Abstract
The mechanisms underlying the evolution of morphology are poorly understood1,2. Distantly related taxa sometimes exhibit correlations between morphological differences and patterns of gene expression3−8, but such comparisons cannot establish how mechanisms evolve to generate diverse morphologies. Answers to these questions require resolution of the nature of developmental evolution within and between closely related species. Here I show how the detailed regulation of the Hox gene Ultrabithorax patterns trichomes on the posterior femur of the second leg in Drosophila melanogaster, and that evolution of Ultrabithorax has contributed to divergence of this feature among closely related species. The cis-regulatory regions of Ultrabithorax, and not the protein itself, appear to have evolved. This study provides experimental evidence that cis-regulatory evolution is one way in which conserved proteins have promoted morphological diversity1.
In most species of the genus Drosophila, non-sensory microtrichiae, or trichomes, cover much of the posterior second femur, leaving a patch of naked cuticle near the proximal end (Fig. 1). The distribution of this naked cuticle varies between, and to some extent within, species. Of the three species studied here, D. melanogaster has a small naked patch, its sister species D. simulans has a larger patch, and the more distantly related D. virilis has no naked cuticle (Fig. 1).
Figure 1.
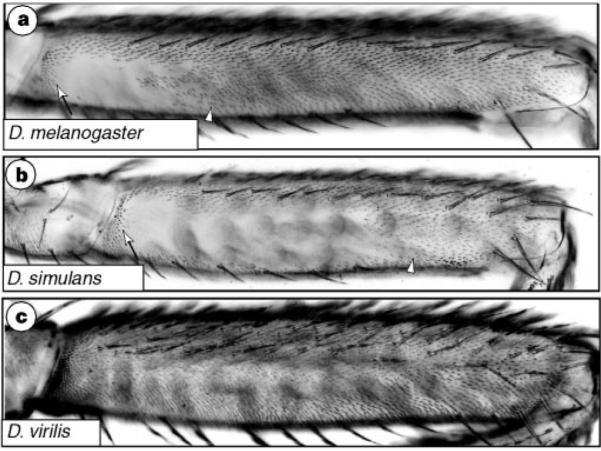
Trichome patterns on the posterior second femur vary among Drosophila species, a, D. melanogaster Oregon-R strain (mean naked cuticle length/femur length ± s.d., 0.31 ± 0.0035). b, D. simulans Tsimbazaza strain (0.61 ± 0.022). c, D. virilis Novosibirsk strain. The naked cuticle length was measured between the proximal (arrow) and maximum distal (arrowhead) extent of naked curticle.
In D. melanogaster, Ultrabithorax (Ubx) patterns unique morphological features from the second thoracic to the seventh abdominal segment9–13. I tested the requirement of Ubx in patterning trichomes on the posterior second femur by generating clones of cells that lacked the ability to produce Ubx protein. When these clones were produced in the proximal patch of naked cuticle they differentiated trichomes; thus, Ubx is needed to repress trichomes in this region (Fig. 2a).
Figure 2.
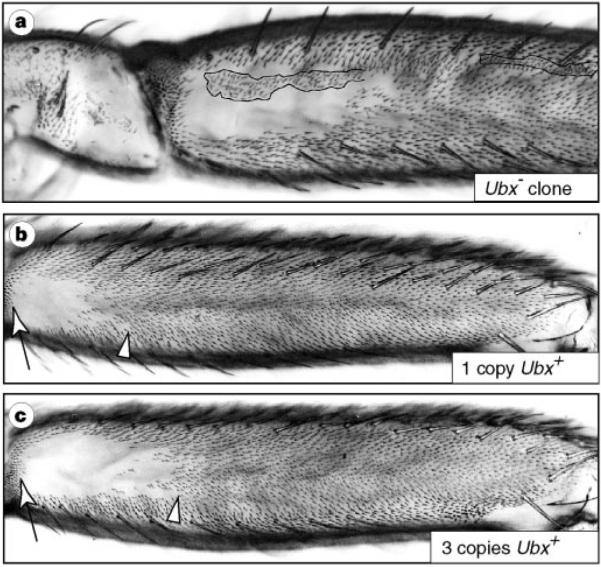
Ubx represses trichomes in the proximal naked cuticle in a dosage-dependent manner in D. melanogaster. a, A multiple-wing-hairs marked clone of Ubx− cells (outlined) differentiated trichomes within the patch of naked cuticle. b, c, Ubx dosage altered the distribution of trichomes. Offspring from the cross Df(3R)P9/Dp(3R)P5 × st pp e11 with one functional copy (b) and three functional copies (c) of Ubx are shown. (Mean naked cuticle length/femur length ± s.d.: Df(3R)P9/st pp e11 = 0.17 ± 0.014 versus Dp(3R)P5/st pp e11 = 0.25 ± 0.014; t = 7.98, d.f. = 5, P = 0.0005.) Arrows and arrowheads delineate the extent of naked cuticle.
Three experiments indicate that the detailed expression pattern of Ubx is required to generate the specific morphology of a naked patch of cuticle. First, flies carrying three copies of the Ubx locus had significantly more naked cuticle than sibling flies carrying one copy (Fig. 2b, c). Second, expression of uniform, high levels of Ubx protein during pupal development repressed trichomes on most of the posterior second femur (Fig. 3). Maximal repression occurred between 20 and 28 hours after puparium formation (APF). Trichomes were repressed in a proximal to distal direction, so that expression before 18 hours APF or at lower levels (results not shown) repressed proximal, but not distal, trichomes. Finally, Ubx protein is expressed in a proximal–distal gradient (Fig. 4a, b), with high levels proximally. Studies of the third leg support the hypothesis that high levels of Ubx repress trichomes. Clonal analysis showed that Ubx is required to repress trichomes on the posterior third leg (results not shown). In all three Drosophila species, most of the posterior third femur lacks trichomes and Ubx is expressed at high levels (Fig. 4a, c, e, h). The amount of expression in the posterior third femur is similar to that in the proximal patch of the posterior second femur, indicating that Ubx expression at or near these levels may repress posterior femur trichomes.
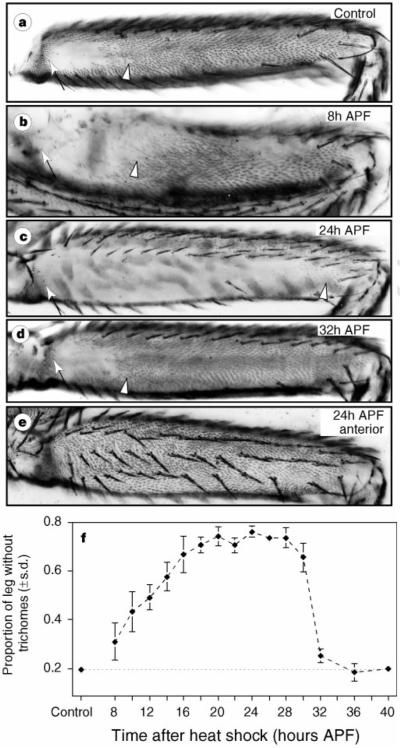
Figure 3.
Uniform expression of Ubx in D. melanogaster represses triohomes on the posterior second femur during a short temporal window, a, Control flies. b, c, Files heat-shocked at 8 h APF (b) and 24 h APF (c) showed progressively larger patches of naked cuticle, d, Heat shocks at 32 h APF had little effect, e, Ectopic Ubx failed to repress anterior femur trichomes. f, Ectopic Ubx repressed posterior second femur trichomes most efficiently between 18 and 28 h APF. (After heat shock, increased amounts of Ubx protein remained detectable for 7 h (results not shown). This long perdurance, combined with the sharp drop in sensitivity to Ubx after 30 h APF, indicates that cells may respond to Ubx level during a shorter time window than is seen here.) Arrows and arrowheads in a–d delineate the extent of naked cuticle.
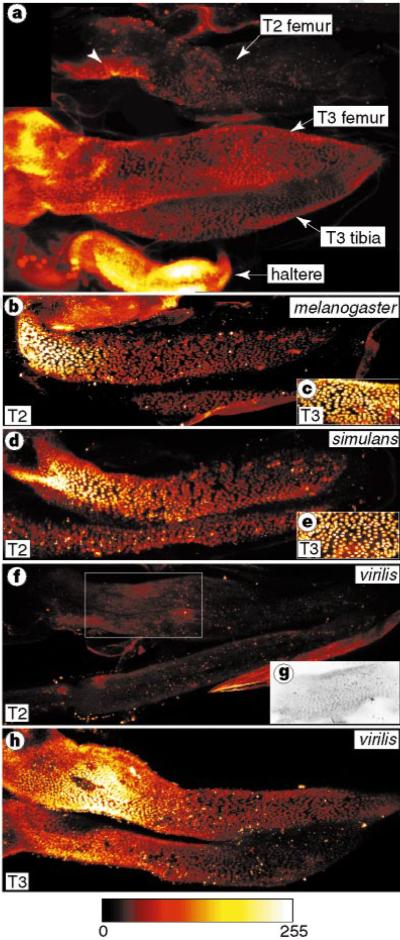
Figure 4.
Distribution of Ubx protein in the posterior femurs of Drosophila species. a, Protein expression in the second (T2) and third (T3) leg and haltere in D. melanogaster. Proximal is to the left and the tibias fold back under the femurs. The second leg is twisted, providing an oblique view of the proximal femur (arrowhead) where Ubx is expressed at levels similar to those in the third femur. b–e, D. melanogaster (b) and D. simulans (d) show a proximal to distal Ubx expression gradient in T2. The approximate expression levels in the T3 femurs are shown for D. melanogaster (c) and D. simulans (e). f–h, Ubx expression in D. virilis is not visible in the second femur (f) at confocal settings that reveal high levels in the third femur (h). The boxed region of f, scanned at maximal sensitivity, shows that Ubx is expressed at low levels (g). Except in g, images were false-coloured with the scale shown below. Expression levels cannot be compared between b, d and f.
Patterns of Ubx expression correlate broadly with interspecific variation in trichome patterning. Ubx expression appeared to be similar in the two sister species D. melanogaster and D. simulans (Fig. 4b, d), both of which have a patch of naked cuticle. However, D. virilis, which has no naked cuticle (Fig. 1), expressed Ubx at levels far below those seen in the posterior third leg (Fig. 4f–h).
Existing methods of visualizing protein expression may be inadequate to detect the potentially small differences in the Ubx expression gradient between D. melanogaster and D. simulans. However, trichome production, which is a binary outcome of the amount of Ubx expression, may provide a more sensitive assay for differences in species' alleles. I therefore used a complementation test, in which a single functional Ubx allele from each species was tested in identical hybrid backgrounds. This approach bypassed the practical difficulties of analysis of F2 generations of these species14.
To minimize the variation that arises from segregating modifier loci in existing stocks of Ubx mutants (results not shown), I generated new null alleles of Ubx in inbred lines of each species. Files heterozygous for each new Ubx mutation were then crossed to the original inbred line of the other species (Fig. 5). F1 offspring carrying a null Ubx allele from one species had a wild-type allele from the other and should be identical hybrids at all other loci. These F1 classes differed significantly from each other in the predicted direction; flies carrying the D. melanogaster wild-type allele of Ubx (Fig. 5c) had a smaller naked patch than flies carrying the D. simulans wild-type allele (Fig. 5d, t = 11.4, degrees of freedom (d.f.) = 43, P < 0.0001). Therefore, the Ubx locus has evolved differently in these species so that the D. simulans allele produces a larger naked patch than the D. melanogaster allele. These conclusions are supported by complementation tests done using previously existing Ubx alleles (results not shown). The presumptive Ubx protein in D. simulans is identical to the D. melanogaster protein (Table 1), indicating that cis-regulation of the Ubx protein may have evolved15.
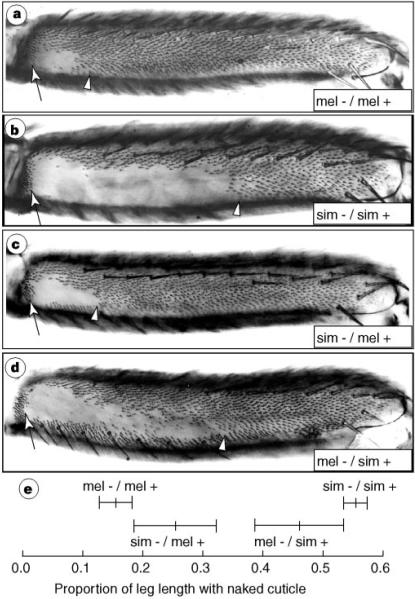
Figure 5.
The Ubx alleles from D. melanogaster and D. simulans produce different triohome patterns in identical hybrid backgrounds, a, b, The posterior second femurs from Ubx heterozygous flies produced from parental strains of D. melanogaster (a) and D. simulans (b) in which new Ubx alleles had been induced. c, d, The posterior second femurs of the Ubx heterozygous F1, offspring of the cross sim − /sim + × mel + /mel+ (c) and mel − /mel + × sim + /sim+ (d). e, The mean proportion of naked cuticle ± s.d. for the four genotypes. Arrows and arrowheads delineate the extent of naked cuticle, mel−, D.melanogaster Ubx null allele; mel+, D. melanogaster Ubx wild-type allele; sim−, D. simulans Ubx null allele; sim+, D. simulans Ubx wild-type allele.
Table 1.
Nucleotide divergence between D. melanogaster and D. simulans for Ubx exons and flanking regions
| Coding |
Flanking |
|||
|---|---|---|---|---|
| Differences* | Total sites | Differences | Total sites | |
| Exon 1 | 11 | 766 | 1 | 53 |
| Exon 2 | 0 | 51 | 5 (+ 1 insert/deletion)† | 167 |
| Exon 3 | 0 | 51 | 1 | 59 |
| Exon 4 | 5 | 302 | 6 | 96 |
| Total | 16 (1.4%) | 1,170 | 13 (3.5%) (+1 insert/deletion) | 375 |
All nucleotide differences in the coding regions were synonymous substitutions at third-base-pair positions.
One 7-base-pair insertion/deletion was found downstream of exon 2.
Ubx is not the only locus that influences this trait, because the hybrids were significantly different from the parental classes that carried the same wild-type allele of Ubx (Fig. 5; D. melanogaster null allele plus wild-type allele versus D. simulans null allele plus D. melanogaster wild-type allele: t = −8.2, d.f. = 58, P < 0.0001; D. simulans null plus wild-type alleles versus D. melanogaster null allele plus D. simulans wild-type allele: t = −6.0, d.f. = 25, P < 0.0001). The hybrids with a D. simulans wild-type allele also exhibited a dorsal–ventral pattern of trichome distribution (Fig. 5d): the ventral naked cuticle was longer than the dorsal cuticle. Therefore, at least one other gene, which is possibly involved in dorsal–ventral leg patterning, has contributed to the evolution of this trait.
This study illustrates one mechanism that contributes to a single difference between fly species. There are about 150,000 species of flies, all using roughly the same set of conserved genes to generate their morphology. How does such diversity arise from a conserved set of proteins? One possible answer, illustrated by my results, is that conserved proteins exhibit complex spatiotemporal regulation and that every element of this pattern is susceptible to subtle evolutionary manipulation1,6–8,15–17.
Methods
Clonal analysis
Two clonal analysis18 experiments were performed. First, I crossed males carrying the alleles mwh jv st red sbd2 Ubx12.5 e11 ro ca/TM1 to females carrying the alleles f36a;M(3)w f+87/TM3. Legs of male offspring without balancer chromosomes were studied for f36a bristles. Second, I crossed mwh jv st red sbd2 Ubx12.5 e11 ro ca/TM1 males to Dp(3:3)S462, Cbx1 Ubx1/TM6B females. Legs of offspring without balancers were studied for mwh trichomes. Larvae were subjected to X-rays (1,000 rad) at 24–72 h after egg-laying.
Ectopic Ubx expression
White prepupae from the cross st ppe11 × HSUbx – 1a (ref. 19) were aged at 25 °C. Separate samples were heat-shocked (at 37 °C) for 1 h at 2-h intervals from 0–48 h APE Samples heat-shocked at 0–6 h APF and some heat-shocked at 8 h APF failed to differentiate cuticle, whereas all others developed to pharate adults but failed to eclose. Most anterior femur trichomes were not repressed, indicating that neither ectopic Ubx expression nor the heat shock itself interfered with the ability of cells to differentiate trichomes.
Antibody staining
White prepupae were aged at 25 °C for 20–30 h for D. melanogaster and D. simulans, and for 38–44 h for D. virilis. Dissected pupal legs were stained using standard techniques20, except that the antibodies were applied at 4 °C for at least 12 h each. The two monoclonal antibodies Fp3.38 (for D. melanogaster and D. simulans) and Fp6.87 (for D. virilis) were used together with a fluorescein isothiocyanate (FITC)-conjugated anti-mouse secondary antibody to detect Ubx protein20,21. Both of the monoclonal antibodies produced comparable results when tested in D. melanogaster (results not shown). Images were captured with a BioRad MRC-1024 confocal microscope.
Generation of new Ubx alleles
New alleles of Ubx were induced in the st pp e11 D. melanogaster and Tsimbazaza D. simulans stocks by X-ray irradiation of 3-day-old virgin males with 4 krad. These males were mated with virgin 3-day-old females. Offspring were screened for enlarged halteres. One of 3,600 D. melanogaster and nine of 8,500 D. simulans flies had enlarged halteres, of which one and two flies of each species, respectively, were fertile. Only alleles that failed to complement Ubx, showed the appropriate null phenotype9, and did not produce Ubx protein (as assessed by antibody staining) were used for the interspecific test.
DNA sequencing
The four Ubx exons were amplified by the polymerase chain reaction (PCR) from genomic DNA extracted from the Tsimbazaza strain of D. simulans, with the following primers: exon 1, 5′-GCCCGTCTCAGACGG AGCAC-3′ and 5′-TGGGATTTCGGGGGACTTTCAG-3′; exon 2, 5′-CCCTA CCACCAGATCCCCACGTACCC-3′ and 5′-GCCCCATTGATTCATGAATTT AGCACACC-3′; exon 3, 5′-TGAGGCATAATGACGTTCCTGGAC-3′ and 5′-CATAGGCAAAACTATAGGCTAAGGGTTTAC-3′; exon 4, 5′-ATGTATGTA TTTCGTCGATGCAGGTC-3′ and 5′-CCAATCCCACATACACCCTAC-3′. PCR fragments were cloned into pCRII (Invitrogen) and four clones of each exon were sequenced using Dye Terminator Cycle sequencing (Perkin Elmer).
Acknowledgements
I thank M. Akam for his patronage; N. Brown, J. Castelli-Gair and J. De Celis for fly tutelage; J. De Celis, E. Lewis, J. Roote and M. Ashburner for stocks; B. Yen and E. Sucena for technical assistance; and M. Akam, M. Ashburner, A. Gonzalez-Reyes, C. Mirth, L. Partridge and E. Sucena for comments. This work was supported by Churchill College, the Wellcome Trust and the BBSRC.
References
- 1.Carroll SB. Homeotic genes and the evolution of arthropods and chordates. Nature. 1995;376:479–485. doi: 10.1038/376479a0. [DOI] [PubMed] [Google Scholar]
- 2.Gerhart J, Kirschner M. Cells, Embryos and Evolution. Blackwell; Malden, Massachusetts: 1997. [Google Scholar]
- 3.Warren RW, Nagy L, Selegue J, Gates J, Carroll S. Evolution of homeotic gene regulation and function in flies and butterflies. Nature. 1994;372:458–461. doi: 10.1038/372458a0. [DOI] [PubMed] [Google Scholar]
- 4.Akam M. Hox genes and the evolution of diverse body plans. Phil. Trans. R. Soc. Lond. B. 1995;349:313–319. doi: 10.1098/rstb.1995.0119. [DOI] [PubMed] [Google Scholar]
- 5.Averof M, Akam M. Hox genes and the diversification of insect and crustacean body plans. Nature. 1995;376:420–423. doi: 10.1038/376420a0. [DOI] [PubMed] [Google Scholar]
- 6.Averof M, Patel NH. Crustacean appendage evolution associated with changes in Hox gene expression. Nature. 1997;388:682–686. doi: 10.1038/41786. [DOI] [PubMed] [Google Scholar]
- 7.Grenier JK, Garber TL, Warren R, Whitington PM, Carroll S. Evolution of the entire arthropod Hox gene set predated the origin and radiation of the onychophoran/arthropod clade. Curr. Biol. 1997;7:547–553. doi: 10.1016/s0960-9822(06)00253-3. [DOI] [PubMed] [Google Scholar]
- 8.Lowe CJ, Wray GA. Radical alterations in the roles of homeobox genes during echinoderm evolution. Nature. 1997;389:718–721. doi: 10.1038/39580. [DOI] [PubMed] [Google Scholar]
- 9.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 10.Morata G, Kerridge S. Sequential functions of the bithorax complex of Drosophila. Nature. 1981;290:778–781. doi: 10.1038/290778a0. [DOI] [PubMed] [Google Scholar]
- 11.Sanchez-Herrero E, Casanova J, Kerridge S, Morata G. Anatomy and function of the bithorax complex of Drosophila. Cold Spring Harb. Symp. Quant. Biol. 1985;50:165–172. doi: 10.1101/sqb.1985.050.01.022. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez-Reyes A, Morata G. The developmental effect of overexpressing a Ubx product in Drosophila embryos is dependent on its interactions with other homeotic products. Cell. 1990;61:515–522. doi: 10.1016/0092-8674(90)90533-k. [DOI] [PubMed] [Google Scholar]
- 13.Castelli-Gair J, Akam M. How the Hox gene Ultrabithorax specifies two different segments: the significance of spatial and temporal regulation within metameres. Development. 1995;121:2973–2982. doi: 10.1242/dev.121.9.2973. [DOI] [PubMed] [Google Scholar]
- 14.Davis AW, et al. Rescue of hybrid sterility in crosses between D. melanogaster and D. simulans. Nature. 1996;380:157–159. doi: 10.1038/380157a0. [DOI] [PubMed] [Google Scholar]
- 15.Gibson G, Hogness DS. Effect of polymorphism in the Drosophila regulatory gene Ultrabithorax on homeotic stability. Science. 1996;271:200–203. doi: 10.1126/science.271.5246.200. [DOI] [PubMed] [Google Scholar]
- 16.Akam M. Hox genes, homeosis and the evolution of segment identity; no need for hopeless monsters. Int. J. Dev. Biol. 1998;42:445–451. [PubMed] [Google Scholar]
- 17.Castelli-Gair J. Implications of the spatial and temporal regulation of Hox genes on development and evolution. Int. J. Dev. Biol. 1998;42:437–444. [PubMed] [Google Scholar]
- 18.Morata G, Ripoll P. Minutes: mutants of Drosophila autonomously affecting cell division rate. Dev. Biol. 1975;42:211–221. doi: 10.1016/0012-1606(75)90330-9. [DOI] [PubMed] [Google Scholar]
- 19.Mann RS, Hogness DS. Functional dissection of Ultrabithorax proteins in D. melanogaster. Cell. 1990;60:597–610. doi: 10.1016/0092-8674(90)90663-y. [DOI] [PubMed] [Google Scholar]
- 20.Kelsh R, Weinzieri ROJ, White RAH, Akam M. Homeotic gene expression in the locust Schistocerca: an antibody that detects conserved epitopes in Ultrabithorax and Abdominal-A proteins. Dev. Genet. 1994;15:19–31. doi: 10.1002/dvg.1020150104. [DOI] [PubMed] [Google Scholar]
- 21.White RAH, Wilcox M. Protein products of the bithorax complex in Drosophila. Cell. 1984;39:163–171. doi: 10.1016/0092-8674(84)90202-2. [DOI] [PubMed] [Google Scholar]