Abstract
Interleukin-12 (IL-12) is a critical cytokine representing the link between the cellular and humoral branches of host immune defense apparatus. IL-12-induced cytotoxic lymphocyte (CTL) development is a central mechanism in immune responses against intracellular infectious agents as well as malignant growth. However, the molecular basis of tumor-specific CTL responses mediated by IL-12 remains poorly defined. In this study, we addressed this issue in a comprehensive manner to probe into IL-12-induced anti-tumor responses by global gene expression profiling of mRNA expression in CD8+T cells in a transplantable syngeneic mouse mammary carcinoma model treated or not with recombinant IL-12. A strong tumor regression was induced by the IL-12 treatment. An introspection of differential gene expression at an early stage of the IL-12-initiated CTL activation reveals interesting genes and molecular pathways that may account for the marked tumor regression, and is likely to provide a rich source of potential targets for further research and development of effective therapeutic modalities.
Keywords: interleukin-12, cytotoxic T lymphocyte, TS/A, mammary carcinoma, microarray
Introduction
The emergence of the concept of cancer immunosurveillance conceived by Paul Ehrlich at the beginning of 20th century and formalized by Burnet and Thomas in 1957, the severe challenges it faced in the 1970s and 80s (1–5), and its renaissance in the last decade brought about by a large body of mouse (6–11) and human (12–17) studies in immunodeficient settings have helped solidify the belief that components of the immune system, such as lymphocytes (T, NK, NKT) and cytokines (IFN-γ, perforin, IL-12) are critically involved in controlling primary tumor development. The refinement of this concept and its extension to “immunoediting” has been put forward by R.D. Schreiber and colleagues to describe the dual host-protecting and tumor-sculpting actions of the immune system that not only impede but also mark neoplastic alterations for immune elimination (18, 19).
Tumors often possess a number of potential recognition sites for immunologic effector cells, which, in theory, could make them susceptible to immune surveillance. Nevertheless, most of such tumors grow progressively in their natural hosts or syngeneic recipients, without being controlled effectively by the immune system. The lack of apparent immunogenicity of tumors in situ might be due to special properties of the tumor cells, e.g., lack of costimulatory molecules, down-regulation of MHC molecules, or production of immunosuppressive factors (20, 21), or due to intrinsic tolerance mechanisms of the immune system (22).
The immune response against cancer cell is complex, involving the interaction of many different cell types and cellular products. However, it is well established cytotoxic T lymphocytes (CTLs) constitute one of the most important anti-tumor immunity (23, 24). CTLs recognize, via their clonal T cell receptors, specific antigenic peptides that are derived from tumors, especially solid ones including carcinomas and sarcomas (25, 26), and presented to them by antigen-presenting cells such as dendritic cells (DCs). These CTLs survey the body and lyse malignant cells expressing the relevant surface antigens. This mechanism of recognition and killing is analogous in many ways to viral infections of peripheral tissues for which CTL is the main effector of adaptive immunity controlling the spread of the infection (27).
In recent years, immunotherapies have been rekindled that attempt to either mark the tumor by upregulating the surface antigens for enhanced interaction with immune effector cells (21, 28), or by directly activating DC, T and NK cells for their heightened “scouting” capacity and increased cytolytic potency. IL-12 is a factor belonging to the latter class (29). IL-12 is a pivotal cytokine representing the link between the cellular and humoral branches of host immune defense apparatus. IL-12 is a heterodimer produced primarily by macrophages and DCs in both innate and adaptive immune responses. It is a key factor in the induction of T cell-dependent and independent activation of macrophages, NK cells, generation of T helper type 1 (Th1) cells and CTLs, induction of opsonizing and complement-fixing antibodies, and resistance to intracellular infections (30). IL-12 has powerful anti-tumor and anti-metastatic activities against many murine tumors as well as human tumors (31). The potent anti-tumor activities of IL-12 are mediated through similar mechanisms that are used by IL-12 against infectious agents, i.e., via the activation of NK cells for the bulk of the non-antigen specific clearance, and activation of CTL and CD4 for tumor-specific elimination and long-term immunity. Numerous recent studies across a wide range of experimental tumor model systems as well as in human cancers have unequivocally confirmed the efficacy of IL-12 as a potent inducer of CTL-mediated anti-tumor immunity (32–47), thus strongly justifying further exploration of IL-12 and CTL in cancer immunotherapy. Furthermore, in a recent study, Lee et al. reported for the first time that IL-12 could inhibit activation-induced CD8+T cell death by downregulating Fas ligand and up-regulating cellular FLIPs, followed by suppressing activation of caspases 8 and 3, thus providing a survival signal for sustained CTL responses (48).
TS/A is an aggressive and poorly immunogenic cell line established from a moderately differentiated mammary adenocarcinoma that arose spontaneously in a multiparous BALB/c mouse (49). It grows progressively, kills both nu/nu and syngeneic mice, and gives rise to lung metastases. It expresses MHC class I (H-2Dd, H-2Kd), but not class II molecules, secretes G-CSF, GM-CSF, TGF-β, basic fibroblast growth factor (FGF) and vascular endothelial growth factor (VEGF), and does not stimulate a syngeneic antitumor response in vivo, nor in mixed lymphocyte-tumor cell cultures (50). IL-12 administered to TS/A tumor-bearing mice via various routes at different stages of tumor establishment has been shown to cause the rejection of the tumor in wild type BALB/c mice through a CD8+T-lymphocyte-dependent reaction associated with macrophage infiltration, vessel damage, and necrosis. Protective immune memory is also established following IL-12 treatment (51).
Genome-wide gene expression profiling allows the viewing, with molecular accuracy, of biological processes as a whole. The mRNA expression of tens of thousand of genes in response to a given signal can be monitored simultaneously using massively parallel DNA microarray technology. The vast amount of data generated can be organized, refined and extrapolated with the aid of bioinformatics tools to reveal a comprehensive, highly concerted, and logical molecular program underlying the process of cellular differentiation and activation (52, 53). Coordinate gene expression takes place among products of the genes that function in a common differentiation program or in the same physiological response pathway. Recent applications of this powerful technology to the analysis of the immune system have uncovered many potential novel pathways and key players in normal as well as pathological immune responses, which are likely to have a great impact on our understanding of autoimmunity, immune deficiencies, and cancers of immune cells (54–58).
The aim of this study was to obtain a high resolution, dynamic signature of a tumor-specific CTL response at every step during its development after activation in lymphoid tissues. The approach was to use microarray to monitor gene expression in the CD8+T cell compartment in the spleen of mice given TS/A tumor cells and recombinant IL-12 over a period of four weeks spanning primary activation, effector phase and memory formation. The choice of TS/A for this study was based on three considerations: (i) these tumor cells expresses MHC class I; (ii) immunotherapy with GM-CSF and IL-2 has demonstrated a strong tumor-specific CTL response and protective immunity (59); (iii) IL-12 administration is effective against these tumors by a CD8+T- or NK-dependent mechanism. Therefore, the TS/A model is highly relevant to studying tumor-specific CTL responses regulated by IL-12. We performed Affymetrix microarrays using RNA samples isolated from purified splenic CD8+T cells of TS/A tumor-bearing mice.
Materials and Methods
Mice
Female BALB/c mice (6–8 week old) were purchased from the Jackson Laboratory (Bar Harbor, Maine). All mice were housed at the Weill Medical College of Cornell University Animal Facilities in accordance with the Principles of Animal Care (NIH publication no. 85-23, revised 1985). Mice bearing TS/A tumors were all sacrificed no later than Day 35 due to the morbidity caused by large tumor size and strong metastasis.
Tumor implantation, size measurement
TS/A mammary carcinoma cells (1 × 105) were injected subcutaneously into the abdominal mammary gland area of recipient mice in 0.1 ml of a single-cell suspension in phosphate buffered saline (PBS) on Day 0. The dose of tumor implantation was empirically determined to give rise to tumors of ~10 mm in diameter in untreated wild type mice in 28 days. Primary tumors were measured using electronic calipers every 3–4 days. Reported measurements are the square root of the product of two perpendicular diameters.
IL-12 treatment
Recombinant murine IL-12 was provided by Genetics Institute (Cambridge, Massachusetts). IL-12 treatment was given by intraperitoneal injection at 1 μg per mouse every other day starting on Day 7 until the end of each experiment unless other wise described. This regimen of IL-12 was well tolerated with no signs of toxicity.
Purification of splenic CD8+T lymphocytes
BALB/c mice spleen cells were prepared by using 40 μm cell strainer (BD Falcon, Bedford, MA), and the red blood cells were lysed with ACK buffer (60), and then washed with RPMI 1640 media with 10% FBS three times. The cells were resuspended at a concentration of 2 × 107/ml in cold PRMI 1640/10% FBS media, and then Dynabeads (CELLection mouse CD8 kit, DYNAL, Lake Success, NY), were added (25 μl containing 107 beads/2 × 107 spleen cells) using the magnetic particle concentrator (Dynal MPC) to positively select the mouse spleen CD8+T cells. Following capturing of the bead-linked CDS8+T cells, the bead-releasing step was omitted before RNA isolation.
Microarray experiment
The high-density oligonucleotide microarray system of Affymetrix (Santa Clara, California), murine Genome U74A Array version 2 containing 12,488 genes, was used. Total RNA was isolated from freshly isolated spleens of all surviving mice on Day 7 (before IL-12 treatment), and Day 14 (one week following the initial IL-12 injection). RNA samples of each mouse within each experiment group were pooled from 3–4 mice. 10 μg total RNA was used to synthesize cDNA using Superscript cDNA synthesis kit (Invitrogen, Carlsbad, California) with a primer containing oligo (dT) and T7 RNA polymerase promoter sequences. Double-stranded cDNA was then purified by phase lock gel (Eppendorf, Westbury, NY) with phenol/chloroform extraction. The purified cDNA was used as a template to generate biotinylated cRNA using the Bioarray High Yield RNA Transcript Labeling Kit (Enzo Biochem), and then biotinylated cRNAs were fragmented and hybridized to Affymetrix Test 3 chips (Affymetrix Inc., Santa Clara, CA). All RNA samples passed quality control (ratio of 3′ to 5′ < 3), then the samples were hybridized to the Murine Genome Array U74Av2 array which contains 12,488 well-substantiated mouse genes. After overnight hybridization, the arrays were washed, stained with streptavidin-phycoerythrin (Molecular Probes, Eugene, OR) on the GeneChip Fluidics Station (Affymetrix), and scanned according to the standard Affymetrix protocol.
Microarray Data collection and analysis
Affymetrix GeneChip 5.0 was used as the image acquisition software for the U74Av2 chips. The signal, which represents the intensity of each gene, was extracted from the image. The target intensity value from each chip was scaled to 250. Data normalization, log transformation, and statistical analysis were performed with GeneSpring software (Silicon Genetics, Redwood City, CA). Array data were globally normalized in two steps. Firstly, all of the measurements on each chip were divided by the 50th percentile value (per-chip normalization). Secondly, each gene was normalized to the baseline value of the control samples (per-gene normalization).
Statistical tests
Tumor growth data to be compared were first subjected to normality test. Where the samples studied were normally distributed, statistical comparisons were performed using the Students’ t test. Where the samples deviated from normality, a nonparametric, Mann Whitney Rank-Sum test was used for comparisons. Statistical analyses were performed using SigmaStat software. For all experiments, the mean and the SD are depicted.
Results and Discussion
IL-12 induces anti-tumor activities in vivo
To assess the effects of IL-12 in tumor regression, TS/A mammary carcinoma was initiated by s.c. injection of TS/A cells into mice on the syngeneic BALB/c background. Recombinant mouse IL-12 (rmIL-12) was given i.p. starting on Day 7 post tumor injection to mice when the primary tumor had grown to ~ 4–5 mm in diameter. The timing of IL-12 administration was based on potential therapeutic considerations to mimic clinical situations in which breast cancer patients do not get therapy until the presence of malignant growth in the breast has been identified by mammogram or other means. As shown in Figure 1, by Day 35 (four weeks following the initial IL-12 injection), there was a strong regression in the TS/A tumor growth in mice given IL-12 treatment. The results are consistent with published data (51) showing great efficacy of IL-12 as a therapeutic agent in this model.
Figure 1.
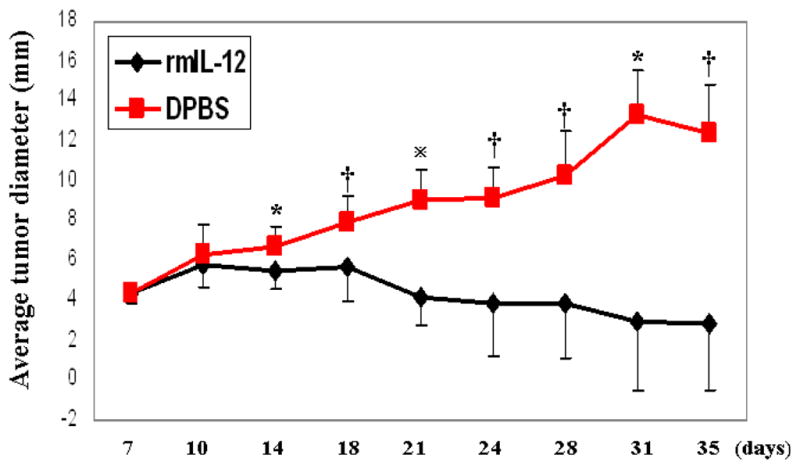
TS/A tumor growth in syngeneic mice. TS/A cells were injected subcutaneously in the middle of the right flank of BALB/c mice with 0.1 ml of a single-cell suspension containing the indicated number of TS/A cells. Tumor growth was monitored every 3–4 days and size measured with a caliper. Each data point is comprised of 4–11 mice. Error bars represent standard deviation. *p < 0.05; †p < 0.01;  p < 0.0001.
p < 0.0001.
Genome-wide analysis of gene expression in splenic CD8+ lymphocytes
Our main interest in this study was to investigate the molecular mechanism(s) by which IL-12 activates CTL against the developing TS/A tumor. The strategy we chose to achieve this objective was to use DNA microarray to comprehensively survey the gene expression in the CD8+T cell populations in the spleen of mice treated or not with IL-12 over the entire phase of tumor growth or regression, with the hope to identify molecular targets that may play critical roles in mediating IL-12’s anti-tumor activity by activating CTL. As a first step towards obtaining a detailed description of the molecular events taking place in the CD8+ T cells in mice treated or not with rmIL-12, we performed global gene expression analysis of the two groups of tumors on Day 14 (7 days following the initial IL-12 injection) using the Affymetrix oligonucleotide microarray system (Murine Genome U74A Array version 2 containing 12,488 genes). To reduce variations between individual mice, RNA samples were pooled from all mice within each group for the microarray analysis. We applied this technology to the search of genes that undergo altered expression in CD8+ splenic T cells in vivo following IL-12 therapy in an attempt to identify the downstream targets of IL-12 in this particular lymphocyte compartment. RNA samples were prepared from the two experimental groups shown in Figure 1 for comparison of differential gene expression. This microarray experiment (4–11 mice from each group) yielded a large amount of data, which was processed through the GeneSpring software for (i) data normalization (bias correction), (ii) data transformation (to ensure normal distribution), and (iii) gene filtering to identify specific genes that are expressed differentially by using appropriate statistic tools. The group of PBS-treated wild type mice was set as the baseline (with an expression value of 1.0) to which the IL-12-treated group was compared. Most of the genes show no altered expression. The vast majority of the genes were either present equally or absent in both samples. A small number of genes manifested changes in expression to varying degrees. These are summarized in Table 1a and 1b as IL-12-induced genes and IL-12-inhibited genes, respectively.
Table 1.
Genes differentially regulated by IL-12. Genes are selected based on the following criteria: (1) They must be expressed more than 2 times than the respective control sample (PBS-treated) to qualify as IL-12-induced genes (Table 1a), or 2 times less than the control to qualify as IL-12-inhibited genes (Table 1b); (2) Their expression detection must be statistically valid as present with detection p < 0.05; Genbank accession numbers are given in the second column. The numbers in column 3 are relative fold of expression over the control (PBS-treated group).
| Table 1a. Genes induced by IL-12 (117 genes). | |||||
|---|---|---|---|---|---|
| Gene and description | Genbank accession | Fold induction | Gene and description | Genbank accession | Fold induction |
| Aldehyde dehydrogenase (ALDH2) | U07235 | 2.01 | Ig rearranged k-chain mRNA, clone AN08K. | M19911 | 4.35 |
| Aldehyde dehydrogenase 2, mitochondrial | AI647493 | 2.24 | Ig variable light chain. | X88903 | 2.05 |
| Alpha fetoprotein | AV037200 | 3.03 | Ig Vkappa-HNK20 | X82688 | 3.43 |
| AMY, Noe1, OlfA, Pancortin, Pancortin3 | D78265 | 4.86 | IgA | J00475 | 18.97 |
| Annexin A2 | M14044 | 2.2 | IgA | J00475 | 3 |
| Annexin A3 | AJ001633 | 2.34 | IgA V-D-J-heavy chain | X94418 | 4.2 |
| Anti-DNA light chain IgM, antibody 363p.168 | U55576 | 3.1 | IgG variable region. | Z22111 | 2.53 |
| Antigen identified by monoclonal antibody Ki 67 | X82786 | 2.23 | Igh | AF042086 | 2.65 |
| Atf3, leucine zipper transcription factor LRG-21 | U19118 | 3.79 | Igh-6 | X94422 | 2.93 |
| Balb/c neutrophil elastase gene, exons 4 and 5 | U04962 | 3.1 | Igk-V28 | U62386 | 3.63 |
| Bmk, Hck-1 | J03023 | 2.82 | IL-1receptor antagonist | L32838 | 3.13 |
| Carbonic anhydrase I (CAI) | M32452 | 23.21 | Immediate early response 3 | X67644 | 2.62 |
| Carboxypeptidase A3, mast cell | AV172041 | 10.9 | Immunoglobulin heavy chain variable gene | X16740 | 5.85 |
| CB17 SCID immunoglobulin heavy chain V region | U23095 | 2.43 | Immunoglobulin kappa chain variable 28 (V28) | Z70661 | 3.68 |
| CD24a antigen | M58661 | 2.81 | Immunoglobulin kappa chain variable 28 (V28) | U48716 | 3.49 |
| CDR3 region; Ig heavy chain gene | AF042798 | 3.77 | Immunoglobulin superfamily, member 4 | AF061260 | 2.78 |
| Cell division cycle 25 homolog C (S. cerevisiae) | L16926 | 3.56 | Immunoglobulin superfamily, member 4 | AF061260 | 2.03 |
| c-Fes | X12616 | 2.14 | Interferon inducible protein 10 (IP-10) | M33266 | 5.18 |
| Chemokine (C-C) receptor 1 | U29678 | 2.36 | Interferon-inducible GTPase | AA914345 | 5.26 |
| Chemokine (C-C) receptor 5 | AV370035 | 2.22 | Interferon-inducible GTPase | AJ007971 | 4.28 |
| Chitinase 3-like 3 | M94584 | 2.17 | Interleukin 1 beta | M15131 | 2.21 |
| Complement component 4 (within H-2S) | X06454 | 5.44 | Interleukin 6 (IL-6) | X54542 | 20.27 |
| Cpa3, carboxypeptidase A | J05118 | 7.37 | Killer cell lectin-like receptor, Ly49b | U10304 | 2.77 |
| Ctla2b | X15592 | 4.45 | Kinesin-like 1 | AJ223293 | 2.88 |
| Ctsg, serine proteinase | X70057 | 14.48 | Lipo 1, lipocortin I | M69260 | 3.3 |
| C-type lectin, superfamily member 6 | AJ133533 | 2.55 | Lipoprotein lipase | AA726364 | 5.86 |
| Cx26, connexin | M81445 | 2.77 | Ly-6G.1 | X70920 | 7.16 |
| Emr1, F4/80 | X93328 | 11.56 | Mitotic checkpoint protein kinase (Bub1) | AF002823 | 2.26 |
| Entpd1, ecto-apyrase CD39 | AF037366 | 2.91 | Myeloperoxidase | X15313 | 9.08 |
| Epcr, endothelial cell activated protein C | L39017 | 2.58 | N-acylsphingosine amidohydrolase 1 | AW124297 | 2.14 |
| Epstein-Barr virus induced gene 3 | AF013114 | 2.14 | Nfe2, basic leucine zipper transcription factor | L09600 | 2.65 |
| EST | AI317217 | 12.66 | NKG2-D (Nkg2d) | AF054819 | 3.31 |
| EST | X67210 | 6.95 | NP-1, neuropilin | D50086 | 2.87 |
| EST | X67210 | 5.7 | paraoxonase-3 (Pon3) | L76193 | 2.32 |
| EST | AL078630 | 5.51 | PLA2, Calcium-dependent phospholipids binding protein | M72394 | 2.79 |
| EST | AW259499 | 3.3 | Plasma glutamate carboxypeptidase | AF009513 | 3.84 |
| EST | AI854793 | 3.16 | precursor of C and V-D-J regions from 7B6.8 | D14625 | 3.44 |
| EST | AI842940 | 2.98 | Primary response gene B94 | L24118 | 9.04 |
| EST | AW215456 | 2.65 | Proteinase 3, myeloblastin | U43525 | 6.34 |
| EST | AI841689 | 2.42 | Ptpn13, protein tyrosine phosphatase | D83966 | 17.04 |
| EST | AW123773 | 2.29 | PYT, MPS1L1, putative esk kinase | M86377 | 4.04 |
| EST | AA590345 | 2.17 | Ra175c | AB021966 | 2.2 |
| EST, similar to 202 interferon-activatable protein | AV229143 | 4.11 | Rab6, kinesin-like | Y09632 | 2.3 |
| EST, similar to gb:M33308 vinculin (human) | AI462105 | 2.43 | rhom-2 | M64360 | 2.88 |
| EST, similar to gb:X52634 tlm oncogene | AI504305 | 4.16 | S100 calcium binding protein A1 | AF087687 | 2.84 |
| Fc receptor, IgE, high affinity I | J05018 | 4.24 | Secreted phosphoprotein 1 | X13986 | 5.6 |
| Fcgr1 | M31314 | 3.89 | Sid23 | AB025406 | 2.08 |
| Formyl peptide receptor, related sequence 2 | AF071180 | 6.03 | Small inducible cytokine A3 | J04491 | 3.61 |
| germline immunoglobulin V(H)II gene H18 | X02468 | 2.27 | Spi-1, PU.1 | L03215 | 2.27 |
| Hex (Prh) | AB017132 | 2.07 | spi2/eb1 | M64085 | 3.38 |
| Histidine decarboxylase cluster | X57437 | 2.79 | ST2L | D13695 | 6.63 |
| ICE, Il1bc, Caspase-1 | L28095 | 2.4 | Stx3, syntaxin 3A | D29797 | 3.42 |
| Ier5, immediate early response 5 gene | AF079528 | 3.94 | Stx3, syntaxin 3D-2 | D38375 | 15.65 |
| Ifi202, Ifbip-1 | M31418 | 4.07 | TCRγ-V4 | X00697 | 13.42 |
| Ig B cell antigen receptor gene | L28059 | 7.61 | Tiap, IAP repeat | AB013819 | 2.08 |
| Ig B cell antigen receptor gene | L28060 | 4.06 | UPase, UdRPase, uridine phosphorylase | D44464 | 11.84 |
| Ig g-3 V-D-J region | D14625 | 5.47 | Vh186.2/Jh2 | AF065324 | 2.98 |
| Ig heavy chain 6 (heavy chain of IgM) | X94420 | 4.07 | Vk10c | AF029261 | 3 |
| Ig heavy chain variable region precursor, gene | AF036737 | 3.99 | |||
| Table 1b. IL-12-inhibited genes (169 genes). | |||||
|---|---|---|---|---|---|
| Gene and description | Genbank accession | Fold inhibition | Gene and description | Genbank accession | Fold inhibition |
| ALL 1-fused gene from chromosome 1q | U95498 | 2.93 | f8a, factor VIII-associated protein | M83118 | 2.5 |
| Angiotensin converting enzyme | J04946 | 2.07 | Fatty acid amide hydrolase gene | AF098009 | 2.18 |
| Antigenic determinant of rec-A protein | X58472 | 2.01 | Fc receptor, IgG, alpha chain transporter | L17022 | 5.59 |
| Arp1, alpha-Arp1 | AB010297 | 2.12 | Feminization 1 homolog a (C. elegans) | AI836048 | 2.07 |
| ATPase, H+ transporting, lysosomal (vacuolar proton pump), subunit 1 | AI646638 | 3.21 | FK506 binding protein 4 (59 kDa) | X17069 | 2.2 |
| BAP, Bap37 | AC002397 | 2.02 | galK, galactokinase | AB027012 | 2.26 |
| Bat-4 | L76155 | 2.14 | General control of amino acid synthesis-like 2 (yeast) | AW049299 | 2.07 |
| Bromodomain-containing 4 | AI838366 | 2.44 | Glns-ps1, outative introless glutamine synthetase | M60803 | 2.55 |
| BSP1, Nrpn, mGk-8, Prss19, TADG14 | D30785 | 2.24 | Gna11, guanine nucleotide binding protein | U37413 | 2.83 |
| Bystin-like | AI132491 | 2.04 | GR, glucagon receptor | L38613 | 2.08 |
| Cannabinoid receptor 2 (macrophage) | X86405 | 2.43 | Granzyme G | J02872 | 2.04 |
| Casein kinase II, alpha 1 related sequence 4 | U51866 | 2.23 | Guanosine diphosphate (GDP) dissociation inhibitor 2 | U07951 | 5.16 |
| Cd28 | M34563 | 2.01 | Histocompatibility 2, L region | AI326621 | 3.13 |
| CD3 antigen, delta polypeptide | X02339 | 2.01 | Histone cell cycle regulation defective homolog A (S. cerevisiae) | AW125193 | 2.72 |
| CD6 antigen | U12434 | 2.09 | Hnrpa1, alternative splicing modulator | U65316 | 3.15 |
| Cerebellar degeneration-related 2 | U88588 | 2.99 | hsp40 | AB028272 | 2.76 |
| Chaperonin subunit 5 (epsilon) | AV170770 | 2.24 | hsp-E7I | L40406 | 2.8 |
| Chemokine (C-X-C) receptor 4 | Z80112 | 2.42 | IGFBP-4, insulin like growth factor binding protein 4 | X76066 | 2.1 |
| Chromobox homolog 1 (Drosophila HP1 beta) | X56690 | 20.49 | IL-4 receptor (IL4R) | M27960 | 2.21 |
| CIS, cytokine SH2-containing protein | D89613 | 2.25 | IL-7 receptor (IL7R) | M29697 | 2.66 |
| Cleavage and polyadenylation specificity factor 3 | AI849311 | 2.56 | Insulin-like growth factor I receptor | AF056187 | 2.09 |
| Cmah, CMP-N-acetylneuraminic acid hydroxylase | D21826 | 2.02 | Intercellular adhesion molecule 2 | X65493 | 2.4 |
| Ctps2, CTP synthetase homolog | U49385 | 2.29 | K+ intermediate/small conductance Ca2+-activated channel, subfamily N, member 4 | AF042487 | 2.36 |
| CYL2, D-type cyclin | M83749 | 2.5 | Kallikrein 8 | AV145185 | 2.42 |
| Defender against cell death 1 | AV099898 | 2.16 | Keratin complex 1, acidic, gene 10 | V00830 | 3.38 |
| DNA polymerase epsilon, subunit 2 | AF036898 | 2.21 | Kruppel-like factor 2 (LKLF) | U25096 | 2.3 |
| DNA segment, Chr 2, ERATO Doi 391, expressed | AW121160 | 2.15 | LEF-1 | D16503 | 2.29 |
| DnaJ (Hsp40) homolog, subfamily B, member 10 | AI843164 | 2.63 | LEF-1 | D16503 | 2.02 |
| EBI 1, G-protein coupled receptor | L31580 | 2.13 | Leucine zipper-EF-hand containing transmembrane protein 1 | AI851685 | 2.62 |
| Epoxide hydrolase 1, microsomal | U89491 | 2.52 | lpC1, G protein-coupled receptor EDG6 | AJ006074 | 2.28 |
| EST | AV356018 | 6.02 | Lymphocyte protein tyrosine kinase | AV314529 | 2.1 |
| EST | AA673252 | 4.96 | Max dimerization protein 4 | U32395 | 2.49 |
| EST | AW061161 | 2.76 | Melanocyte proliferating gene 1 | AI842612 | 2 |
| EST | AI843106 | 2.48 | MEN1, menin | AB023401 | 2.36 |
| EST | AI854141 | 2.46 | Mesoderm development candiate 2 | AW045534 | 2.38 |
| EST | AW011716 | 2.44 | Mitochondrial ribosomal protein L53 | AI854607 | 2.09 |
| EST | AV274270 | 2.44 | MMET-1, granzyme M | AB015728 | 2.07 |
| EST | AV164757 | 2.44 | Mouse ORF | M37030 | 2.49 |
| EST | AV117844 | 2.43 | mRECK, metastasis and invasion | AB006960 | 2.39 |
| EST | AW259411 | 2.42 | Ndr2 | AB033921 | 3.19 |
| EST | AI842128 | 2.4 | Npm3 | U64450 | 2.08 |
| EST | AI834777 | 2.38 | P4ha1, prolyl 4-hydroxylase alpha(I)-subunit | U16162 | 2.36 |
| EST | AI846549 | 2.38 | PAC1, tyrosine-threonine dual specificity phosphatase | U09268 | 2.01 |
| EST | AA795923 | 2.36 | Period homolog (Drosophila) | AF022992 | 2.05 |
| EST | AW049142 | 2.36 | Pkcz, protein kinase C zeta | M94632 | 2.61 |
| EST | AW050018 | 2.33 | Polymerase, gamma | U53584 | 2.05 |
| EST | AV352777 | 2.33 | Polyomavirus enhancer activator 3 | X63190 | 4.55 |
| EST | AA798971 | 2.32 | Polypyrimidine tract binding protein | AV274525 | 2.55 |
| EST | AI849939 | 2.28 | Polypyrimidine tract binding protein 2 | AI119718 | 2.3 |
| EST | AI843155 | 2.27 | Pore forming protein, perforin | X12760 | 2.56 |
| EST | C79210 | 2.25 | Praja1, similar to neurodegeneration associated protein 1 | U06944 | 3.21 |
| EST | AI847879 | 2.24 | Praja1, similar to rat neurodegeneration associated protein 1 | U06944 | 2.09 |
| EST | AW121399 | 2.23 | Protein kinase C, zeta | AV367375 | 2.4 |
| EST | AW124529 | 2.23 | Protein typrotein tyrosine kinase 9-like (A6-related protein) | Y17808 | 2.2 |
| EST | AV171460 | 2.22 | RAB23, member RAS oncogene family | Z22821 | 2.25 |
| EST | AA718040 | 2.21 | rab6 | L40934 | 2.04 |
| EST | AW050015 | 2.2 | Rbm14 | X52102 | 2.09 |
| EST | AW060927 | 2.19 | Rearranged T-cell receptor beta V14/D1.1/J2.3 gene segment | X03278 | 2.41 |
| EST | AW125116 | 2.18 | RFC1 | L23755 | 2.05 |
| EST | AW046470 | 2.18 | Rpl10 | AV105022 | 2.24 |
| EST | AW123921 | 2.17 | s11-6, zinc finger protein | AB020542 | 2.14 |
| EST | AW061306 | 2.17 | Sepiapterin reductase | AI530375 | 2.24 |
| EST | AV369210 | 2.17 | Sex comb on midleg-like 1 (Drosophila) | AI853225 | 2.8 |
| EST | AI845814 | 2.16 | Sgne1, neuroendocrine protein 7B2 | X15830 | 2.19 |
| EST | AA867778 | 2.16 | SHP2 interacting transmembrane adaptor | AJ236881 | 2.87 |
| EST | AF110520 | 2.15 | Solute carrier family 16 (monocarboxylic acid transporters), member 1 | AF058055 | 2.04 |
| EST | AA168476 | 2.14 | Srm, spermidine synthase pseudogene 2 | Z80833 | 2.37 |
| EST | AW208513 | 2.13 | TATA box binding protein (Tbp)-associated factor, RNA polymerase I, C | Y09974 | 2.12 |
| EST | AW049326 | 2.13 | Tcra-V8 | X06307 | 2.12 |
| EST | AV084635 | 2.13 | Tcrz, T cell receptor zeta chain | J04967 | 2.07 |
| EST | AV314618 | 2.13 | Thromboxane A2 receptor | D10849 | 2.44 |
| EST | AI527477 | 2.11 | Thy1 | M12379 | 2.08 |
| EST | AW060526 | 2.1 | Thymoma viral proto-oncogene 2 (Akt2) | U22445 | 2.42 |
| EST | AI007117 | 2.1 | Tob, Trob | D78382 | 2.02 |
| EST | AI854358 | 2.09 | Translocase of inner mitochondrial membrane 22 homolog (yeast) | AA760359 | 2.08 |
| EST | AI854144 | 2.07 | TRAP, acid phosphatase type 5 gene | M99054 | 2.3 |
| EST | AI846994 | 2.07 | Ubiquitin-conjugating enzyme E2D 2 | AV171056 | 2.33 |
| EST | AA733372 | 2.05 | Ubiquitin-conjugating enzyme E2H | U19854 | 2.21 |
| EST | AA250414 | 2.05 | UDP-N-acetyl-alpha-D-galactosamine | U18975 | 2.3 |
| EST | AI850991 | 2.04 | Upregulated by 1,25-dihydroxyvitamin D-3 | AI839138 | 2.1 |
| EST | AW123267 | 2.02 | Zfp-35, zinc finger proetin | M36146 | 3.53 |
| EST | AI596360 | 2.01 | Zinc finger protein 161 | AI447619 | 2.11 |
| EST | AA688761 | 2.01 | Zinc finger protein 54 | AF080070 | 2.24 |
| EST | AV28333 | 2.01 | Zinc finger protein 94 | U46187 | 2.92 |
Although the CD8+T cell response activated by IL-12 analyzed in this study was a total one instead of being specific for given target molecules in the TS/A tumor because of the lack of identified tumor antigens, interesting information regarding the CTL response to a developing tumor can still be gleaned and extrapolated from the microarray data. On the side of the induced genes by IL-12, many of them were quite as expected. Genes that are involved in proteolysis, cytotoxic killing, and cell migration were found, e.g., several types of carboxypeptidases, myeloperoxidase, proteinase 3, neutrophil elastase, caspase-1 (IL-1-converting enzyme, ICE), NK killer receptor NKG2-D, Ly49b, IFN-γ-induced GTPases and chemokines (IP-10, macrophage inflammatory protein MIP), chemokine receptors (CCR1 and CCR5), etc.
One surprising finding is the strong induction of IL-6 by IL-12. IL-6 is a multifunctional cytokine that regulates cell growth, differentiation, and cell survival in the immune system. These two cytokines are not known to interact and cross-talk. In some malignant tumors, tumor-infiltrating lymphocytes (TIL) secrete IL-6 (61, 62). IL-6 completely antagonizes the immunosuppressive effects of TGF-β1 on T cell proliferation in eyes with endotoxin-induced uveitis (63). Combined IL-2 and IL-6 gene therapy, by liposome-mediated intratumor delivery to mice bearing B16F10 melanoma, significantly enhances the CTL and NK activity of splenocytes and TILs (64). TIL secretion of IL-6 antagonizes tumor-derived TGF-β1 and restores the lymphokine-activated killing activity against canine transmissible venereal tumor (CTVT) (65). In light of these new findings and our own data, the role of IL-6 in tumor-specific CTL responses and the interaction between IL-12 and IL-6 should be explored.
A much less appreciated aspect of IL-12 immunobiology is the flip side of the coin, i.e., its inhibitory activities. Among the genes that were inhibited by IL-12 in the CD8+T cells are more novelties. Take for example, guanosine diphosphate (GDP) dissociation inhibitor 2 (GDI-2). The GDP dissociation inhibitors (GDIs) represent an important class of regulatory proteins in the functional cycle and recycling of Rab GTPases. Accumulated evidence over the past several years points to a functional role for the Rab/YPT1/SEC4 gene subfamily of p21 ras-like small GTP-binding proteins in the mechanisms regulating membrane trafficking in yeast and mammalian cells. Reported data are consistent with the notion that Rabs are essential in each step of vesicular transport, including vesicle formation, vesicle docking, and membrane fusion (66). Among the three presently known members of the GDI protein class (GDI-1, also known as RabGDI or GDIα, GDI-2, and GDIβ) (67–70), GDI-1, the best-studied member, appears crucial for progression through the membrane/cytosol localization cycle and the recycling of all Rabs. Because GDIs display a high degree of homology (86% and 96% identity between GDI-2 and GDI-1, or GDIβ, respectively), it is largely accepted, yet not proven, that they exhibit a redundant function. Although several lines of biochemical and morphological evidence suggest distinct functional roles for GDI-1 and GDI-2 in the context of living cells (71–73), no specificity toward an individual Rab or a subset of Rab proteins has been documented. Thus, GDI-2 displays a general activity to release Rabs from membranes (74). Although the involvement of Rabs in cancer is presently unknown, their regulation by GDIs could be an interesting direction in immune response to developing cancer.
Polyomavirus enhancer activator 3 (PEA3) is the prototypical member of one major subgroup of the superfamily of Ets-related transcription factors which, at the present time, is composed of three members, PEA3 (also known as E1AF or ETV4) (75), ER81/ETV1 (76) and ERM/ETV5 (77). These three transcription factors are > 95% identical in the Ets-domain and > 85% in the transactivation acidic domain (78). Several candidate target genes of the PEA3 transcription factor have been reported based largely on the occurrence of Ets-binding sites in their upstream regulatory regions. A large proportion of these genes encode matrix proteases (MMP-2, -7, -9, -13, -14 and -19, uPA) and some of their inhibitors (TIMP-1), whose dysregulated expression has been associated with the invasive potential of tumor cells (79–84). Expression of PEA3 has been positively linked to MMP2, NRG1 and CGB expression in breast tumorigenesis (85). It would be interesting to establish the role of PEA3 in IL-12-mediated anti-tumor CTL response.
IL-12-induced CTL development is a central theme in the immune responses against intracellular infectious agents as well as against malignant growth, united by a common mechanism regulated by IL-12. The microarray data collected at various stages of a tumor-specific CTL development induced by IL-12 represent a comprehensive molecular survey of these important differentiation events in a biologically dynamic and kinetic manner. With the aid of sophisticated bioinformatic tools and experimental verification, we will be able to uncover previously unknown pathways and identify potential targets critical for CTL activation in response to tumor development. These novel pathways and targets will be invaluable to our efforts to understand the process of anti-tumor responses effected by the immune system.
Acknowledgments
S. Cao was supported by a fellowship from the Susan G. Komen Breast Cancer Foundation.
Abbreviation
- CTL
cytotoxic T lymphocyte
- DC
dendritic cell
- FGF
fibroblast growth factor
- VEGF
vascular endothelial growth factor
- GDP
guanosine diphosphate
- CTVT
canine transmissible venereal tumor
- PEA3
polyomavirus enhancer activator 3
- MIP
macrophage inflammatory protein
References
- 1.Stutman O. Tumor development after 3-methylcholanthrene in immunologically deficient athymic-nude mice. Science. 1974;183:534–536. doi: 10.1126/science.183.4124.534. [DOI] [PubMed] [Google Scholar]
- 2.Stutman O. Chemical carcinogenesis in nude mice: comparison between nude mice from homozygous matings and heterozygous matings and effect of age and carcinogen dose. J Natl Cancer Inst. 1979;62:353–358. [PubMed] [Google Scholar]
- 3.Outzen HC, Custer RP, Eaton GJ, Prehn RT. Spontaneous and induced tumor incidence in germfree “nude” mice. J Reticuloendothel Soc. 1975;17:1–9. [PubMed] [Google Scholar]
- 4.Rygaard J, Povlsen CO. Is immunological surveillance not a cell-mediated immune function? Transplantation. 1974;17:135–136. [PubMed] [Google Scholar]
- 5.Rygaard J, Povlsen CO. The mouse mutant nude does not develop spontaneous tumours. An argument against immunological surveillance. Acta Pathol Microbiol Scand [B] Microbiol Immunol. 1974;82:99–106. doi: 10.1111/j.1699-0463.1974.tb02299.x. [DOI] [PubMed] [Google Scholar]
- 6.Engel AM, Svane IM, Mouritsen S, Rygaard J, Clausen J, Werdelin O. Methylcholanthrene-induced sarcomas in nude mice have short induction times and relatively low levels of surface MHC class I expression. Apmis. 1996;104:629–639. doi: 10.1111/j.1699-0463.1996.tb04923.x. [DOI] [PubMed] [Google Scholar]
- 7.Engel AM, Svane IM, Rygaard J, Werdelin O. MCA sarcomas induced in scid mice are more immunogenic than MCA sarcomas induced in congenic, immunocompetent mice. Scand J Immunol. 1997;45:463–470. doi: 10.1046/j.1365-3083.1997.d01-419.x. [DOI] [PubMed] [Google Scholar]
- 8.Dighe AS, Richards E, Old LJ, Schreiber RD. Enhanced in vivo growth and resistance to rejection of tumor cells expressing dominant negative IFN-γ receptors. Immunity. 1994;1:447–456. doi: 10.1016/1074-7613(94)90087-6. [DOI] [PubMed] [Google Scholar]
- 9.Kaplan DH, Shankaran V, Dighe AS, et al. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc Natl Acad Sci U S A. 1998;95:7556–7561. doi: 10.1073/pnas.95.13.7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Street SE, Cretney E, Smyth MJ. Perforin and interferon-γ activities independently control tumor initiation, growth, and metastasis. Blood. 2001;97:192–197. doi: 10.1182/blood.v97.1.192. [DOI] [PubMed] [Google Scholar]
- 11.Street SE, Trapani JA, MacGregor D, Smyth MJ. Suppression of lymphoma and epithelial malignancies effected by interferon gamma. J Exp Med. 2002;196:129–134. doi: 10.1084/jem.20020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Penn I. Malignant melanoma in organ allograft recipients. Transplantation. 1996;61:274–278. doi: 10.1097/00007890-199601270-00019. [DOI] [PubMed] [Google Scholar]
- 13.Clemente CG, Mihm MC, Jr, Bufalino R, Zurrida S, Collini P, Cascinelli N. Prognostic value of tumor infiltrating lymphocytes in the vertical growth phase of primary cutaneous melanoma. Cancer. 1996;77:1303–1310. doi: 10.1002/(SICI)1097-0142(19960401)77:7<1303::AID-CNCR12>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 14.Mihm MC, Jr, Clemente CG, Cascinelli N. Tumor infiltrating lymphocytes in lymph node melanoma metastases: a histopathologic prognostic indicator and an expression of local immune response. Lab Invest. 1996;74:43–47. [PubMed] [Google Scholar]
- 15.Rilke F, Colnaghi MI, Cascinelli N, et al. Prognostic significance of HER-2/neu expression in breast cancer and its relationship to other prognostic factors. Int J Cancer. 1991;49:44–49. doi: 10.1002/ijc.2910490109. [DOI] [PubMed] [Google Scholar]
- 16.Lipponen PK, Eskelinen MJ, Jauhiainen K, Harju E, Terho R. Tumour infiltrating lymphocytes as an independent prognostic factor in transitional cell bladder cancer. Eur J Cancer. 1992;29A:69–75. doi: 10.1016/0959-8049(93)90579-5. [DOI] [PubMed] [Google Scholar]
- 17.Naito Y, Saito K, Shiiba K, et al. CD8+T cells infiltrated within cancer cell nests as a prognostic factor in human colorectal cancer. Cancer Res. 1998;58:3491–3494. [PubMed] [Google Scholar]
- 18.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3:991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 19.Ikeda H, Old LJ, Schreiber RD. The roles of IFN-γ in protection against tumor development and cancer immuno-editing. Cytokine Growth Factor Rev. 2002;13:95–109. doi: 10.1016/s1359-6101(01)00038-7. [DOI] [PubMed] [Google Scholar]
- 20.Browning MJ, Bodmer WF. MHC antigens and cancer: implications for T-cell surveillance. Curr Opin Immunol. 1992;4:613–618. doi: 10.1016/0952-7915(92)90036-e. [DOI] [PubMed] [Google Scholar]
- 21.Chen L, Ashe S, Brady WA, et al. Costimulation of antitumor immunity by the B7 counterreceptor for the T lymphocyte molecules CD28 and CTLA-4. Cell. 1992;71:1093–1102. doi: 10.1016/s0092-8674(05)80059-5. [DOI] [PubMed] [Google Scholar]
- 22.Naor D. Suppressor cells: permitters and promoters of malignancy? Adv Cancer Res. 1979;29:45–125. doi: 10.1016/s0065-230x(08)60846-5. [DOI] [PubMed] [Google Scholar]
- 23.Greenberg PD. Adoptive T cell therapy of tumors: mechanisms operative in the recognition and elimination of tumor cells. Adv Immunol. 1991;49:281–355. doi: 10.1016/s0065-2776(08)60778-6. [DOI] [PubMed] [Google Scholar]
- 24.Robins RA. T-cell responses at the host: tumour interface. Biochim Biophys Acta. 1986;865:289–305. doi: 10.1016/0304-419x(86)90019-3. [DOI] [PubMed] [Google Scholar]
- 25.Boon T, Coulie PG, Van den Eynde B. Tumor antigens recognized by T cells. Immunol Today. 1997;18:267–268. doi: 10.1016/s0167-5699(97)80020-5. [DOI] [PubMed] [Google Scholar]
- 26.Old LJ. Tumor immunology: the first century. Curr Opin Immunol. 1992;4:603–607. doi: 10.1016/0952-7915(92)90034-c. [DOI] [PubMed] [Google Scholar]
- 27.Ochsenbein AF. Principles of tumor immunosurveillance and implications for immunotherapy. Cancer Gene Ther. 2002;9:1043–1055. doi: 10.1038/sj.cgt.7700540. [DOI] [PubMed] [Google Scholar]
- 28.Townsend SE, Allison JP. Tumor rejection after direct costimulation of CD8+T cells by B7-transfected melanoma cells. Science. 1993;259:368–370. doi: 10.1126/science.7678351. [DOI] [PubMed] [Google Scholar]
- 29.Coughlin CM, Salhany KE, Gee MS, et al. Tumor cell responses to IFN-γ affect tumorigenicity and response to IL-12 therapy and antiangiogenesis. Immunity. 1998;9:25–34. doi: 10.1016/s1074-7613(00)80585-3. [DOI] [PubMed] [Google Scholar]
- 30.Trinchieri G. Interleukin-12: a cytokine at the interface of inflammation and immunity. Adv Immunol. 1998;70:83–243. doi: 10.1016/s0065-2776(08)60387-9. [DOI] [PubMed] [Google Scholar]
- 31.Trinchieri G, Scott P. Interleukin-12: basic principles and clinical applications. Curr Top Microbiol Immunol. 1999;238:57–78. doi: 10.1007/978-3-662-09709-0_4. [DOI] [PubMed] [Google Scholar]
- 32.Gong J, Koido S, Chen D, et al. Immunization against murine multiple myeloma with fusions of dendritic and plasmacytoma cells is potentiated by interleukin 12. Blood. 2002;99:2512–2517. doi: 10.1182/blood.v99.7.2512. [DOI] [PubMed] [Google Scholar]
- 33.Gri G, Chiodoni C, Gallo E, Stoppacciaro A, Liew FY, Colombo MP. Antitumor effect of interleukin (IL)-12 in the absence of endogenous IFN-γ: a role for intrinsic tumor immunogenicity and IL-15. Cancer Res. 2002;62:4390–4397. [PubMed] [Google Scholar]
- 34.Guo J, Wang B, Zhang M, et al. Macrophage-derived chemokine gene transfer results in tumor regression in murine lung carcinoma model through efficient induction of antitumor immunity. Gene Ther. 2002;9:793–803. doi: 10.1038/sj.gt.3301688. [DOI] [PubMed] [Google Scholar]
- 35.Asada H, Kishida T, Hirai H, et al. Significant antitumor effects obtained by autologous tumor cell vaccine engineered to secrete interleukin (IL)-12 and IL-18 by means of the EBV/lipoplex. Mol Ther. 2002;5:609–616. doi: 10.1006/mthe.2002.0587. [DOI] [PubMed] [Google Scholar]
- 36.Chen YM, Tsai CM, Whang-Peng J, Perng RP. Double signal stimulation was required for full recovery of the autologous tumor-killing effect of effusion-associated lymphocytes. Chest. 2002;122:1421–1427. doi: 10.1378/chest.122.4.1421. [DOI] [PubMed] [Google Scholar]
- 37.Chaperot L, Manches O, Mi JQ, et al. Differentiation of anti-tumour cytotoxic T lymphocytes from autologous peripheral blood lymphocytes in non-Hodgkin’s lymphomas. Br J Haematol. 2002;119:425–431. doi: 10.1046/j.1365-2141.2002.03885.x. [DOI] [PubMed] [Google Scholar]
- 38.Derre L, Corvaisier M, Pandolfino MC, Diez E, Jotereau F, Gervois N. Expression of CD94/NKG2-A on human T lymphocytes is induced by IL-12: implications for adoptive immunotherapy. J Immunol. 2002;168:4864–4870. doi: 10.4049/jimmunol.168.10.4864. [DOI] [PubMed] [Google Scholar]
- 39.O’Sullivan BJ, Thomas R. CD40 ligation conditions dendritic cell antigen-presenting function through sustained activation of NF-κB. J Immunol. 2002;168:5491–5498. doi: 10.4049/jimmunol.168.11.5491. [DOI] [PubMed] [Google Scholar]
- 40.Wajchman J, Simmons WJ, Klein A, Koneru M, Ponzio NM. Interleukin-12-induced cytotoxicity against syngeneic B cell lymphomas of SJL/J mice. Leuk Res. 2002;26:577–590. doi: 10.1016/s0145-2126(01)00179-5. [DOI] [PubMed] [Google Scholar]
- 41.Furumoto K, Mori A, Yamasaki S, et al. Interleukin-12-gene transduction makes DCs from tumor-bearing mice an effective inducer of tumor-specific immunity in a peritoneal dissemination model. Immunol Lett. 2002;83:13–20. doi: 10.1016/s0165-2478(02)00071-8. [DOI] [PubMed] [Google Scholar]
- 42.Mazda O. Improvement of nonviral gene therapy by Epstein-Barr virus (EBV)-based plasmid vectors. Curr Gene Ther. 2002;2:379–392. doi: 10.2174/1566523023347814. [DOI] [PubMed] [Google Scholar]
- 43.Saudemont A, Buffenoir G, Denys A, et al. Gene transfer of CD154 and IL12 cDNA induces an anti-leukemic immunity in a murine model of acute leukemia. Leukemia. 2002;16:1637–1644. doi: 10.1038/sj.leu.2402590. [DOI] [PubMed] [Google Scholar]
- 44.Strohlein MA, Grutzner KU, Schildberg FW, Heiss MM. Induction of cytotoxicity against autologous tumour cells by interleukin-12: evidence for intrinsic anti-tumor immune capacity in curatively resected gastrointestinal tumour patients. Cancer Immunol Immunother. 2002;51:505–512. doi: 10.1007/s00262-002-0310-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pan PY, Zang Y, Weber K, Meseck ML, Chen SH. OX40 ligation enhances primary and memory cytotoxic T lymphocyte responses in an immunotherapy for hepatic colon metastases. Mol Ther. 2002;6:528–536. doi: 10.1006/mthe.2002.0699. [DOI] [PubMed] [Google Scholar]
- 46.Monzavi-Karbassi B, Shamloo S, Kieber-Emmons M, et al. Priming characteristics of peptide mimotopes of carbohydrate antigens. Vaccine. 2003;21:753–760. doi: 10.1016/s0264-410x(02)00703-x. [DOI] [PubMed] [Google Scholar]
- 47.Vegh Z, Mazumder A. Generation of tumor cell lysate-loaded dendritic cells preprogrammed for IL-12 production and augmented T cell response. Cancer Immunol Immunother. 2003;52:67–79. doi: 10.1007/s00262-002-0338-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee SW, Park Y, Yoo JK, Choi SY, Sung YC. Inhibition of TCR-Induced CD8 T Cell Death by IL-12: Regulation of Fas Ligand and Cellular FLIP Expression and Caspase Activation by IL-12. J Immunol. 2003;170:2456–2460. doi: 10.4049/jimmunol.170.5.2456. [DOI] [PubMed] [Google Scholar]
- 49.Nanni P, de Giovanni C, Lollini PL, Nicoletti G, Prodi G. TS/A: a new metastasizing cell line from a BALB/c spontaneous mammary adenocarcinoma. Clin Exp Metastasis. 1983;1:373–380. doi: 10.1007/BF00121199. [DOI] [PubMed] [Google Scholar]
- 50.Giovarelli M, Santoni A, Forni G. Alloantigen-activated lymphocytes from mice bearing a spontaneous “nonimmunogenic” adenocarcinoma inhibit its growth in vivo by recruiting host immunoreactivity. J Immunol. 1985;135:3596–3603. [PubMed] [Google Scholar]
- 51.Cavallo F, Signorelli P, Giovarelli M, et al. Antitumor efficacy of adenocarcinoma cells engineered to produce interleukin 12 (IL-12) or other cytokines compared with exogenous IL-12. J Natl Cancer Inst. 1997;89:1049–1058. doi: 10.1093/jnci/89.14.1049. [DOI] [PubMed] [Google Scholar]
- 52.Southern EM, Maskos U, Elder JK. Analyzing and comparing nucleic acid sequences by hybridization to arrays of oligonucleotides: evaluation using experimental models. Genomics. 1992;13:1008–1017. doi: 10.1016/0888-7543(92)90014-j. [DOI] [PubMed] [Google Scholar]
- 53.Chee M, Yang R, Hubbell E, et al. Accessing genetic information with high-density DNA arrays. Science. 1996;274:610–614. doi: 10.1126/science.274.5287.610. [DOI] [PubMed] [Google Scholar]
- 54.DeRisi J, Penland L, Brown PO, et al. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 55.DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 56.Heller RA, Schena M, Chai A, et al. Discovery and analysis of inflammatory disease-related genes using cDNA microarrays. Proc Natl Acad Sci U S A. 1997;94:2150–2155. doi: 10.1073/pnas.94.6.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strausberg RL, Dahl CA, Klausner RD. New opportunities for uncovering the molecular basis of cancer. Nat Genet. 1997;15(Spec No):415–416. doi: 10.1038/ng0497supp-415. [DOI] [PubMed] [Google Scholar]
- 58.Chen H, Centola M, Altschul SF, Metzger H. Characterization of gene expression in resting and activated mast cells. J Exp Med. 1998;188:1657–1668. doi: 10.1084/jem.188.9.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brockstedt DG, Diagana M, Zhang Y, et al. Development of anti-tumor immunity against a non-immunogenic mammary carcinoma through in vivo somatic GM-CSF, IL-2, and HSVtk combination gene therapy. Mol Ther. 2002;6:627–636. [PubMed] [Google Scholar]
- 60.Gately MK, Chizzonite R, Presky DH. Measurement of human and mouse interleukin-12. In: Coligan JE, Kruisbeek AM, Margulies DH, Shevach EM, Strober W, editors. Current Protocols in Immunology. New York, NY: John Wiley & Sons; 1995. pp. 16.11–16.15. [Google Scholar]
- 61.Kharkevitch DD, Seito D, Balch GC, Maeda T, Balch CM, Itoh K. Characterization of autologous tumor-specific T-helper 2 cells in tumor-infiltrating lymphocytes from a patient with metastatic melanoma. Int J Cancer. 1994;58:317–323. doi: 10.1002/ijc.2910580302. [DOI] [PubMed] [Google Scholar]
- 62.Ortegel JW, Staren ED, Faber LP, Warren WH, Braun DP. Modulation of tumor-infiltrating lymphocyte cytolytic activity against human non-small cell lung cancer. Lung Cancer. 2002;36:17–25. doi: 10.1016/s0169-5002(01)00472-x. [DOI] [PubMed] [Google Scholar]
- 63.Ohta K, Yamagami S, Taylor AW, Streilein JW. IL-6 antagonizes TGF-β and abolishes immune privilege in eyes with endotoxin-induced uveitis. Invest Ophthalmol Vis Sci. 2000;41:2591–2599. [PubMed] [Google Scholar]
- 64.Cao X, Wang Q, Ju DW, Tao Q, Wang J. Efficient inducation of local and systemic antitumor immune response by liposome-mediated intratumoral co-transfer of interleukin-2 gene and interleukin-6 gene. J Exp Clin Cancer Res. 1999;18:191–200. [PubMed] [Google Scholar]
- 65.Hsiao YW, Liao KW, Hung SW, Chu RM. Tumor-infiltrating lymphocyte secretion of IL-6 antagonizes tumor-derived TGF-β1 and restores the lymphokine-activated killing activity. J Immunol. 2004;172:1508–1514. doi: 10.4049/jimmunol.172.3.1508. [DOI] [PubMed] [Google Scholar]
- 66.Schimmoller F, Simon I, Pfeffer SR. Rab GTPases, directors of vesicle docking. J Biol Chem. 1998;273:22161–22164. doi: 10.1074/jbc.273.35.22161. [DOI] [PubMed] [Google Scholar]
- 67.Matsui Y, Kikuchi A, Araki S, et al. Molecular cloning and characterization of a novel type of regulatory protein (GDI) for smg p25A, a ras p21-like GTP-binding protein. Mol Cell Biol. 1990;10:4116–4122. doi: 10.1128/mcb.10.8.4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nishimura N, Nakamura H, Takai Y, Sano K. Molecular cloning and characterization of two rab GDI species from rat brain: brain-specific and ubiquitous types. J Biol Chem. 1994;269:14191–14198. [PubMed] [Google Scholar]
- 69.Janoueix-Lerosey I, Jollivet F, Camonis J, Marche PN, Goud B. Two-hybrid system screen with the small GTP-binding protein Rab6. Identification of a novel mouse GDP dissociation inhibitor isoform and two other potential partners of Rab6. J Biol Chem. 1995;270:14801–14808. doi: 10.1074/jbc.270.24.14801. [DOI] [PubMed] [Google Scholar]
- 70.Shisheva A, Sudhof TC, Czech MP. Cloning, characterization, and expression of a novel GDP dissociation inhibitor isoform from skeletal muscle. Mol Cell Biol. 1994;14:3459–3468. doi: 10.1128/mcb.14.5.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shisheva A, Czech MP. Association of cytosolic Rab4 with GDI isoforms in insulin-sensitive 3T3-L1 adipocytes. Biochemistry. 1997;36:6564–6570. doi: 10.1021/bi970202g. [DOI] [PubMed] [Google Scholar]
- 72.Shisheva A, Doxsey SJ, Buxton JM, Czech MP. Pericentriolar targeting of GDP-dissociation inhibitor isoform 2. Eur J Cell Biol. 1995;68:143–158. [PubMed] [Google Scholar]
- 73.Shisheva A, Buxton J, Czech MP. Differential intracellular localizations of GDP dissociation inhibitor isoforms. Insulin-dependent redistribution of GDP dissociation inhibitor-2 in 3T3-L1 adipocytes. J Biol Chem. 1994;269:23865–23868. [PubMed] [Google Scholar]
- 74.Shisheva A, Chinni SR, DeMarco C. General role of GDP dissociation inhibitor 2 in membrane release of Rab proteins: modulations of its functional interactions by in vitro and in vivo structural modifications. Biochemistry. 1999;38:11711–11721. doi: 10.1021/bi990200r. [DOI] [PubMed] [Google Scholar]
- 75.Xin JH, Cowie A, Lachance P, Hassell JA. Molecular cloning and characterization of PEA3, a new member of the Ets oncogene family that is differentially expressed in mouse embryonic cells. Genes Dev. 1992;6:481–496. doi: 10.1101/gad.6.3.481. [DOI] [PubMed] [Google Scholar]
- 76.Brown TA, McKnight SL. Specificities of protein-protein and protein-DNA interaction of GABP alpha and two newly defined ets-related proteins. Genes Dev. 1992;6:2502–2512. doi: 10.1101/gad.6.12b.2502. [DOI] [PubMed] [Google Scholar]
- 77.Monte D, Baert JL, Defossez PA, de Launoit Y, Stehelin D. Molecular cloning and characterization of human ERM, a new member of the Ets family closely related to mouse PEA3 and ER81 transcription factors. Oncogene. 1994;9:1397–1406. [PubMed] [Google Scholar]
- 78.de Launoit Y, Baert JL, Chotteau A, et al. Structure-function relationships of the PEA3 group of Ets-related transcription factors. Biochem Mol Med. 1997;61:127–135. doi: 10.1006/bmme.1997.2605. [DOI] [PubMed] [Google Scholar]
- 79.Sharrocks AD, Brown AL, Ling Y, Yates PR. The ETS-domain transcription factor family. Int J Biochem Cell Biol. 1997;29:1371–1387. doi: 10.1016/s1357-2725(97)00086-1. [DOI] [PubMed] [Google Scholar]
- 80.Clark IM, Rowan AD, Edwards DR, et al. Transcriptional activity of the human tissue inhibitor of metalloproteinases 1 (TIMP-1) gene in fibroblasts involves elements in the promoter, exon 1 and intron 1. Biochem J. 1997;324 (Pt 2):611–617. doi: 10.1042/bj3240611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Crawford HC, Fingleton B, Gustavson MD, et al. The PEA3 subfamily of Ets transcription factors synergizes with beta-catenin-LEF-1 to activate matrilysin transcription in intestinal tumors. Mol Cell Biol. 2001;21:1370–1383. doi: 10.1128/MCB.21.4.1370-1383.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Evans CP, Stapp EC, Dall’Era MA, Juarez J, Yang JC. Regulation of u-PA gene expression in human prostate cancer. Int J Cancer. 2001;94:390–395. doi: 10.1002/ijc.1469. [DOI] [PubMed] [Google Scholar]
- 83.Higashino F, Yoshida K, Noumi T, Seiki M, Fujinaga K. Ets-related protein E1A-F can activate three different matrix metalloproteinase gene promoters. Oncogene. 1995;10:1461–1463. [PubMed] [Google Scholar]
- 84.Pendas AM, Balbin M, Llano E, Jimenez MG, Lopez-Otin C. Structural analysis and promoter characterization of the human collagenase-3 gene (MMP13) Genomics. 1997;40:222–233. doi: 10.1006/geno.1996.4554. [DOI] [PubMed] [Google Scholar]
- 85.Bieche I, Tozlu S, Girault I, et al. Expression of PEA3/E1AF/ETV4, an Ets-related transcription factor, in breast tumors: positive links to MMP2, NRG1 and CGB expression. Carcinogenesis. 2004;25:405–411. doi: 10.1093/carcin/bgh024. [DOI] [PubMed] [Google Scholar]


