Abstract
CD74, a Type II membrane glycoprotein and MHC class II chaperone involved in antigen processing, is normally expressed by cells associated with the immune system. CD74 also forms heterodimers with CD44 to generate receptors to macrophage migration inhibitory factor (MIF), a proinflammatory cytokine. Following targeted Alb-Cre-mediated deletion of Ikkβ in IkkβΔhep mice (IkkβF/F:Alb-Cre, a strain highly susceptible to chemically-induced hepatotoxicity and hepatocarcinogenesis), CD74 is expressed abundantly by adult hepatocytes throughout liver acini, albeit more intensely in midzonal-to-centrilobular regions. By comparison, CD74 expression is not observed in IkkβF/F hepatocytes, nor is it augmented in the livers of Ikkβ+/+:Alb-Cre mice; CD74 is barely detectable in cultured embryonic fibroblasts from Ikkβ-/- mice. Microarray profiling shows that constitutive CD74 expression in IkkβΔhep hepatocytes is accompanied by significantly augmented expression of CD44 and key genes associated with antigen processing and host defense, including MHC class II I-Aα, I-Aβ, and I-Eβ chains, CIITA and CD86. Taken together, these observations suggest that IkkβΔhep hepatocytes might express functional capacities for class II-restricted antigen presentation and heightened responsiveness to MIF-signaling, and also suggest further roles for intrahepatocellular IKKβ in the suppression or inactivation of molecules normally associated with the formation and differentiation of cells of the immune system.
Keywords: Hepatocytes, Ikkβ deletion, CD74, MHC Class II
Introduction
CD74 is recognized as the invariant chain chaperone (Ii) of MHC class II processing (Bertolino and Rabourdin-Combe, 1996). Its molecular sizes range between 31-45 kDa; the most abundant mouse isoforms, splice variants p31 and p41, are post-translationally modified with chondroitin sulfate side-chains to generate Type II membrane glycoproteins with 30 aa cytoplasmic tails (Bertolino and Rabourdin-Combe, 1996; Stumptner-Cuvelette and Benaroch, 2002). CD74 is expressed throughout the immune system by B cells, activated T cells, dendritic cells, monocytes and macrophages (Bertolino and Rabourdin-Combe, 1996; Calandra and Roger, 2003; Faure-André et al., 2008; Leng and Bucala, 2006; Lue et al., 2006; Stumptner-Cuvelette and Benaroch, 2002). In the liver, CD74 is expressed constitutively in dendritic and Kupffer cells (Momburg et al., 1986), and in resident hepatic stellate cells (HSCs) (Maubach et al., 2007); however, except for one isolated report of a scattered sub-population of CD74-positive hepatocyte clusters (containing 1 to 5 cells) in ♀ B10.BR H-2k mice (Momburg et al., 1986), CD74 is not expressed in hepatocytes. CD74 is also a component of heterodimeric CD74/CD44 cell surface receptors (Leng and Bucala, 2006; Lou et al., 2006) which mediate mitogenic and proinflammatory effects of macrophage migration inhibitory factor (MIF) (Calandra and Roger, 2003; Leng and Bucala, 2006; Lue et al., 2006), a 12.5 kDa cytokine secreted by a variety of T and B cells, epithelial cells, and liver Kupffer cells (Calandra and Roger, 2003; Kobayashi et al., 1999; Lan, 2008; Leng and Bucala, 2006; Lue et al., 2006). MIF exhibits intrinsic enzymatic activities; induces cyclin D1 expression, mitogenesis and NO production; and plays major roles in the pathogenesis of inflammatory disease (Javeed et al., 2008; Leng and Bucala, 2006; Swant et al., 2005). MIF expression is causally related to chronic hepatitis B virus (HBV) infection (Zhang et al., 2005), fulminant alcoholic hepatitis (Kumagi et al., 2001) and Bacille-Calmette-Guerin-primed lipopolysaccharide (LPS)-induced T cell-mediated liver failure (BLTLF) (Kobayashi et al., 1999). Anti-MIF antisera block BLTLF (Kobayashi et al., 1999) and attenuate experimentally-induced murine colitis (de Jong et al., 2001); anti-sense MIF cDNA attenuates LPS-induced liver injury (Iwaki et al., 2003); and MIF knockout mice are protected from concanavalin A (ConA), LPS- and acetaminophen-induced liver injuries (Bourdi et al., 2002; Bozza et al., 1999; Nakajima et al., 2006). CD74 also is cleaved by regulated intracellular proteolysis (RIP) to form a 10-kDa cytoplasmic polypeptide fragment which migrates into the nucleus and activates nuclear factor kappa B (NF-κB) expression (Becker-Herman et al., 2005). However, the relationship between CD74 and NF-κB in liver disease remains unclear.
To understand how NF-κB, a transcription factor that plays anti-inflammatory and protective roles in hepatotoxic injury (Heyninck et al., 2003) and mediates liver disease, we have investigated an hepatocyte-targeted mouse strain (IkkβΔhep) with Cre recombinase-mediated deletions of IκB kinase β (Ikkβ) exon 3 (Maeda et al., 2003). IKKβ, a subunit of the IκB Kinase (IKK) complex, is required for activation of NF-κB (Ghosh and Karin, 2002; Heyninck et al., 2003). IkkβΔhep mice exhibit defective activation of NF-κB and survival genes, and enhanced susceptibility to hepatotoxins (Maeda et al., 2003, 2005). Following ConA- or LPS+Galactosamine-induced inflammation, IkkβΔhep mice exhibit heightened susceptibility to liver failure, resulting partly from tumor necrosis factor (TNF)α-activation of TNF receptor 2 (TNFR2; Maeda et al., 2003), and from increased reactive oxygen species (ROS) formation. ROS inhibits mitogen activated protein kinase (MAPK) phosphatases and sustains c-JUN nuclear kinase 1 (JNK1) activation (Kamata et al., 2005; Maeda et al., 2003). Following treatment with diethylnitrosamine (DEN), IkkβΔhep mice exhibit increased susceptibility to hepatocellular carcinoma (HCC), resulting partly from cytokine-driven NF-κB activation-independent compensatory hepatocyte proliferation (Laurent et al., 2005; Maeda et al., 2005); from sustained JNK1 activation, accompanied by necrotic hepatocellular interleukin (IL)-1α release which activates IL-1 receptors in Kupffer cells and IL-6 secretion (Sakurai et al., 2008); and, as described most recently, from intrinsic IkkβΔhep hepatocellular growth advantages (Koch et al., 2009).
Neither CD74 nor MIF has been linked to the pathophysiological responses of IkkβΔhep hepatocytes, or to Ikkβ-deletion in other cells in other IKKβ-deficient mice (Chen et al., 2006; Malato et al., 2008). However, CD74 is required for major histocompatibility (MHC) class II antigen processing (Bertolino and Rabourdin-Combe, 1996); and MIF has wide-ranging effects on hepatic inflammation, infection and tumor progression (Bourdi et al., 2002; Bozza et al., 1999; Calandra and Roger, 2003; de Jong et al., 2001; Iwaki et al., 2003; Nakajima et al., 2006; Zhang et al., 2005). In addition, because the proinflammatory effects elicited by the CD74/CD44 MIF receptor, like those by TNFR2 (Maeda et al., 2003), require sustained kinase activation (Leng and Bucala, 2006), we investigated constitutive CD74 expression in IkkβΔhep and IkkβF/F mice.
Surprisingly, we find that IkkβΔhep hepatocytes express CD74, whereas IkkβF/F hepatocytes do not. In addition, as assessed by microarray profiling, the hepatocellular CD74-positive phenotype is accompanied by the expression of large numbers of genes normally associated with immune system processing and host defense mechanisms, including MHC class II I-Aα, I-Aβ, and I-Eβ chains, CD44, the transcriptional coactivator of MHC class II expression CIITA (Ting and Trowsdale, 2002), and coactivator CD86. These observations suggest that targeted Ikkβ deletion confers several unexpected and unusual immunologic phenotypes in hepatocytes which, if functionally active, might have significant implications for both the pathophysiology of liver disease and the regulation of animal cell differentiation.
Materials and Methods
Animals and cell culture
C57BL/6, Ikkβ+/+:Alb-Cre, IkkβF/F and IkkβF/F:Alb-Cre (referred to as IkkβΔhep) mice were bred at UCSD (La Jolla, CA), maintained and sacrificed according to NIH guidelines (Koch et al., 2009; Maeda et al., 2003). Protocols used in all animal studies were approved by the UCSD IACUC animal ethics committee. Mouse embryonic fibroblasts (MEFs) from Ikkβ-/- embryos were cultured under standard conditions (Chen et al., 2006).
Isolation and analyses of primary hepatocytes, tissues and macromolecules
Freshly isolated hepatocytes and liver tissues were harvested from adult mice; DNA, RNA and proteins were isolated from hepatocytes, liver tissues and MEFs as described elsewhere (Chen et al, 2006; Koch et al., 2009; Maeda et al., 2003). Ikkβ deletion and genotyping tests were performed by polymerase chain reaction (PCR) using DNA from tails, livers and cultured hepatocytes and MEFs (Chen et al, 2006; Koch et al., 2009; Maeda et al., 2003). RNA and DNA were sized by gel-electrophoresis (Koch et al., 2009).
Microarray profiling
Animals were sacrificed and tissues harvested at 5-6 weeks of age. RNA samples were labeled and hybridized by standard procedures (Feng et al., 2007) using two Illumina WG-6 V2 mouse bead-chips (San Diego, CA, cat. #BD 201-0202) in the UCSD BIOGEM Core (http://microarrays.ucsd.edu/). Each chip carried 6 microarrays housing 47,000 oligonucleotide targets representing ∼5,000 genes/array (see: Fig. 1). Data were normalized and grouped (using the gene ontology databases from the Gene Ontology Consortium [www.genego.org] including pathways from KEGG, Biocarta, and Proteinlounge databases), and analyzed for statistical significance (Feng et al., 2007). Quantitative variation among data samples for individual genes was <1-2%. The original data (see Tables 1 and 2) have been entered into the NCBI GEO Repository (http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GSE15476); a sample of selected data in Table 1 has been presented elsewhere (Koch and Leffert, 2010). For GenBank details and probes employed for these analyses, see the Illumina Reference Chip file, MouseWG-6_V2_0_R2_11278593_A (http://www.switchtoi.com/annotationfiles.ilmn).
Figure 1.
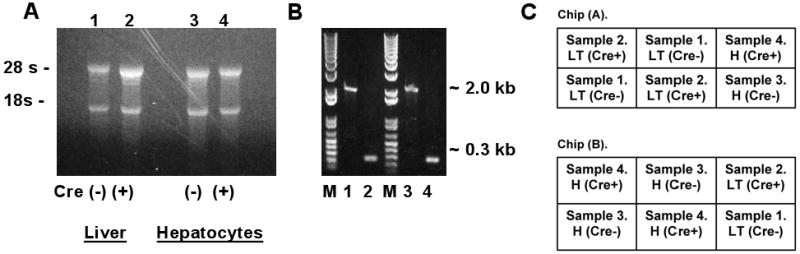
Analysis of RNA and DNA from male Cre(-) IkkβF/F and Cre(+) IkkβΔhep mouse livers and freshly isolated hepatocytes used for microarray profiling. (A) EtBr-stained RNA-sizing gel. (B) Deletion analysis. EtBr-stained DNA-sizing gel. Genotyping for deletion confirmation was performed by PCR using standard primers (Koch et al., 2009). M, size markers (bps); lanes #1-#4 as in panel (A); Cre(-) ∼2-kb, Cre(+) ∼0.3-kb. (C) Hybridization format: chips 1 (A) and 2 (B). Each RNA sample (liver tissue [LT], or isolated hepatocytes [H]) was obtained from a separate mouse: Samples #1 and #2, Cre(-)- and Cre(+)-tissues; #3 and #4, Cre(-)- and Cre(+)-isolated hepatocytes. Each sample was hybridized 3×, on 3 separate sectors across the 2 chips, as diagrammed.
Table 1.
Partial results of microarray profiling: Freshly Isolated Liver Tissues.*
| IMMUNE SYSTEM PROCESS p = 6.46685868E-28 | ||||
|---|---|---|---|---|
| Rank | Locus | GenBank Acc.# | Description | E** |
| 2 | 14961 | NM_207105.1 | Histocompatibility 2, class II antigen A, beta 1 (H2-Ab1) | -8.86 |
| 3 | 14969 | NM_010382.1 | Histocompatibility 2, class II antigen E beta (H2-Eb1) | -8.65 |
| 4 | 16149 | NM_010545.2 | CD74 antigen (invariant polypeptide of major histocompatibility complex) | -8.37 |
| 5 | 14960 | NM_010378.2 | Histocompatibility 2, class II antigen A, alpha (H2-Aa) | -6.21 |
| 6 | 14963 | NM_008199.1 | Histocompatibility 2, blastocyst (H2-Bl) | +5.72 |
| 8 | 14998 | NM_010386.2 | Histocompatibility 2, class II, locus DMa (H2-DMa) | -5.11 |
| DEFENSE RESPONSE p = 2.07679921E-27 | ||||
|---|---|---|---|---|
| Rank | Locus | GenBank Acc.# | Description | E** |
| 4 | 16149 | NM_010545.2 | CD74 antigen (invariant polypeptide of major histocompatibility complex) | -8.37 |
| 6 | 14963 | NM_008199.1 | Histocompatibility 2, blastocyst (H2-Bl) | +5.72 |
| 10 | 17329 | NM_008599 | Chemokine (C-X-C motif) ligand 9 (Cxcl9) | -4.97 |
| 18 | 15019 | NM_023124 | Histocompatibility 2, Q region locus 8 (H2-Q8) | -3.82 |
| 20 | 20210 | NM_011315 | Serum amyloid A 3 (Saa3) | -3.74 |
| 29 | 55985 | NM_018866.1 | Chemokine (C-X-C motif) ligand 13 (Cxcl13) | -3.31 |
| 30 | 20304 | NM_013653.1 | Chemokine (C-C motif) ligand 5 (Ccl5) | -3.29 |
| 34 | 12262 | NM_007574.1 | Complement component 1, q subcomponent, C chain (C1qc) | -3.08 |
| 37 | 17105 | NM_017372 | Lysozyme (Lyzs) | -2.9 |
| 42 | 12260 | NM_009777.1 | Complement component 1, q subcomponent, beta polypeptide (C1qb) | -2.85 |
| 43 | 17069 | NM_008529.2 | Lymphocyte antigen 6 complex, locus E (Ly6e) | -2.82 |
| 53 | 14127 | NM_010185.2 | Fc receptor, IgE, high affinity I, gamma polypeptide (Fcer1g) | -2.55 |
| 67 | 24088 | NM_011905.2 | Toll-like receptor 2 (Tlr2) | -2.31 |
| 72 | 21356 | NM_009318.1 | TAP binding protein (Tapbp) | -2.29 |
| 74 | 15945 | NM_021274.1 | Chemokine (C-X-C motif) ligand 10 (Cxcl10) | -2.29 |
| 78 | 17110 | NM_013590.2 | Lysozyme (Lyzs) | -2.25 |
| 82 | 12505 | AK045226 | CD44 antigen (Cd44) | -2.22 |
Selected expression comparisons between markers from IkkβF/F and IkkβΔhep freshly isolated liver tissues are listed; the markers with these comparatively most disparate expression levels are clustered within two functional groups.
Enrichment, E**, as measured by relative microarray chip hybridization signals, is defined as follows: a negative (−) E value signifies that IkkβΔhep > IkkβF/F, and a positive (+) E value signifies that IkkβF/F > IkkβΔhep (because the initial comparative chip data were expressed as X-fold enrichment of hybridization markers from IkkβF/F compared to those from IkkβΔhep RNA samples).
Table 2.
Partial results of microarray profiling: Freshly Isolated Hepatocytes.*
| DEFENSE RESPONSE p = 1.38089039E-21 | |||||
|---|---|---|---|---|---|
| Rank | Locus | GenBank Acc.# | Description | E** | |
| 1 | 16149 | NM_010545.2 | CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated) (Cd74) | -38.31 | |
| 19 | 15953 | NM_008330.1 | Interferon gamma inducible protein 47 (Ifi47) | -7.15 | |
| 23 | 15944 | NM_008326.1 | Immunity-related GTPase family, M (Irgm) | -6.51 | |
| 24 | 17329 | NM_008599 | Chemokine (C-X-C motif) ligand 9 (Cxcl9) | -6.51 | |
| 27 | 93671 | NM_053094.1 | CD163 antigen (Cd163) | +5.7 | |
| 34 | 21356 | NM_001025313.1 | TAP binding protein (Tapbp) | -5.23 | |
| 45 | 17105 | NM_017372 | Lysozyme (Lyzs) | -4.5 | |
| 53 | 12505 | AK045226 | CD44 antigen (Cd44) | -4.25 | |
| 55 | 15024 | NM_010395.5 | Histocompatibility 2, T region locus 10 (H2-T10) | -4.17 | |
| 64 | 21354 | NM_013683.1 | Transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) (Tap1) | -3.96 | |
| 65 | 24088 | NM_011905.2 | Toll-like receptor 2 (Tlr2) | -3.96 | |
| 66 | 12260 | NM_009777.1 | Complement component 1, q subcomponent, beta polypeptide (C1qb) | -3.96 | |
| 67 | 12774 | NM_009917.2 | Chemokine (C-C motif) receptor 5 (Ccr5) | -3.92 | |
| 69 | 17110 | NM_013590.2 | Lysozyme (Lyzs) | -3.87 | |
| IMMUNE SYSTEM PROCESS p = 1.17768030E-18 | |||||
|---|---|---|---|---|---|
| Rank | Locus | GenBank Acc.# | Description | E** | |
| 1 | 16149 | NM_010545.2 | CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated) (Cd74) | -38.31 | |
| 3 | 14961 | NM_207105.1 | Histocompatibility 2, class II antigen A, beta 1 (H2-Ab1) | -22.48 | |
| 4 | 14969 | NM_010382.1 | Histocompatibility 2, class II antigen E beta (H2-Eb1) | -18.24 | |
| 6 | 14998 | NM_010386 | Histocompatibility 2, class II, locus DMa (H2-DMa) | -15.82 | |
| 7 | 14960 | NM_010378.2 | Histocompatibility 2, class II antigen A, alpha (H2-Aa) | -14.39 | |
| 8 | 21822 | NM_011579.2 | T-cell specific GTPase (Tgtp) | -13.26 | |
| 9 | 14468 | NM_010259.1 | Guanylate nucleotide binding protein 1 (Gbp1) | -11.94 | |
| 11 | 14999 | NM_010387.2 | Histocompatibility 2, class II, locus Mb1 (H2-DMb1) | -10.6 | |
| 13 | 15000 | NM_010388 | Histocompatibility 2, class II, locus Mb2 (H2-DMb2) | -9.36 | |
| 14 | 55932 | NM_018734.2 | Guanylate nucleotide binding protein 3 (Gbp3) | -8.96 | |
| 20 | 14469 | NM_010260.1 | Guanylate nucleotide binding protein 2 (Gbp2) | -7.02 | |
| 24 | 17329 | NM_008599 | Chemokine (C-X-C motif) ligand 9 (Cxcl9) | -6.51 | |
| 30 | 60533 | NM_021893.2 | CD274 antigen (Cd274) | -5.54 | |
| 33 | 16912 | NM_013585.1 | Proteasome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2) (Psmb9) | -5.31 | |
| 34 | 21356 | NM_001025313.1 | TAP binding protein (Tapbp) | -5.23 | |
| 49 | 16913 | NM_010724 | Proteasome (prosome, macropain) subunit, beta type 8 (large multifunctional peptidase 7) (Psmb8) | -4.38 | |
| 56 | 16362 | NM_008390.1 | Interferon regulatory factor 1 (Irf1) | -4.16 | |
| 64 | 21354 | NM_013683.1 | Transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) (Tap1) | -3.96 | |
| 65 | 24088 | NM_011905.2 | Toll-like receptor 2 (Tlr2) | -3.96 | |
Selected expression comparisons between markers from IkkβF/F and IkkβΔhep freshly isolated hepatocytes are listed; the markers with these comparatively most disparate expression levels are clustered within two functional groups.
Enrichment, E**, as measured by relative microarray chip hybridization signals, is defined as follows: a negative (−) E value signifies that IkkβΔhep > IkkβF/F, and a positive (+) E value signifies that IkkβF/F > IkkβΔhep (because the initial comparative chip data were expressed as X-fold enrichment of hybridization markers from IkkβF/F compared to those from IkkβΔhep RNA samples).
Western blots
Animals were sacrificed and tissues harvested at 58 and 48 days (see Figures 2A [lanes 3-6] and 2B [lanes 1-4], both panels, respectively), and at 125 days (see Figure 2B [lanes 5 and 6], both panels) of age. Liver extracts (30 μg proteins/lane) and cultured MEF extracts (40 μg) were analyzed on non-reducing prefabricated 10% SDS-PAGE gels (Invitrogen, Carlsbad, CA; Koch et al., 2009). Proteins were transferred to nitrocellulose membranes using an Xcell II blot module (Invitrogen, Carlsbad, CA). Membranes were incubated overnight with either primary (‘1°’) rat anti-mouse CD74 antibody (clone In-1, BD Biosciences Pharmingen, San Diego, CA, catalog (cat.) #555317) or with matched rat IgG2b,κ isotype control (BD Biosciences Pharmingen, San Diego, CA, cat. #553986), washed, incubated 2 hr with secondary (‘2°’) horse radish peroxidase (HRP)-conjugated goat anti-rat IgG (H+L) (Pierce, ThermoScientific, Rockford, IL, cat. #31470), washed, and incubated 1 hr with ECL-Western-Blotting substrate according to the supplier's instructions (Pierce, ThermoScientific, Rockford, IL, cat. #32106). Loading control blots using identical extracts were performed with anti-GRP94 (gp96) antibody (Santa Cruz Biotechnologies, Santa Cruz, CA, cat. #sc-71182). Chemiluminescence was visualized by 5-30 sec exposure to Amersham ECL-Hyperfilm (GE Healthcare Biosciences, Piscataway, NJ, cat. #28-9068-36). Results of Western blots were obtained from different extracts from two independent experiments.
Figure 2.
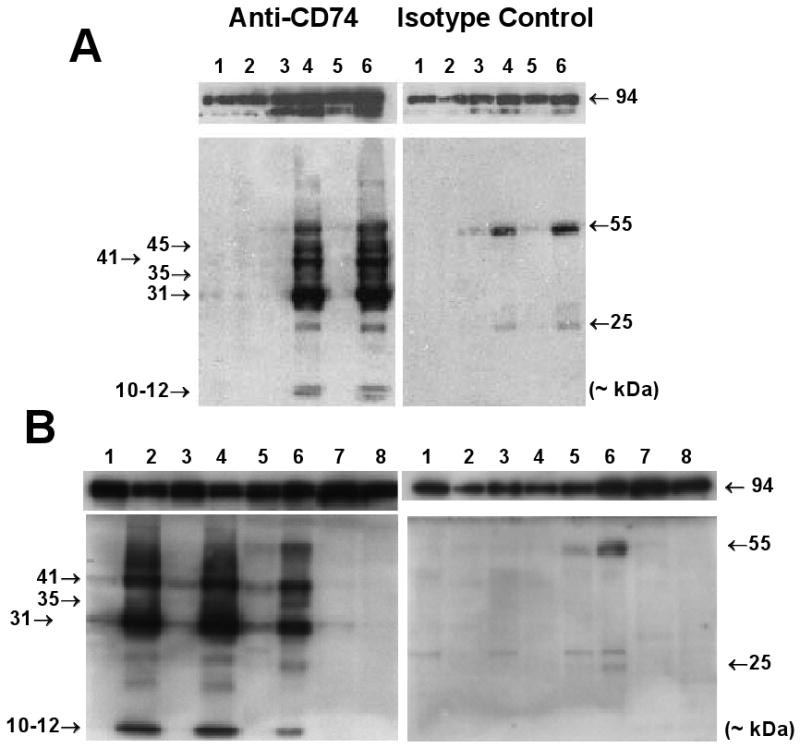
Detection of CD74 in cell extracts on Western Blots. (A) IkkβF/F and IkkβΔhep mouse livers, and cultured MEFs (each lane from a separate mouse). Lanes: 1 (wildtype MEFs), 2 (Ikkβ-/- MEFs), 3 (♂IkkβF/F liver), 4 (♂IkkβΔhep liver), 5 (♀IkkβF/F liver), 6 (♀IkkβΔhep liver). Non-specific ∼25 kDa and ∼55 kDa bands in isotype control panels (right upper and lower) likely reflect non-specific cross-reactions with serum or reagent light and heavy IgG chains. (B) Ikkβ+/+:Alb-Cre mouse livers and spleens, and cultured MEFs (each lane from a separate mouse). Lanes: 1 (♂ #1, Alb-Cre liver), 2 (♂ #1, Alb-Cre spleen), 3 (♂ #2, Alb-Cre liver), 4 (♂ #2, Alb-Cre spleen), 5 (♂IkkβF/F liver), 6 (♂IkkβΔhep liver), 7 (Ikkβ-/- MEFs), 8 (wildtype MEFs). Loading controls (both panels) showed comparative intensities of 94-96 kDa bands in all lanes.
Immunohistochemistry
Animals were sacrificed and liver tissues (portions of the right lobe) harvested at 58 days of age. Thin sections (5 μm) of paraffin blocks of fixed tissues (4% paraformaldehyde in PBS, 24 hr at 4 °C), and routine hematoxylin and eosin (H&E) stained sections were prepared by standard automated procedures in the UCSD Cancer Center Histology Core (http://cancer.ucsd.edu/Research/Shared/histology.asp). For immunohistochemical staining, endogenous peroxidase activity was blocked in deparaffinized sections with 3% H2O2. Sections were pretreated with Retrievagen A* according to the supplier's protocol (BD Pharmingen, San Diego, CA, cat. #550524), followed by 10% fetal bovine serum (Gibco Invitrogen, Carlsbad, CA) to block non-specific binding. Slides were washed, and incubated with 1° anti-CD74 antibody or with matched isotype control rat IgG2b,κ antibody at final concentrations of 5 μg/mL for 1 hr at 21°C. Slides were washed, treated with biotinylated goat 2° anti-rat Ig (BD Pharmingen, San Diego CA, cat. #51-7605KC) for 30 min at 21 °C, washed and treated with Streptravidin-HRP (BD Pharmingen, San Diego CA, cat. #551013) for 30 min at 21 °C. After further washing, the slides were incubated with diaminobenzidine solution (BD Pharmingen, San Diego CA, cat. #551013) for 0.5-5 min (or, as needed), washed, counterstained in Meyer's hematoxylin, dehydrated and coverslipped. Digital images and information (available on request) were acquired with NIS-Elements F 2.30 software, by Nikon DS-Ri1 camera, from Nikon Diaphot with 20× (Fig. 3) or 4× (Fig. 4) objectives. Independent immunohistochemical staining experiments utilizing sections from at least two animals of each genotype were performed multiple times.
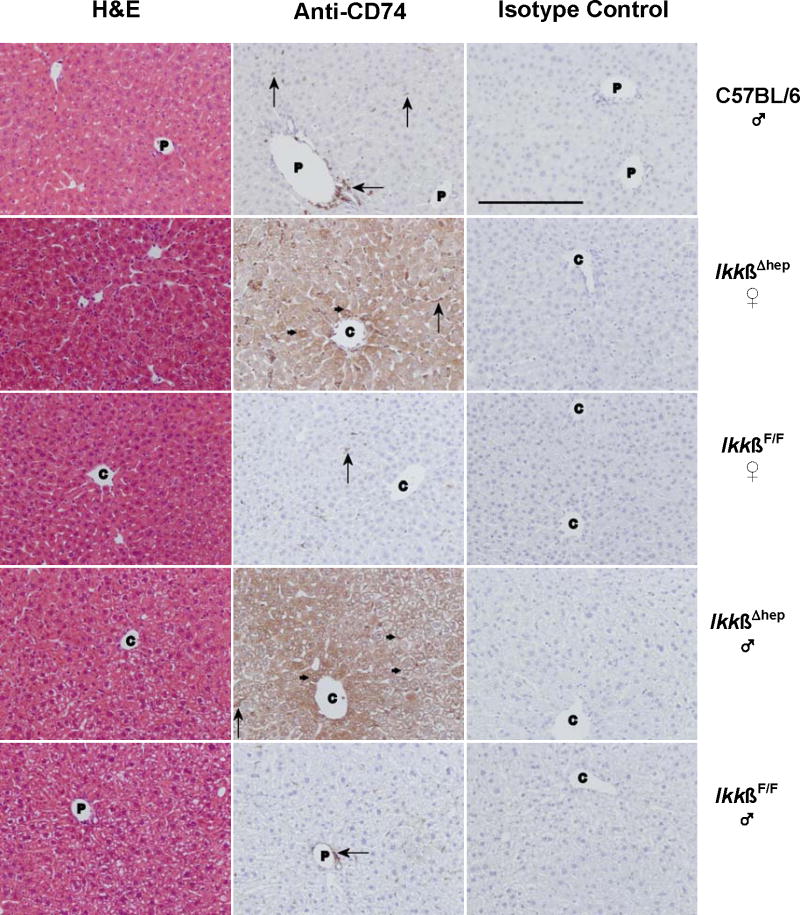
Figure 3.
Histochemical studies of CD74 in livers from C57BL/6 ♂, and IkkβF/F and IkkβΔhep ♀ and ♂ mice. Photomicrographs of liver tissue sections stained with H&E, anti-CD74 antibody, or isotype control antibody are shown in the left-, middle-, and right-hand columns, respectively. The sections are from individual C57BL/6 (row 1), IkkβΔhep ♀ (row 2), IkkβF/F ♀ (row 3), IkkβΔhep ♂ (row 4), and IkkβF/F ♂ (row 5) mice. Annotations indicate hepatocyte membrane (small →) and NPC (large →) staining; C, central vein; P, portal triad. (The unannotated H&E stained IkkβΔhep ♀ liver section appears to approach a central region.) Specific CD74 staining is present in hepatocytes from IkkβΔhep mice but not in hepatocytes from C57BL/6 and IkkβF/F mice. Tissue-specific CD74 staining is not seen in sections treated with isotype control antibody. Objective magnification = 20× (the solid bar, lower left, in the C57BL/6 isotype control panel = 200 μm; the scale bar is identical for experimental panels).
Figure 4.
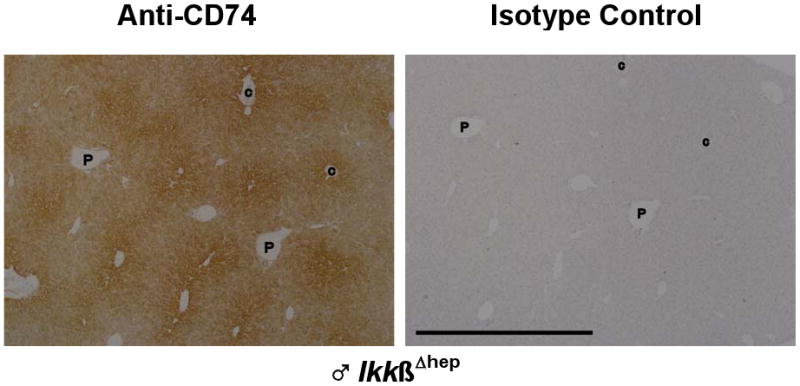
Enriched CD74 staining in IkkβΔhep hepatocytes in midzonal-to-centrilobular regions. Serial sections from a ♂ IkkβΔhep liver, stained with either anti-CD74 or isotype control antibodies as described in the text and in Fig. 3, are here placed in register. Low objective magnification (4×) was used to illustrate the zonal gradient of staining intensity. Hepatocytes proximal to central veins (C) are more intensely stained than hepatocytes proximal to portal triads (P). At lower magnification, scattered NPC staining is more difficult to visualize. The few notations are placed within the same vessels of both serial sections to facilitate registration of the two panels. The solid bar, lower left, in the isotype control panel = 1000 μm (the scale bar is identical for the experimental panel).
Results
Microarray profiling shows upregulation of immune phenotypic expression in IkkβΔhep hepatocytes
Liver tissue and isolated hepatocytes from both genotypes (four separate samples) were compared to see whether profiling differences would be more or less enriched in hepatocytes compared to total liver. RNA sizing and DNA genotyping were performed prior to profiling studies. Intact RNA of high quality (Fig. 1A), and PCR products consistent with wild type and Ikkβ-deleted genomes were observed (Fig. 1B) (Maeda et al., 2003). Microarray hybridizations were performed in triplicate with formatting as diagrammed in Figure 1C.
Key findings in the largest expression group (‘Biological Process’) are shown for freshly isolated livers and freshly isolated hepatocytes in Tables 1 and 2, respectively. Two functional sub-families topped both lists at very high levels of statistical significance (albeit in reverse order): liver (immune system process, p ∼6.5 × 10-28; defense response, p ∼2 × 10-27); hepatocytes (defense response, p = 1.4 × 10-21; immune system process, p ∼1.2 × 10-18). Examination of the 6-19 genes topping these lists showed that all were expressed at relatively higher levels in IkkβΔhep samples, except for two which were higher in IkkβF/F liver tissue (histocompatibility 2, blastocyst H2-Bl [both sub-families]) and one which was apparently higher in IkkβF/F hepatocytes (CD163 antigen [defense response]).
Enhanced CD74 expression in IkkβΔhep samples ranked highest throughout; CD74 was enriched ∼38-fold in isolated IkkβΔhep hepatocytes, and ∼8-fold in IkkβΔhep liver tissue (a 5-fold enrichment following hepatocyte isolation; see both Tables 1 and 2). Profiling also indicated that additional immune process and defense genes were significantly upregulated >4-10-fold in IkkβΔhep hepatocytes, including MHC class II I-Aα, I-Aβ, and I-Eβ chains, CD44, Cxcl9, guanylate nucleotide binding proteins 1-3, TAP, Tlr2, interferon gamma (IFN-γ) inducible protein 47 and IFN-γ regulatory factor 1 (see both Tables 1 and 2). Direct inspection of the microarray data also revealed augmented expression levels of IkkβΔhep hepatocyte CIITA, STAT1, cathepsin S, co-activator CD86 and programmed death ligand-1 (PD-L1) (> 6-, 5.4-, 3.6-, 2- and 5.5-fold, respectively), all significantly greater than isolated liver tissue. As shown in Table 3, the available data from microarray comparisons between IkkβΔhep hepatocytes and Ikkβ-/- MEFs revealed few similarities (Chen et al., 2006).
Table 3.
Microarray profiling: Partial comparisons between mouse embryo fibroblasts (MEFs, Ikkβ-/- vs. wild type MEFs) and freshly isolated hepatocytes (IkkβΔhep vs. IkkβF/F).
| Gene | MEFs (Ikkβ-/- vs. wt) |
Hepatocytes (IkkβΔhep vs. IkkβF/F) |
GenBank Acc. # | Function |
|---|---|---|---|---|
| X-fold enrichment* | ||||
| CARP | +156.3 | -1.21 | AF041847 | Cardiac ankyrin repeat protein, binding to myopalladin |
| Versican | +9.7 | NR | D45889 | Anti-cell adhesive molecules |
| α-Actin | +2.6 | +1.28 | X13297 | Cytoskeleton, stress fibers |
| Prothymosin β4 | +2.34 | -1.98 | U38967 | Prevents actin polymerization |
| S100A4 | +2.2 | NR | M36579 | Tumor metastasis, calcium-binding protein |
| Tropomyosin 2 | +2.0 | -1.47 | M22479 | Actin-binding protein |
| IGFBP2 | -134 | -1.79 | X81580 | Inhibitor of cell mitogenesis |
| TIMP-2 | -5.8 | -2.32 | X62622 | Inhibitor of matrix metalloproteinases |
| TIMP-3 | -4.0 | -1.99 | U26437 | Inhibitor of matrix metalloproteinases |
| VCAM | -2.9 | +5.62 | M84487 | Vascular cell adhesion molecule |
| Integrin β5 | -2.0 | -1.2 | AF022110 | Cell adhesion |
X-fold enrichment values for MEFs were taken from Chen et al., 2006. The X-fold enrichment value is here in Table 3 defined as the relative comparison of RNA hybridization expression of MEF Ikkβ-/- to MEF Ikkβ+/+, or of IkkβΔhep to IkkβF/F such that a positive (+) value signifies that MEF Ikkβ-/- > MEF Ikkβ+/+, or that IkkβΔhep > IkkβF/F; and, a negative (−) value signifies that MEF Ikkβ+/+ > MEF Ikkβ-/-, or that IkkβF/F > IkkβΔhep. Note that this definition is here the reverse of that in Tables 1 and 2, in order to establish congruence with the E values previously published for the MEF Ikkβ-/- and Ikkβ+/+ pair.
Ectopic expression of CD74 proteins is upregulated in IkkβΔhep hepatocyte extracts
Four specific bands (Mr ∼ 31, 35, 41 kDa, and a 10-12 kDa doublet) were detected in IkkβΔhep liver extracts by antibody specific to cytosolic CD74 epitopes (Figs. 2A and 2B [left panels]). CD74-specific bands were not seen in similar extracts probed with isotype control antibody (Figs. 2A and 2B [right panels]). Band intensities were similar in ♂ and ♀ samples; the relative intensities of p31 were ∼2.5-fold higher than p41 as expected (Bertolino and Rabourdin-Combe, 1996). In contrast, some (p31 and p41) but not all (p35, p45 and the 10-12 kDa doublet) CD74-specific bands were detectable at significantly lower levels in IkkβF/F liver extracts (Figs. 2A and 2B [left panels]); these bands were likely due to CD74 expressed by non-parenchymal cells (NPCs) in the intact livers (Fig. 3). The finding of CD74-specific expression in IkkβΔhep liver extracts was not an artifact of albumin promoter-driven Cre expression, as CD74 expression levels in liver extracts from Ikkβ+/+:Alb-Cre mice were similar to those from IkkβF/F mice, with no detectable p35, p45, or 10-12 kDa doublet (Fig. 2B).
As expected from profiling (Chen et al., 2006), CD74 was undetectable in wildtype MEF extracts whereas extracts from Ikkβ-/- MEFs variably expressed very low levels of p31 and p41 bands but undetectable levels of p35 or the 10-12 kDa doublet (Figs. 2A and 2B [left panels]). The significance of these observations is currently unclear.
Ectopic and specific expression of CD74 is observed in situ in IkkβΔhep hepatocytes
To localize hepatic CD74 expression, livers from ♂ C57BL/6, and ♂ and ♀ IkkβF/F and IkkβΔhep mice were analyzed by routine histology and immunohistochemistry. In all cases, the same 1° antibody used for Western blots was employed; with respect to IkkβF/F and IkkβΔhep mice, portions of the same tissue samples used for Western blots (Fig. 2A) were fixed for histochemistry (Figs. 3 and 4).
Normal histology was observed by H&E staining of C57BL/6 liver (Fig. 3, row 1). As expected (Maubach et al., 2007; Momburg et al, 1986), specific CD74 staining was observed in NPCs bordering a portal triad, and also scattered sparsely throughout the liver. In contrast, specific staining was not observed either in hepatocytes, or in cells bordering the central vein, or in sections incubated with isotype control antibody (Fig. 3, row 1). The faint CD74-positive p31 and p41 bands on Western blots of liver extracts from Ikkβ+/+:Alb-Cre and IkkβF/F mice (Figs. 2A and 2B) may reflect NPC staining. Further work is needed for precise identification of NPCs, whether dendritic, HSC, Kupffer, sinusoidal endothelial and/or biliary ductular epithelial cells, which are responsible for this CD74-positive staining.
As shown in Fig. 3 (rows 2-5), H&E staining showed no morphological differences between IkkβΔhep, and control IkkβF/F, ♀ or ♂ livers. However, intense CD74 staining was observed in virtually all IkkβΔhep hepatocytes throughout liver acini and regardless of gender. Specific CD74 staining in IkkβΔhep hepatocytes was associated with cytoplasmic and with membrane sites. In contrast, although scattered periportal and acinar staining was observed in NPCs, specific CD74 staining was undetectable in IkkβF/F hepatocytes. No CD74 staining was observed in any liver sections incubated with isotype control antibody (Fig. 3) or in the absence of 1° antibody (data not shown).
By observation of liver sections at lower magnification (Fig. 4), specific CD74 staining in IkkβΔhep livers appeared to be more intense in hepatocytes bordering central veins, and to decrease slightly and gradually in intensity from centrilobular to periportal zones.
Discussion
Immunohistochemical, microarray profiling, and Western blot findings show ectopic and gender-independent expression of major, minor and putatively processed CD74 isoforms in hepatocytes deleted in Ikkβ (IkkβΔhep). Immunohistochemistry suggests that antibody-specific CD74 expression occurs in all hepatocytes, as might be expected from Ikkβ-deletion analyses indicating no undeleted product in a population of isolated IkkβΔhep hepatocytes (Maeda et al., 2003; Koch et al, 2009); however, the isoform(s) responsible for the observed immunochemical reactivities have not been specifically identified. The prominent 35 kDa band in IkkβΔhep liver extracts might represent another CD74 murine isoform, so far unreported; the nature of the 45 kDa band observed in younger but not older mice (compare Fig. 2A with Fig. 2B) is unclear. The 10-12 kDa doublet, also observed in spleens from Ikkβ+/+:Alb-Cre mice (Fig. 2B [left panel]), might be a form of RIP-processed CD74 (Becker-Herman et al., 2005); if functional, this might account for residual NF-κB activity in IkkβΔhep hepatocytes (Maeda et al., 2003).
The occurrence of multiple isoforms complicates interpretation of immunohistochemical observation of enriched midzonal-to-centrilobular localization of hepatocellular CD74, since site-enrichment could be due either to increased expression of all or selected isoforms, or to increased availability of antigenic sites of one or more equally abundant isoforms. Alternatively, owing to the relatively short paraformaldehyde fixation time, and to subsequent variably effective antigen retrieval steps, zonal staining intensity differences might reflect differential fixation of antigenic sites accessible to the anti-CD74 antibody employed. Whereas longer fixation times (> 24 hr) clearly reduce antigen detection (data not shown), zonal differences were still visible. Therefore, because the zonal differentiation of hepatocytes is responsible for many functional attributes of liver under both normal, and pathological conditions, further experiments are needed to clarify these observations as well as to eliminate other possible fixation artifacts.
Although the upregulation of CD74 expression in hepatocytes appears to be a consequence of targeted hepatocellular deletion of Ikkβ in IkkβΔhep mice, Ikkβ deletion alone is not a uniformly sufficient condition of CD74 upregulation, because Ikkβ-/- MEFs express only barely detectable levels of CD74, as revealed by Western blots (Fig. 2), and none by RNA profiling (Chen et al., 2006). While the mechanisms responsible for ectopic hepatocellular CD74 expression remain to be determined, they do not appear to be random or isolated. This conclusion is supported by liver micorarray profiling results which reveal, with very high statistical significance, that augmented CD74 expression in IkkβΔhep hepatocytes is concomitant with enriched expression of two large families of immune system-related genes, among them CD44, MHC class II I-Aα, I-Aβ, and I-Eβ chains, CIITA and PD-L1 mRNAs, the functions of which are involved in antigen processing, host defense and liver tolerance (Tiegs and Lohse, 2009). This enrichment is selectively greater in isolated hepatocytes compared to that in unfractionated liver extracts. Although further work is needed to show the translation of these mRNAs directly and to characterize the cellular locations of these putatively translated proteins, these additional findings are surprising and intriguing because the expression of all five of these molecules, like CD74, is usually restricted to lymphoid (Abbas et al., 2010; Rafi-Janajreh et al., 1998) and NPCs (Tiegs and Lohse, 2009).
The collective, and significantly augmented, expression of CD74 and these two gene families in IkkβΔhep hepatocytes suggests several interrelated hypotheses about ectopic immunologic functions and susceptibility to HCC in these mice, as discussed elsewhere (Koch and Leffert, 2010).
Acknowledgments
This work was supported by NIH (CA113602, AI067354) and the Superfund Basic Research Program (P42ES010337). Breeding colonies were started with mice generously supplied by M. Karin (UCSD), whom we also thank for the suggestion to examine Ikkβ+/+:Alb-Cre mice. We thank K. Juson, M. Zhang and A. Elbehti for technical assistance; Anne Chang (UCSD, Karin Laboratory) for wildtype and Ikkβ-/- MEFs and Ikkβ+/+:Alb-Cre mice; and R. Sasik (UCSD BIOGEM Core) for statistical analyses and organization of microarray profiling data.
Abbreviations
- Alb
refers to albumin-specific gene promotor
- BLTLF
Bacille-Calmette-Guerin-primed lipopolysaccharide-induced T cell-mediated acute liver failure
- ConA
lectin concanavalin A
- CIITA
transcriptional coactivator of MHC class II expression
- Cre
cre recombinase
- DEN
diethylnitrosamine
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- H&E
hematoxylin and eosin
- HRP
horse radish peroxidase
- HSC
hepatic stellate cell
- IFN-γ
interferon γ
- Ii
invariant chain CD74 chaperone
- IKK
IκB complex
- IKKβ
protein kinase subunit of IKK
- IkkβF/F
mouse strain carrying Cre-recombinase sites (F/F) spanning Ikkβ exon 3
- IkkβΔhep
mouse strain IkkβF/F:Alb-Cre, specifically deleted in hepatocyte Ikkβ
- IL
interleukin
- JNK
c-JUN nuclear kinase
- LPS
lipopolysaccharide
- MAPK
mitogen-activated protein kinase
- MEFs
mouse embryonic fibroblasts
- MHC
major histocompatibility
- MIF
macrophage migration inhibitory factor
- NPC
non-parenchymal cell
- NF-κB
nuclear factor kappa B
- PD-L1
programmed death ligand
- RIP
regulated intramembrane proteolysis
- ROS
reactive oxygen species
- STAT
signal transducers and activator of transcription
- TNF
tumor necrosis factor
- TNFR2
tumor necrosis factor receptor 2
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbas AK, Lichtman AH, Pillai S. Cellular and Molecular Immunology. Updated 6th. Philadelphia: Saunders Elsevier; 2010. [Google Scholar]
- Becker-Herman S, Arie G, Medvedovsky H, Kerem A, Shachar I. CD74 is a member of the regulated intramembrane proteolysis-processed protein family. Mol Biol Cell. 2005;11:5061–9. doi: 10.1091/mbc.E05-04-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolino P, Rabourdin-Combe C. The MHC class II-associated invariant chain: a molecule with multiple roles in MHC class II biosynthesis and antigen presentation to CD4+ T cells. Crit Rev Immunol. 1996;16:359–79. [PubMed] [Google Scholar]
- Bozza M, Satoskar AR, Lin G, Lu B, Humbles AA, Gerard C, et al. Targeted disruption of migration inhibitory factor gene reveals its critical role in sepsis. J Exp Med. 1999;189:341–6. doi: 10.1084/jem.189.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdi M, Reilly TP, Elkahloun AG, George JW, Pohl LR. Macrophage migration inhibitory factor in drug induced liver injury: a role in susceptibility and stress responsiveness. Biochem Biophys Res Commun. 2002;294:225–30. doi: 10.1016/S0006-291X(02)00466-7. [DOI] [PubMed] [Google Scholar]
- Calandra T, Roger T. Macrophage migration inhibitory factor: a regulator of innate immunity. Nature Reviews Immunology. 2003;3:791–800. doi: 10.1038/nri1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Lu Y, Castranova V, Li Z, Karin M. Loss of Ikkbeta promotes migration and proliferation of mouse embryo fibroblast cells. J Biol Chem. 2006;281:37142–9. doi: 10.1074/jbc.M603631200. [DOI] [PubMed] [Google Scholar]
- de Jong YP, Abadia-Molina AC, Satoskar AR, Clarke K, Rietdijk ST, Faubion WA, et al. Development of chronic colitis is dependent on the cytokine MIF. Nat Immunol. 2001;2:1061–6. doi: 10.1038/ni720. [DOI] [PubMed] [Google Scholar]
- Faure-André G, Vargas P, Yuseff MI, Heuzé M, Diaz J, Lankar D, et al. Regulation of dendritic cell migration by CD74, the MHC class II-associated invariant chain. Science. 2008;322:1705–10. doi: 10.1126/science.1159894. [DOI] [PubMed] [Google Scholar]
- Feng Z, Davis DP, Sásik R, Patel HH, Drummond JC, Patel PM. Pathway and gene ontology based analysis of gene expression in a rat model of cerebral ischemic tolerance. Brain Res. 2007;1177:103–23. doi: 10.1016/j.brainres.2007.07.047. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Karin M. Missing pieces in the NF-κB puzzle. Cell. 2002;109:S81–S96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- Heyninck K, Wullaert A, Beyaert R. Nuclear factor-κB plays a central role in tumour necrosis factor-mediated liver disease. Biochem Pharmacol. 2003;66:1409–15. doi: 10.1016/s0006-2952(03)00491-x. [DOI] [PubMed] [Google Scholar]
- Iwaki T, Sugimura M, Nishihira J, Matsuura T, Kobayashi T, Kanayama N. Recombinant adenovirus vector bearing antisense macrophage migration inhibitory factor cDNA prevents acute lipopolysaccharide induced liver failure in mice. Lab Invest. 2003;83:561–70. doi: 10.1097/01.lab.0000062857.26210.ef. [DOI] [PubMed] [Google Scholar]
- Javeed A, Zhao Y, Zhao Y. Macrophage-migration inhibitory factor: role in inflammatory diseases and graft rejection. Inflamm Res. 2008;57:45–50. doi: 10.1007/s00011-007-7110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamata H, Honda S, Maeda S, Chang L, Hirata H, Karin M. Reactive oxygen species promote TNFalpha induced death and sustained JNK activation by inhibiting MAP kinase phosphatases. Cell. 2005;120:649–61. doi: 10.1016/j.cell.2004.12.041. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Nishihira J, Watanabe S, Todo S. Prevention of lethal acute hepatic failure by antimacrophage migration inhibitory factor antibody in mice treated with bacille Calmette-Guerin and lipopolysaccharide. Hepatology. 1999;29:1752–9. doi: 10.1002/hep.510290610. [DOI] [PubMed] [Google Scholar]
- Koch KS, Leffert HL. Hypothesis: Targeted Ikkβ deletion upregulates MIF signaling responsiveness and MHC class II expression in mouse hepatocytes. Hepatic Medicine: Evidence and Research. 2010 doi: 10.2147/HMER.S7208. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch KS, Maeda S, He G, Karin M, Leffert HL. Targeted deletion of hepatocyte Ikkβ confers growth advantages. Biochem Biophys Res Communic. 2009;380:349–54. doi: 10.1016/j.bbrc.2009.01.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagi T, Akbar F, Horiike N, Onji M. Increased serum levels of macrophage migration inhibitory factor in alcoholic liver diseases and their expression in liver tissues. Clin Biochem. 2001;34:189–93. doi: 10.1016/s0009-9120(01)00214-4. [DOI] [PubMed] [Google Scholar]
- Lan HY. Role of macrophage migration inhibition factor in kidney disease. Nephron Exp Nephrol. 2008;109:e79–e83. doi: 10.1159/000145463. [DOI] [PubMed] [Google Scholar]
- Laurent S, Horsmans Y, Stärkel P, Leclercq I, Sempoux C, Lambotte L. Disrupted NF-κB activation after partial hepatectomy does not impair hepatocyte proliferation in rats. World J Gastroenterol. 2005;11:7345–50. doi: 10.3748/wjg.v11.i46.7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng L, Bucala R. Insight into the biology of macrophage migration inhibitory factor (MIF) revealed by the cloning of its cell surface receptor. Cell Res. 2006;16:162–8. doi: 10.1038/sj.cr.7310022. [DOI] [PubMed] [Google Scholar]
- Lue H, Kapurniotu A, Fingerle-Rowson G, Roger T, Leng L, Thiele M, et al. Rapid and transient activation of the ERK MAPK signaling pathway by macrophage migration inhibitory factor (MIF) and dependence on JAB1/CSN5 and Src kinase activity. Cell Signal. 2006;18:688–703. doi: 10.1016/j.cellsig.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Maeda S, Chang L, Li ZW, Luo JL, Leffert H, Karin M. IKKβ is required for prevention of apoptosis mediated by cell-bound but not by circulating TNFα. Immunity. 2003;19:725–37. doi: 10.1016/s1074-7613(03)00301-7. [DOI] [PubMed] [Google Scholar]
- Maeda S, Kamata H, Luo JL, Leffert H, Karin M. IKKβ couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell. 2005;121:977–90. doi: 10.1016/j.cell.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Malato Y, Sander LE, Liedtke C, Al-Masaoudi M, Tacke F, Trautwein C, et al. Hepatocyte-specific inhibitor-of-κB-kinase deletion triggers the innate immune response and promotes earlier cell proliferation during liver regeneration. Hepatology. 2008;47:2036–50. doi: 10.1002/hep.22264. [DOI] [PubMed] [Google Scholar]
- Maubach G, Lim MC, Kumar S, Zhuo L. Expression and upregulation of cathepsin S and other early molecules required for antigen presentation in activated hepatic stellate cells upon IFN-gamma treatment. Biochim Biophys Acta. 2007;1773:219–31. doi: 10.1016/j.bbamcr.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Momburg F, Koch N, Möller P, Moldenhauer G, Butcher GW, Hämmerling GJ. Differential expression of Ia and Ia-associated invariant chain in mouse tissues after in vivo treatment with IFN-gamma. J Immunol. 1986;136:940–8. [PubMed] [Google Scholar]
- Nakajima H, Takagi H, Horiguchi N, Toyoda M, Kanda D, Otsuka T, et al. Lack of macrophage migration inhibitory factor protects mice against concanavalin A-induced liver injury. Liver Int. 2006;26:346–51. doi: 10.1111/j.1478-3231.2005.01216.x. [DOI] [PubMed] [Google Scholar]
- Rafi-Janajreh AQ, Nagarkatti PS, Nagarkatti M. Role of CD44 in CTL and NK cell activity. Front Biosci. 1998;3:d665–71. doi: 10.2741/a311. [DOI] [PubMed] [Google Scholar]
- Sakurai T, He G, Matsuzawa A, Yu GY, Maeda S, Hardiman G, et al. Hepatocyte necrosis induced by oxidative stress and IL-1α release mediate carcinogen-induced compensatory proliferation and liver tumorigenesis. Cancer Cell. 2008;14:156–65. doi: 10.1016/j.ccr.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumptner-Cuvelette P, Benaroch P. Multiple roles of the invariant chain in MHC class II function. Biochim Biophys Acta. 2002;1542:1–13. doi: 10.1016/s0167-4889(01)00166-5. [DOI] [PubMed] [Google Scholar]
- Swant JD, Rendon BE, Symons M, Mitchell RA. Rho GTPase-dependent signaling is required for macrophage migration inhibitory factor-mediated expression of cyclin D1. J Biol Chem. 2005;280:23066–72. doi: 10.1074/jbc.M500636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiegs G, Lohse AW. Immune tolerance: What is unique about the liver. J Autoimmun. 2009 Aug 28; doi: 10.1016/j.jaut.2009.08.008. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Ting JP, Trowsdale J. Genetic control of MHC class II expression. Cell. 2002;109:S21–33. doi: 10.1016/s0092-8674(02)00696-7. [DOI] [PubMed] [Google Scholar]
- Zhang HY, Nanji AA, Luk JM, Huang XR, Lo CM, Chen YX, et al. Macrophage migration inhibitory factor expression correlates with inflammatory changes in human chronic hepatitis B infection. Liver Int. 2005;25:571–9. doi: 10.1111/j.1478-3231.2005.01047.x. [DOI] [PubMed] [Google Scholar]