Abstract
Background
Agrobacterium tumefaciens has long been known to transform plant tissue in nature as part of its infection process. This natural mechanism has been utilised over the last few decades in laboratories world wide to genetically manipulate many species of plants. More recently this technology has been successfully applied to non-plant organisms in the laboratory, including fungi, where the plant wound hormone acetosyringone, an inducer of transformation, is supplied exogenously. In the natural environment it is possible that Agrobacterium and fungi may encounter each other at plant wound sites, where acetosyringone would be present, raising the possibility of natural gene transfer from bacterium to fungus.
Methodology/Principal Findings
We investigate this hypothesis through the development of experiments designed to replicate such a situation at a plant wound site. A. tumefaciens harbouring the plasmid pCAMDsRed was co-cultivated with the common plant pathogenic fungus Verticillium albo-atrum on a range of wounded plant tissues. Fungal transformants were obtained from co-cultivation on a range of plant tissue types, demonstrating that plant tissue provides sufficient vir gene inducers to allow A. tumefaciens to transform fungi in planta.
Conclusions/Significance
This work raises interesting questions about whether A. tumefaciens may be able to transform organisms other than plants in nature, or indeed should be considered during GM risk assessments, with further investigations required to determine whether this phenomenon has already occurred in nature.
Introduction
Agrobacterium tumefaciens is a specialized plant pathogen that has evolved a complex mechanism for transforming many hundreds of plant species in order to provide a unique chemical and environmental habitat for itself. The transfer of DNA from Agrobacterium species to plants (which in the case of A. tumefaciens results in production of crown galls) represents the only accepted example of common natural trans-kingdom gene transfer. Virulent A. tumefaciens strains possess an extrachromosomal tumour inducing (Ti) plasmid, typically ∼200 kb in size. This plasmid contains a number of features important to the transformation process, one of which is the region to be transferred, called T-DNA (transferred DNA). Through the detection of phenolic compounds produced by a wounded plant, the bacterium's virulence system is activated and T-DNA is transferred to the nuclear genome of the plant. T-DNA is delimited by a left and right border each consisting of 25 bp repeat sequences, and it is the presence of these borders, not the sequence between them that is crucial for transfer [1]. This remarkable feature has led to the development of A. tumefaciens as a vital tool for genetic engineering [2], [3]. In theory any piece of DNA can be placed between the borders and will be transferred, upon infection, to the plant. However, it should be noted that in nature the Ti plasmids of A. tumefaciens, while diverse in nucleotide sequence, all contain a relatively-conserved set of genes that code for a specific set of amino acids required by the pathogen.
Bundock et al., [4] demonstrated that the repertoire of organisms amenable to ATMT could be extended if the phenolic wound hormone acetosyringone was added in vitro to induce the virulence pathway. They reported successful A. tumefaciens-mediated transformation (ATMT) in the laboratory of the yeast Saccharomyces cerevisiae [4], with transformation only possible in the presence of acetosyringone. Experiments investigating the effect of various vir mutants confirmed that the mechanism for transfer uses the same vir machinery as for plant transformation [5], [6]. Following the publication of de Groot et al., [5], an ever increasing number of fungal species are proving amenable to ATMT in the laboratory [7], [8], [9], [10], [11], [12], [13], [14]. This discovery that non-plant species could also be amenable to ATMT in the laboratory demonstrates that the barriers to HGT via A. tumefaciens are not as strict as was first thought. Indeed it has even been reported that A. tumefaciens is able to transform human cells in the laboratory [15]. The A. tumefaciens virulence system has evolved to respond to phenolic wound hormones such as acetosyringone, and is able to transform a range of fungal species if acetosyringone is provided in vitro. As A. tumefaciens is a soil-dwelling pathogen that often infects plants through wound sites, it is conceivable that it could encounter numerous species of microorganisms at such an environmental niche, including plant pathogenic fungi also utilizing this method of plant entry. Such wound sites are likely to be exuding wound hormones such as acetosyringone, so the bacteria are likely to be primed for T-DNA transfer. This paper investigates the possibility that ATMT of fungi could be happening at such wound sites in nature. The wilt-causing fungus Verticillium albo-atrum is a strong candidate for encountering Agrobacterium in planta, as it has a similar wide host range, infecting root and crown. It has a well characterised disease cycle, is amenable to culture in the laboratory, and has recently been transformed via ATMT [16]. Perhaps most importantly, V. albo-atrum cannot be transformed by Agrobacterium in the absence of acetosyringone [16], therefore if Agrobacterium and V. albo-atrum are present together on plant tissue and transformation does occur, the acetosyringone will have been plant derived.
Materials and Methods
Fungal and bacterial strains
A. tumefaciens strain LBA1126 containing the binary vector pCAMDsRed [12] was used in all experiments. pCAMDsRed contains the DsRed (Express) gene under the control of the Aspergillus nidulans gpdA promoter and the trpC terminator, and the hygromycin B resistance gene (hph) under the control of a trpC promoter. A. tumefaciens cultures were prepared as in Knight et al., [16], with the modifcation that following 48 hour incubation in LB, cultures were diluted with sterile water to an OD600 nm of 0.25, instead of being induced with acetosyringone in Induction Medium.
Conidia from wild type V. albo-atrum strain 1974, (Warwick-HRI) were harvested from potato dextrose agar (PDA) plates and diluted to a concentration of 108 spores.ml−1 in sterile water.
Preparation of plant tissues
Potato tubers (Maris Piper) and carrots (Scarlet Nantes) were aseptically peeled, sliced (approximately 30 mm diameter and 5 mm thickness) and surface sterilised in a 10% sodium hypochlorite solution. Tissue was then either placed on plates containing Murashige and Skoog (MS) agar [17] +1.5% sucrose with or without 200 µM acetosyringone, or was placed in petri dishes in the absence of any media.
Tobacco plantlets (Nicotiana tabacum var. White Burley) were grown in sterile magenta pots containing MS agar supplemented with 1.5% sucrose. Sections of leaf (average area 120 mm2) were removed and inoculated with bacterial/fungal suspension before 3–5 sections were placed on each MS+1.5% sucrose plate.
Tobacco plants (N. tabacum var. White Burley) approximately 8 weeks old had all leaves removed and the stem cut into sections approximately 5 cm long. These were then surface sterilised as described above. One section of stem was placed in each petri dish, in the absence of any media and a sterile scalpel used to score the surface.
Inoculation of plant material with A. tumefaciens and V. albo-atrum
V. albo-atrum conidial suspension (ca. 108 spores.ml−1) was mixed with an equal volume of A. tumefaciens culture (an OD600 nm of 0.25–0.30) before being inoculated onto plant tissue. For potato slice experiments, either 40 µl or 200 µl was pipetted onto the surface, for carrot slices the volume was 40 µl. For tobacco stem experiments, 100 µl suspension was inoculated onto the surface of stems. Leaf experiments involved placing leaf sections in a 1∶1 suspension of conidia and A. tumefaciens for 5 minutes. Control plates for each experiment comprised plant material inoculated solely with wild type V. albo-atrum.
Plates were left at room temperature for a minimum of 8 days and a maximum of 42 days.
Harvesting of samples
A range of methods were attempted with the early experiments (potato and carrot) in order to successfully isolate putative transformants from plant tissue. These included using an inoculating loop to disrupt the previously inoculated surface of the plant tissue in the presence of ∼2 ml sterile water. This liquid was then pippetted onto selection plates (PDA+50 µg.ml−1 hygromycin B and 75 µg.ml−1 Timentin). Alternatively, plant material was removed from the co-cultivation plate and homogenised in an autoclaved pestle and mortar, before either being added to molten selection media, or spread over selection media plates. In the case of experiments with tobacco stems, stem sections were cut in half longitudinally before being placed (cut side down) onto selection plates.
For potato slices, transformants were obtained using one of three methods: by homogenising the plant material in a pestle and mortar and either spreading the suspension directly on selection plates (PDA+50 µg.ml−1 hygromycin B and 75 µg.ml−1 Timentin) or by adding the suspension to molten selection media and pouring into a plate, or by disrupting the plant tissue using a sterile inoculating loop and ∼2 ml sterile water and adding the suspension to molten selection media before pouring into a plate.
For tobacco leaf experiments, plant tissue was homogenised in pestle and mortars before being spread over selection plates. In the case of experiments with tobacco stems, stem sections were cut in half longitudinally before being placed (cut side down) onto selection plates.
Microscopy
Putative transformants were screened for DsRed expression in situ and following subculture onto fresh selection plates using a DM-LB Leica microscope fitted with a red-shifted TRITC filter (excitation 545/30 nm, emission 620/60 nm) [18], [19].
DNA extraction and analysis
Following repeated subculturing onto selection plates to obtain single colonies, fungal genomic DNA was extracted essentially as described in Keon and Hargreaves [20] (see [21]). PCR amplification was then conducted on genomic DNA using primers HygF (5′-GCGTGGATATGTCCTGCGGG-3′) and HygR (5′-CCATACAAGCCAACCACGG-3′) to produce a 598 bp fragment of the hph gene [22]. For confirmation of presence of the DsRed gene, primers DsRedF (5′-AGGACGTCATCAAGGAGTTC-3′) and DsRedR (5′-CAGCCCATAGTCTTCTTCTG-3′) resulted in the amplification of a 414 bp fragment. Species specific ITS primers - VaaF (5′-CCGCCGGTACATCAGTCTCTTTATTTATAC-3′) and VaaR (5′- GTGCTGCGGGACTCCGATGCGAGCTGTAAT-3′) that amplify a ∼300 bp ITS region were used to confirm fungal DNA was of V. albo-atrum origin [23].
To determine T-DNA copy number, Southern analysis was performed by digesting 10 µg genomic DNA with XbaI, before being separated via electrophoresis and transferred to Hybond N+ membrane. Standard protocols [24] were used for hybridisation using a [α-32P] dCTP labeled DsRed PCR product.
Results
Transformation of V. albo-atrum by A. tumefaciens on carrot and potato tissues
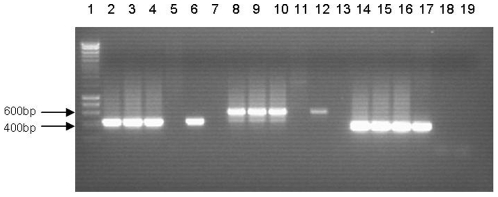
Potato tubers and carrots are both natural hosts of V. albo-atrum [25] and are easy to obtain and sterilise. The first transformants were obtained from control experiments, with co-cultivation with A. tumefaciens and V. albo-atrum on a potato slice, where acetosyringone was present in the MS co-cultivation medium. This demonstrated that the selection conditions worked, and that the plant tissue in this system was not in any way inhibitory to the ATMT of fungi. Following this initial success, ‘more natural’ experiments were conducted, whereby no exogenous acetosyringone was provided other than that produced by the wounded plant tissue. From these conditions, 3 transformants, 2 from potato slices (17 plates, 1 slice per plate), and 1 from carrot (15 plates, 1 slice per plate) were isolated. All putative transformants were subcultured onto fresh selection plates to confirm hygromycin B resistance, and screened using microscopy for DsRed expression. Transformants maintained hygromycin B resistance and DsRed expression following repeated subculturing in both the presence and absence of selection suggesting that stable transformation had occurred. No hygromycin B resistant, DsRed expressing colonies were obtained in any of the controls, where just fungal material was inoculated onto plant tissue in the absence of A. tumefaciens. PCR was performed on the transformants generated on potato disc, which confirmed presence of both the hph and DsRed genes in all transformants (see Figure 1), and Southern analysis demonstrated that as with conventional ATMT of V. albo-atrum [16] T-DNA integrated randomly into genomic DNA, and as single copy insertions (Figure 2).
Figure 1. PCR analysis of 3 V. albo-atrum transformants obtained from ATMT experiments on potato.
Lane 1, DNA Hyperladder I (Bioline), lanes 2–7, PCR with DsRed primers; 3 transformants (2–4), wild type V. albo-atrum (5), pCAMDsRed (6), water (7). Lanes 8–13, PCR with hygromycin B primers; 3 transformants (8–10), wild type V. albo-atrum (11), pCAMDsRed (12), water (13). Lanes 14–19, PCR with wild type V. albo-atrum ITS primers; 3 transformants (14–16), wild type V. albo-atrum (17), pCAMDsRed (18), water (19).
Figure 2. Autoradiograph showing Southern analysis of 3 transformants obtained from potato slice hosts.
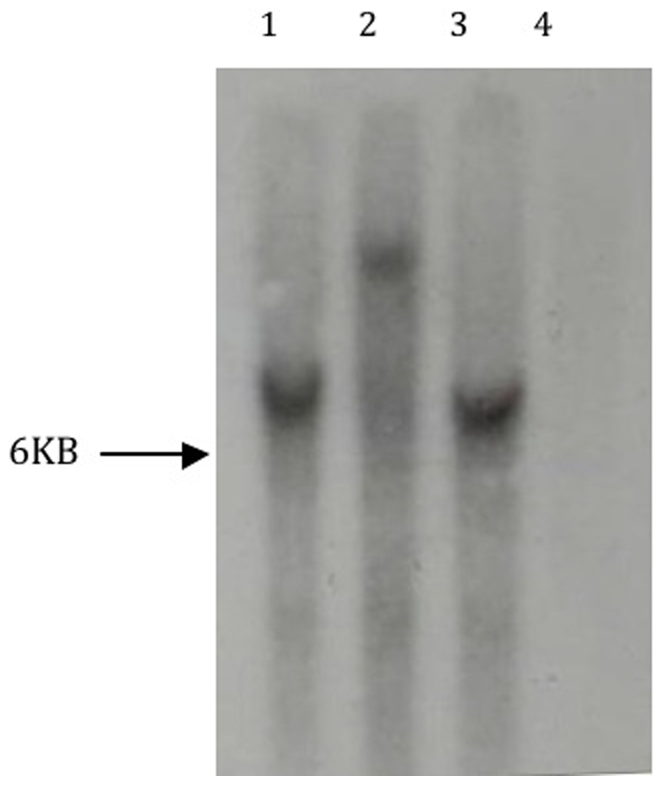
10 µg genomic DNA was digested with XbaI and probed with the 414 bp fragment of the DsRed gene obtained via PCR of pCAMDsRed using primers DsRed forward and reverse. Lanes 1–3, transformants, lane 4, wild type V. albo-atrum.
Transformation of V. albo-atrum by A. tumefaciens on tobacco stem and leaf tissues
Following successful isolation of fungal transformants from potato and carrot slices, tobacco leaves were investigated as another example of a semi-natural damaged area upon which ATMT of a fungus may occur in the absence of exogenously provided acetosyringone. A total of 14 transformants were obtained from four replicated experiments (comprising 42 plates, 3–5 leaf pieces per plate). In all four experiments the only source of acetosyringone was the plant tissue and A. tumefaciens had not been pre-induced. Control plates containing just plant tissue inoculated with V. albo-atrum never resulted in hygromycin B resistant DsRed expressing transformants. Putative fungal transformants were screened via DsRed microscopy and repeated subculturing on selection plates, before a subset of 10 were shown to be transformed by PCR for both DsRed and hygromycin and Southern analysis (data not shown).
Finally, following successful ATMT of V. albo-atrum on tobacco leaves in the absence of exogenous acetosyringone, further experiments even more representative of a ‘natural’ situation for ATMT of V. albo-atrum were conducted. A. tumefaciens was co-cultivated with V. albo-atrum on and within tobacco stems in the absence of any medium on the plates. The lack of agar medium on the plate means that the fungus and bacteria are confined within/on the dying stem section for survival, and any acetosyringone produced could not diffuse into the medium.
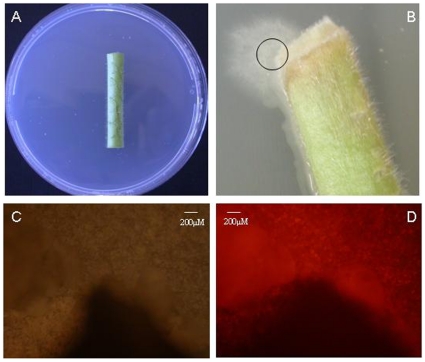
Co-cultivation of A. tumefaciens with V. albo-atrum conidia on tobacco stems (Fig. 3A) resulted in the isolation of 10 fungal transformants in total from two independent experiments (31 plates in total, 1 stem per plate). Transformants were visualised in situ (Figure 3B,C and D) with all demonstrating strong fluorescence before being subcultured to fresh selection plates, and again confirmed positive via PCR analysis (data not shown). No transformants were observed on control plates.
Figure 3. ATMT of V. albo-atrum on stems.
Stem section prior to inoculation (Panel A), (Panel B) stem section showing a transformed V. albo-atrum colony emerging from the stem onto the selection medium ten days after transfer to selection (stems were co-cultivated on no media for 12–22 days prior to selection), (Panel C) white light microscopy of plant/PDA interface circled in B, (Panel D) the same region viewed under the DsRed TRITC filter.
Discussion
It is well established that the plant pathogenic bacterium A. tumefaciens transforms plants in nature, producing crown galls as a result of the transfer of T-DNA to the plant genome [26]. This natural form of genetic modification has been adapted in laboratories over the last 20 years to enable the insertion of almost any foreign gene into plants [27]. More recently it has been found that non-plant organisms such as fungi, and even human cells can be transformed via ATMT in the laboratory. We investigated whether ATMT of non-plant organisms could happen in planta, with induction of A. tumefaciens virulence system being provided by damaged plant tissue. The plant pathogenic fungus V. albo-atrum was chosen as an initial test system because such fungi have the potential to co-exist on plant surfaces with the possibility of colonising microenvironments such as stems and crown galls, with V. albo-atrum and A. tumefaciens commonly causing disease on the same plant species such as hops, rose and raspberry. V. albo-atrum has also recently been transformed via ATMT in the laboratory [16] further confirming its suitability for such studies.
In the case of fungi at least, transformation using A. tumefaciens appears to require the addition of acetosyringone [9], [10], [12], [16]. We recently demonstrated this to also be true in the case of V. albo-atrum [16] with no transformants being produced if acetosyringone was not provided exogenously. Initial experiments on plant tissue therefore included acetosyringone provided in the co-cultivation medium. The production of transformants from such conditions demonstrated that selection conditions were suitable, and that plant material was not in any way inhibitory to ATMT of fungi.
This led to the development of further experiments designed to mimic a more natural situation, with the only source of acetosyringone being the damaged plant tissue. Consequently, V. albo-atrum transformants were successfully obtained following co-cultivation of A. tumefaciens and V. albo-atrum on carrot and potato slices, tobacco leaves and stems. These reproducible findings appear to be the first evidence that A. tumefaciens is capable of transforming a fungus in a semi-natural environment, and suggests that in principle at least, this could also be possible in nature.
Whilst transformation is less efficient than in conventional ATMT of V. albo-atrum [16], the relative ease (following optimisation) with which fungal transformants were obtained from a range of plant material indicates that there is a possibility that this could be occurring in nature.
This study focused on ATMT of V. albo-atrum in planta, however there are numerous other fungi that A. tumefaciens could also encounter at wound sites in nature (for example Fusarium oxysporum) that could have been used in this study, and which may also be amenable to ‘semi-natural’ ATMT. The isolation of fungal transformants following co-cultivation on several types of plant tissue – carrot, potato and tobacco – may also indicate that not only is ATMT of V. albo-atrum possible on different parts of a plant, but also on a range of different host plants.
This work therefore raises interesting questions about whether the host range of A. tumefaciens in nature is greater than just plants. It is possible that evidence of such events could be looked for retrospectively in the increasing number of genome sequences becoming available. Machida et al., [28] reported the finding of two genes of possible Agrobacterium origin in the genome sequence of Aspergillus oryzae, although as they resemble chromosomal genes rather than T-DNA the mechanism for such transfer is unclear. Nevertheless, there have been reports of similar transfers of Agrobacterium chromosomal DNA being transferred to plants in the absence of any obvious T-DNA signatures [29]. Therefore such hypothetical transfer events warrant further investigation.
If this is indeed a possible natural form of horizontal gene flow previously unappreciated it is interesting purely from an evolutionary point of view. However, the biological significance of any such gene transfers may depend on whether the transferred DNA confers any selective advantage to the organism in its natural environment.
In addition, the results may well have implications for the risk assessment of GM plants generated via Agrobacterium-mediated transformation, as Agrobacterium can survive within plant tissue through transformation and tissue culture and can therefore be found within regenerated transgenic plants [30], [31]. Much has been made of the fact that the elimination of Agrobacterium is essential to prevent this as a possible route of gene escape possibly from Agrobacterium to other bacteria, but this study shows that the encounter between Agrobacterium and a plant pathogenic fungus on a plant surface can lead to gene flow in a new, and to-date, under investigated way.
Indeed, many susceptible crop plants are thought to be infected by Agrobacterium primarily during propagation, especially perennial crops that are grafted (e.g., rose), and it is clear that more and more GM perennial crops are being generated and grown each year.
In conclusion, we have shown that a range of plant tissues are capable of inducing A. tumefaciens to transfer T-DNA to the plant pathogenic fungus V. albo-atrum in a range of semi-natural experiments reminiscent of plant wound/damage sites. This suggests that A. tumefaciens may be able to transform non-plant organisms such as fungi in nature, the implications of which are unknown.
Acknowledgments
This work was carried out under DEFRA Licence PHL 247A/6355.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by a Natural Environment Research Council (NERC) PhD studentship awarded to CJK. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Tzfira T, Citovsky V. The Agrobacterium-Plant Cell Interaction. Taking Biology Lessons from a Bug Plant Physiol. 2003;133:943–947. doi: 10.1104/pp.103.032821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gelvin SB. Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol Mol Biol Rev. 2003;67:16–37. doi: 10.1128/MMBR.67.1.16-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCullen CA, Binns AN. Agrobacterium tumefaciens and Plant Cell Interactions and Activities Required for Interkingdom Macromolecular Transfer. Annu Rev Cell Dev Biol. 2006;22:101–127. doi: 10.1146/annurev.cellbio.22.011105.102022. [DOI] [PubMed] [Google Scholar]
- 4.Bundock P, den Dulk-Ras A, Beijersbergen A, Hooykaas PJ. Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J. 1995;14:3206–14. doi: 10.1002/j.1460-2075.1995.tb07323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Groot MJ, Bundock P, Hooykaas PJ, Beijersbergen AG. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat Biotechnol. 1998;16:839–42. doi: 10.1038/nbt0998-839. [DOI] [PubMed] [Google Scholar]
- 6.Michielse CB, Ram AF, Hooykaas PJ, van den Hondel CA. Agrobacterium-mediated transformation of Aspergillus awamori in the absence of full-length VirD2, VirC2, or VirE2 leads to insertion of aberrant T-DNA structures. J Bacteriol. 2004;186:2038–45. doi: 10.1128/JB.186.7.2038-2045.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X, Stone M, Schlagnhaufer C, Romaine CP. A Fruiting Body Tissue Method for Efficient Agrobacterium-Mediated Transformation of Agaricus bisporus. Appl Environ Microbiol. 2000;66:4510–4513. doi: 10.1128/aem.66.10.4510-4513.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Covert SF, Kapoor P, Lee M-h, Briley A, Nairn CJ. Agrobacterium tumefaciens-mediated transformation of Fusarium circinatum. Mycol. 2001;Res105:259–264. [Google Scholar]
- 9.Mullins ED, Chen X, Romaine P, Raina R, Geiser DM, et al. Agrobacterium-mediated transformation of Fusarium oxysporum: An efficient tool for insertional mutagenesis and gene transfer. Phytopathology. 2001;91:173–180. doi: 10.1094/PHYTO.2001.91.2.173. [DOI] [PubMed] [Google Scholar]
- 10.Amey RC, Athey-Pollard A, Burns C, Mills PR, Bailey A, et al. PEG-mediated and Agrobacterium-mediated transformation in the mycopathogen Verticillium fungicola. Mycol Res. 2002;106:4–11. [Google Scholar]
- 11.dos Reis MC, Pelegrinelli Fungaro MH, Delgado Duarte RT, Furlaneto L, Furlaneto MC. Agrobacterium tumefaciens-mediated genetic transformation of the entomopathogenic fungus Beauveria bassiana. J Microbiol Methods. 2004;58:197–202. doi: 10.1016/j.mimet.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 12.Eckert M, Maguire K, Urban M, Foster S, Fitt B, et al. Agrobacterium tumefaciens-mediated transformation of Leptosphaeria spp. and Oculimacula spp. with the reef coral gene DsRed and the jellyfish gene gfp. FEMS Microbiol Lett. 2005;253:67–74. doi: 10.1016/j.femsle.2005.09.041. [DOI] [PubMed] [Google Scholar]
- 13.Burns C, Leach KM, Elliott TJ, Challen MP, Foster GD, et al. Evaluation of Agrobacterium-mediated transformation of Agaricus bisporus using a range of promoters linked to hygromycin resistance. Mol Biotechnol. 2006;32:129–138. doi: 10.1385/mb:32:2:129. [DOI] [PubMed] [Google Scholar]
- 14.Kilaru S, Collins CM, Hartley AJ, Bailey AM, Foster GD. Establishing molecular tools for genetic manipulation of the pleuromutilin producing fungus Clitopilus passeckerianus. Appl Environ Microbiol. 2009a;75:7196–7204. doi: 10.1128/AEM.01151-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunik T, Tzfira T, Kapulnik Y, Gafni Y, Dingwall C, et al. Genetic transformation of HeLa cells by Agrobacterium. Proc Natl Acad Sci USA. 2001;98:1871–1876. doi: 10.1073/pnas.041327598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knight CJ, Bailey AM, Foster GD. Agrobacterium-mediated transformation of the plant pathogenic fungus Verticillium albo-atrum. Journal of Plant Pathology. 2009;91:637–642. [Google Scholar]
- 17.Murashige T, Skoog F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- 18.Patel RM, Heneghan MN, van Kan JA, Bailey AM, Foster GD. The pOT vector system: Improving ease of transgene expression in Botrytis cinerea. J Gen Appl Microbiol. 2008;54:367–376. doi: 10.2323/jgam.54.367. [DOI] [PubMed] [Google Scholar]
- 19.Collins C, Heneghan M, Kilaru S, Bailey A, Foster GD. Improvement of the Coprinopsis cinereus Molecular Toolkit using new construct design and additional marker genes. J Microbiol Method. 2010 doi: 10.1016/j.mimet.2010.05.007. In Press. [DOI] [PubMed] [Google Scholar]
- 20.Keon J, Hargreaves J. Isolation and heterologous expression of a gene encoding 4-hydroxyphenylpyruvate dioxygenase from the wheat leaf-spot pathogen, Mycosphaerella graminicola. FEMS Microbiol Lett. 1998;161:337–343. doi: 10.1111/j.1574-6968.1998.tb12966.x. [DOI] [PubMed] [Google Scholar]
- 21.Hartley AJ, De Mattos-Shipley K, Collins CM, Kilaru S, Foster GD, et al. Investigating Pleuromutilin Producing Clitopilus Species and related Basidiomycetes. FEMS Microbiol Lett. 2009;297:24–30. doi: 10.1111/j.1574-6968.2009.01656.x. [DOI] [PubMed] [Google Scholar]
- 22.Kilaru S, Collins CM, Hartley AJ, Burns C, Foster GD, et al. Investigating dominant selection markers for Coprinopsis cinerea a carboxin: resistance system and re-evaluation of hygromycin & phleomycin resistance vectors. Curr Genet. 2009b;55:543–550. doi: 10.1007/s00294-009-0266-6. [DOI] [PubMed] [Google Scholar]
- 23.Nazar RN, Hu X, Schmidt J, Culham D, Robb J. Potential Use of PCR-Amplified Ribosomal Intergenic Sequences in the Detection and Differentiation of Verticillium Wilt Pathogens. Physiol Mol Plant Pathol. 1991;39:1–11. [Google Scholar]
- 24.Sambrook J, Fritsch EF, Maniatis T. New York: Cold Spring Harbour Laboratory Press; 1989. Molecular Cloning - A Laboratory Manual, 2nd Edition. [Google Scholar]
- 25.Pegg GF, Brady BL. Verticillium Wilts, CABI Publishing 2002.
- 26.Chilton MD, Drummond MH, Merio DJ, Sciaky D, Montoya AL, et al. Stable incorporation of plasmid DNA into higher plant cells: the molecular basis of crown gall tumorigenesis. Cell. 1977;11:263–71. doi: 10.1016/0092-8674(77)90043-5. [DOI] [PubMed] [Google Scholar]
- 27.Gelvin SB. Agricultural biotechnology: Gene exchange by design. Nature. 2005;433:583–584. doi: 10.1038/433583a. [DOI] [PubMed] [Google Scholar]
- 28.Machida M, Asai K, Sano M, Tanaka T, Kumagai T, et al. Genome sequencing and analysis of Aspergillus oryzae. Nature. 2005;438:1157–1161. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- 29.Ulker B, Li Y, Rosso MG, Logemann E, Somssich IE, et al. T-DNA-mediated transfer of Agrobacterium tumefaciens chromosomal DNA into plants. Nat Biotech. 2008;26:1015–1017. doi: 10.1038/nbt.1491. [DOI] [PubMed] [Google Scholar]
- 30.Barrett C, Cobb E, McNicol R, Lyon G. A risk assessment study of plant genetic transformation using Agrobacterium and implications for analysis of transgenic plants. Plant Cell Tiss Org. 1997;47:135–144. [Google Scholar]
- 31.Yang M, Mi D, Ewald D, Wang Y, Liang H, et al. Survival and escape of Agrobacterium tumefaciens in triploid hybrid lines of Chinese white poplar transformed with two insect-resistant genes. Acta Ecologica Sinica. 2006;26:3555–3561. [Google Scholar]