Abstract
Although aging is a ubiquitous process that prevails in all organisms, the mechanisms governing both the rate of decline in functionality and the age of onset remain elusive. A profound constitutively upregulated cytoprotective response is commonly observed in naturally long-lived species and experimental models of extensions to lifespan (e.g., genetically-altered and/or experimentally manipulated organisms), as indicated by enhanced resistance to stress and upregulated downstream components of the cytoprotective nuclear factor erythroid 2-related factor 2 (Nrf2)-signaling pathway. The transcription factor Nrf2 is constitutively expressed in all tissues, although levels may vary among organs, with the key detoxification organs (kidney and liver) exhibiting highest levels. Nrf2 may be further induced by cellular stressors including endogenous reactive-oxygen species or exogenous electrophiles. The Nrf2-signaling pathway mediates multiple avenues of cytoprotection by activating the transcription of more than 200 genes that are crucial in the metabolism of drugs and toxins, protection against oxidative stress and inflammation, as well as playing an integral role in stability of proteins and in the removal of damaged proteins via proteasomal degradation or autophagy. Nrf2 interacts with other important cell regulators such as tumor suppressor protein 53 (p53) and nuclear factor-kappa beta (NF-κB) and through their combined interactions is the guardian of healthspan, protecting against many age-related diseases including cancer and neurodegeneration. We hypothesize that this signaling pathway plays a critical role in the determination of species longevity and that this pathway may indeed be the master regulator of the aging process.
Introduction
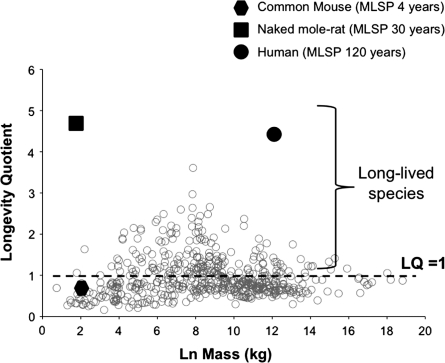
Understanding the mechanisms employed in nature during the evolution of species with disparate longevities is a fundamental focus for comparative research into the biology of aging. Long-lived species are defined as those species that live at least twice as long as expected on the basis of their body size [i.e., longevity quotient (LQ) observed lifespan divided by that predicted by body size = 2; Fig. 1]. Using this definition, prolonged longevity with concomitant attenuated rates of aging, has evolved repeatedly in mammals with long-lived species (e.g., bats, naked mole-rats, and humans) reported in many mammalian taxa. Among the rodents, the naked mole-rat, a mouse-sized hystricognath, stands out as being exceptionally long-lived (Buffenstein 2005), with a maximum lifespan of 30 years and a similar longevity quotient to that of humans (Buffenstein 2008). However, several other rodent species in multiple families live at least twice as long as expected on the basis of body size. By comparing species of similar size, we are able to elucidate both common and novel mechanisms naturally involved in aging processes. This approach may also identify common mechanisms for abrogating aging, as well as those traits that are unique to a particular species or phyla. Few studies in aging research have employed a comparative approach to address the elusive mechanisms involved in divergent rates of aging. This approach, though, through the use of non-traditional animal models, is poised to make pivotal inroads by identifying pathways that are altered in long-lived species, leading to a better understanding of aging mechanisms that indeed may highlight new directions for aging research that may be particularly pertinent to human aging.
Fig. 1.
Lifespan is predicted by body size. The natural log (Ln) of body size in mammals and birds has long been known to correlate allometrically with maximum species lifespan (MLSP) as modified from Sacher (1959) and updated with new species longevity data. This relationship may be described by the equation MLSP = 3.34(Mass)0.193 (from Hulbert et al. 2007). The LQ is the ratio of actual longevity to the expected longevity from this regression of longevity on body size of non-flying, eutherian mammals. Species that live twice as long as expected on the basis of this relationship are considered long-lived and may have evolved mechanisms that resist the aging process. We hypothesize that the cytoprotective Nrf2-signalling pathway is upregulated in long-lived species, conferring enhanced resilience, and resistance to the aging process.
Aging is an inherently blatant process, easily distinguishable in all mammals. The decline in individual fitness and quality of health, coupled with increased difficulty in overcoming illness or physical stresses, leads to permanent loss of function and increased mortality in the elderly. These aging traits are generally thought to be due to a gradual loss of genomic, proteomic, and metabolic integrity. However, despite considerable research endeavor, the mechanisms that contribute to and characterize the aging process remain obscure. It often has been suggested that aging is a multi-causal process linked to multiple molecular and cellular causes of damage (Rattan 2006; Hayflick 2007; Kirkwood 2008). However, current proximate theories of aging are generally focused on a single causative factor of aging. For example, the most widely accepted theory of aging is that it is due to oxidative stress. First proposed by Denham Harman in 1956, this theory postulates that reactive oxygen species (ROS), inevitable byproducts of aerobic respiration, inflict indiscriminate oxidative modification of all cellular components leading to damage that cannot always be completely neutralized by antioxidants and/or repaired (Harman 1956). Damage then can accrue with age leading to the gradual decline in cellular integrity and functionality. Furthermore, ROS levels may increase in damaged or aged mitochondria, causing even higher levels of ROS production and increased levels of subsequent damage. Despite the intuitive logic and vast support for this theory (Barja et al. 1994; Beckman and Ames 1998; Sohal et al. 2002; Droge and Schipper 2007), a causal link between oxidative stress and the rate of aging still has not been clearly established, and in recent times a considerable pool of data incompatible with predictions of this theory have led many to question its validity (Sanz et al. 2006; Muller et al. 2007; Buffenstein et al. 2008; Pérez et al. 2009; Lapointe and Hekimi 2010). Many long-lived rodents, reptiles, bats, and birds (Hamilton et al. 2001; Brunet Rossinni 2004; Andziak and Buffenstein 2006; Furtado-Filho et al. 2007; Wilhelm Filho et al. 2007; Buffenstein et al. 2008), exhibit high levels of oxidative damage, even in young individuals, and these data are not commensurate with their longevity. Furthermore, data from transgenic manipulations of antioxidant levels (Huang et al. 2000; Van Remmen et al. 2003; Pérez et al. 2009) are also contrary to the basic premises of the oxidative-stress theory of aging, for the life spans of these animals are not influenced by either the levels of antioxidant expression and/or accrued oxidative damage. This august theory of aging, while definitively implicated in certain age-associated diseased states, is now openly challenged by many, including renowned scientists who have spent the great majority of their research career focusing on this mechanism of aging (Howes 2006; Sanz et al. 2006; Bonawitz and Shadel 2007; Kregel and Zhang 2007; Buffenstein et al. 2008; Fukui and Moraes 2008; Gems and Doonan 2009; Pérez et al. 2009; Salmon et al. 2009b). Clearly, the oxidative stress theory of aging has been over simplified and requires some modification.
A common denominator exists between the many naturally long-lived species and the transgenic models that exhibit extended longevity and offer some support for the oxidative-stress theory of aging; namely skin fibroblasts from long-living animals show greater levels of cytoprotection and are more resistant to oxidative and chemical insults (Ogburn et al. 1998; Kapahi et al. 1999; Salmon et al. 2005; Harper et al. 2006; Mele et al. 2006; Salmon et al. 2008). The strong relationship between species longevity and cellular resistance to oxidative insults has been reported in a wide variety of organisms across the animal kingdom (Kapahi et al. 1999; Salmon et al. 2005; Labinskyy et al. 2006; Harper et al. 2007). Cellular stress resistance, however, extends to numerous other non-oxidative stressors. These include heat, heavy metals that are not involved in redox reactions, chemotherapeutic agents, dietary alterations, and xenobiotics. This pronounced difference between short-lived and long-lived species most likely represents an evolutionary trade-off between diverting energy and resources into somatic maintenance (and thereby contributing to the survival of the individual) versus investments in growth and reproduction, thereby ensuring the rapid attainment of sexual maturity and the long-term survival of the species (Kirkwood et al. 2000).
An alternate explanation may be that cellular protection against oxidative insults is just one component of a multi-factorial cytoprotective pathway that protects against a plethora of cellular stressors by activating a diverse suite of defenses, including synthesis of molecular chaperones, components of cell cycle surveillance and protein degradation as well as antioxidants, to maintain the cellular, protein, and genomic integrity observed in long-lived species (Pérez et al. 2009; Salmon et al. 2009b). One such pathway is mediated by the transcription factor nuclear factor erythroid 2-related factor 2 (Nrf2) (Motohashi and Yamamoto 2004). Nrf2 (Fig. 2) is a master regulator of a diverse array of more than 200 cytoprotective genes encoding proteins that neutralize and detoxify both endogenous and environmental toxins, regulate factors in cell cycle and growth, and facilitate the maintenance of a high quality proteome (Lee et al. 2003; Liu et al. 2007). Given that Nrf2 controls a broad sweep of cytoprotective mechanisms, this signaling pathway may substantially contribute to the multi-factorial phenotype associated with the aging process. Furthermore, this could explain why some data reportedly support the oxidative-stress theory of aging whereas other data are incompatible with this theory; for example the full spectrum of positive and negative correlations and no correlation at all between antioxidant activities and lifespan (Perez-Campo et al. 1993; Sohal et al. 1993; Andziak et al. 2005; Pamplona and Barja 2007; Wilhelm Filho et al. 2007). Those data that appear to be irreconcilable with this theory, such as the reported high levels of oxidative damage in long-lived birds (Hamilton et al. 2001), vampire bats (Ferreira-Cravo et al. 2007), and naked mole-rats (Andziak and Buffenstein 2006) may be explained by a constitutive upregulation of other compensatory components of the cytoprotective Nrf2 signaling pathway (such as molecular chaperones, upregulation of cell cycle surveillance mechanisms, and components of quality control of proteins).
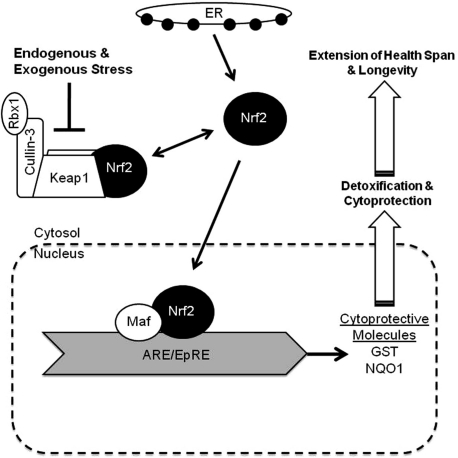
Fig. 2.
The Nrf2 cytoprotective signaling pathway. This pathway regulates transcription of a variety of cellular defenses that bolster an organism’s response to stresses normally encountered during metabolism as well as protect against unexpected cellular insults from environmental toxins. Following its biosynthesis in the endoplasmic reticulum, Nrf2 may translocate directly into the nucleus (as indicated by the arrow) where it transactivates its target genes, thereby contributing to their constitutive expression. Steady state levels of Nrf2 are determined primarily through modulation of its continuous proteasomal degradation following ubiquitination by Cullin-3/Rbx1. Under homeostatic conditions Nrf2 is bound in the cytoplasm to Keap1 an E3 ubiquitin ligase substrate adaptor that targets Nrf2 for degradation. Under stressful conditions, conformational changes in Keap1 block the ubiquitylation of Nrf2, increasing both the half-life of Nrf2 and the size of the free Nrf2 pool, allowing more Nrf2 to translocate into the nucleus, bind to the ARE/EpRE and induce expression of a multitude of cytoprotective enzymes, including GSTs and NQO1 and those that increase GSH biosynthesis. Not surprisingly, depletion and deregulation of this pathway and or downstream components is implicated in diverse pathological conditions, many of which show increased incidence with age (e.g., diabetes, cancer, and neurodegeneration). We predict that this pathway is constitutively upregulated in longer-lived species.
The Nrf2-signaling pathway
Nrf2 is a member of the Cap ‘n’ Collar (CNC) subfamily of transcription factors that contain a basic-region leucine zipper (bZIP) DNA binding domain at its C-terminus (Hayes and McMahon 2001; Aleksunes and Manautou 2007; Kensler et al. 2007; Cavin et al. 2008). The CNC family includes the closely related factors Nrf1, Nrf3 and p45 NF-E2 in addition to Nrf2 (Motohashi et al. 2002; Blank 2008). Both Nrf1 and Nrf2 are ubiquitously expressed in all tissues and show similar binding to the antioxidant/electrophilic response element (ARE/EpRE) (Bishop and Guarente 2007) and whilst they regulate some common genes, Nrf1 differs from Nrf2 in that it is targeted to the endoplasmic reticulum (Zhang et al. 2006). However, knock-out studies reveal that Nrf1 and Nrf2 have distinct phenotypes and different roles in the activation of ARE-dependent genes (Leung et al. 2003; Ohtsuji et al. 2008). The function of Nrf3 has yet to be truly elucidated, but its expression is restricted to placenta and liver and it possesses relatively weak transactivation activity (Sankaranarayanan and Jaiswal 2004; Zhang et al. 2009). Evidence suggests that while Nrf1 and Nrf2 are both constitutively active, Nrf2 is principally activated by pro-oxidants and electrophiles.
Under homeostatic, non-stressful conditions, the levels of Nrf2 are restricted through binding to Kelch-like ECH-associated protein 1 (Keap1) by a ‘hinge-and-latch’ mechanism, otherwise called a ‘two-site tethering’ mechanism, and is targeted for polyubiquitination and subsequent degradation by the proteasome via interactions with Keap1 and Cullin 3 based-E3/Rbx1 ligase complex (Tong et al. 2006; McMahon et al. 2006) (Fig. 2). Under these basal conditions, the half-life of Nrf2 remains short (∼15 min). Upon a stressful insult to the organism or cell itself, cysteine residues (C151, C273, and C288) in the intervening region of Keap1 are oxidized or chemically modified by oxidants or electrophiles preventing the ubiquitylation of Nrf2, thus increasing its half-life to ∼60 min. Nrf2 then rapidly moves into the nucleus and forms a heterodimer with other transcription factors such as small musculoaponeurotic fibrosarcoma proteins (Mafs) that bind to the ARE/EpRE, initiating the transcription of a diverse array of cytoprotective molecules and enzymes (Thimmulappa et al. 2002; Kwak et al. 2003), and thereby controlling cellular detoxification as well as those processes involved in maintenance of stability and integrity of proteins (Fig. 2).
Nrf2-mediated cytoprotection
Nrf2 plays a central role in cytoprotection, by detoxifying and eliminating ROS, xenobiotics and electrophilic carcinogens, as well as removing damaged proteins and organelles (McMahon et al. 2001; Kwak et al. 2003; Cho et al. 2005). Importantly, Nrf2 regulates the synthesis of glutathione (GSH), as well as enzymes involved in GSH homeostasis, namely glutamate-cysteine ligase catalytic (GCLC) and glutamate-cysteine modifier (GCLM) subunits that combine to form the GCL heterodimer, which catalyses the rate-limiting enzymes in GSH biosynthesis, and glutathione reductase 1 (GR), which reduces the oxidized glutathione disulfide (GSSG) and thereby recycles GSH. Nrf2 also mediates induction of several other classes of antioxidant proteins [e.g., thioredoxin, peroxiredoxin, sulfiredoxin, ferritin, metallothionein, and heme oxygenase 1 (HO-1)] and mediates induction of phase I and phase II drug-metabolizing enzymes [e.g., aldo–keto reductases (AKRs), glutathione S-transferases (GSTs), and NAD(P)H:quinone oxidoreductase 1 (NQO1)].
Perhaps the most important role of Nrf2 in the response to toxins is the regulation of GSH biosynthesis and detoxification through direct regulation of GCL and NQO1. GSH production augments the capacity of cells to scavenge ROS and upon conjugation with xenobiotics, decreases their hydrophobic nature and increases their elimination via MRP efflux pumps (Hayes et al. 2005; Meijerman et al. 2008). NQO1 inhibits the formation of free radicals by the redox-cycling of quinones and also stabilizes p53, which may lead to better regulation of the cell cycle under genotoxic stress (Nioi and Hayes 2004).
GST is an enzyme essential for the detoxification of organic hyperoxides and α,β-unsaturated aldehydes and is also an important component in the regulation of GSH, of which Nrf2 also plays a direct role. GSH is considered to be an abundant (present in mM concentrations in tissues) and an important intracellular, non-protein sulfhydryl compound (Meister and Anderson 1983; Franco et al. 2007; Wu et al. 2009) as GSH is involved in the reduction and detoxification of ROS, electrophiles, and xenobiotics (Hayes et al. 2005). GSH also participates in limiting the deleterious actions of NO and inhibits peroxynitrite formation and NO-induced neuronal apoptosis, and as such is an important neuroprotectant (Vargas et al. 2006). GSH also serves as a large and important reservoir of cysteine, a cofactor in metabolism (e.g., formaldehyde dehydrogenase), hormone biosynthesis and is the conjugating substrate involved in GST-mediated detoxification (Jones 2008).
The importance of Nrf2 in many cytoprotective activities has been clearly illustrated through the use of Nrf2 knock-out (Nrf2–/–) mice or their fibroblasts under stressful conditions (Enomoto et al. 2001; Chanas et al. 2002; Gong and Cederbaum 2006). Under normal laboratory conditions, Nrf2–/– mice show very few phenotypic differences from wild-type (Nrf2+/+) mice (Yanagawa et al. 2004). However, these knock-out animals are more susceptible than are their wild-type equivalents to pro-oxidant stimuli such as hyperoxia, lipopolysaccharide (LPS), cigarette smoke, diesel exhaust fumes, and ultraviolet irradiation (UVA and UVB), as well as numerous chemicals such as butylated hydroxytoluene and benzo[a]pyrene (Cho et al. 2002; Rangasamy et al. 2004; Hirota et al. 2005; Thimmulappa et al. 2006a; Reddy et al. 2007). Cytoprotective effects of Nrf2 are evident in all tissues studied to date, with reports of increased susceptibility to many cancers (Ramos-Gomez et al. 2001; Iida et al. 2004; Xu et al. 2006; Kensler et al. 2007; Osburn and Kensler 2008), neurodegeneration (Shih et al. 2005; Burton et al. 2006), lung disease (Cho et al. 2004; Rangasamy et al. 2005), impaired liver and gastrointestinal function (Enomoto et al. 2001; Khor et al. 2008) and inflammation (Rangasamy et al. 2005; Thimmulappa et al. 2006b; Osburn et al. 2007) in Nrf2−/− mice. Indeed, even the tumor-protective effects of dietary restriction are dependent upon the presence of Nrf2 and are virtually ablated in calorically restricted Nrf2−/− mice (Pearson et al. 2008). Because an abundance of cytoprotective molecules would be beneficial for any organism, we believe that these factors would be selected for in longer-lived species, better protecting them against cytotoxicity and thereby contributing to longer healthspans.
The many roles of the Nrf2-signaling pathway
The Nrf2 signaling pathway is emerging as a critical and principal factor in the protection against cancer, neurodegeneration, and inflammation (Hayes and McMahon 2001; Droge and Schipper 2007; Nair et al. 2007). Because of its prominence in a large variety of age-associated diseases and processes, we propose that Nrf2 plays a pivotal role in longevity and in determination of healthspan (Fig. 3).
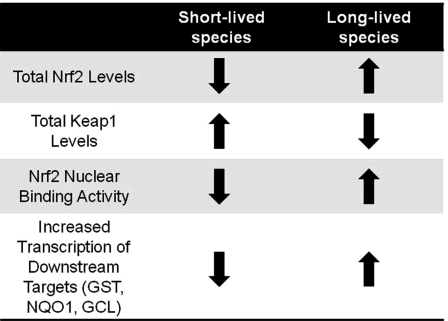
Fig. 3.
Increased cytoprotection is a key contributor to longevity. We predict that marked differences among species in Nrf2 protein levels and concomitant signaling exist and that they are commensurate with species longevity. In this regard we predict that naturally or genetically-altered long-lived species have more free Nrf2 that is localized in the nucleus, resulting in greater Nrf2 binding to the ARE and thus in an increase in the transcription of cytoprotective target genes such as GST and NQO1, which show greater activity and that this enhanced cellular protection contributes to species longevity. We hypothesize that shorter-lived species, in contrast, show low constitutive levels of Nrf2 with concomitant lower primed defenses against cellular insults and are thus more susceptible to toxins and accumulated damage.
Detoxification
Many diverse types of stress continuously challenge cells of all organisms. These include endogenous stressors (e.g., ROS, hydroperoxides, carbonyls, and quinones) and environmental stressors (e.g. heat, UV irradiation, air pollutants, food toxins, heavy metals, drugs, and bacterial or viral infections). Although agents like ROS are predictably and continuously produced as metabolic byproducts, other stressors may be more capricious, resulting from acute exposure to toxins in food, pollutants, or even infections. These different types of stress, independent of origin, can cause modifications of cellular protein and DNA, possibly leading to the decline in function of cells and tissues and, in extreme cases, resulting in death of the organism. The role of Nrf2 has been mainly studied with regards to the metabolism of toxins including acetaminophen, butylated hydroxytoluene, bleomycin, and 7,12-dimethylbenz(a)anthracene (Copple et al. 2008). Nrf2 is the primary responder to such toxins and upregulates expression of several compounds and enzymes (NQO1, GSH, GST) that promote the efficient neutralization, conjugation and concomitant elimination of the toxins.
Stability and turnover of proteins
Changes in the stability and degradation of proteins have been argued to lead to age-related conformational changes in proteins with a propensity for aggregation. This correlation has been observed in several neurodegenerative diseases (Grune et al. 2004; Gregersen et al. 2006) including Alzheimer’s disease. Maintenance of proteasome activity is required for cell survival, and loss of proteasomal capacity may further exacerbate cellular stress due to the accrual of misfolded or damaged proteins. When ineffectively removed and degraded, damaged proteins can form toxic aggregates (Glickman and Ciechanover 2002). Not surprisingly, failure to properly maintain stability and turnover of proteins has been hypothesized to be a causal factor in the aging process (Grune 2000; Herczenik and Gebbink 2008; Pérez et al. 2009).
Homeostasis of proteins is maintained in part by ubiquitin-mediated targeting of misfolded proteins for both 26S proteasomal degradation and autophagy. However, during oxidative stress, both have been shown to play a minor role in the degradation of oxidized proteins (Shringarpure et al. 2001). Instead, the 19S subunits dissociate from the 26S proteasome, and oxidatively modified proteins are directly targeted to the 20S catalytic cylindrical core of the proteasome (Grune and Davies 1997).
Microrrays have shown that Nrf2 also influences expression of molecular chaperones, subunits of the 26S and 20S proteasome, and the sequestosome 1 (p62), a key component in autophagy (Kwak et al. 2003; Korashy and El-Kadi 2006; Hayes and McMahon 2009), thereby controlling mechanisms that enable folding of proteins and the removal of misfolded and ubiquitinated proteins as well as damaged organelles (Lane 1992; Nioi and Hayes 2004; Gamet-Payrastre 2006). Kapeta et al. (2010) have demonstrated a role of Nrf2 and proteasome function is regulating replicative senescence of human cells. Induction of Nrf2 preserves proteasome function in human fibroblasts and extends replicative lifespan up to 65% by reducing the levels of ROS and oxidized proteins. This finding supports our premise of the protective role of Nrf2 in regulating longevity (Fig. 3) as observed in one model of cellular aging.
Long-lived species and mutant-mouse models appear to share features that contribute to enhanced protein stability that may be regulated in part by elevated levels of Nrf2. Several researchers have demonstrated that long-lived bats and naked mole-rats possess proteins that are resistant to unfolding, low levels of ubiquitinated-proteins, and moderate to higher levels of 20S proteasomal specific activity, compared to shorter-lived bats and mice (Pérez et al. 2009; Salmon et al. 2009a); all these traits are indicators of enhanced stability of proteins and maintenance of protein quality. These observations may be explained if long-lived species have elevated levels of Nrf2 that would become active during times of oxidative stress and act on target genes to dissipate that stress and degrade misfolded proteins with a 20S proteasome complement of greater specific activity.
Cellular integrity
Nrf2 also interacts with p53, the tumor suppressor transcription factor widely regarded as the guardian of the genome. p53 regulates cellular pathways of response to stress that may lead to cell-cycle arrest, repair of DNA, cellular senescence and apoptosis. p53 also transcriptionally activates p21 under toxic conditions, allowing p21 to exert its function as an inhibitor of the cell cycle. Alternatively p53 can activate a variety of apoptotic genes, leading to cell death (Shen 2001). Chen et al. (2009) have demonstrated that p21 increases Nrf2 levels by interfering with Keap1-directed Nrf2 ubiquitination and degradation. Thus, p21 widens the survival functions of p53 under conditions of low to moderate stress through an interaction with the Nrf2 pathway by protecting the cell from cytotoxins and may be another mechanism by which p21 suppresses tumors. If toxin levels exceed a certain threshold, p21 may induce cell cycle arrest to allow time for DNA repair. Should damage exceed the repair capabilities of the organism, apoptosis is induced (Villeneuve et al. 2009). Thus, the p21-Nrf2-dependent and p21-dependent pathways of cell-cycle arrest may represent two separate, but reinforcing, cytoprotective mechanisms mediated by p53 and Nrf2 all at the level of p21. It is not known whether Nrf2 signaling is constitutively elevated in different types of cells that may accumulate p21 in response to stress.
An interesting question that arises from the influence of p21 on Nrf2 activity is whether cells of long-lived and short-lived organisms have different thresholds for switching from p21-induced upregulation of Nrf2-dependent cell-survival mechanisms to that of p53/p21-induced cell death. In contrast, p53 has been shown to antagonize Nrf2 binding at ARE-containing promoters, suggesting that at high levels of stress and/or irreparable damage the balance shifts to a pro-apoptotic state, reinforced by attenuated activitiy of Nrf2 and increased levels of ROS. Given the difference in cellular resistance to cytotoxins among species of disparate longevity, we hypothesize that cells of long-lived species have higher thresholds of stress tolerance, favoring the upregulation of Nrf2-dependent cytoprotection, and only as a last resort, when damage is excessive and/or irreparable, suppressing Nrf2 responses and thereby inducing apoptosis. The decision whether p53 selects induction of growth arrest or apoptosis appears to be defined by the availability of the survival signal. When survival signals dominate, p53 activation leads to cessation of the cell cycle and facilitates repair. Conversely, when survival signals are inadequate, p53 will drive apoptosis (Ashcroft et al. 2000; Sears 2002). Long-lived species may maintain a more stable environment, which coordinates more survival signals through p53 activation, which biases the system to enter pathways of repair or cell-cycle arrest. Nrf2 target genes, such as NQO1, have been shown to bind and stabilize p53. This could lead to a mutually reinforcing feedback loop regulating cell-cycle arrest or apoptosis, depending on the degree of ROS and the number of damaged macromolecules.
Thus, there are numerous interacting points between the p53 and Nrf2 pathways that are critically dependent upon the severity of stress and upon the kinetics of the Nrf2 response, either of which may determine survival or cell death.
Inflammation
Recent data also reveal that Nrf2 signaling plays an important role in reducing the inflammatory response. Nrf2 also represses multiple pro-inflammatory genes, including tumor necrosis factor α (TNFα), and the interleukins (IL-1β, IL-6) as well as cell adhesion molecules, prostaglandin metabolites, matrix-metalloproteinases, and inducible nitric oxide synthase (Kim et al. 2009). This is thought to be primarily through its ability to antagonize NF-κB (Chen et al. 2006; Jin et al. 2008). For example lipopolysaccharide (LPS) induction of NF-κB can be attenuated by chemical activators (e.g., curcumin or sulforaphane) of Nrf2 (Jeong et al. 2004). The enzyme, HO-1 has prominent anti-inflammatory activity. Importantly, HO-1 is up-regulated by Nrf2 and this is likely to modulate innate immunity, inflammation, and wound healing (Srisook et al. 2005; Patil et al. 2008). There is also considerable cross-talk between the Nrf2 pathway and inflammatory signaling. NF-κB has been reported to directly repress Nrf2 signaling at the transcriptional level. Moreover, NF-κB may also impede Nrf2 signaling through the induction of hypoacetylation through the recruitment of histone deacetylase 3 (HDAC) and by competitively binding to the transcription coactivator-binding protein (CBP) (Liu and Shen 2008).
Nrf2-induced repression of the inflammatory system has widespread implications for many diseased states. Chronic inflammation is implicated in the etiology of several degenerative human diseases and promotes the pathogenesis and progression of autoimmune disease, asthma, neurodegeneration (e.g., Parkinson’s and Alzheimer’s disease), pulmonary fibrosis, osteoarthritis, colitis, renal failure, cardiovascular disease, atherosclerosis, and cancer (for review see Kim et al. 2009). For example, the down-regulation of pro-inflammatory factors appears to have a profound beneficial effect on cardiovascular function and has made Nrf2 a novel target in the maintenance of cardiac health. Nrf2 upregulation has been shown to protect arterial endothelial cells from inflammation (Zakkar et al. 2009) and attenuate a variety of factors associated with cardiovascular disease (Ungvari et al. 2008). Sulforaphane, an activator of Nrf2, has been shown to protect against ischemic injury via the upregulation of phase-II detoxification enzymes as well as HO-1 (Piao et al. 2009). The protective antioxidant enzyme HO-1 also mediates mitochondrial biogenesis in cardiac tissue (Piantadosi et al. 2008). The Nrf2 cytoprotective pathway is crucial in protecting the cardiovascular system against oxidative damage by ROS (Zhu et al. 2008).
Similarly, the drugs used in treatment for rheumatoid arthritis, a chronic inflammatory autoimmune disease that causes progressive damage to joints, are known to activate the Nrf2-signaling pathway and upregulate the expression of HO-1 and GCL. Upregulation of Nrf2 by Auranofin in rheumatoid arthritis, and by asthma medications may abrogate the accumulation of prostaglandin through the inhibition of cyclooxygenase-2 (Kim et al. 2009).
Chronic neuroinflammation is commonly associated with neurodegenerative diseases (Rojo et al. 2010) and is attributed to the uncontrolled defensive microglial activity. Microglia promote healing of the brain by removing pathogens and cellular debris. Damaged neurons release chemical stimuli that trigger the expansion of the microglial pool in response to inflammatory stress and this in turn promotes further neuronal death, chronic neuroinflammation and neurodegeneration (Kim et al. 2009; Rojo et al. 2010).
Neurodegeneration
The role of Nrf2 in the protection against neurodegenerative diseases and maintenance of cognitive function is being aggressively pursued. Nrf2 is required for adequate healing after a traumatic injury to the brain and is now regarded as an important therapeutic agent in a variety of age-associated neurodegenerative disorders (Jin et al. 2009). Neuronal cell death, commonly associated with several disorders, including Alzheimer’s and Parkinson’s disease, is decreased in vitro following induction of Nrf2 signaling in neurons (Johnson et al. 2008). In addition, upregulation of the Nrf2 signaling pathway in astrocytes protects neighboring neurons from cytotoxic threats (Kraft et al. 2004). Similarly, in both Parkinson’s and Alzheimer’s disease high levels of oxidative stress may be countered by an upregulation of Nrf2 signaling and concomitant enhanced levels of HO-1, NQO1, and other downstream targets involved in GSH metabolism (Cuadrado et al. 2009). Laboratory mice show an age-related decline in Nrf2 levels and downstream signaling components in the nervous system (Duan et al. 2009) and this is thought to possibly contribute to their age-related cognitive decline.
Cancer
Chronic inflammation may also lead to spontaneous neoplasia (Ahmad et al. 2009). Nrf2 has been shown to have both positive and negative effects on the development and treatment of cancer (Lau et al. 2008). Tumor cell lines isolated and profiled from human patients have indicated that many tumors have adapted to exploit the cytoprotective actions of Nrf2 both in vivo and in vitro through mutations of Keap1 and Nrf2, which lead to the constitutive upregulation and permanent activation of Nrf2-signaling to enhance the tolerance of the cancer cells to toxins and thereby limit the efficacy of chemotherapeutic agents (Wang et al. 2008). It is not currently known at what stage of tumorigenesis these somatic mutations contribute most to the tumorigenic phenotype. Chemotherapeutic-resistant tumor lines also often upregulate GSH biosynthetic and drug-metabolizing enzyme pathways making them difficult to treat in vivo. It has been shown that upregulation of GSH biosynthetic and GSH-dependent enzymes is associated with resistance to drugs (McLellan and Wolf 1999) and we now recognize that this is likely to be due to dysregulation of Nrf2 (Hayes and McMahon 2009).
Potential benefits may be realized by studying how long-lived species prevent these mutations and maintain control of the regulation of the Nrf2-Keap1 pathway. The chemo-preventative effects of the Nrf2-signaling pathway, the anti-tumorigenic properties of the Nrf2-signaling pathway and its widespread presence throughout the organism are most likely an integral component of cancer resistance and augmented longevity. In addition, learning how to regulate the Nrf2-Keap1 pathway may lead to more effective treatments for human cancers.
Other roles of Nrf2
Although the roles of Nrf2-signaling are being thoroughly studied in fields of cancer biology and neurodegeneration, the importance of Nrf2 cytoprotection is emerging in other research areas as well. Nrf2 upregulation has also been hypothesized to play a role in global antioxidant defense during arousal from hibernation in various species. In squirrels, constitutive Nrf2 levels, as well as components downstream of Nrf2, are increased throughout hibernation but return to normal levels after resuming normal activity (Morin et al. 2008). Nrf2 also mediates protection against damage from oxidative stress during dehydration in the African clawed frog (Malik and Storey 2009). These fascinating studies performed in different species illustrate the diverse yet profound physiological effects Nrf2 can exert in biology.
Because Nrf2 cytoprotection is implicated in so many areas of research, compounds that activate this signaling pathway are of great interest to biomedical scientists. Many natural compounds, particularly phytochemicals from edible plants, have already been identified as stimulants of this pathway including sulforaphane (present in broccoli), resveratrol (a flavenoid from grape skin), and curcumin (the key component of turmeric) (Lee and Surh 2005).
Activation of Nrf2 confers protection against oxidative stressors, electrophiles, pro-inflammatory agents and situations that lead to accumulation of damaged proteins and other molecules. Given this myriad of cytoprotective regulatory functions, Nrf2 is most likely an important component of enhanced stress resistance in long-lived species, long-lived transgenic mouse models (e.g., dwarf mice), as well as those species subjected to life-extending environmental manipulations (e.g., caloric restriction). Novel therapeutics, mainly dietary polyphenols, which elicit beneficial effects through the activation of Nrf2 also modulate cellular signaling processes that are important in the defense against inflammation, neurodegeneration, and cancer.
The Nrf2-signaling pathway and aging
The paradigm of hormesis has been used to explain the ability of long-lived species to handle stress and thereby increase longevity (Masoro 2007). Indeed caloric restriction is viewed as a mild stress that induces protective cellular mechanisms and these most likely are dependent upon the Nrf2-ARE cytoprotective cell-signaling pathway. The hormetic profile of Nrf2 action has been recently reviewed from an evolutionary perspective (Maher and Yamimoto 2010). These authors make the point that there is a narrow range of Nrf2 activity that provides an optimal ability of an organism to detoxify stressors. We hypothesize that long-lived species constitutively have a primed Nrf2 pathway ready to respond immediately to cell stressors and thereby minimize exposure to substances that may induce cell damage. These organisms may also have a broader range of set points before Nrf2 levels result in the unmasking of unfavorable states. Evidence in support of this premise comes from constitutive expression of ARE-regulated genes (as observed in the Keap1 knockout mice) or improper expression of ARE-regulated genes which may give rise to deleterious effects observed in disease states (e.g., multidrug resistance phenotype in cancer cells). There is mounting evidence across evolutionarily distant species that Nrf2-ARE-dependent components are associated with both longevity and extension of healthspan. Nrf2-dependent enzymes involved in detoxification and GSH metabolism are increased in multiple murine models of the extension of lifespan, including calorically restricted mice, methionine-restricted mice, and dwarf mice under non-stressful conditions (Brown-Borg and Rakoczy 2005; Amador-Noguez et al. 2007). Enhanced GSH has also been implicated as a potential mechanism in cellular resistance to drug-induced apoptosis (O'Brien and Tew 1996). Depletion of GSH alters cellular resistance to electrophiles and hydroperoxides (Higgins et al. 2009) and reduced GSH bioavailability has been implicated in aging and associated age-related diseases (Maher 2005; Droge and Schipper 2007).
Similarly, flies that overexpress the GCL rate-limiting enzyme responsible for GSH synthesis in neural tissue, and have as a consequence higher GSH levels in their central nervous sytem, live 50% longer than do wild type flies (Orr et al. 2005). Longer-living mutants of Caenorhabditis elegans, with alterations in insulin/IGF-1-like signaling, have been found to up-regulate GSH biosynthetic enzymes as well as phase I and phase II drug-metabolizing enzymes including NQO1 and GST (McElwee et al. 2004; Jasper 2008; Tullet et al. 2008), and overexpression of GSTs increases worm median lifespan (Ayyadevara et al. 2005a;. 2005b). Conversely, it is well known that short-lived rats and mice possess a diminished thiol status as they age, and that this is associated with decreased Nrf2 activity (Suh et al. 2004).
We have found that, like experimental mouse models of extended longevity (Brown-Borg and Rakoczy 2005), long-lived rodents such as naked mole-rats and white-footed mice (Peromyscus leucopus) exhibit constitutive elevation of both GSH and GST. As discussed above, raised GSH confers considerable protection, firstly by serving as a redox buffer, as well as a co-substrate required to neutralize and eliminate ROS and electrophiles (Hayes and Strange 1995; Maher 2005). Based on work in the mouse (McMahon et al. 2001), it seems probable that these increases in GSH-dependent enzymes in long-lived rodents are directed by Nrf2. Furthermore, we have found that both young (2-year-old) and old (26-year-old) naked mole-rats show similarly low levels of ubiquitinated protein, indicative of highly efficient removal of damaged proteins (Pérez et al. 2009). As Nrf2 regulates α and β subunits of the 26S proteasome, along with p62, a protein that promotes protein aggregate removal by autophagy (Bjorkoy et al. 2006; Nakaso et al. 2006; Du et al. 2009), it seems likely that up-regulation of the ARE-gene battery may, at least in part, account for the failure to accumulate ubiquitinated protein. We therefore speculate that long-lived species have a constitutively active Nrf2 cytoprotective pathway. Consistent with this notion, we have found that under normal physiological conditions the levels of Nrf2 protein are between three-fold and eight-fold higher in the various organs and fibroblasts of naked mole-rats than in those of mice, and that the white-footed mouse shows intermediate values. The molecular basis for this apparent constitutive upregulation and activation of Nrf2 in naked mole-rats is not known, nor is the consequence of its activation clear. We expect that augmented Nrf2 activity contributes significantly to the extraordinary longevity of this species and hypothesize that this is a common feature in long-lived mammals.
Interestingly, one of the two transcription factors implicated in longevity in C. elegans is Skn-1, a CNC-type protein that is related to mammalian Nrf2 (An et al. 2005; Tullet et al. 2008), which is required for longevity induced by dietary restriction in C. elegans (Bishop and Guarente 2007). Support for the notion that Nrf2 has the capacity to increase longevity also comes from Drosophila in which deletion of one Keap1 allele increases expression of GSTD1, resulting in resistance to paraquat, and longevity in male flies (Sykiotis and Bohmann 2008). Over-expression of downstream components of the Nrf2 signaling pathway extends Drosophila’s lifespan (Orr et al. 2005) and long-lived C. elegans models also exhibit upregulated expression of Skn-1-dependent enzymes (Jasper 2008). Conversely, when the homologue of Nrf2 is knocked out in both flies and worms, the extension of lifespan associated with caloric restriction is abrogated (Bishop and Guarente 2007). Taken together, these studies suggest that the Nrf2-ARE pathway (or its evolutionary equivalent in invertebrates) is commonly involved in paradigms of prolonged longevity in the mouse, worm, and fly.
Collectively, these data, further support our hypothesis that Nrf2 directly plays a role in the determination of both longevity and healthspan. Despite the well-documented relationship between Nrf2 and cellular protective mechanisms relevant to aging, surprisingly very few studies have evaluated the role of Nrf2 in mediating rates of aging and longevity. However, these indirect data outlined above, support our premise that the protective role of Nrf2 against toxins and tumorigenesis actively determines longevity and healthspan (Fig. 3). Many of the single gene mutations that have been shown to modulate aging could induce their effects through manipulation of this pathway and it thus may be a source of novel hypothesis testing in comparative aging research in this direction. Mechanistic dissection of this pathway and its regulatory role in aging will determine the validity of those hypotheses and may ultimately lead to intervention studies culminating in an extension of life and prolonged good health in humans.
Funding
American Federation for Aging Research (to R.B.), the Glenn Foundation for Medical Research grant (to R.B.), NIH/NIA (to R.B.), Cancer Research–UK (to J.D.H.) and the Association for International Cancer Research (to J.D.H.).
References
- Ahmad AB, Wang Z, Kong D, Majumdar AP, Sarkar FH. Aging and inflammation: etiological culprits of cancer. Curr Aging Sci. 2009;2:174–86. doi: 10.2174/1874609810902030174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleksunes LM, Manautou JE. Emerging role of Nrf2 in protecting against hepatic and gastrointestinal disease. Toxic Path. 2007;35:459–73. doi: 10.1080/01926230701311344. [DOI] [PubMed] [Google Scholar]
- Amador-Noguez D, Dean A, Huang WD, Setchell K, Moore D, Darlington G. Alterations in xenobiotic metabolism in the long-lived Little mice. Aging Cell. 2007;6:453–70. doi: 10.1111/j.1474-9726.2007.00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An JH, Vranas K, Lucke M, Inoue H, Hisamoto N, Matsumoto K, Blackwell TK. Regulation of the Caenorhabditis elegans oxidative stress defense protein SKN-1 by glycogen synthase kinase-3. Proc Natl Acad Sci USA. 2005;102:16275–80. doi: 10.1073/pnas.0508105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andziak B, Buffenstein R. Disparate patterns of age-related changes in lipid peroxidation in long-lived naked mole-rats and shorter-lived mice. Aging Cell. 2006;5:525–32. doi: 10.1111/j.1474-9726.2006.00246.x. [DOI] [PubMed] [Google Scholar]
- Andziak B, O'Connor TP, Buffenstein R. Antioxidants do not explain the disparate longevity between mice and the longest-living rodent, the naked mole-rat. Mech Ageing Dev. 2005;126:1206–12. doi: 10.1016/j.mad.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Ashcroft MT, Taya Y, Vousden KH. Stress signals utilize multiple pathways to stabilize p53. Mol Cell Biol. 2000;20:3224–33. doi: 10.1128/mcb.20.9.3224-3233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyadevara S, Dandapat A, Singh SP, Benes H, Zimniak L, Shmookler Reis RJ, Zimniak P. Lifespan extension in hypomorphic daf-2 mutants of Caenorhabditis elegans is partially mediated by glutathione transferase CeGSTP2-2. Aging Cell. 2005a;4:299–307. doi: 10.1111/j.1474-9726.2005.00172.x. [DOI] [PubMed] [Google Scholar]
- Ayyadevara S, Engle MR, Singh SP, Dandapat A, Lichti CF, Benes H, Shmookler Reis RJ, Liebau E, Zimniak P. Lifespan and stress resistance of Caenorhabditis elegans are increased by expression of glutathione transferases capable of metabolizing the lipid peroxidation product 4-hydroxynonenal. Aging Cell. 2005b;4:257–71. doi: 10.1111/j.1474-9726.2005.00168.x. [DOI] [PubMed] [Google Scholar]
- Barja G, Cadenas S, Rojas C, Lopeztorres M, Perez-Campo R. A decrease of free-radical production near-critical targets as a cause of maximum longevity in animals. Comp Biochem Physiol. 1994;108:501–12. doi: 10.1016/0305-0491(94)90103-1. [DOI] [PubMed] [Google Scholar]
- Beckman KB, Ames BN. The free radical theory of aging matures. Physiol Rev. 1998;78:547–81. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–9. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Johansen T. p62/SQSTM1: a missing link between protein aggregates and the autophagy machinery. Autophagy. 2006;2:138–9. doi: 10.4161/auto.2.2.2405. [DOI] [PubMed] [Google Scholar]
- Blank V. Small Maf proteins in mammalian gene control: mere dimerization partners or dynamic transcriptional regulators? J Mol Biol. 2008;376:913–25. doi: 10.1016/j.jmb.2007.11.074. [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Shadel GS. Rethinking the mitochondrial theory of aging - the role of mitochondrial gene expression in lifespan determination. Cell Cycle. 2007;6:1574–8. doi: 10.4161/cc.6.13.4457. [DOI] [PubMed] [Google Scholar]
- Brown-Borg HM, Rakoczy SG. Glutathione metabolism in long-living Ames dwarf mice. Exp Geront. 2005;40:115–20. doi: 10.1016/j.exger.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Brunet Rossinni AK. Testing the free radical theory of aging in bats. Ann N Y Acad Sci. 2004;1019:506–8. doi: 10.1196/annals.1297.093. [DOI] [PubMed] [Google Scholar]
- Buffenstein R. The naked mole-rat? A new long-living model for human aging research. J Gerontol A. 2005;60:1369–77. doi: 10.1093/gerona/60.11.1369. [DOI] [PubMed] [Google Scholar]
- Buffenstein R. Negligible senescence in the longest living rodent, the naked mole-rat: insights from a successfully aging species. J Comp Physiol B. 2008;178:439–45. doi: 10.1007/s00360-007-0237-5. [DOI] [PubMed] [Google Scholar]
- Buffenstein R, Edrey YH, Yang T, Mele J. The oxidative stress theory of aging: embattled or invincible? Insights from non-traditional model organisms. Age. 2008;30:99–109. doi: 10.1007/s11357-008-9058-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton NC, Kensler TW, Guilarte TR. In vivo modulation of the Parkinsonian phenotype by Nrf2. Neurotoxicol. 2006;27:1094–100. doi: 10.1016/j.neuro.2006.07.019. [DOI] [PubMed] [Google Scholar]
- Cavin C, Marin-Kuan M, Langouet S, Bezencon C, Guignard G, Verguet C, Piguet D, Holzhauser D, Cornaz R, Schilter B. Induction of Nrf2-mediated cellular defenses and alteration of phase I activities as mechanisms of chemoprotective effects of coffee in the liver. Food Chem Toxicol. 2008;46:1239–48. doi: 10.1016/j.fct.2007.09.099. [DOI] [PubMed] [Google Scholar]
- Chanas SA, et al. Loss of the Nrf2 transcription factor causes a marked reduction in constitutive and inducible expression of the glutathione S-transferase Gsta1, Gsta2, Gstm1, Gstm2, Gstm3 and Gstm4 genes in the livers of male and female mice. Biochemical J. 2002;365:405–16. doi: 10.1042/BJ20020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WM, Sun Z, Wang XJ, Jiang T, Huang ZP, Fang DY, Zhang DD. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol Cell. 2009;34:663–73. doi: 10.1016/j.molcel.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XL, Dodd G, Thomas S, Zhang XL, Wasserman MA, Rovin BH, Kunsch C. Activation of Nrf2/ARE pathway protects endothelial cells from oxidant injury and inhibits inflammatory gene expression. AJP. 2006;290:H1862–70. doi: 10.1152/ajpheart.00651.2005. [DOI] [PubMed] [Google Scholar]
- Cho HY, Jedlicka AE, Reddy SP, Kensler TW, Yamamoto M, Zhang LY, Kleeberger SR. Role of NRF2 in protection against hyperoxic lung injury in mice. Am J Resp Cell Mol Bio. 2002;26:175–82. doi: 10.1165/ajrcmb.26.2.4501. [DOI] [PubMed] [Google Scholar]
- Cho HY, Reddy SP, Debiase A, Yamamoto M, Kleeberger SR. Gene expression profiling of NRF2-mediated protection against oxidative injury. Free Radic Biol Med. 2005;38:325–43. doi: 10.1016/j.freeradbiomed.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Cho HY, Reddy SP, Yamamoto M, Kleeberger SR. The transcription factor NRF2 protects against pulmonary fibrosis. FASEB J. 2004;18:1258–60. doi: 10.1096/fj.03-1127fje. [DOI] [PubMed] [Google Scholar]
- Copple IM, Goldring CE, Kitteringham NR, Park BK. The Nrf2-Keapl defence pathway: role in protection against drug-induced toxicity. Toxicology. 2008;246:24–33. doi: 10.1016/j.tox.2007.10.029. [DOI] [PubMed] [Google Scholar]
- Cuadrado A, Moreno-Murciano P, Pedraza-Chaverri J. The transcription factor Nrf2 as a new therapeutic target in Parkinson's disease. Exp Opin Therap Targets. 2009;13:319–29. doi: 10.1517/13543780802716501. [DOI] [PubMed] [Google Scholar]
- Droge W, Schipper HM. Oxidative stress and aberrant signaling in aging and cognitive decline. Aging Cell. 2007;6:361–70. doi: 10.1111/j.1474-9726.2007.00294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Wooten MC, Gearing M, Wooten MW. Age-associated oxidative damage to the p62 promoter: implications for Alzheimer disease. Free Rad Bio Med. 2009;46:492–501. doi: 10.1016/j.freeradbiomed.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan WS, Zhang RY, Guo YS, Jiang YF, Huang YL, Jiang H, Li CY. Nrf2 activity is lost in the spinal cord and its astrocytes of aged mice. In Vitro Cell Dev Biol Anim. 2009;45:388–97. doi: 10.1007/s11626-009-9194-5. [DOI] [PubMed] [Google Scholar]
- Enomoto A, Itoh K, Nagayoshi E, Haruta J, Kimura T, O'Connor T, Harada T, Yamamoto M. High sensitivity of Nrf2 knockout mice to acetaminophen hepatotoxicity associated with decreased expression of ARE-regulated drug metabolizing enzymes and antioxidant genes. Tox Sci. 2001;59:169–77. doi: 10.1093/toxsci/59.1.169. [DOI] [PubMed] [Google Scholar]
- Ferreira-Cravo MW, Andrade RG, Drew K, Hermes-Lima M. Physiological oxidative stress in the animal world. Comp Biochem Phys A. 2007;148:S63–4. [Google Scholar]
- Franco R, Schoneveld OJ, Pappa A, Panayiotidis MI. The central role of glutathione in the pathophysiology of human diseases. Arch Physiol Biochem. 2007;113:234–58. doi: 10.1080/13813450701661198. [DOI] [PubMed] [Google Scholar]
- Fukui H, Moraes CT. The mitochondrial impairment, oxidative stress and neurodegeneration connection: reality or just an attractive hypothesis? Trends in Neurosci. 2008;31:251–6. doi: 10.1016/j.tins.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furtado-Filho OV, Polcheira C, Machado DP, Mourao G, Hermes-Lima M. Selected oxidative stress markers in a South American crocodilian species. Comp Biochem Physiol C. 2007;146:241–54. doi: 10.1016/j.cbpc.2006.11.017. [DOI] [PubMed] [Google Scholar]
- Gamet-Payrastre L. Signaling pathways and intracellular targets of sulforaphane mediating cell cycle arrest and apoptosis. Curr Cancer Drug Targets. 2006;6:135–45. doi: 10.2174/156800906776056509. [DOI] [PubMed] [Google Scholar]
- Gems D, Doonan R. Antioxidant defense and aging in C. elegans Is the oxidative damage theory of aging wrong? Cell Cycle. 2009;8:1681–7. doi: 10.4161/cc.8.11.8595. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Gong PF, Cederbaum AI. Nrf2 is increased by CYP2E1 in rodent liver and HepG2 cells and protects against oxidative stress caused by CYP2E1. Hepatol. 2006;43:144–53. doi: 10.1002/hep.21004. [DOI] [PubMed] [Google Scholar]
- Gregersen N, Bross P, Vang S, Christensen JH. Protein misfolding and human disease. Ann Rev Gen Human Genetics. 2006;7:103–24. doi: 10.1146/annurev.genom.7.080505.115737. [DOI] [PubMed] [Google Scholar]
- Grune T. Oxidative stress, aging and the proteasomal system. Biogeron. 2000;1:31–40. doi: 10.1023/a:1010037908060. [DOI] [PubMed] [Google Scholar]
- Grune T, Davies KJA. Breakdown of oxidized proteins as a part of secondary antioxidant defenses in mammalian cells. Biofactors. 1997;6:165–72. doi: 10.1002/biof.5520060210. [DOI] [PubMed] [Google Scholar]
- Grune T, Jung T, Merker K, Davies KJA. Decreased proteolysis caused by protein aggregates, inclusion bodies, plaques, lipofuscin, ceroid, and ‘aggresomes’ during oxidative stress, aging, and disease. Int J Biochem Cell Biol. 2004;36:2519–30. doi: 10.1016/j.biocel.2004.04.020. [DOI] [PubMed] [Google Scholar]
- Hamilton ML, Guo Z, Fuller CD, Van Remmen H, Ward WF, Austad SN, Troyer DA, Thompson I, Richardson A. A reliable assessment of 8-oxo-2-deoxyguanosine levels in nuclear and mitochondrial DNA using the sodium iodide method to isolate DNA. Nucleic Acids Res. 2001;29:2117–26. doi: 10.1093/nar/29.10.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JM, Salmon AB, Chang YY, Bonkowski M, Bartke A, Miller RA. Stress resistance and aging: influence of genes and nutrition. Mech Ageing Dev. 2006;127:687–94. doi: 10.1016/j.mad.2006.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JM, Salmon AB, Leiser SF, Galecki AT, Miller RA. Skin-derived fibroblasts from long-lived species are resistant to some, but not all, lethal stresses and to the mitochondrial inhibitor rotenone. Aging Cell. 2007;6:1–13. doi: 10.1111/j.1474-9726.2006.00255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Ann Rev Pharm Tox. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- Hayes JD, McMahon M. Molecular basis for the contribution of the antioxidant responsive element to cancer chemoprevention. Cancer Lett. 2001;174:103–13. doi: 10.1016/s0304-3835(01)00695-4. [DOI] [PubMed] [Google Scholar]
- Hayes JD, McMahon M. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem Sci. 2009;34:176–88. doi: 10.1016/j.tibs.2008.12.008. [DOI] [PubMed] [Google Scholar]
- Hayes JD, Strange RC. Invited commentary potential contribution of the glutathione-S-transferase supergene family to resistance to oxidative stress. Free Rad Res. 1995;22:193–207. doi: 10.3109/10715769509147539. [DOI] [PubMed] [Google Scholar]
- Hayflick L. Entropy explains aging, genetic determinism explains longevity, and undefined terminology explains misunderstanding both. PLOS Genet. 2007;3:2351–4. doi: 10.1371/journal.pgen.0030220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herczenik E, Gebbink M. Molecular and cellular aspects of protein misfolding and disease. FASEB J. 2008;22:2115–33. doi: 10.1096/fj.07-099671. [DOI] [PubMed] [Google Scholar]
- Higgins LG, Kelleher MO, Eggleston IM, Itoh K, Yamamato M, Hayes JD. Transcription factor Nrf2 mediates an adaptive response to sulforaphane that protects fibroblasts in vitro against the cytotoxic effects of electrophiles, peroxides and redox-cycling agents. Tox App Pharm. 2009;237:267–80. doi: 10.1016/j.taap.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Hirota A, Kawachi Y, Itoh K, Nakamura Y, Xu XZ, Banno T, Takahashi T, Yamamoto M, Otsuka F. Ultraviolet A irradiation induces NF-E2-related factor 2 activation in dermal fibroblasts: protective role in UVA-induced apoptosis. J Invest Derm. 2005;124:825–32. doi: 10.1111/j.0022-202X.2005.23670.x. [DOI] [PubMed] [Google Scholar]
- Howes RM. The free radical fantasy: a panoply of paradoxes. Ann N Y Acad Sci. 2006;1067:22–6. doi: 10.1196/annals.1354.004. [DOI] [PubMed] [Google Scholar]
- Huang HC, Nguyen T, Pickett CB. Regulation of the antioxidant response element by protein kinase C-mediated phosphorylation of NF-E2-related factor 2. Proc Natl Acad Sci USA. 2000;97:12475–80. doi: 10.1073/pnas.220418997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulbert AJ, Paplona R, Buffenstein R, Buttemer WA. Life and death: Metabolic rate, membrane composition, and life span of animals. Physiol Rev. 2007;87:1175–213. doi: 10.1152/physrev.00047.2006. [DOI] [PubMed] [Google Scholar]
- Iida K, Itoh K, Kumagai Y, Oyasu R, Hattori K, Kawai K, Shimazui T, Akaza H, Yamamoto M. Nrf2 is essential for the chemopreventive efficacy of oltipraz against urinary bladder carcinogenesis. Cancer Res. 2004;64:6424–31. doi: 10.1158/0008-5472.CAN-04-1906. [DOI] [PubMed] [Google Scholar]
- Jasper H. SKNy worms and long life. Cell. 2008;132:915–6. doi: 10.1016/j.cell.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Jeong WK, Hu R, Kong AN. Modulatory properties of various natural chemopreventive agents on the activation of NF-ka. Pharm Res. 2004;21:661–70. doi: 10.1023/b:pham.0000022413.43212.cf. [DOI] [PubMed] [Google Scholar]
- Jin W, Wang H, Yan W, Xu L, Wang X, Zhao X, Yang X, Chen G, Ji Y. Disruption of Nrf2 enhances upregulation of nuclear factor-kappaB activity, proinflammatory cytokines, and intercellular adhesion molecule-1 in the brain after traumatic brain injury. Mediators Inflamm. 2008;2008:725174. doi: 10.1155/2008/725174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Wang HD, Ji Y, Zhu L, Yan W, Qiao L, Yin HX. Genetic ablation of Nrf2 enhances susceptibility to acute lung injury after traumatic brain injury in mice. Exp Bio and Med. 2009;234:181–9. doi: 10.3181/0807-RM-232. [DOI] [PubMed] [Google Scholar]
- Johnson JA, Johnson DA, Kraft AD, Calkins MJ, Jakel RJ, Vargas MR, Chen PC. The Nrf2-ARE pathway an indicator and modulator of oxidative stress in neurodegeneration. Ann NY Acad Sci. 2008;1147:61–9. doi: 10.1196/annals.1427.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DP. Radical-free biology of oxidative stress. AJP. 2008;295:C849–68. doi: 10.1152/ajpcell.00283.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapahi P, Boulton ME, Kirkwood TBL. Positive correlation between mammalian life span and cellular resistance to stress. Free Rad Bio Med. 1999;26:495–500. doi: 10.1016/s0891-5849(98)00323-2. [DOI] [PubMed] [Google Scholar]
- Kapeta S, Chondrogianni N, Gonos ES. Nuclear erythroid factor 2 (Nrf2) mediated proteasome activation delays senescence in human fibroblasts. J Biol Chem. 2010;285:8171–84. doi: 10.1074/jbc.M109.031575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensler TW, Wakabayash N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Ann Rev of Pharm Tox. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- Khor TO, et al. Increased susceptibility of nrf2 knockout mice to colitis-associated colorectal cancer. Cancer Prev Res. 2008;1:187–91. doi: 10.1158/1940-6207.CAPR-08-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Cha YN, Surh YJ. A protective role of nuclear factor-erythroid 2-related factor-2 (Nrf2) in inflammatory disorders. Mutat Res. 2009 doi: 10.1016/j.mrfmmm.2009.09.007. Epub ahead of print: September 30. PMID:19799917. [DOI] [PubMed] [Google Scholar]
- Kirkwood TBL. A systematic look at an old problem. Nature. 2008;451:644–7. doi: 10.1038/451644a. [DOI] [PubMed] [Google Scholar]
- Kirkwood TL, Kapahi P, Shanley DP. Evolution, stress, and longevity. J Anat. 2000;197(Pt 4):587–90. doi: 10.1046/j.1469-7580.2000.19740587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korashy HM, El-Kadi AOS. Transcriptional regulation of the NAD(P)H: quinone oxidoreductase 1 and glutathione S-transferase Ya genes by mercury, lead, and copper. Drug Metab Dispos. 2006;34:152–65. doi: 10.1124/dmd.105.005397. [DOI] [PubMed] [Google Scholar]
- Kraft AD, Johnson DA, Johnson JA. Nuclear factor E2-related factor 2-dependent antioxidant response element activation by tert-butylhydroquinone and sulforaphane occurring preferentially in astrocytes conditions neurons against oxidative insult. J Neurosci. 2004;24:1101–12. doi: 10.1523/JNEUROSCI.3817-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kregel KC, Zhang HJ. An integrated view of oxidative stress in aging: basic mechanisms, functional effects, and pathological considerations. AJP. 2007;292:R18–36. doi: 10.1152/ajpregu.00327.2006. [DOI] [PubMed] [Google Scholar]
- Kwak MK, Itoh K, Yamamoto M, Sutter TR, Kensler TW. Role of transcription factor Nrf2 in the induction of hepatic phase 2 and antioxidative enzymes in vivo by the cancer chemoprotective agent, 3H-1,2-dithiole-3-thione. Mol Med. 2001;7:135–45. [PMC free article] [PubMed] [Google Scholar]
- Kwak MK, Wakabayashi N, Itoh K, Motohashi H, Yamamoto M, Kensler TW. Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway–identification of novel gene clusters for cell survival. J Biol Chem. 2003;278:8135–45. doi: 10.1074/jbc.M211898200. [DOI] [PubMed] [Google Scholar]
- Labinskyy N, Csiszar A, Orosz Z, Smith K, Rivera A, Buffenstein R, Ungvari Z. Comparison of endothelial function, O(2)− and H2O2 production, and vascular oxidative stress resistance between the longest-living rodent, the naked mole rat, and mice. AJP. 2006;291:H2698–704. doi: 10.1152/ajpheart.00534.2006. [DOI] [PubMed] [Google Scholar]
- Lane DP. Cancer and p53, guardian of the genome. Nature. 1992;358:15–6. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- Lapointe J, Hekimi S. When a theory of aging ages badly. Cell Mol Life Sci. 2010;67:1–8. doi: 10.1007/s00018-009-0138-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau AV, Sun Z, Wong PK, Zhang DD. Dual roles of Nrf2 in cancer. Pharm Res. 2008;58:262–70. doi: 10.1016/j.phrs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Calkins MJ, Chan KM, Kan YW, Johnson JA. Identification of the NF-E2-related factor-2-dependent genes conferring protection against oxidative stress in primary cortical astrocytes using oligonucleotide microarray analysis. J Biol Chem. 2003;278:12029–38. doi: 10.1074/jbc.M211558200. [DOI] [PubMed] [Google Scholar]
- Lee JS, Surh YJ. Nrf2 as a novel molecular target for chemoprevention. Cancer Let. 2005;224:171–84. doi: 10.1016/j.canlet.2004.09.042. [DOI] [PubMed] [Google Scholar]
- Leung L, Kwong M, Hou S, Lee C, Chan JY. Deficiency of the Nrf1 and Nrf2 transcription factors results in early embryonic lethality and severe oxidative stress. J Biol Chem. 2003;278:48021–9. doi: 10.1074/jbc.M308439200. [DOI] [PubMed] [Google Scholar]
- Liu J, Shen X. NF-kappa B/p65 antagonizes Nrf2-ARE pathway by depriving CBP from Nrf2 and facilitating recruitment of HDAC3 to MafK. Biochem Biophys Acta Mol Cell Res. 2008;1783:713–27. doi: 10.1016/j.bbamcr.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Liu XM, Peyton KJ, Ensenat D, Wang H, Hannink M, Alam J, Durante W. Nitric oxide stimulates heme oxygenase-1 gene transcription via the Nrf2/ARE complex to promote vascular smooth muscle cell survival. Cardiovas Res. 2007;75:381–9. doi: 10.1016/j.cardiores.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher J, Yamamoto M. The rise of antioxidant signaling – the evolution and hormetic actions of Nrf2. Toxicol Appl Pharmacol. 2010;244:4–15. doi: 10.1016/j.taap.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Maher P. The effects of stress and aging on glutathione metabolism. Ageing Res Rev. 2005;4:288–314. doi: 10.1016/j.arr.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Malik AI, Storey KB. Activation of antioxidant defense during dehydration stress in the African clawed frog. Gene. 2009;442:99–107. doi: 10.1016/j.gene.2009.04.007. [DOI] [PubMed] [Google Scholar]
- Masoro EJ. The role of hormesis in life extension by dietary restriction. Interdiscip Top Gerontol. 2007;35:1–17. doi: 10.1159/000096552. [DOI] [PubMed] [Google Scholar]
- McElwee JJ, Schuster E, Blanc E, Thomas JH, Gems D. Shared transcriptional signature in Caenorhabditis elegans dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J Biol Chem. 2004;279:44533–43. doi: 10.1074/jbc.M406207200. [DOI] [PubMed] [Google Scholar]
- McLellan LI, Wolf CR. Glutathione and glutathione-dependent enzymes in cancer drug resistance. Drug Resistance Updates. 1999;2:153–64. doi: 10.1054/drup.1999.0083. [DOI] [PubMed] [Google Scholar]
- McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a “tethering” mechanism: a two-site interaction model for the Nrf2-Keap1 complex. J Biol Chem. 2006;281:24756–68. doi: 10.1074/jbc.M601119200. [DOI] [PubMed] [Google Scholar]
- McMahon M, Itoh K, Yamamoto M, Chanas SA, Henderson CJ, McLellan LI, Wolf CR, Cavin C, Hayes JD. The cap ‘n’ collar basic leucine zipper transcription factor Nrf2 (NF-E2 p45-related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer Res. 2001;61:3299–307. [PubMed] [Google Scholar]
- Meijerman I, Beijnen JH, Schellens JHM. Combined action and regulation of phase II enzymes and multidrug resistance proteins in multidrug resistance in cancer. Cancer Treatment Rev. 2008;34:505–20. doi: 10.1016/j.ctrv.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Meister A, Anderson ME. Glutathione. Annu Rev Biochem. 1983;52:711–60. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- Mele J, Van Remmen H, Vijg J, Richardson A. Characterization of transgenic mice that overexpress both copper zinc superoxide dismutase and catalase. Antiox Redox Sig. 2006;8:628–38. doi: 10.1089/ars.2006.8.628. [DOI] [PubMed] [Google Scholar]
- Morin P, Ni ZL, McMullen DC, Storey KB. Expression of Nrf2 and its downstream gene targets in hibernating 13-lined ground squirrels, Spermophilus tridecemlineatus. Mol Cell Biochem. 2008;312:121–9. doi: 10.1007/s11010-008-9727-3. [DOI] [PubMed] [Google Scholar]
- Motohashi H, O'Connor T, Katsuoka F, Engel JD, Yamamoto M. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene. 2002;294:1–12. doi: 10.1016/s0378-1119(02)00788-6. [DOI] [PubMed] [Google Scholar]
- Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends in Molec Med. 2004;10:549–57. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Muller FL, Lustgarten MS, Jang Y, Richardson A, Van Remmen H. Trends in oxidative aging theories. Free Rad Biology and Med. 2007;43:477–503. doi: 10.1016/j.freeradbiomed.2007.03.034. [DOI] [PubMed] [Google Scholar]
- Nair S, Li WG, Kong ANT. Natural dietary anti-cancer chemopreventive compounds: redox-mediated differential signaling mechanisms in cytoprotection of normal cells versus cytotoxicity in tumor cells. Acta Pharma Sinica. 2007;28:459–72. doi: 10.1111/j.1745-7254.2007.00549.x. [DOI] [PubMed] [Google Scholar]
- Nakaso K, Nakamura C, Sato H, Imamura K, Takeshima T, Nakashima K. Novel cytoprotective mechanism of anti-parkinsonian drug deprenyl: PI3K and Nrf2-derived induction of antioxidative proteins. Biochem Biophys Res Commun. 2006;339:915–22. doi: 10.1016/j.bbrc.2005.11.095. [DOI] [PubMed] [Google Scholar]
- Nioi P, Hayes JD. Contribution of NAD(P)H :quinone oxidoreductase 1 to protection against carcinogenesis, and regulation of its gene by the Nrf2 basic-region leucine zipper and the arylhydrocarbon receptor basic helix-loop-helix transcription factors. Mutat Res-Fund Molec Mech of Mutagen. 2004;555:149–71. doi: 10.1016/j.mrfmmm.2004.05.023. [DOI] [PubMed] [Google Scholar]
- O'Brien ML, Tew KD. Glutathione and related enzymes in multidrug resistance. Eur J Cancer. 1996;32A:967–78. doi: 10.1016/0959-8049(96)00051-2. [DOI] [PubMed] [Google Scholar]
- Ogburn CE, Austad SN, Holmes DJ, Kiklevich JV, Gollahon K, Rabinovitch PS, Martin GM. Cultured renal epithelial cells from birds and mice: enhanced resistance of avian cells to oxidative stress and DNA damage. J Geron A. 1998;53:B287–92. doi: 10.1093/gerona/53a.4.b287. [DOI] [PubMed] [Google Scholar]
- Ohtsuji M, Katsuoka F, Kobayashi A, Aburatani H, Hayes JD, Yamamoto M. Nrf1 and Nrf2 play distinct roles in activation of antioxidant response element-dependent genes. J Biol Chem. 2008;283:33554–62. doi: 10.1074/jbc.M804597200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr WC, Radyuk SN, Prabhudesai L, Toroser D, Benes JJ, Luchak JM, Mockett RJ, Rebrin I, Hubbard JG, Sohal RS. Overexpression of glutamate-cysteine ligase extends life span in Drosophila melanogaster. J Biol Chem. 2005;280:37331–8. doi: 10.1074/jbc.M508272200. [DOI] [PubMed] [Google Scholar]
- Osburn WO, Karim B, Dolan PM, Liu G, Yamamoto M, Huso DL, Kensler TW. Increased colonic inflammatory injury and formation of aberrant crypt foci in Nrf2-deficient mice upon dextran sulfate treatment. Int J Cancer. 2007;121:1883–91. doi: 10.1002/ijc.22943. [DOI] [PubMed] [Google Scholar]
- Osburn WO, Kensler TW. Nrf2 signaling: an adaptive response pathway for protection against environmental toxic insults. Mutat Res. 2008;659:31–9. doi: 10.1016/j.mrrev.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamplona G, Barja G. Highly resistant macromolecular components and low rate of generation of endogenous damage: two key traits of longevity. Ageing Res Rev. 2007;6:189–210. doi: 10.1016/j.arr.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Patil L, Cullaro G, Gotlinger KH, Dunn MW, Schwartzman ML. Heme oxygenase-1 induction attenuates corneal inflammation and accelerates wound healing after epithelial injury. Invest Opthal and Vis Sci. 2008;49:3379–86. doi: 10.1167/iovs.07-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson KJ, et al. Nrf2 mediates cancer protection but not prolongevity induced by caloric restriction. Proc Natl Acad Sci USA. 2008;105:2325–30. doi: 10.1073/pnas.0712162105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez V, et al. Protein stability and resistance to oxidative stress are determinants of longevity in the longest-living rodent, the naked mole-rat. Proc Natl Acad Sci USA. 2009;106:3059–64. doi: 10.1073/pnas.0809620106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Campo RL, Rojas C, Cadenas S, Barja G. Comparative-study of free-radicals in vertebrates. 1 Antioxidant enzymes. Comp Biochem and Physio B. 1993;105:749–55. doi: 10.1016/0305-0491(93)90116-m. [DOI] [PubMed] [Google Scholar]
- Piantadosi CA, Carraway MS, Babiker A, Suliman HB. Heme oxygenase-1 regulates cardiac mitochondrial biogenesis via nrf2-mediated transcriptional control of nuclear respiratory factor-1. Circ Res. 2008;103:1232–60. doi: 10.1161/01.RES.0000338597.71702.ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao CS GS, Lee GH, Kim DS, Park BH, Chae SW, Chae HJ, Kim SH. Sulforaphane protects ischemic injury of hearts through antioxidant pathway and mitochondrial K(ATP) channels. Pharmacol Res. 2010;61:342–8. doi: 10.1016/j.phrs.2009.11.009. [DOI] [PubMed] [Google Scholar]
- Ramos-Gomez M, Kwak MK, Dolan PM, Itoh K, Yamamoto M, Talalay P, Kensler TW. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc Natl Acad Sci USA. 2001;98:3410–5. doi: 10.1073/pnas.051618798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangasamy T, Cho CY, Thimmulappa RK, Zhen L, Srisuma SS, Kensler TW, Yamamoto M, Petrache I, Tuder RM, Biswal S. Genetic ablation of Nrf2 enhances susceptibility to cigarette smoke-induced emphysema in mice. J Clin Invest. 2004;114:1248–59. doi: 10.1172/JCI21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangasamy T, et al. Disruption of Nrf2 enhances susceptibility to severe airway inflammation and asthma in mice. J Exp Med. 2005;202:47–59. doi: 10.1084/jem.20050538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattan SIS. Theories of biological aging: genes, proteins, and free radicals. Free Rad Res. 2006;40:1230–8. doi: 10.1080/10715760600911303. [DOI] [PubMed] [Google Scholar]
- Reddy NM, Kleeberger SR, Cho HY, Yamamoto M, Kensler TW, Biswal S, Reddy SP. Deficiency in Nrf2-GSH signaling impairs type II cell growth and enhances sensitivity to oxidants. Am J Respir Cell Mol Biol. 2007;37:3–8. doi: 10.1165/rcmb.2007-0004RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo AI, Innamorato NG, Martín-Moreno AM, De Ceballos ML, Yamamoto M, Cuadrado A. Nrf2 regulates migroglial dynamics and neuroinflammation in experimental Parkinson's disease. Glia. 2010;58:588–98. doi: 10.1002/glia.20947. [DOI] [PubMed] [Google Scholar]
- Sacher GA. Relation of lifespan to brain weight and body weight in mammals. Ciba foundation Colloquia on Ageing. 1959;5:115–16. [Google Scholar]
- Salmon AB, Akha AAS, Buffenstein R, Miller RA. Fibroblasts from naked mole-rats are resistant to multiple forms of cell injury, but sensitive to peroxide, ultraviolet light, and endoplasmic reticulum stress. J Geron A. 2008;63:232–41. doi: 10.1093/gerona/63.3.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon AB, Leonard S, Masamsetti V, Pierce A, Podlutsky AJ, Podlutskaya N, Richardson A, Austad SN, Chaudhuri AR. The long lifespan of two bat species is correlated with resistance to protein oxidation and enhanced protein homeostasis. FASEB J. 2009a;23:2317–26. doi: 10.1096/fj.08-122523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon AB, Murakami S, Bartke A, Kopchick J, Yasumura K, Miller RA. Fibroblast cell lines from young adult mice of long-lived mutant strains are resistant to multiple forms of stress. AJP. 2005;289:E23–9. doi: 10.1152/ajpendo.00575.2004. [DOI] [PubMed] [Google Scholar]
- Salmon AB, Perez VI, Bokov A, Jernigan A, Kim G, Zhao H, Levine RL, Richardson A. Lack of methionine sulfoxide reductase A in mice increases sensitivity to oxidative stress but does not diminish life span. FASEB J. 2009b;23:3601–8. doi: 10.1096/fj.08-127415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaranarayanan K, Jaiswal AK. Nrf3 negatively regulates antioxidant-response element-mediated expression and antioxidant induction of NAD(P)H :quinone oxidoreductase1 gene. J Biol Chem. 2004;279:50810–7. doi: 10.1074/jbc.M404984200. [DOI] [PubMed] [Google Scholar]
- Sanz A, Pamplona R, Barja G. Is the mitochondrial free radical theory of aging intact? Antiox Redox Sig. 2006;8:582–99. doi: 10.1089/ars.2006.8.582. [DOI] [PubMed] [Google Scholar]
- Sears RN. Signaling networks that link cell proliferation and cell fate. J Biol Chem. 2002;277:11617–20. doi: 10.1074/jbc.R100063200. [DOI] [PubMed] [Google Scholar]
- Shen Y. p53-dependent apoptosis pathways. Adv Cancer Res. 2001;82:55–84. doi: 10.1016/s0065-230x(01)82002-9. [DOI] [PubMed] [Google Scholar]
- Shih AY, Li P, Murphy TH. A small-molecule-inducible Nrf2-mediated antioxidant response provides effective prophylaxis against cerebral ischemia in vivo. J Neurosci. 2005;25:10321–35. doi: 10.1523/JNEUROSCI.4014-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shringarpure R, Grune T, Davies KJA. Protein oxidation and 20S proteasome-dependent proteolysis in mammalian cells. Cell Mol Life Sci. 2001;58:1442–50. doi: 10.1007/PL00000787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal RA, Dubey A, Orr WC. Protein oxidative damage is associated with life expectancy of housefiles. Proc Natl Acad Sci USA. 1993;90:7255–9. doi: 10.1073/pnas.90.15.7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal RS, Mockett RJ, Orr WC. Mechanisms of aging: an appraisal of the oxidative stress hypothesis. Free Rad Biol Med. 2002;33:575–86. doi: 10.1016/s0891-5849(02)00886-9. [DOI] [PubMed] [Google Scholar]
- Srisook K, Kim C, Cha YN. Molecular mechanisms involved in enhancing HO-1 expression: de-repression by heme and activation by Nrf2, the “one-two” punch. Antiox Redox Sig. 2005;7:1674–87. doi: 10.1089/ars.2005.7.1674. [DOI] [PubMed] [Google Scholar]
- Suh JH, Shenvi SV, Dixon BM, Liu HL, Jaiswal AK, Liu RM, Hagen TM. Decline in transcriptional activity of Nrf2 causes age-related loss of glutathione synthesis, which is reversible with lipoic acid. Proc Natl Acad Sci USA. 2004;101:3381–6. doi: 10.1073/pnas.0400282101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykiotis GP, Bohmann D. Keapl/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev Cell. 2008;14:76–85. doi: 10.1016/j.devcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimmulappa RK, Lee H, Rangasamy T, Reddy SP, Yamamoto M, Kensler TW, Biswal S. Nrf2 is a critical regulator of the innate immune response and survival during experimental sepsis. J Clin Invest. 2006a;116:984–95. doi: 10.1172/JCI25790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimmulappa RK, Mai KH, Srisuma S, Kensler TW, Yamamato M, Biswal S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62:5196–203. [PubMed] [Google Scholar]
- Thimmulappa RK, Scollick C, Traore K, Yates M, Trush MA, Liby KT, Sporn MB, Yamamoto M, Kensler TW, Biswal S. Nrf2-dependent protection from LPS induced inflammatory response and mortality by CDDO-Imidazolide. Biochem Biophys Res Commun. 2006b;351:883–9. doi: 10.1016/j.bbrc.2006.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong KI, Kobayashi A, Katsuoka F, Yamamoto M. Two-site substrate recognition model for the Keap1-Nrf2 system: a hinge and latch mechanism. Biol Chem. 2006;387:1311–20. doi: 10.1515/BC.2006.164. [DOI] [PubMed] [Google Scholar]
- Tullet JMA, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, Blackwell TK. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–38. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungvari Z, Buffenstein R, Austad SN, Podlutsky A, Kaley G, Csiszar A. Oxidative stress in vascular senescence: lessons from successfully aging species. Front Biosci. 2008;13:5056–70. doi: 10.2741/3064. [DOI] [PubMed] [Google Scholar]
- Van Remmen H, et al. Life-long reduction in MnSOD activity results in increased DNA damage and higher incidence of cancer but does not accelerate aging. Physiol Genom. 2003;16:29–37. doi: 10.1152/physiolgenomics.00122.2003. [DOI] [PubMed] [Google Scholar]
- Vargas MR, Pehar M, Cassina P, Beckman JS, Barbeito L. Increased glutathione biosynthesis by Nrf2 activation in astrocytes prevents p75(NTR)-dependent motor neuron apoptosis. J Neurochem. 2006;97:687–96. doi: 10.1111/j.1471-4159.2006.03742.x. [DOI] [PubMed] [Google Scholar]
- Villeneuve NF, Sun Z, Chen WM, Zhang DD. Nrf2 and p21 regulate the fine balance between life and death by controlling ROS levels. Cell Cycle. 2009;8:3255–6. doi: 10.4161/cc.8.20.9565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ, et al. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis. 2008;29:1235–43. doi: 10.1093/carcin/bgn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm Filho D, Althoff SL, Dafre AL, Boveris A. Antioxidant defenses, longevity and ecophysiology of South American bats. Comp Biochem Physiol C. 2007;146:214–20. doi: 10.1016/j.cbpc.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Wu H, McBride TJ, Isanhart JP, Cox SB, Hooper MJ. Responses of glutamate cysteine ligase and glutathione to oxidants in deer mice (Peromyscus maniculatus) Ecotoxicol Environ Saf. 2009;72:1572–8. doi: 10.1016/j.ecoenv.2009.02.008. [DOI] [PubMed] [Google Scholar]
- Xu CJ, Huang MT, Shen GX, Yuan XL, Lin W, Khor TO, Conney AH, Kong ANT. Inhibition of 7,12-dimethylbenz(a)anthracene-induced skin tumorigenesis in C57BL/6 mice by sulforaphane is mediated by nuclear factor E2-related factor 2. Cancer Res. 2006;66:8293–6. doi: 10.1158/0008-5472.CAN-06-0300. [DOI] [PubMed] [Google Scholar]
- Yanagawa T, Itoh K, Uwayama J, Shibata Y, Yamaguchi A, Sano T, Ishii T, Yoshida H, Yamamoto M. Nrf2 deficiency causes tooth decolourization due to iron transport disorder in enamel organ. Genes Cells. 2004;9:641–51. doi: 10.1111/j.1356-9597.2004.00753.x. [DOI] [PubMed] [Google Scholar]
- Zakkar M, et al. Activation of Nrf2 in endothelial cells protects arteries from exhibiting a proinflammatory state. Art Thromb Vasc Biol. 2009;29:1851–7. doi: 10.1161/ATVBAHA.109.193375. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Crouch DH, Yamamoto M, Hayes JD. Negative regulation of the Nrf1 transcription factor by its N-terminal domain is independent of Keap1: Nrf1, but not Nrf2, is targeted to the endoplasmic reticulum. Biochem J. 2006;399:373–85. doi: 10.1042/BJ20060725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kobayashi A, Yamamoto M, Hayes JD. The Nrf3 transcription factor is a membrane-bound glycoprotein targeted to the endoplasmic reticulum through its N-terminal homology box 1 sequence. J Biol chem. 2009;284:3195–210. doi: 10.1074/jbc.M805337200. [DOI] [PubMed] [Google Scholar]
- Zhu H, Jia ZQ, Misra BR, Zhang L, Cao ZX, Yamamoto M, Trush MA, Misra HP, Li YB. Nuclear factor E2-related factor 2-dependent myocardiac cytoprotection against oxidative and electrophilic stress. Cardiovas Toxicol. 2008;8:71–85. doi: 10.1007/s12012-008-9016-0. [DOI] [PubMed] [Google Scholar]