct link between hnRNP L function and human lung cancer.
hnRNP L regulates caspase-9 isoform expression in NSCLC
Caspase-9 is an apoptosis-initiating caspase that is known to have two distinct protein isoforms generated as a result of alternative pre-mRNA splicing (Figure 2 and ref. 8). Caspase-9a mRNA contains variable exons 3, 4, 5, and 6 and encodes the active proapoptotic caspase, while caspase-9b mRNA lacks the four variable exons and encodes an antiapoptotic protein (9). A number of studies have demonstrated the importance of splice variants associated with the etiology of lung cancers, prime examples of which include CD44 and BCLX mRNAs (10). In the case of BCLX mRNA, higher levels of the longer antiapoptotic BCLXL mRNA have been found in both NSCLC and small cell lung cancer cells; whereas differential inclusion/exclusion of variable exons 3 and 6 in the CD44 pre-mRNA has been shown to have prognostic implications in lung carcinomas (10). In this issue of the JCI, Goehe et al. (7) have added to the list of lung cancer–specific splice variants, using RT-PCR and immunoblot assays to demonstrate that the ratio of caspase-9a/9b is lower in a large number of NSCLC cells as well as in pure populations of lung adenocarcinomas. The increase in levels of caspase-9b mRNA and protein levels in a lung adenocarcinoma cell line (A549) resulted in decreased caspase activity. Importantly, using caspase-9b overexpression and shRNA-mediated knockdown experiments, the authors directly demonstrated that caspase-9b promotes anchorage-independent growth and tumor induction. These experiments highlight how two mRNA isoforms of a given gene can encode proteins that have opposing physiological functions and can differentially impact cellular growth.
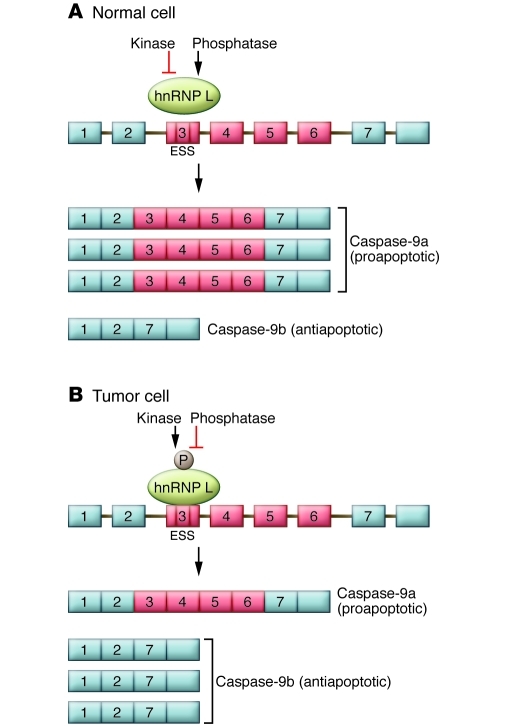
Figure 2. Model for the regulation of caspase-9 mRNA expression by hnRNP L.
The caspase-9 pre-mRNA contains four variable exons (exons 3, 4, 5, and 6) that are differentially included or excluded to generate the longer caspase-9a or shorter caspase-9b mRNAs. (A) The work of Goehe et al. (7) indicates that in normal cells, a higher ratio of caspase-9a/9b mRNA is present, which leads to increased levels of the proapoptotic caspase-9a protein isoform. (B) In tumor cells, the four variable exons are skipped, leading to a lower ratio of caspase-9a/9b mRNA, which leads to increased levels of the antiapoptotic caspase-9b protein isoform. Goehe et al. show that regulation of the alternative RNA processing pattern of the caspase-9 mRNA is achieved by the differential phosphorylation of the hnRNP L protein. hnRNP L protein is largely unphosphorylated in normal cells, while in NSCLC cells, hnRNP L is phosphorylated at specific serine, threonine, and tyrosine residues. The differential phosphorylation pattern of hnRNP L could be regulated by cell type–specific kinases and/or phosphatases. Phosphorylation of hnRNP L Ser52, specifically in tumor cells, regulates the interaction of hnRNP L with caspase-9 RNA and regulates differential RNA processing of this transcript, as discussed in the text. ESS, exonic splicing silencer.
In order to address the molecular mechanism governing the alternative splicing pattern of caspase-9, Goehe and colleagues used a minigene assay comprising the variable exons 3, 4, 5, and 6 and flanking intronic sequences (7). Using this assay, they showed that mutation of a purine-rich sequence in exon 3 of the caspase-9 pre-mRNA decreased the abundance of the 9b isoform. Using an RNA affinity approach, hnRNP L was found to be a major protein associated specifically with the exon 3 element in nuclear extract from NSCLC cell lines. Reduction of hnRNP L protein levels, using RNAi technology combined with overexpression studies, provided direct confirmation that hnRNP L promoted the expression of the caspase-9b mRNA isoform and altered tumorigenicity. In contrast, other hnRNP family members that bound adjacent sequences in exon 3 had no effect on isoform expression. These studies link hnRNP L specifically to the regulation of caspase-9 expression.
Intriguingly, control experiments in nontransformed lung epithelial cell lines revealed that the activity of hnRNP L in regulating caspase-9 was specific for NSCLC cells (7). This differential activity of hnRNP L correlated with increased phosphorylation of hnRNP L on specific serine, threonine, and tyrosine residues in the NSCLC cell lines as compared with control cells. Among these residues, phosphorylation of hnRNP L on serine 52 was specifically required for regulating the alternative splicing of caspase-9 pre-mRNA. Furthermore, similar to expression of caspase-9b, expression of phospho–hnRNP L, but not that of the serine 52 to alanine mutant, was found to induce anchorage-independent growth in the NSCLC cell line. This is an elegant example of how hnRNP activity can be regulated, with profound physiologic consequences, by individual phosphorylation events.
hnRNPs in the cause and detection of cancer
The data presented in this issue of the JCI by Goehe and colleagues (7) have important implications for the understanding of tumor biology as well as RNA processing mechanisms. From a tumor biology standpoint, this study highlights and adds to the theme of splicing regulators acting as oncogenes or as biomarkers for cancers (11). Research in other laboratories has shown that members of the hnRNP family, such as hnRNP A1, hnRNP A2, hnRNP B1, and polypyrimidine tract binding protein 1 (PTB), are frequently overexpressed in different types of tumors, including lung cancer (12–14); however, changes in the phosphorylation status of such proteins has not previously been reported in cancer cells. It will be interesting to survey the lung tumor samples used by Goehe et al. for the phosphorylated form of hnRNP L and to further examine the incidence of this posttranslational modification in other types of tumors. Such a systematic analysis would help determine whether the phosphorylated form of hnRNP L can serve as a biomarker for lung or other types of cancers.
The other question that remains to be addressed is whether there are additional mRNA targets of the phosphorylated form of hnRNP L that can cause or promote tumors. Multiple targets of hnRNP L have been identified in normal cells (15). Further, hnRNPs, such as PTB, hnRNP A1, and hnRNP A2, have been recently shown to regulate the splicing pattern of pyruvate kinase mRNA to promote aerobic glycolysis in cancer cells (13, 14). Determining whether Ser52 phosphorylated hnRNP L also regulates pyruvate kinase splicing or whether any other targets of hnRNP L influence tumor growth will be essential to understanding the breadth of the contribution of hnRNP L to cancer. However, even identification of the functional significance of caspase-9b upregulation in tumors opens the possibility of developing drugs that can inhibit the expression of this antiapoptotic isoform. Notably, both a splicing-modulating small molecule (emetine) and antisense oligonucleotide technology are in preclinical development for the modulation of Bcl-xL expression as an anticancer therapy (10).
Regulation of hnRNP L function
From a splicing mechanism perspective, the data presented by Goehe et al. (7) also highlight several interesting questions. The sequence of the regulatory element to which hnRNP L was shown to associate in caspase-9 exon 3 (AGGGGA) differs markedly from the canonical C/A-rich high-affinity binding site for hnRNP L (16, 17). It is possible that phosphorylation of hnRNP L at Ser52 alters the binding specificity of the protein, so as to expand its regulatory repertoire. However, it is also possible that hnRNP L binds indirectly to the purine-rich sequence in exon 3, since all of the binding assays performed by Goehe et al. were done in the presence of nuclear extract. Even in the case of indirect binding, phosphorylation of hnRNP L on Ser52 could regulate its protein-protein interaction properties to help recruit hnRNP L to the caspase RNA. This would be consistent with other examples in the field; other studies have found that phosphorylation of splicing regulators, such as alternative-splicing factor 1 (ASF, also known as SF2), regulates protein-protein interactions and consequently splicing of target mRNAs (18). Obviously, determining the biochemical nature of the interaction between hnRNP L and caspase-9 will be essential to further uncovering the mechanistic details of caspase-9 regulation.
Perhaps an even more pressing question is to determine exactly which step of the processing of caspase-9 is controlled by hnRNP L. Given the vast abundance of alternative splicing, it is tempting to assume that hnRNP L controls the splicing of caspase-9 pre-mRNA. While there is extensive precedent for hnRNP L functioning as a splicing repressor, it is difficult to envision a model in which binding of hnRNP L at a single site in exon 3 causes the concomitant skipping of exons 4, 5, and 6. It is possible that hnRNP L only causes skipping of the 35-nt exon 3, thereby altering the subsequent reading frame such that the resulting message is destroyed by nonsense-mediated decay (NMD) (1). Alternatively, hnRNP L could regulate caspase-9 mRNA isoform expression by preferentially destabilizing the caspase-9a mRNA that contains the exon 3 regulatory sequence. Such a role for hnRNP L in mRNA stability has been shown in the case of VEGF and eNOS mRNAs (17, 19). Clearly, further experiments are required to differentiate among these mechanistic possibilities.
Finally, the fact that the phosphorylation status of hnRNP L is different in tumor versus normal tissues raises the obvious question regarding the kinases and phosphatases that govern this cell type–specific modification. Regulation of alternative splicing by cellular signaling pathways is an emerging theme, and data from several laboratories have established several kinases that modulate the activities of splicing regulators (20). Identification of the enzymes that control the phosphorylation status of hnRNP L would both add to the understanding of the regulation of hnRNPs and potentially also provide additional novel targets for the therapeutic manipulation of hnRNP L function in splicing, stability, and/or tumorigenesis.
Acknowledgments
Ganesh Shankarling and Kristen W. Lynch are supported by NIH grants R01 GM067719 and GM084034 to K.W. Lynch.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(11):3798–3801. doi:10.1172/JCI45037
See the related article beginning on page 3923.
References
- 1.Moore MJ, Proudfoot NJ. Pre–mRNA processing reaches back to transcription and ahead to translation. . Cell. 2009;136(4):688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456(7221):470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high–throughput sequencing. Nat Genet. 2008;40(12):1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 4.Han SP, Tang YH, Smith R. Functional diversity of the hnRNPs: past, present and perspectives. . Biochem J. 2010;430(3):379–392. doi: 10.1042/BJ20100396. [DOI] [PubMed] [Google Scholar]
- 5.Black DL. Mechanisms of alternative pre–messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 6.Faustino NA, Cooper TA. Pre–mRNA splicing and human disease. Genes Dev. 2003;17(4):419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 7.Goehe RW, et al. hnRNP L regulates the tumorigenic capacity of lung cancer xenografts in mice via caspase-9 pre-mRNA processing. J Clin Invest. 2010;120(11):3923–3939. doi: 10.1172/JCI43552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seol DW, Billiar TR. A caspase–9 variant missing the catalytic site is an endogenous inhibitor of apoptosis. J Biol Chem. 1999;274(4):2072–2076. doi: 10.1074/jbc.274.4.2072. [DOI] [PubMed] [Google Scholar]
- 9.Philchenkov A, Zavelevich M, Kroczak TJ, Los M. Caspases and cancer: mechanisms of inactivation and new treatment modalities. Exp Oncol. 2004;26(2):82–97. [PubMed] [Google Scholar]
- 10.Pio R, Montuenga LM. Alternative splicing in lung cancer. J Thorac Oncol. 2009;4(6):674–678. doi: 10.1097/JTO.0b013e3181a520dc. [DOI] [PubMed] [Google Scholar]
- 11.Skotheim RI, Nees M. Alternative splicing in cancer: noise, functional, or systematic? Int J Biochem Cell Biol. 2007;39(7–8):1432–1449. doi: 10.1016/j.biocel.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 12.Boukakis G, Patrinou-Georgoula M, Lekarakou M, Valavanis C, Guialis A. Deregulated expression of hnRNP A/B proteins in human non–small cell lung cancer: parallel assessment of protein and mRNA levels in paired tumor/non–tumor tissues. BMC Cancer. 2010;10:434. doi: 10.1186/1471-2407-10-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clower CV, Chatterjee D, Wang Z, Cantley LC, Vander Heiden MG, Krainer AR. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc Natl Acad Sci U S A. 2010;107(5):1894–1899. doi: 10.1073/pnas.0914845107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c–Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463(7279):364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hung LH, Heiner M, Hui J, Schreiner S, Benes V, Bindereif A. Diverse roles of hnRNP L in mammalian mRNA processing: a combined microarray and RNAi analysis. RNA. 2008;14(2):284–296. doi: 10.1261/rna.725208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hui J, Reither G, Bindereif A. Novel functional role of CA repeats and hnRNP L in RNA stability. RNA. 2003;9(8):931–936. doi: 10.1261/rna.5660803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hui J, Stangl K, Lane WS, Bindereif A. HnRNP L stimulates splicing of the eNOS gene by binding to variable–length CA repeats. Nat Struct Biol. 2003;10(1):33–37. doi: 10.1038/nsb875. [DOI] [PubMed] [Google Scholar]
- 18.Xiao SH, Manley JL. Phosphorylation of the ASF/SF2 RS domain affects both protein–protein and protein–RNA interactions and is necessary for splicing. Genes Dev. 1997;11(3):334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- 19.Shih SC, Claffey KP. Regulation of human vascular endothelial growth factor mRNA stability in hypoxia by heterogeneous nuclear ribonucleoprotein L. J Biol Chem. 1999;274(3):1359–1365. doi: 10.1074/jbc.274.3.1359. [DOI] [PubMed] [Google Scholar]
- 20.Lynch KW. Regulation of alternative splicing by signal transduction pathways. Adv Exp Med Biol. 2007;623:161–174. doi: 10.1007/978-0-387-77374-2_10. [DOI] [PubMed] [Google Scholar]