Abstract
Background
The new field of paleomicrobiology allows past outbreaks to be identified by testing dental pulp of human remains with PCR.
Methods
We identified a mass grave in Douai, France dating from the early XVIIIth century. This city was besieged during the European war of Spanish succession. We tested dental pulp from 1192 teeth (including 40 from Douai) by quantitative PCR (qPCR) for R. prowazekii and B. quintana. We also used ultra-sensitive suicide PCR to detect R. prowazekii and genotyped positive samples.
Results and Discussion
In the Douai remains, we identified one case of B. quintana infection (by qPCR) and R. prowazekii (by suicide PCR) in 6/21 individuals (29%). The R. prowazekii was genotype B, a genotype previously found in a Spanish isolate obtained in the first part of the XXth century.
Conclusion
Louse-borne outbreaks were raging during the XVIIIth century; our results support the hypothesis that typhus was imported into Europe by Spanish soldiers from America.
Introduction
As mentioned by Zinsser, infectious illness has killed more soldiers during war than weapons [1]. The current genetic tools make it possible in a certain number of cases to identify the probable causes of epidemics [2]. The combination of an anthropological approach that identifies burials from catastrophes with a molecular approach that makes it possible to identify the genes of bacteria in dental pulp has developed recently into the framework of a new speciality called paleomicrobiology [3]. Thanks to these elements, we found that the Justinian plague, like the great plague of the Middle Ages, was due to Yersinia pestis Orientalis [4]–[6]. We also identified the presence of Bartonella quintana in very ancient samples [7]. More recently, we showed that some of the soldiers of the Grand Army that died in Vilnius after the passage of the Bérézina river died of diseases transmitted by lice: Bartonella quintana and Rickettsia prowazekii [8].
Recently, a mass grave dating back to the 18th century was explored in Douai, France. The city of Douai was besieged from 1710 to 1712, being successively occupied by the French and then the Dutch and then retaken by French in 1712 during the war of Spanish succession (Figure 1). These events were in the framework of a generalised European war, with France and Spain opposing the other nations, the battle being carried out on the French side by Louis XIV “Le Grand Monarque.” The investigation of this mass grave showed that few skeletons presented lesions compatible with weapon wounds. This led to the assumption that a certain number of these skeletons were caused by an epidemic that occurred during the siege. Indeed, the medical condition of the men was very bad in spite of their young age. This could be consistent with an epidemic of diseases transmitted by lice.
Figure 1. Representation of the bi siege of Douai in an Almanach.
Materials and Methods
Source of the materials
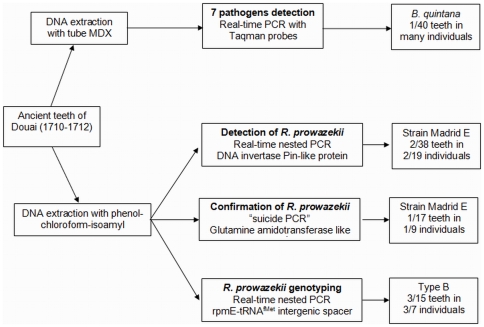
Douai is a village located in northern France. It was disputed between France and the Great Alliance of La Haye between 1702 and 1712 [9], [10]. The site of the street Martin-du-Nord was discovered during building construction in 1981 (Figure 2). A total of twelve multiple burials and four individual graves have been uncovered at this site. The graves appeared to be scattered over the plot in various directions (Figure 3), and the individuals were deposited head-to-foot into exiguous pits, a characteristic of disaster graves linked to sudden and massive mortality. This type of burial can also occur during an epidemic [11], [12]. The demographic profile of young males as well as historical documents suggested a military installation. Five individuals with evidence of traumatic injuries were buried in a single pit. The cause of death could not be attributed to trauma in the other individuals, and 21 such individuals were studied herein, from whom a total of 55 teeth were used for the molecular investigations [13], [14] (Figure 4).
Figure 2. General view of the burial site of Douai.
Figure 3. Featuring multiple burials discovered in Douai.
Figure 4. Summary of the materials and methods used in this study.
High-throughput detection of pathogens
Dental pulp recovered as previously described [4] was incubated overnight at 56°C with 600 µL ATL buffer and 50 µL proteinase K. DNA was then extracted by using QIAamp Media MDx Bacterio-pulverisation in the BioRobot® MDx workstation in a final 100 µL volume (Qiagen GmbH, Hilden, Germany). Seven PCR primer pairs and seven probes were designed for the specific detection of Bacillus anthracis (anthrax), Borrelia recurrentis (louse-borne relapsing fever), Bartonella quintana (trench fever), Rickettsia prowazekii (epidemic typhus), Salmonella enterica Typhi (typhoid fever), Poxvirus (smallpox) and Yersinia pestis (plague) (Table 1). Real-time PCR amplification was performed using the QuantiTech Probe PCR Kit (Qiagen) and a 7900HT Fast Real-Time PCR System (Applied Biosystem, Courtaboeuf, France). Each well of a 384-well plate was filled with 10 µL Mix Quantitech, 2 µL sterile water, 2 µL of 2 pmol/µL probe, 0.5 µL forward primer (10 pmol/µL), 0.5 µL reverse primer (10 pmol/µL) and 5 µL DNA. Amplification consisted of 15-min activation at 95°C followed by 50 cycles of 30-sec denaturation at 95°C and 1-min hybridisation at 60°C. In every plate, two wells containing sterile water and two wells containing DNA extracted from dental pulp collected from skeletons devoid of anthropologic evidence of infection were used as negative controls.
Table 1. Primers for molecular detection of all pathogens into 1192 acient teeth.
| Desired specificity | Gene | Name | Probe sonde and primers | Sequence |
| Bacillus anthracis(anthrax) | pag | Bant_pag_P | 6 FAM- TAC CGC AAA TTC AAG AAA CAA CTG C -TAMRA | 94 bp |
| Bant_pag_F | 5′- AGG CTC GAA CTG GAG TGA A -3′ | |||
| Bant_pag_R | 5′- CCG CCT TTC TAC CAG ATT T -3′ | |||
| Borrelia recurrentis(louse-borne relapsing fever) | unknown | Brec_P | 6 FAM- CTG CTG CTC CTT TAA CCA CAG GAG CA -TAMRA | 111 bp |
| Brec_F | 5′- TCA ACT GTT TTT CTT ATT GCC ACA -3′ | |||
| Brec_R | 5′- TCC TTA TGT TGG TTA TGG GAT TGA -3′ | |||
| Bartonella quintana(Trench fever) | ITS | Barto ITS_P | 6 FAM- GCG CGC GCT TGA TAA GCG TG -TAMRA | 102 bp |
| Barto ITS_F | 5′- GAT GCC GGG GAA GGT TTT C -3′ | |||
| Barto ITS_R | 5′- GCC TGG GAG GAC TTG AAC CT -3′ | |||
| Rickettsia prowazekii(typhus) | ompB | Rpr_ompB_P | 6 FAM- CGG TGG TGT TAA TGC TGC GTT ACA ACA -TAMRA | 134 bp |
| Rpr_ompB_F | 5′- AAT GCT CTT GCA GCT GGT TCT -3′ | |||
| Rpr_ompB_R | 5′- TCG AGT GCT AAT ATT TTT GAA GCA -3′ | |||
| Salmonella Typhi(typhoid fever) | unknown | Styp_put_P | 6 FAM- GCT TTT TGT GAA GCA ACG CTG GCA -TAMRA | 138 bp |
| Styp_put_F | 5′- CTC CAT GCT GCG ACC TCA AA -3′ | |||
| Styp_put_R | 5′- TTC ATC CTG GTC CGG TGT CT -3′ | |||
| Poxvirus(smallpox) | HA | Var_HA_P | 6 FAM- AAG ATC ATA CAG TCA CAG ACA CTG T -TAMRA | 100 bp |
| Var_HA_F | 5′- GAC KTC SGG ACC AAT TAC TA -3′ | |||
| Var_HA_R | 5′- TTG ATT TAG TAG TGA CAA TTT CA -3′ | |||
| Yersinia pestis(plague) | pla | Yper_PLA_P | 6 FAM- TCC CGA AAG GAG TGC GGG TAA TAG G -TAMRA | 98 bp |
| Yper_PLA_F | 5′- ATG GAG CTT ATA CCG GAA AC -3′ | |||
| Yper_PLA_R | 5′- GCG ATA CTG GCC TGC AAG -3′ |
Suicide PCR detection of R. prowazekii
The Douai specimens were further examined with a suicide nested PCR protocol after conventional phenol chloroform DNA extract [4]. The program PerlPrimer version 1.1.6 was used to design PCR primers. The first suicide nested-PCR targeted a 206-base pair fragment of the Rickettsia prowazekii DNA invertase Pin-like protein (ORF0698; gi|3861237|emb|AJ235273.1|) by combining external primers RpDet-F1 5′-GTTGGATATATAAGGGTTTC-3′ and RpDet-R2 5′-CCGAGTCTATCTAATTTCCA-3′ and internal primers RpDet-F3 5′-ATGATCGTCAAGTGTTCGAT-3′ and RpDet-R4 5′-TAGACAGTCGCCATCTTGTA-3′; the final expected PCR product was 152 bp (Table 2). This region had never been amplified in our laboratory. The first round PCR mix contained 1.6 µL MgCl2, 0.5 µL bovine serum albumin (BSA), 2 µL Master Mix (Light Cycle® FastStart DNA Master Sybr Green, Roche Applied Science, France), 1 µL of a 10 µM solution of RpDet-F1, 1 µL of a 10 µM solution of RpDet-R2, 8.9 µL sterile water and 5 µL DNA (experimental tube) or 5 µL sterile water (negative control tube). The first round of amplification was done using a LightCycler™ apparatus (Roche Diagnostics) and the following conditions: 10-min activation at 95°C followed by 45 cycles of 15-sec denaturation at 95°C, 20-sec hybridisation at 55°C and 25-sec elongation at 72°C. PCR products were recovered by centrifugation of capillaries at 380 g for 2 minutes. The nested PCR was performed in the capillary containing 1.6 µL MgCl2, 0.5 µL BSA, 2 µL Master Mix, 1 µL of 10 µM solution of RpDet-F3; 1 µL of 10 µM solution of RpDet-R4; 8.9 µL sterile water and 5 µL first round PCR product. The second amplification in the LightCycler™ apparatus used the following conditions: activation at 95°C for 10 minutes and 45 cycles of denaturation at 95°C for 15 seconds, hybridisation at 57°C for 20 seconds, elongation at 72°C for 25 seconds. Nested PCR products were detected on a 2% agarose gel (Invitrogen™, Paisley, Scotland) in the presence of molecular weight marker VI (Boehringer Mannheim, Germany).
Table 2. Primers for detection and genotyping of R. prowazekii into ancient teeth of Douai.
| PCR | Name | Primers | Tm | Sequence zise | Reference | |
| Detection ofR. prowazekii | Real-time PCR | RpDet-F1RpDet-R2 | 5′-GTTGGATATATAAGGGTTTC-3′ 5′-CCGAGTCTATCTAATTTCCA-3 ′ | 55°C | 206 bp | rpr_ORF0698 site-specific recombinases, DNA invertase Pin-like protein |
| Nested PCR | RpDet-F3RpDet-R4 | 5′-ATGATCGTCAAGTGTTCGAT-3 ′ 5′-TAGACAGTCGCCATCTTGTA-3′ | 57°C | 152 bp | ||
| Confirmation ofR. prowazekii | PCR standard | Rpro-F1Rpro-R1 | 5′-ACTGTTATTACCGATCTTGCCA-3′ 5′-TGGTTGATGCTAGGTTATTTGG-3′ | 58°C | 187 bp | rpr_ORF0700, glutamine amidotransferase-like protein |
| Nested PCR | Rpro-F11Rpro-R11 | 5′-GTATTAAGAATTTGATGCCACCA-3′ 5′-GTTATTAGTCCAAATGACGTGAA-3′ | 62°C | 130 bp | ||
| R. prowazekiigenotyping | Real-time PCR | rpmE-F1rpmE-R2 | 5′-CCGGAAATGTAGTAAATCAATC-3′ 5′-CTGAGAATTTAAAGATTTATCTG-3 ′ | 59°C | 210 bp | Yong Zhu et al., 2005 rpmE-tRNAfMet intergenic spacer |
| Nested PCR | rpmE-F3rpmE-R4 | 5′-CTTTCGATAGCAAGAAAGAAGC-3 ′ 5′-CAGAGTATTAGTAGACGATACG 3′ | 62°C | 115 bp |
A second suicide nested-PCR targeted a 187-base pair fragment of the R. prowazekii glutamine amidotransferase-like protein (rpr_ORF0700; gi|3861237|emb|AJ235273.1|) by combining external primers Rpro-F1 5′-ACTGTTATTACCGATCTTGCCA-3′ and Rpro-R1 5′-TGGTTGATGCTAGGTTATTTGG-3′ and internal primers Rpro-F11 5′-GTATTAAGAATTTGATGCCACCA-3′and Rpro-R11 5′-GTTATTAGTCCAAATGACGTGAA-3′; the final expected PCR product was 130 bp (Table 2). This region had never been amplified in our laboratory. The first round of PCR was performed using a HotSartTaq DNA Polymerase Kit (Qiagen) with 0.8 µL MgCl2, 0.2 µL HotStart Taq, 2.5 µL 10X PCR buffer, 2.5 µL dNTP, 0.5 µL BSA, 0.5 µL of a 10 µM solution of each Rpro-F1 and Rpro-R1, 12.5 µL sterile water and 5 µL DNA (experimental tube) or 5 µL sterile water (negative control tube). The first round of amplification was done in an ABI GeneAmp™ 2700 thermocycler (Applied Biosystems, CA, USA) under the following conditions: 10-min activation at 95°C and 45 cycles of 30-sec denaturation at 95°C, 45-sec hybridisation at 58°C and 90-sec elongation at 72°C. The PCR products were purified using the Millipore plate protocol and suspended into 40 µL water. The second round of PCR was performed using 0.8 µL MgCl2, 0.2 µL HotStart Taq, 2.5 µL 10X PCR buffer, 2.5 µL dNTP, 0.5 µL BSA, 0.5 µL Rpro-F11, 0.5 µL Rpro-R11, 12.5 µL sterile water and 5 µL first round PCR product. The second amplification was performed using the following conditions: 10-min activation at 95°C and 45 cycles of 30-sec denaturation at 95°C, 45-sec hybridisation at 62°C and 90-sec elongation at 72°C. Nested PCR products were detected on a 2% agarose gel (Invitrogen™, Paisley, Scotland) in the presence molecular weight marker VI (Boehringer Mannheim, Germany).
Nested PCR products were purified using the Millipore plate protocol and suspended in 50 µL water. The sequencing reaction was carried out in a tube containing 3 µL Big Dye Terminator, 0.5 µL forward or reverse primer, 3.5 µL purified PCR product and 3 µL sterile water using the following conditions: activation at 95°C for 5 minutes followed by 25 cycles consisting of 30-sec denaturation at 96°C, 20-sec hybridisation at 55°C, 4-min elongation at 60°C and 7-min extension at 15°C. The sequencing products were purified on Séphadex® G50 5% gel and analysed with the ABI PRISM 3100 Genetic Analyser (HITACHI). The sequences were read and corrected by using the software ChromasPro version 1.34 and then aligned with BLAST to compare the sequences available in GenBank.
Genotyping of R. prowazekii
The software PerlPrimer version 1.1.6 was used to design two pairs of primers targeting a 210-bp sequence (external primers: rpmE-F1 5′-CCGGAAATGTAGTAAATCAATC-3′ and rpmE-R2 5′-CTGAGAATTTAAAGATTTATCTG-3′) and a 115-bp sequence (internal primes: rpmE-F3 5′-CTTTCGATAGCAAGAAAGAAGC-3′ and rpmE-R4 5′-CAGAGTATTAGTAGACGATACG 3′) on the rpmE-tRNAfMet intergenic spacer (gi|56967982|gb|AY695449.1|) (Yong Zhu et al., 2005) (Table 2). Genotyping was performed by mixing 1.6 µL MgCl2, 0.5 µL BSA, 2 µL Master Mix, 1 µL of 10 µM solution of rpmE-F1; 1 µL of 10 µM solution of rpmE-R2, 8.9 µL sterile water and 5 µL DNA (experimental tube) or 5 µL sterile water (negative control tube) in a Stratagene plate. The first round of amplification was performed in a Stratagene™ thermocycler (Agilent Technologies Company) using the following conditions: activation at 95°C for 10 minutes, 45 cycles of denaturation at 95°C for 30 seconds, hybridisation at 59°C for 30 seconds, elongation at 72° C for 1 minute. Nested PCR was realised in a Stratagene plate containing 1.6 µL MgCl2, 0.5 µL BSA, 2 µL Master Mix, 1 µL of 10 µM solution of rpmE-F3, 1 µL of 10 µM solution of rpmE-R4, 8.9 µL sterile water and 5 µL first round PCR product. The second amplification in the Stratagene™ thermocycler was performed using the following conditions: activation at 95°C for 10 minutes followed by 45 cycles consisting of denaturation at 95°C for 30 seconds, hybridisation at 62°C for 30 seconds and elongation at 72°C for 1 minute. The nested PCR products were detected on a 2% agarose gel (Invitrogen™) in the presence of molecular weight marker VI (Boehringer Mannheim). The PCR products were sequenced as described above.
Prevention of DNA contamination
all manipulations of ancient materials were done in two successive laboratories where R. prowazekii had never been previously amplified. Each step was conducted in a separate room under a hood with air-capture. All instruments were sterilised and used only once. No positive control was included, and one negative control consisting of sterile water was used for each three or five samples.
Results
High-throughput detection of pathogens
A total of 1.192 dental pulp specimens collected from several burial sites in France were analysed by high throughput detection, including 40 specimens collected in Douai tested blindly. Whereas the negative controls remained negative, high throughput real-time PCR detected B. quintana DNA in 1/40 dental pulp specimens, and no other pathogen was detected in these 40 specimens. We then attempted to detect R. prowazekii using a more sensitive technique.
Molecular detection of R. prowazekii
In all of the experiments, the negative controls remained negative. The first suicide real-time nested PCR detected R. prowazekii DNA in 2/38 (5.3%) ancient teeth collected from 2/19 (10.5%) different individuals (coded as A6-4 and A10-19). Sequence alignment yielded 100% sequence similarity with the reference R. prowazekii strain Madrid E (Genbank accession emb|AJ235273.1|RPXX04). As for the second suicide PCR, it detected R. prowazekii DNA in 1/17 (5.9%) dental pulp specimens collected from 1/9 (11%) different individuals (coded as 126/220-39) (Figure 5).
Figure 5. Summary of R. prowazekii detection and genotyping results.
Genotyping of R. prowazekii
In the genotyping, the negative controls remained negative while we amplified the R. prowazekii rpmE/tRNAfMet intergenic spacer sequence in 3/15 (20%) ancient teeth collected from 3/7 (43%) different individuals (coded as 075-24, 077-27, 1074-35). All PCR products yielded an identical sequence exhibiting 100% sequence similarity to R. prowazekii type B, which is characterised by a T to C substitution at position 111 (Figure 6). Merging all of the PCR results yielded positive detection of R. prowazekii DNA in 6/55 (11%) teeth collected from 6/21 (28.6%) individuals.
Figure 6. Result of Genbank alignments for genotyping.
Grey boxes indicate the locations of PCR primers.
Discussion
The results presented herein were interpreted as authentic. Indeed, we selected teeth that had a closed apex and that were free of dental caries and traumatic lesions to minimise any risk of external contamination of the dental pulp. For the first time, we used a high throughput paleomicrobiological approach to screen 1192 teeth for seven infectious agents (8,344 tests). This technique is less sensitive than suicide PCR but is well adapted to testing numerous samples. Our work was carried out in a laboratory where R. prowazekii had never been worked on, nor had R. prowazekii DNA been extracted. All operations were carried out under a hood with air-capture using sterilised instruments that were used only once. All the negative controls remained negative. We obtained an original sequence that was consistently found in several teeth collected in several individuals, its uniqueness thus giving much confidence. After blindly detecting B. quintana in one tooth from Douai, we decided to employ more sensitive techniques to recover R. prowazekii from other teeth from this site.
In this study, we observed that not all dental pulp specimens collected from a single individual yielded a positive PCR product. Such variability in the positivity of PCR-based detection has been previously observed in detecting Yersinia pestis DNA [4]. Specific R. prowazekii sequences were detected in 6/55 (11%) teeth collected in 6/21 (28.6%) individuals; this prevalence is significantly higher (P<0.05, test χ2) than that previously reported in the Napoleon's Grand Army study, which detected R. prowazekii in 4/72 (5.6%) teeth collected from 3/35 (8.6%) soldiers [15]. As we also found an individual infected by B. quintana, we suspect the circulation of louse-borne diseases in this place, but we did not recover Borrelia recurrentis DNA.
In the present study, we also took advantage of the dental pulp to detect and genotype 300-year-old R. prowazekii DNA. Dental pulp is a specialised conjunctive tissue occupying the central position in the tooth, where it is protected from the external environment by the dentine. We previously demonstrated that blood-borne bacteria could be detected in dental pulp both by culture and by molecular detection of specific DNA sequences [16]–[18]. In addition, dental pulp is the only conjunctive tissue that can persist for several thousand years after the degradation of other tissues because it is very well preserved within the dentine and enamel, which are the hardest tissues of the human body [4]. In 2004, Tran H Lam et al. showed that PCR amplifying a fragment of 286 base pairs of the 16S rRNA gene could identify several bacteria in the pulp of ancient teeth dated to the 17th century [19]. It is difficult to amplify fragments of DNA more than 300 base pairs from old samples [4], [6]. For these reasons, we chose a fragment of 115–210 base pairs for amplification. Nested PCR, which is effected by two successive PCR reactions with two pairs of external-internal primers, can increase PCR's sensitivity and specificity considerably. With the combination of the benefits of real-time PCR [20] and nested PCR, we have successfully amplified an ancient bacterial DNA sequence from the 18th century and identified it as R. prowazekii genotype B. It was later shown that real-time PCR is a rapid, specific and sensitive method for detecting R. prowazekii in blood [20], [21]. Sequence analysis of the rpmE/tRNAfMet spacer in 15 modern R. prowazekii DNA samples found three genotypes: A, B and C. Genotype B had been sequenced in the avirulent R. prowazekii Madrid E strain (derived from a Spanish isolate [22]), in its virulent revertant R. prowazekii Evir in two blood isolates from Russia and Algeria, and in four lice collected in Rwanda and Burundi [23]. We herein demonstrated that this genotype was already present in 18th-century Europe.
Despite the fact that typhus epidemics has been depicted in historical sources for two millennia or so [24], only few demonstrations of R. prowazekii issued from ancient materials have been provided. One individual diagnosed with fatal epidemic typhus in Maryland in 1901 was demonstrated by using immunohistological detection ninety years later to have died of Rocky Mountain spotted fever caused by Rickettsia rickettsii [25]. In another report, sequence-based analysis of remains of Napoleon's Grand Army soldiers yielded molecular evidence for R. prowazekii and B. quintana in an estimated one-third of the individuals [26]. The data herein reported remind us that epidemic typhus, a disease now confined to relatively limited geographic areas, had a broad geographic range of prevalence only a few centuries ago, being one of the plagues reported in historical descriptions (Table 3) [27]. In the past, several typhus epidemics have been described in central Africa [28]. Today, cases of typhus are still described in Peru [29] and in industrial cities of Russia [30], Algeria [31] and France [32]. Further studies applying the techniques herein described to the remains of individuals in America, Europe and Africa may help to paint a clearer picture of the evolution and spread of epidemic typhus in connection with human history.
Table 3. Dental pulp: a source for the paleomicrobiology of ancient epidemics.
| Epidemic | Site - date | Materials and Methods | Results | Reference |
| Plague | Lambesc – 1590 and Marseille – 1722 (France) | Dental pulp, DNA amplification by PCR, gene RpoB (133-bp) and gene pla (300-bp) | Y. pestis detected in 6/12 teeth | [3] |
| Saint-Côme and Saint-Damien (Montpellier) – 14th century (France) | Dental pulp, “suicide” PCR, gene pla (148-bp) | Y. pestis detected in 20/23 teeth | [5] | |
| Sens: 5th–6th century, Dreux: 12th–14th century and Monpellier – 1348 (France) | Dental pulp, DNA amplification by PCR, spacer YP | Y. pestis strain Orientalis detected in 7/11 individuals | [31] | |
| Aschheim – 6th century (Upper Bavaria) | Dental pulp, “suicide” PCR, gene pla (148-bp) | Y. pestis detected in 2/6 teeth | [32] | |
| Vienne: 7th–9th century, Martigues: 1720–1721 and Marseille – 1722 | Dental pulp, suicide-nested PCR, gene glpD (191-bp) | Y. pestis strain Orientalis detected in 5/46 teeth | [33] | |
| Lambesc – 1590, Saint-Pierre: 1628–1632, Draguignan: 1649–1650, Martigues: 1720–1721, Berre l'Etang: 1720–1721, Marseille – 1722 (France) | Dental pulp and spongy bone, immuno-detection by RDT, F1 antigen | Y. pestis detected in 19/28 individuals | [34] | |
| Typhoid fever | Athens: 430–426 BC (Greek) | Dental pulp, “suicide” PCR, gene osmC-clyA (322-bp) and gene narC (360-bp) | S. enterica Typhi detected in 3/3 teeth | [35] |
| Rocky Mountainspotted fever | Maryland – 1901 (USA) | Immunohistology detection | Detection of R. rickettsii | [22] |
| Cat-cratch disease | Compiègne – 16th century, Montbéliard – 14th century and Paris – 13th century | Dental pulp of cats, nested PCR, gene groEL (269 – bp) and gene Pap31 (164 – bp) | B. henselae detected in 3/135 teeth of cats | [36] |
| Trench fever | Roaix: 2100–2200 BC and Peyraoutes: 2230–1950 BC (France) | Dental pulp, nested PCR, gene hemin-binding protein-E (283-bp) and gene groEL (269-bp) | B. quintana detected in 1/12 teeth | [6] |
| Typhus and trench fever | Vilnius – 1812 (Lithuania) | Lice and dental pulp, DNA amplification by PCR, gene dnaA (141–279 bp) and gene hbpE (282–429 bp) | R. prowazekii detected in 4/72 teeth, B. quintana detected in 7/72 teeth and 3/5 lice | [7] |
| Typhus and trench fever | Douai: 1710–1712 (France) | Dental pulp, real-time PCR and “suicide PCR”, gene ITS (102-bp) and gene DNA invertase Pin-like protein (152–206 bp), gene glutamine amidotransferase-like protein (130–187 bp), rpmE-tRNAfMet intergenic spacer (115-210 bp) | B. quintana detected in 1/40 teeth and R. prowazekii strain Madrid E type B detected in 6/55 teeth | Present work |
Taken together, the results reported herein show clearly that an epidemic disease transmitted by lice prevailed during the long siege of Douai. Diseases transmitted by body lice were probably extremely frequent in the past. When sanitary arrangements are degraded, lice are likely to quickly expand and the population of lice can increase by 10% per day [33] when the conditions of hygiene are met [29]. Under these conditions, it is easy to imagine a B. quintana epidemic persisting in Europe for an extremely long time. The oldest trace of infection of humans by this bacterium goes back 4.000 years [34]. Epidemic typhus appeared in Europe later. The majority of authors suggest that it was introduced by the Spanish returning from America [35]. The war in Douai was that of the Spanish succession, and one can propose that it was imported by Spanish soldiers. It is interesting therefore to observe that the genotype B found here is identical to that of a Spanish isolate from the beginning of the XXth century in Spain. The first descriptions go back to Fracastor, and early in the 16th century a number of epidemics compatible with the diagnosis of typhus were reported. However, at the beginning of the 18th century, the clinical individualisation of epidemic typhus, like that of trench fever of the trenches, was not yet carried out. The first definition of typhus was provided by Boissier de Sauvages in 1772 [36].
In conclusion, the molecular diagnosis of past epidemic infections related to the teeth made it possible to identify the first outbreak of epidemic typhus in the 18th century in the context of a pan-European Great War. Working together, molecular biologists, dentists and anthropologists have explored burials from catastrophes to identify the prevailing epidemics in past centuries.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: No current external funding received for this study.
References
- 1.Zinsser H. Londres: Broadway House; 1935. Rats, lice, and history.301 [Google Scholar]
- 2.Drancourt M, Raoult D. Palaeomicrobiology: current issues and perspectives. Nat Rev Microbiol. 2005;3:23–35. doi: 10.1038/nrmicro1063. [DOI] [PubMed] [Google Scholar]
- 3.Drancourt M, Raoult D. Palaeomicrobiology: current issues and perspectives. Nat Rev Microbiol. 2005;3:23–35. doi: 10.1038/nrmicro1063. [DOI] [PubMed] [Google Scholar]
- 4.Drancourt M, Aboudharam G, Signoli M, Dutour O, Raoult D. Detection of 400-year-old Yersinia pestis DNA in human dental pulp: an approach to the diagnosis of ancient septicemia. Proc Natl Acad Sci USA. 1998;95:12637–12640. doi: 10.1073/pnas.95.21.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drancourt M, Raoult D. Molecular detection of Yersinia pestis in dental pulp. Microbiology. 2004;150:263–264. doi: 10.1099/mic.0.26885-0. [DOI] [PubMed] [Google Scholar]
- 6.Raoult D, Aboudharam G, Crubezy E, Larrouy G, Ludes B, et al. Molecular identification by “suicide PCR” of Yersinia pestis as the agent of Medieval Black Death. Proc Natl Acad Sci U S A. 2000;97:12800–12803. doi: 10.1073/pnas.220225197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drancourt M, Tran-Hung L, Courtin J, Lumley H, Raoult D. Bartonella quintana in a 4000-year-old human tooth. J Infect Dis. 2005;191:607–611. doi: 10.1086/427041. [DOI] [PubMed] [Google Scholar]
- 8.Raoult D, Dutour O, Houhamdi L, Jankauskas R, Fournier PE, et al. Evidence for louse-transmitted diseases in soldiers of Napoleon's Grand Army in Vilnius. J Infect Dis. 2006;193:112–120. doi: 10.1086/498534. [DOI] [PubMed] [Google Scholar]
- 9.Hardy de Perini E, Majesté A, Bouchardeau L. Les batailles françaises, Les armées sous l'ancien régime, 1700 à 1789. Châteauroux-Flammarion (Paris) 1906 [Google Scholar]
- 10.Corvisier A. Les Hommes, la guerre et la mort. Economica. 1985:453. [Google Scholar]
- 11.Raoult D, Dutour O, Houhamdi L, Jankauskas R, Fournier PE, et al. Evidence for louse-transmitted diseases in soldiers of Napoleon's Grand Army in Vilnius. J Infect Dis. 2006;193:112–120. doi: 10.1086/498534. [DOI] [PubMed] [Google Scholar]
- 12.Rigeade C, Signoli M. Les sépultures de catastrophe: une gestion originale des cadavres en temps de crises démographiques. Sociologie et Santé. 2005;117-131 [Google Scholar]
- 13.Signoli M, Ardagna Y, Adalian P, Devriendt W, Lalys L, et al. Discovery of a mass grave of Napoleonic period in Lithuania (1812, Vilnius). C R Palevol. 2004;3:219–227. [Google Scholar]
- 14.Rigeade C, Willot JP, Demolon P, Rabino Massa E, et al. Approche anthropologique de sépultures de catastrophe du XVIIIe siècle (rue Martin-du-Nord, Douai, France). C R Palevol. 2006;5:901–907. [Google Scholar]
- 15.Raoult D, Dutour O, Houhamdi L, Jankauskas R, Fournier PE, et al. Evidence for louse-transmitted diseases in soldiers of Napoleon's Grand Army in Vilnius. J Infect Dis. 2006;193:112–120. doi: 10.1086/498534. [DOI] [PubMed] [Google Scholar]
- 16.Aboudharam G, La Scola B, Raoult D, Drancourt M. Detection of Coxiella burnetii DNA in dental pulp during experimental bacteremia. Microb Pathog. 2000;28:249–254. doi: 10.1006/mpat.1999.0343. [DOI] [PubMed] [Google Scholar]
- 17.Aboudharam G, Drancourt M, Raoult D. Culture of C. burnetii from the dental pulp of experimentally infected guinea pigs. Microb Pathog. 2004;36:349–350. doi: 10.1016/j.micpath.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 18.Aboudharam G, Fournier PE, Drancourt M, Raoult D, et al. Molecular detection of Bartonella quintana DNA in the dental pulp of a homeless patient. Eur J Clin Microbiol Infect Dis. 2004;23:920–922. doi: 10.1007/s10096-004-1244-z. [DOI] [PubMed] [Google Scholar]
- 19.Tran-Hung L, Tran-Thi N, Aboudharam G, Raoult D, Drancourt M. A new method to extract dental pulp DNA: application to universal detection of bacteria. PLoS ONE. 2007;2:e1062. doi: 10.1371/journal.pone.0001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Svraka S, Rolain JM, Bechah Y, Gatabazi J, Raoult D. Rickettsia prowazekii and real-time Polymerase Chain Reaction. Emerg Infect Dis. 2006;12:428–432. doi: 10.3201/eid1203.050888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stenos J, Graves SR, Unsworth NB. A highly sensitive and specific real-time PCR assay for the detection of spotted fever and typhus group Rickettsiae. Am J Trop Med Hyg. 2005;73:1083–1085. [PubMed] [Google Scholar]
- 22.Bechah Y, El Karkouri K, Mediannikov O, Leroy Q, Pelletier N, et al. Genomic, proteomic, and transcriptomic analysis of virulent and avirulent Rickettsia prowazekii reveals its adaptive mutation capabilities. Genome Res. 2010;20:655–663. doi: 10.1101/gr.103564.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu Y, Fournier PE, Ogata H, Raoult D. Multispacer typing of Rickettsia prowazekii enabling epidemiological studies of epidemic typhus. J Clin Microbiol. 2005;43:4708–4712. doi: 10.1128/JCM.43.9.4708-4712.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bechah Y, Capo C, Mege JL, Raoult D. Epidemic typhus. Lancet Infect Dis. 2008;8:417–426. doi: 10.1016/S1473-3099(08)70150-6. [DOI] [PubMed] [Google Scholar]
- 25.Dumler JS. Fatal Rocky Mountain Spotted Fever in Maryland - 1901. J Amer Med Assoc. 1991;265:718. doi: 10.1001/jama.265.6.718. [DOI] [PubMed] [Google Scholar]
- 26.Raoult D, Dutour O, Houhamdi L, Jankauskas R, Fournier PE, et al. Evidence for louse-transmitted diseases in soldiers of Napoleon's Grand Army in Vilnius. J Infect Dis. 2006;193:112–120. doi: 10.1086/498534. [DOI] [PubMed] [Google Scholar]
- 27.Raoult D, Woodward T, Dumler JS. The history of epidemic typhus. Infect Dis Clin North Am. 2004;18:127–140. doi: 10.1016/S0891-5520(03)00093-X. [DOI] [PubMed] [Google Scholar]
- 28.Raoult D, Ndihokubwayo JB, Tissot-Dupont H, Roux V, Faugere B, et al. Outbreak of epidemic typhus associated with trench fever in Burundi. Lancet. 1998;352:353–358. doi: 10.1016/s0140-6736(97)12433-3. [DOI] [PubMed] [Google Scholar]
- 29.Raoult D, Birtles RJ, Montoya M, Perez E, Tissot-Dupont H, et al. Survey of louse-associated diseases among rural Andean communities in Peru: Prevalence of epidemic typhus, trench fever, and relapsing fever. Clin Infect Dis. 1999;29:434–436. doi: 10.1086/520229. [DOI] [PubMed] [Google Scholar]
- 30.Tarasevich I, Rydkina E, Raoult D. Epidemic typhus in Russia. Lancet. 1998;352:1151. doi: 10.1016/S0140-6736(05)79799-3. [DOI] [PubMed] [Google Scholar]
- 31.Mokrani K, Fournier PE, Dalichaouche M, Tebbal S, Aouati A, et al. Reemerging threat of epidemic typhus in algeria. J Clin Microbiol. 2004;42:3898–3900. doi: 10.1128/JCM.42.8.3898-3900.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Badiaga S, Brouqui P, Raoult D. Autochthonous epidemic typhus associated with Bartonella quintana bacteremia in a homeless person. Am J Trop Med Hyg. 2005;72:638–639. [PubMed] [Google Scholar]
- 33.Raoult D, Roux V. The body louse as a vector of reemerging human diseases. Clin Infect Dis. 1999;29:888–911. doi: 10.1086/520454. [DOI] [PubMed] [Google Scholar]
- 34.Drancourt M, Tran-Hung L, Courtin J, Lumley H, Raoult D. Bartonella quintana in a 4000-year-old human tooth. J Infect Dis. 2005;191:607–611. doi: 10.1086/427041. [DOI] [PubMed] [Google Scholar]
- 35.Raoult D, Woodward T, Dumler JS. The history of epidemic typhus. Infect Dis Clin North Am. 2004;18:127–140. doi: 10.1016/S0891-5520(03)00093-X. [DOI] [PubMed] [Google Scholar]
- 36.Boissier de Sauvages F. Lyon: Bruyset JM, Imprimeur, 10 vol; 1772. Nosologie méthodique. [Google Scholar]