Abstract
There has been considerable interest in developing new therapies with adult multipotent progenitor stromal cells or mesenchymal stem cells (MSCs) in organ replacement and repair. To be effectively seeded into scaffolds for therapy, large numbers of cells are needed, but concerns remain regarding their chromatin stability in long-term culture. We therefore expanded four donors of human MSCs (hMSCs) from bone marrow aspirates with a protocol that maintains the cells at low density. MSCs initially proliferated at average doubling times of 24 h and then gradually reached senescence after 8–15 passages (33–55 population doublings) without evidence of immortalization. Comparative genomic hybridization assays of two preparations revealed no abnormalities through 33 population doublings. One preparation had a small amplification of unknown significance in chromosome 7 (7q21:11) after 55 population doublings. Microarray assays demonstrated progressive changes in the transcriptome of the cells. However, the transcriptomes clustered more closely over time within a single passage, rather than with passage number, indicating a partial reversibility of the patterns of gene expression. One of the largest changes was a decrease in mRNA for Sox11, a transcription factor previously identified in neural progenitor cells. Knockdown of Sox11 with siRNA decreased the proliferation and osteogenic differentiation potential of hMSCs. The results suggested that assays for Sox11 may provide a biomarker for early progenitor hMSCs.
Introduction
There has been considerable interest in developing new therapies with the adult stem/progenitor cells, referred to as mesenchymal stem cells or multipotent mesenchymal stromal cells (MSCs). The cells are relatively easy to isolate from bone marrow and several other tissues, and they have the potential for multilineage differentiation in vitro and in vivo.1 The ability to proliferate and generate large quantities of cells in vitro is especially of interest for clinical therapies utilizing the cells alone or seeded on biomimetic scaffolds, most of which require large numbers of cells. A series of publications demonstrated that like most healthy cells from human adult tissues, human MSCs (hMSCs) gradually become senescent as they are expanded in culture.2–4 Several recent reports, however, raised the concern that hMSCs expanded in culture may emerge from senescence and generate cells that are both immortal in culture and tumorigenic in patients.3,5–11 We speculated that culture methodology could play a significant role in the putative tumorigenicity of expanded hMSC cultures. In particular, exposure to high density can substantially and irreversibly alter the characteristics of hMSCs12,13 much as high-density cultures predispose mouse fibroblasts to spontaneous transformation.14
To reexamine changes in hMSCs during expansion in culture, we serially expanded the cells from four preparations of bone marrow in low-density cultures until they reached senescence and characterized the cells in terms of their genomic stability and transcriptome profiles. After substantial expansion in low-density conditions, none of the cultures emerged from senescence, indicating that there was no evidence of immortalization of the cells. Genomic hybridization assays, as the cells approached senescence, detected no chromosomal abnormalities but for one preparation propagated to 55 doublings that contained one minor abnormality of unknown significance. Transcriptome analysis demonstrated that most of the changes occurred within the duration of a single passage rather than as function of expansion. The changes within a passage were partially reversed upon reseeding at low density. A noteworthy exception was a decrease in the mRNA for Sox11 with population doublings. Knockdown of Sox11 with an siRNA decreased both the rate of proliferation and the osteogenic differentiation potential of early passage MSCs.
Materials and Methods
Isolation and culture of hMSCs
hMSCs were obtained from the National Institutes of Health-sponsored Center for the Preparation and Distribution of Adult Stem Cells (www.som.tulane.edu/gene_therapy/distribute.shtml; currently available from msc@medicine.tamhsc.edu). The hMSCs were isolated from 1 to 4 mL of bone marrow aspirates taken from the left and right iliac crest of normal adult donors under local anesthesia and after informed consent under a protocol approved by an Institutional Review Board. Cells from eight separate preparations of bone marrow from eight donors of bone marrow aspirates were used. They were defined by the anonymous numbers assigned to the donor and whether obtained from the right or left iliac crest as donor preparations 89R (male), 220R (male), 240L (male), 260R (female), 281L (male), 5064L (male), 7064L (male), and 7068L (male). Nucleated cells were isolated with a density gradient (Ficoll-Paque; Pharmacia) and resuspended in a complete culture medium (CCM) containing alpha minimal essential medium (GIBCO/BRL), 17% fetal bovine serum lot selected for rapid growth of MSCs (Atlanta Biologicals, Inc.), 100 units/mL penicillin, 100 μg/mL streptomycin, and 2 mM L-glutamine (GIBCO/BRL). All of the nucleated cells (15 to 100 million) were plated in 20 mL CCM in a 175 cm2 culture dish (Nunc) and incubated at 37°C with 5% CO2. After 24 h, nonadherent cells were discarded, and adherent cells were thoroughly washed twice with phosphate-buffered saline (PBS). The cells were incubated in the CCM for 4 to 11 days until ∼70% confluent, harvested with 0.25% trypsin and 1 mM ethylenediaminetetraacetic acid (EDTA) for 5 min at 37°C, and replated at 50 cells/cm2 in the CCM in an intercommunicating system of culture flasks (6300 cm2; Cell Factory; Nunc). The cells were incubated 7 to 10 days until ∼70% confluent, harvested with trypsin/EDTA, suspended at 1 × 106 cells/mL in 5% dimethylsulfoxide and 30% fetal bovine serum, frozen in 1 mL aliquots at −80°C overnight, and stored in liquid nitrogen (passage 1 cells).
To expand the MSCs, a frozen vial was thawed, plated in a 57 cm2 culture dish in the CCM, and incubated for 1 day, to recover viable adherent cells. MSCs were then replated and incubated in the CCM in an intercommunicating system of culture flasks (Cell Factory; Nunc) at 100 cells/cm2 or in culture dishes (Nunc) at 1–1000 cells/cm2. This culture was designated passage 2 (P2) MSCs. Cell morphology was observed, and recorded by photomicrographs with phase-contrast microscopy. For most experiments, the medium was replaced every 3 days, and the cells were serially passaged by lifting with trypsin/EDTA on day 7 and replating at 100 cells/cm2. MSCs for microarray assays were cultured by passaging every 7 days without medium change to eliminate the effect of serum activation within each passage.
Fluorescence-activated cell scanning
MSCs were detached with trypsin/EDTA, suspended in Hank's balanced salt solution (GIBCO/BRL), and assayed by flow cytometric analysis (Cytomics FC 500; Beckman Coulter). The gains and voltages on the photomultiplier tubes were adjusted and standardized daily to maintain constant mean channel. To characterize distinct cell fractions on the basis of forward scatter (FS) and side scatter (SS), we divided gated events into four user-defined quadrants.
Microarray sample preparation
Total RNA was extracted from MSCs (Ambion) and samples for microarrays were prepared according to manufacturer's directions. In brief, 5 μg of total RNA was used to synthesize double-stranded cDNA (Superscript Choice System; GIBCO/BRL). After synthesis, the double-stranded cDNA was purified by phenol/chloroform extraction (Phase Lock Gel; Eppendorf Scientific) and concentrated by ethanol precipitation. In vitro transcription was used to produce biotin-labeled cRNA (BioArray HighYield RNA Transcription Labeling Kit; Enzo Diagnostics). The biotinylated cRNA was then cleaned (RNAeasy Mini Kit; Qiagen), fragmented, and hybridized on the HG-U133 Plus 2.0 microarray chips (Affymetrix). The chips consisted of over 54,000 transcripts, representing over 31,000 human genes. After washing, individual microarray chips were stained with streptavidin-phycoerythrin (Molecular Probes), amplified with biotinylated anti-streptavidin (Vector Laboratories), and scanned for fluorescence (GeneChip Scanner 3000; Affymetrix) using the GeneChip Operating software 1.0 (GCOS; Affymetrix).
Microarray data processing
GCOS software provided the intensities for perfect match and mismatch oligonucleotides, and determined whether genes were present (P), marginal (M), or absent (A). The scanned images were then transferred to the dChip program.15,16 To allow comparisons between different microarrays, one array was chosen as the baseline array against which the other arrays were normalized at the probe intensity level. The dChip program then calculated the model-based expression values using the perfect matches and mismatches. Negative values were assigned a value of 1. Microarray data will be available at the Gene Expression Omnibus database.
Filtering of the data
To prepare for the clustering algorithm, three different protocols were used: (1) no filtering of the transcripts; (2) filtering for genes with a present call in at least 25% of the samples, and for genes with a value >0.4 for the coefficient of variation (standard deviation of expression value for each gene divided by the mean expression value for that gene across all the samples); (3) filtering by analysis of variance for genes differentially expressed between day 2 and 7 with a p-value <0.01 and retaining only the genes that were present in either all four day 2 or all four day 7 samples.
Hierarchical clustering algorithm in dChip
The dChip program was used to standardize the expression values for each gene by linearly adjusting their values across all samples to a mean of zero with a standard deviation of one. Individual genes were then clustered using an algorithm in dChip program that determined the correlation coefficients (r-values) for the normalized expression values (distances between genes were defined as 1 − r). Genes with the shortest distances between them were merged into super-genes, connected in a dendogram by branches with lengths proportional to their genetic distances, and then merged (centroid-linkage). This process was repeated n − 1 times until all genes had been clustered. A similar algorithm was also used to cluster the samples. The standardization and clustering methods follow published procedures.
Real-time reverse transcriptase polymerase chain reaction
To assay DNA damage signaling pathways, total RNA was extracted from MSCs (RNAqueous Kit; Ambion) and converted into first-strand cDNA (RT2 First Strand Kit; SuperArray). Then, the cDNA and RT2 quantitative polymerase chain reaction (PCR) Master Mix was placed into plates containing predispensed gene-specific primer sets (SuperArray). PCR was performed, and relative expression calculated.
For GAPDH (AssayID Hs00266705_g1), Sox4 (AssayID Hs00268388_s1), Sox11(AssayID Hs00846583_s1), and Sox12 (AssayID Hs00272869_s1) gene expression, total RNA was extracted (RNAqueous Kit; Ambion) and ∼100 ng was converted into first strand cDNA (SuperScript III FirstStrand Synthesis Kit; Invitrogen). Approximately 10 ng of cDNA per reaction was amplified by real-time reverse transcriptase (RT)-PCR using TaqMan (Universal PCR Master Mix and Inventoried Primer Probes; Applied Biosystems). For the assays, reactions were incubated at 50°C for 2 min, 95°C for 10 min, and then 40 cycles at 95°C for 15 s followed by 60°C for 1 min. Relative quantity was determined as quantity relative to GAPDH expression value.
Comparative genomic hybridization
Total DNA was isolated from MSCs at passage 2 and 8 from donor 260R and from passage 2, 8, and 12 from donor 281L (Puregene® DNA Purification Kit; Gentra). Proteins were digested with proteinase K solution and removed by salt precipitation. Genomic DNA was precipitated with graded alcohols. Samples were then shipped to the Vanderbilt Microarray Shared Resource Center at Vanderbilt University (Nashville, TN) where comparative genomic hybridization (cGH) was performed (Genome-Wide Human SNP Nsp/Sty 6.0 Kit; Affymetrix) with an array that contained 906,600 single-nucleotide polymorphism probes and 945,826 copy number probes. Passage 8 DNA from donor 260R was hybridized with passage 2 DNA from the same donor. Passage 8 and passage 12 DNA from donor 281L was hybridized separately with passage 8 and passage 12 DNA from the same donor. The data were analyzed by segmentation analysis (Partek Genomics Solution Software; Partek).
Transfection with siRNAi for Sox11
MSCs for transfection experiments were plated at 100 cells/cm2 in the CCM without antibiotics in six-well microtiter plates and incubated for 2 days to about 5% confluency. Cells were transfected with 10 nM siRNA for Sox11 or a high guan-osine cytosine content siRNA-negative control (Invitrogen) mixed with Lipofectamine RNAi MAX reagent (Invitrogen) in the presence of OPTI-minimal essential medium. After 4 h the medium was replaced with the CCM without antibiotics.
Assays for adipogenic and osteogenic differentiation
For adipogenic differentiation, MSCs at ∼70% confluence were incubated in the CCM supplemented with 0.5 μM isobutylmethylxanthine (Sigma-Aldrich), 50 μM indomethacin (Sigma-Aldrich), and 0.5 μM dexamethasone (DEX; R&D Systems) for up to 21 days. The medium was changed every 3 days.
For osteogenic differentiation, MSCs at ∼70% confluence were incubated in the CCM supplemented with 10 nM DEX (R&D Systems), 10 mM beta-glycerol phosphate (BGP; Sigma-Aldrich), and 50 μM L-ascorbic acid-2 phosphate (Sigma-Aldrich) for up to 21 days. To define optimal conditions, the concentrations of BGP and DEX were varied between 0–100 mM and 0–100 nM, respectively. The medium was changed every 3 days.
Quantitative assays of adipogenic and osteogenic differentiation
For the quantitative assays, Oil Red O stock solution (0.5%) was prepared by adding 2.5 g Oil Red O (Sigma-Aldrich) into 500 mL isopropyl alcohol. Fresh Oil Red O stain was prepared before each use by diluting three parts of stock with two parts PBS and filtering through 0.2 μm filter. Alizarin Red stain was prepared fresh before each use by adding 1 g Alizarin Red (Sigma-Aldrich) to 100 mL deionized water, mixing well, and filtering through a 0.2 μm filter.
Plates were washed three times with PBS and then fixed with 4% paraformaldehyde for 20 min at room temperature. After washing with PBS three times, Oil Red O or Alizarin Red stain was added to the plates and left for 5 min at room temperature. Plates were then thoroughly washed with deionized water and left to dry overnight. Samples were observed under light microscopy.
For quantitative assay of adipogenesis, 200 μL isopropyl alcohol was added to the Oil Red O-stained and dried plates. The samples were extracted for about 0.5 min. Absorbance of the extracted dye was then measured by plate reader (Bio-Rad) at 510 nm at 5 min after dilution to a linear range.
For quantitative assay of osteogenesis, cells stained with Alizarin Red (Sigma-Aldrich) were scraped from the plates, transferred to microcentrifuge tubes, and incubated at 85°C for 15 min in 1 mL of 10% (v/v) acetic acid overlaid with 0.5 mL of light mineral oil. The extraction was cooled on ice and then centrifuged at 21,000 g, and 0.5 mL of the supernatant was transferred to a fresh tube containing 10% (v/v) ammonium hydroxide. The solution was then transferred to a 96-well plate and absorbance read at 450 nm on a plate reader (Fluostar Omega; BMG Labtech).17 Kinetic assays for alkaline phosphatase were used for quantitative assays of the early stages of osteogenic commitment. For the determination of alkaline phosphatase activity, unstained and unfixed plates were washed with PBS twice followed by a one wash with reaction buffer containing 100 mM NaCl, 100 mM Tris-HCl, and 1 mM MgCl2 (Sigma-Aldrich) in deionized water at a final pH of 9.0. After aspirating final wash, 1 mL of reaction buffer was added to each of the six wells containing the intact monolayers. Then, 0.5 mL of 1-Step p-nitrophenyl phosphate disodium salt (PNPP; Pierce Biotechnology) was added to each well followed by immediate measurement by spectrophotometer. One-Step PNPP solution was maintained at 4°C until addition to the plates. The increase in absorbance at 405 nm, caused by the alkaline phosphatase-catalyzed conversion of PNPP to nitrophenolate, was measured every 30 s by plate reader (Fluostar Omega; BMG Labtech) for 15 min. The observed rate of reaction was calculated from the linear phase of the kinetic plot and employed as an arbitrary unit of alkaline phosphatase activity.18
Results
Proliferation and morphology of MSCs with passage
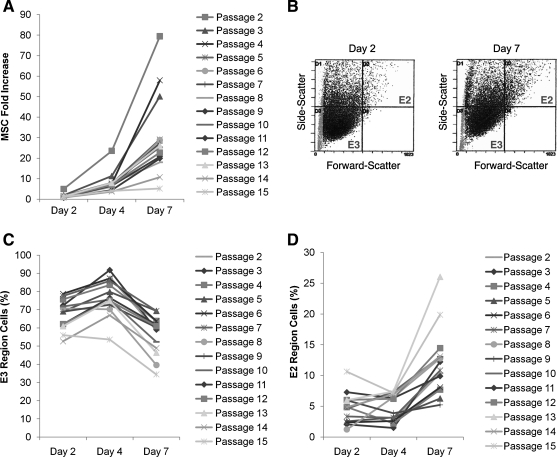
hMSCs from a frozen vial at passage 1 were plated at 100 cells/cm2, expanded for 7 days, and then repeatedly expanded by replating at 100 cells/cm2 and incubating for 7 days. Consistent with previous observations,19,20 the rates of propagation in each sub-culture reflected a lag period in that the cells expanded more slowly between days 2 and 4 than between days 4 and 7 (Fig. 1A). The rates of propagation progressively decreased as the cells were expanded. Also, consistent with previous observations,13 the cells in each sub-culture at day 2 were primarily small rapidly self-replicating MSCs, and the cells at day 7 were primarily larger slowly replicating MSCs (Fig. 1B–E).
FIG. 1.
Expansion of MSCs by replating at low density. MSCs from donor 5064Lp2 were plated at 100 cells/cm2 and replated at day 7 through 15 passages for RNA harvest and cell size determination. (A) MSC proliferation was greatest at early passages and decreased until senescence at passage 15. (B) Representative schema of MSCs analysis by flow cytometry for size and complexity. (C, D) The percentage of smaller, less complex cells (E3) and larger, more complex cells (E2) in culture was determined at days 2, 4, and 7 in each passage. Cell became larger with time in culture and then reverted back to being smaller size when replated at low density. MSCs, mesenchymal stem cells.
Senescence of MSCs
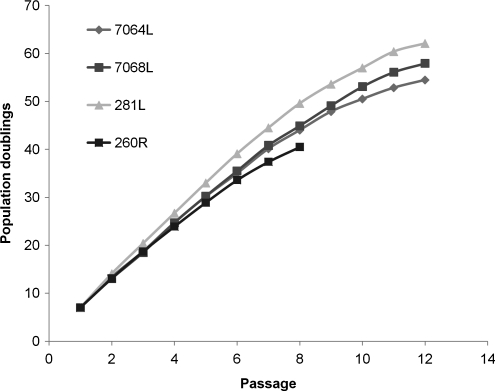
With repeated expansion, the cells entered into senescence (Fig. 2), with a preparation from one donor failing to expand significantly after passage 8 (∼33 population doublings) and three preparations from three donors failing to expand significantly after passage 12 (∼55 population doublings). The four samples were cultured for an additional 3 months with a change in the medium twice per week. The cells remained adherent and viable, but there was no further expansion or appearance of colonies that may have emerged from senescence.
FIG. 2.
Expansion of MSCs to senescence. The four donor preparations (7764L, 7068L, 281L, and 260R) were passed as indicated in Figure 1 until no further propagation of the cells was apparent. As indicated in text, the cells remained viable but there was no further propagation when each of the samples was cultured for an additional 3 months.
DNA damage signaling with passage
As one assay of genomic stability, real-time RT-PCR assays were performed for expression of 19 genes for pathways known to be involved in signaling of DNA damage. Gene expression fold changes were measured at passages 6 and 12 relative to passage 2 for donor 5064. The results indicated than none of the transcripts increased more than threefold (Supplemental Table S1, available online at www.liebertonline.com/ten).
Genomic stability of MSCs with passage
As a further assay of genomic stability, cells from early passage cultures and from cultures that had reached senescence were compared by cGH. Cultures from two separate donor preparations were employed for the assay. MSCs from one of the donor preparations (260R) reached senescence by passage 8 and failed to expand further (Fig. 2). MSCs from the second donor preparation (281L) expanded to passage 12 (∼62 population doublings). Hybridization of passage 8 DNA (about 33 to 42 population doublings) was compared with passage 2 DNA (∼14 population doublings) from the same two donor preparations. The data did not reveal any losses in heterozygosity or variations in copy number. However, an additional comparative hybridization from passage 12 DNA of donor preparation 281L with passage 2 DNA revealed an amplification of about 400 kb at 7q21.11 (Supplemental Fig. S1, available online at www.liebertonline.com/ten). The only gene known to be present in this locus encodes membrane-associated guanylate kinase, WW, and PDZ domain-containing protein 2, MAGI2. The gene interacts with Delta proteins in zebrafish,21 inhibits cell migration and proliferation via PTEN in human hepatocarcinoma cells,22 and has been associated with inflammatory bowel disease.23
Transcriptome changes in MSCs during passaging
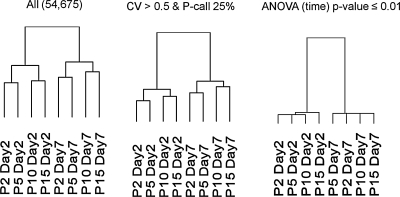
To determine the changes in transcriptome as the cells were expanded, microarray assays were performed on cells from day 2 to 7 cultures. Analysis of the data by hierarchical clustering before filtering revealed a pattern in which day 2 samples clustered together, and day 7 samples clustered together, regardless of population doublings and passage number (Fig. 3). The pattern was more pronounced when the data were filtered by removing genes not present on either day 2 or 7, or by removing genes not changing significantly when analyzed analysis of variance (p-values <0.01). As reported previously,24,25 the transcripts that exhibited the largest changes between day 2 and 7 of a single passage suggested a transition from proliferation to differentiation (Supplemental Table S2, available online at www.liebertonline.com/ten). The transition was observed with each replating of the cells at low density. Genes with the greatest fold changes with expansion are shown in Table 1. Among the genes that decreased with expansion of the cells were genes involved in both proliferation and differentiation. This observation is consistent with previously reported reductions in proliferation and potential to differentiate as the cells are expanded.2,26,27
FIG. 3.
Microarray analysis of MSCs during passage. Microarrays from MSCs at passages 2, 5, 10, and 15 in day 2 and 7 were analyzed by dChip software. Hierarchical clustering was performed without filtering, using all probeset data; filtering for probes present in at least 25% of samples and demonstrating significant changes; and by ANOVA. ANOVA, analysis of variance.
Table 1.
Microarray Data for Transcripts with Largest Changes in Signal Intensities with Expansion of the Cells
| |
Day 2 |
||
|---|---|---|---|
| Gene | P2 versus P5 | P2 versus P10 | P2 versus P15 |
| SRY sex determining region Y-box 11 (Sox11) | 1.2 | 8.4 | 61.8 |
| Proenkephalin | 1.2 | 4.0 | 29.2 |
| Elastase 3A, pancreatic | 1.5 | 3.9 | 27.7 |
| Claudin 4 | 1.8 | 1.5 | 17.6 |
| Neuritin 1 | 1.5 | 1.6 | 15.5 |
| CD24 molecule | 4.3 | 11.8 | 14.8 |
| Cytochrome c oxidase | −1.3 | −1.1 | 12.8 |
| Suppressor of cytokine signaling 2 | 3.4 | 2.9 | 12.4 |
| ADAM metallopeptidase | 2.1 | 7.6 | 11.7 |
| Pleckstrin | 3.5 | 1.7 | 10.6 |
| Fibroblast growth factor 7 | 2.4 | 5.6 | 8.2 |
| Hephaestin | 1.3 | 2.6 | 8.0 |
| Parathyroid hormone-like hormone | 1.5 | 2.1 | 7.8 |
| Suppressor of cytokine signaling 2 | 1.3 | 3.4 | 7.5 |
| Day 7 | |||
|---|---|---|---|
| Fibroblast growth factor 7 | 4.5 | 23.5 | 77.4 |
| Suppressor of cytokine signaling 2 | 6.4 | 11.2 | 64.4 |
| Proenkephalin | 1.9 | 11.0 | 57.4 |
| Fibroblast growth factor receptor 2 | 2.9 | 17.1 | 42.3 |
| SRY sex determining region Y-box 11 (Sox11) | 1.6 | 46.4 | 40.8 |
| Collagen, type XI, alpha 1 | 19.5 | 45.6 | 39.2 |
| Cytochrome c oxidase | 2.0 | 2.2 | 32.2 |
| Fibronectin type III | 10.4 | 47.4 | 29.3 |
| Hephaestin | 2.5 | 8.7 | 25.4 |
| Aggrecan 1 | 4.5 | 25.3 | 22.0 |
| Neuritin 1 | 1.1 | 1.5 | 19.1 |
Sox11 in proliferating and differentiating MSCs
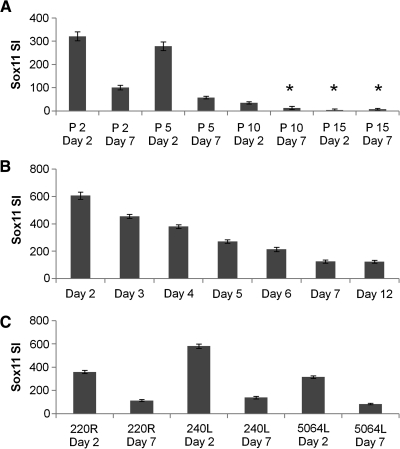
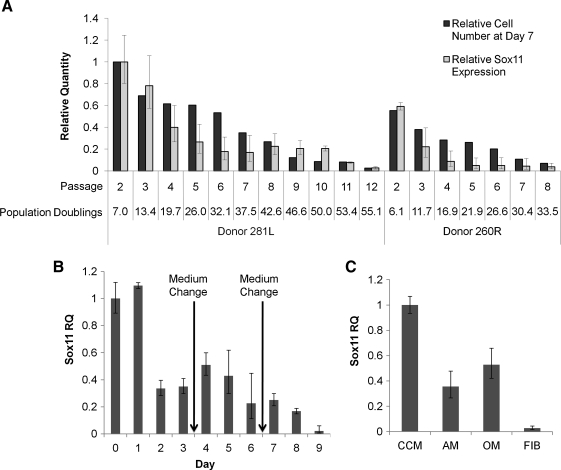
From our microarray results (Table 1), Sox11 was the only nuclear transcription factor detected that was highly downregulated with expansion of the cells. The expression levels decreased progressively with passage of the cells (Fig. 4). Expression decreased with time in culture within each passage (Fig. 4B, C) and increased with replating of the cells at low density (Fig. 4A).
FIG. 4.
Sox11 expression with expansion of MSCs. Sox11 expression signal intensity (SI) was measured by microarray in various samples. (A) MSCs from donor 5064Lp2 were plated at 100 cells/cm2 and replated at day 7 through 15 passages. (B) MSCs from donor 89Rp3 were plated at 100 cells/cm2 and cultured for 12 days without medium change. (C) MSCs from three donors were plated at 100 cells/cm2 and cultured for 7 days without medium change (mean ± standard error for multiple probes as calculated by dChip). Asterisks indicate absence of genes as calculated by dChip algorithm.
To confirm microarray data, two MSC donors were expanded until senescence and the relative quantity of Sox11 mRNA transcripts was determined by real-time RT-PCR. Sox11 expression decreased with each passage in parallel with the decrease in proliferation rate (Fig. 5A). To examine the effects of serum activation on expression of the gene, cells were plated at low density and the medium was changed without replating. Expression of the gene was increased slightly by changing the medium during a given passage of the cells (Fig. 5B) and it decreased 24 h after transfer of the cells to the adpiogenic or osteogenic medium (Fig. 5C). Sox11 transcription was not detectable in donor-matched fibroblast samples tested.
FIG. 5.
Real-time reverse transcriptase-polymerase chain reaction assays for Sox11 expression. (A) MSCs from donors 281Lp2 and 260Rp2 were plated at 100 cells/cm2 and replated at day 7 through 15 passages with medium changed on days 3 and 6. Cell number and RNA isolation were performed at each replating at day 7. (B) MSCs from donor 7068Lp2 were plated at 100 cells/cm2 and cultured for 9 days with medium changed at days 3 and 6. (C) MSCs from donor 7068Lp2 were plated at 100 cells/cm2 and cultured until 70% confluence at day 7 when the CCM, adipogenic medium (AM), or osteogenic medium (OM) was added. Twenty-four hours later RNA was isolated from cultures. Fibroblast (FIB) control cultures are also shown (mean ± 95% confidence intervals for triplicate assays). CCM, complete culture medium; RQ, relative quantity.
Sox11 knockdown in MSCs
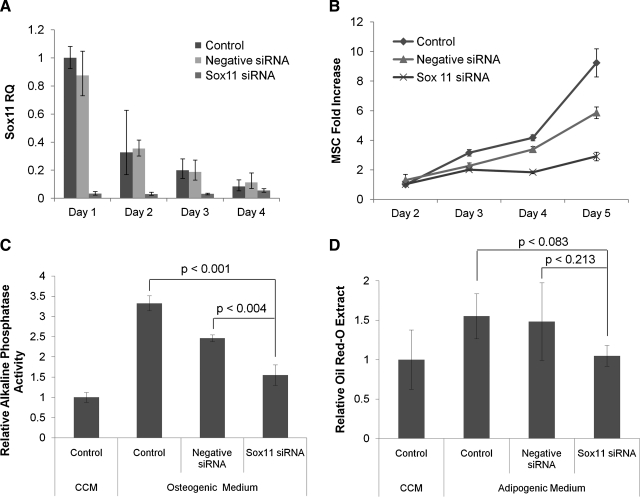
To examine the role of Sox11, MSCs were incubated for 2 days to ∼5% confluence and transfected with siRNA for Sox11 for 4 h. One day later, Sox11 expression was <5% of the control MSCs, but increased thereafter (Fig. 6A). Knockdown of Sox11 decreased the rate of proliferation by 70% after 5 days of the low-density culture, and by 60% after 4 days of higher density culture (Fig. 6B; Supplemental Fig. S2, available online at www.liebertonline.com/ten). The negative control siRNA decreased the rate of proliferation to a lesser extent.
FIG. 6.
Sox11 knockdown with an siRNA. Sox11 expression relative quantity was measured by real-time reverse transcriptase-polymerase chain reaction in MSCs tranfected with an siRNA for Sox11 or negative control sequences. (A) MSCs from donor 7027Lp2 were plated at 100 cells/cm2 and transfected 2 days later. Knockdown efficiency was >95% on day 1 after transfection and decreased thereafter. (B) MSC proliferation was decreased by the knockdown after 4 days. (C) MSCs from donor 7068Lp2 were plated at 100 cells/cm2 and cultured until 70% confluence at day 7 when the CCM or osteogenic medium was added. Relative alkaline phosphatase activity was measured when mineralized matrix began to appear at 6 days after differentiation. (D) MSCs from donor 7068Lp2 were plated at 100 cells/cm2 and cultured until 70% confluence at day 7 when the CCM or adipogenic medium was added. Relative Oil Red O extract was measured when lipid vacuoles began to appear at 9 days after differentiation. CCM, complete culture medium; RQ, relative quantity.
To assay the effects of the Sox11 knockdown on the traditional MSC differentiation lineages,28 we first optimized conditions for osteogenic differentiation of MSCs.18,29–33 As indicated in Supplemental Figure S3 (available online at www.liebertonline.com/ten), we found that concentrations of 10 mM BGP and 10 nM DEX were the most effective. The knockdown of Sox11 significantly decreased osteogenic differentiation as assayed by alkaline phosphatase activity and slightly reduced adipogenic differentiation (Fig. 6C, D).
Discussion
The results presented here demonstrated the changing profile of gene expression as hMSCs were expanded in low-density cultures. All four preparations examined here passed into senescence with expansion. There was no evidence of genomic instability, detected by assaying for expression of 19 genes involved in genomic signaling. Also, there was no evidence of genomic instability by cGH of two samples except for the difficult to interpret amplification of a 400 kb region on chromosome 7 in one preparation after over 50 population doublings. Therefore, the results are consistent with most,4,12,34–36 but not all, reports7,10,37,38 that the genomes of MSCs are stable as they are extensively expanded in culture. Microarray assays of the transcriptomes of the cells presented the unexpected result that day 2 samples clustered together and day 7 samples clustered together regardless of how extensively the cells were expanded. Therefore, the data confirmed and extended previous indications that the changes produced in the cells by expansion in culture are partially reversible if the cultures are lifted before they reach confluence and are replated at low density.39
The microarray data were queried for changes in the levels of gene expression that might be useful in distinguishing early progenitors from more mature MSCs.25,31–34 Significant decreases were observed in the transcripts for several genes, including CD24. However, CD24 was not useful under the conditions employed here because the epitope to the antibody tested (Clone ML5; Beckton Dickinson) was cleaved or internalized by the trypsinization used to lift the cells (unpublished results). The most informative marker detected was the transcript for the nuclear transcription factor Sox11, one of several Sox genes involved in neural crest development.40
Knockdown of Sox11 decreased proliferation of the MSCs and decreased osteogenic differentiation. The levels of the Sox11 transcript decreased progressively as the MSCs expanded, with only slight recovery with each addition of the new culture medium, until it was no longer detectable on highly passaged MSCs. Therefore, it may provide a useful marker to distinguish early progenitors of MSCs from MSCs that are approaching senescence. It may also provide another MSC marker to separate MSCs from other surrounding tissues in vivo, such as CD105, CD73, and CD90.28 Recently, Kubo et al. observed that Sox11 was one of several transcription factors that were downregulated when hMSCs were transferred to a differentiation-inducing medium.41 They observed that knockdowns of the same group of transcription factors inhibited both osteogenic and adipogenic differentiation.
Supplementary Material
Acknowledgment
This work was supported by NCRR/NIH grant P40 RR 17447.
Disclosure Statement
No competing financial interests exist.
References
- 1.Prockop D.J. “Stemness” does not explain the repair of many tissues by mesenchymal stem/multipotent stromal cells (MSCs) Clin Pharmacol Ther. 2007;82:241. doi: 10.1038/sj.clpt.6100313. [DOI] [PubMed] [Google Scholar]
- 2.DiGirolamo C.M. Stokes D. Colter D. Phinney D.G. Class R. Prockop D.J. Propagation and senescence of human marrow stromal cells in culture: a simple colony-forming assay identifies samples with the greatest potential to propagate and differentiate. Br J Haematol. 1999;107:275. doi: 10.1046/j.1365-2141.1999.01715.x. [DOI] [PubMed] [Google Scholar]
- 3.Bernardo M.E. Zaffaroni N. Novara F. Cometa A.M. Avanzini M.A. Moretta A. Montagna D. Maccario R. Villa R. Daidone M.G. Zuffardi O. Locatelli F. Human bone marrow derived mesenchymal stem cells do not undergo transformation after long-term in vitro culture and do not exhibit telomere maintenance mechanisms. Cancer Res. 2007;67:9142. doi: 10.1158/0008-5472.CAN-06-4690. [DOI] [PubMed] [Google Scholar]
- 4.Izadpanah R. Kaushal D. Kriedt C. Tsien F. Patel B. Dufour J. Bunnell B.A. Long-term in vitro expansion alters the biology of adult mesenchymal stem cells. Cancer Res. 2008;68:4229. doi: 10.1158/0008-5472.CAN-07-5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosland G.V. Svendsen A. Torsvik A. Sobala E. McCormack E. Immervoll H. Mysliwietz J. Tonn J.C. Goldbrunner R. Lonning P.E. Bjerkvig R. Schichor C. Long-term cultures of bone marrow-derived human mesenchymal stem cells frequently undergo spontaneous malignant transformation. Cancer Res. 2009;69:5331. doi: 10.1158/0008-5472.CAN-08-4630. [DOI] [PubMed] [Google Scholar]
- 6.Lepperdinger G. Brunauer R. Jamnig A. Laschober G. Kassem M. Controversial issue: is it safe to employ mesenchymal stem cells in cell-based therapies? Exp Gerontol. 2008;43:1018. doi: 10.1016/j.exger.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 7.Rubio D. Garcia S. Paz M.F. De la Cueva T. Lopez-Fernandez L.A. Lloyd A.C. Garcia-Castro J. Bernad A. Molecular characterization of spontaneous mesenchymal stem cell transformation. PLoS ONE. 2008;3:e1398. doi: 10.1371/journal.pone.0001398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rubio D. Garcia S. De la Cueva T. Paz M.F. Lloyd A.C. Bernad A. Garcia-Castro J. Human mesenchymal stem cell transformation is associated with a mesenchymal-epithelial transition. Exp Cell Res. 2008;314:691. doi: 10.1016/j.yexcr.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 9.Lazennec G. Jorgensen C. Concise review: adult multipotent stromal cells and cancer: risk or benefit? Stem Cells. 2008;26:1387. doi: 10.1634/stemcells.2007-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dahl J.A. Duggal S. Coulston N. Millar D. Melki J. Shahdadfar A. Brinchmann J.E. Collas P. Genetic and epigenetic instability of human bone marrow mesenchymal stem cells expanded in autologous serum or fetal bovine serum. Int J Dev Biol. 2008;52:1033. doi: 10.1387/ijdb.082663jd. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Z.X. Guan L.X. Zhang K. Wang S. Cao P.C. Wang Y.H. Wang Z. Dai L.J. Cytogenetic analysis of human bone marrow-derived mesenchymal stem cells passaged in vitro. Cell Biol Int. 2007;31:645. doi: 10.1016/j.cellbi.2006.11.025. [DOI] [PubMed] [Google Scholar]
- 12.Ylostalo J. Bazhanov N. Prockop D.J. Reversible commitment to differentiation by human multipotent stromal cells in single-cell-derived colonies. Exp Hematol. 2008;36:1390. doi: 10.1016/j.exphem.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colter D.C. Sekiya I. Prockop D.J. Identification of a subpopulation of rapidly self-renewing and multipotential adult stem cells in colonies of human marrow stromal cells. Proc Natl Acad Sci USA. 2001;98:7841. doi: 10.1073/pnas.141221698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rubin H. Multistage carcinogenesis in cell culture. Dev Biol (Basel) 2001;106:61. discussion 67, 143–160. [PubMed] [Google Scholar]
- 15.Li C. Automating dChip: toward reproducible sharing of microarray data analysis. BMC Bioinformatics. 2008;9:231. doi: 10.1186/1471-2105-9-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li C. Wong W.H. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gregory C.A. Perry A.S. Reyes E. Conley A. Gunn W.G. Prockop D.J. Dkk-1-derived synthetic peptides and lithium chloride for the control and recovery of adult stem cells from bone marrow. J Biol Chem. 2005;280:2309. doi: 10.1074/jbc.M406275200. [DOI] [PubMed] [Google Scholar]
- 18.Gunn W.G. Conley A. Deininger L. Olson S.D. Prockop D.J. Gregory C.A. A crosstalk between myeloma cells and marrow stromal cells stimulates production of DKK1 and interleukin-6: a potential role in the development of lytic bone disease and tumor progression in multiple myeloma. Stem Cells. 2006;24:986. doi: 10.1634/stemcells.2005-0220. [DOI] [PubMed] [Google Scholar]
- 19.Sekiya I. Vuoristo J.T. Larson B.L. Prockop D.J. In vitro cartilage formation by human adult stem cells from bone marrow stroma defines the sequence of cellular and molecular events during chondrogenesis. Proc Natl Acad Sci USA. 2002;99:4397. doi: 10.1073/pnas.052716199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gregory C.A. Singh H. Perry A.S. Prockop D.J. The Wnt signaling inhibitor dickkopf-1 is required for reentry into the cell cycle of human adult stem cells from bone marrow. J Biol Chem. 2003;278:28067. doi: 10.1074/jbc.M300373200. [DOI] [PubMed] [Google Scholar]
- 21.Wright G.J. Leslie J.D. Ariza-McNaughton L. Lewis J. Delta proteins and MAGI proteins: an interaction of Notch ligands with intracellular scaffolding molecules and its significance for zebrafish development. Development. 2004;131:5659. doi: 10.1242/dev.01417. [DOI] [PubMed] [Google Scholar]
- 22.Hu Y. Li Z. Guo L. Wang L. Zhang L. Cai X. Zhao H. Zha X. MAGI-2 Inhibits cell migration and proliferation via PTEN in human hepatocarcinoma cells. Arch Biochem Biophys. 2007;467:1. doi: 10.1016/j.abb.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 23.McGovern D.P. Taylor K.D. Landers C. Derkowski C. Dutridge D. Dubinsky M. Ippoliti A. Vasiliauskas E. Mei L. Mengesha E. King L. Pressman S. Targan S.R. Rotter J.I. MAGI2 genetic variation and inflammatory bowel disease. Inflamm Bowel Dis. 2009;15:75. doi: 10.1002/ibd.20611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larson B.L. Ylostalo J. Prockop D.J. Human multipotent stromal cells undergo sharp transition from division to development in culture. Stem Cells. 2008;26:193. doi: 10.1634/stemcells.2007-0524. [DOI] [PubMed] [Google Scholar]
- 25.Lee R.H. Seo M.J. Pulin A.A. Gregory C.A. Ylostalo J. Prockop D.J. The CD34-like protein PODXL and {alpha}6-integrin (CD49f ) identify early progenitor MSCs with increased clonogenicity and migration to infarcted heart in mice. Blood. 2009;4:816. doi: 10.1182/blood-2007-12-128702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitney M.J. Lee A. Ylostalo J. Zeitouni S. Tucker A. Gregory C.A. Leukemia inhibitory factor secretion is a predictor and indicator of early progenitor status in adult bone marrow stromal cells. Tissue Eng Part A. 2009;15:33. doi: 10.1089/ten.tea.2007.0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muraglia A. Cancedda R. Quarto R. Clonal mesenchymal progenitors from human bone marrow differentiate in vitro according to a hierarchical model. J Cell Sci. 2000;113(Pt 7):1161. doi: 10.1242/jcs.113.7.1161. [DOI] [PubMed] [Google Scholar]
- 28.Dominici M. Le Blanc K. Mueller I. Slaper-Cortenbach I. Marini F. Krause D. Deans R. Keating A. Prockop D. Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 29.Peister A. Mellad J.A. Larson B.L. Hall B.M. Gibson L.F. Prockop D.J. Adult stem cells from bone marrow (MSCs) isolated from different strains of inbred mice vary in surface epitopes, rates of proliferation, and differentiation potential. Blood. 2004;103:1662. doi: 10.1182/blood-2003-09-3070. [DOI] [PubMed] [Google Scholar]
- 30.Sekiya I. Larson B.L. Smith J.R. Pochampally R. Cui J.G. Prockop D.J. Expansion of human adult stem cells from bone marrow stroma: conditions that maximize the yields of early progenitors and evaluate their quality. Stem Cells. 2002;20:530. doi: 10.1634/stemcells.20-6-530. [DOI] [PubMed] [Google Scholar]
- 31.Smith J.R. Pochampally R. Perry A. Hsu S.C. Prockop D.J. Isolation of a highly clonogenic and multipotential subfraction of adult stem cells from bone marrow stroma. Stem Cells. 2004;22:823. doi: 10.1634/stemcells.22-5-823. [DOI] [PubMed] [Google Scholar]
- 32.Mauney J.R. Kirker-Head C. Abrahamson L. Gronowicz G. Volloch V. Kaplan D.L. Matrix-mediated retention of in vitro osteogenic differentiation potential and in vivo bone-forming capacity by human adult bone marrow-derived mesenchymal stem cells during ex vivo expansion. J Biomed Mater Res A. 2006;79:464. doi: 10.1002/jbm.a.30876. [DOI] [PubMed] [Google Scholar]
- 33.Datta N. Holtorf H.L. Sikavitsas V.I. Jansen J.A. Mikos A.G. Effect of bone extracellular matrix synthesized in vitro on the osteoblastic differentiation of marrow stromal cells. Biomaterials. 2005;26:971. doi: 10.1016/j.biomaterials.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 34.Khoo M.L. Shen B. Tao H. Ma D.D. Long-term serial passage and neuronal differentiation capability of human bone marrow mesenchymal stem cells. Stem Cells Dev. 2008;17:883. doi: 10.1089/scd.2007.0185. [DOI] [PubMed] [Google Scholar]
- 35.Choumerianou D.M. Dimitriou H. Perdikogianni C. Martimianaki G. Riminucci M. Kalmanti M. Study of oncogenic transformation in ex vivo expanded mesenchymal cells, from paediatric bone marrow. Cell Prolif. 2008;41:909. doi: 10.1111/j.1365-2184.2008.00559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barlow S. Brooke G. Chatterjee K. Price G. Pelekanos R. Rossetti T. Doody M. Venter D. Pain S. Gilshenan K. Atkinson K. Comparison of human placenta- and bone marrow-derived multipotent mesenchymal stem cells. Stem Cells Dev. 2008;17:1095. doi: 10.1089/scd.2007.0154. [DOI] [PubMed] [Google Scholar]
- 37.Rubio D. Garcia-Castro J. Martin M.C. de la F.R. Cigudosa J.C. Lloyd A.C. Bernad A. Spontaneous human adult stem cell transformation. Cancer Res. 2005;65:3035. doi: 10.1158/0008-5472.CAN-04-4194. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y. Huso D.L. Harrington J. Kellner J. Jeong D.K. Turney J. McNiece I.K. Outgrowth of a transformed cell population derived from normal human BM mesenchymal stem cell culture. Cytotherapy. 2005;7:509. doi: 10.1080/14653240500363216. [DOI] [PubMed] [Google Scholar]
- 39.Prockop D.J. Repair of tissues by adult stem/progenitor cells (MSCs): controversies, myths, and changing paradigms. Mol Ther. 2009;17:939. doi: 10.1038/mt.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hong C.S. Saint-Jeannet J.P. Sox proteins and neural crest development. Semin Cell Dev Biol. 2005;16:694. doi: 10.1016/j.semcdb.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 41.Kubo H. Shimizu M. Taya Y. Kawamoto T. Michida M. Kaneko E. Igarashi A. Nishimura M. Segoshi K. Shimazu Y. Tsuji K. Aoba T. Kato Y. Identification of mesenchymal stem cell (MSC)-transcription factors by microarray and knockdown analyses, and signature molecule-marked MSC in bone marrow by immunohistochemistry. Genes Cells. 2009;3:407. doi: 10.1111/j.1365-2443.2009.01281.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.