Abstract
In eukaryotic cells, the Rad6/Rad18-dependent monoubiquitination of the proliferating cell nuclear antigen (PCNA) plays an essential role in the switching between replication and translesion DNA synthesis (TLS). The DNA polymerase Polη binds to PCNA via a consensus C-terminal PCNA-interacting protein (PIP) motif. It also specifically interacts with monoubiquitinated PCNA thanks to a recently identified ubiquitin-binding domain (UBZ). To investigate whether the TLS activity of Polη is always coupled to PCNA monoubiquitination, we monitor the ability of cell-free extracts to perform DNA synthesis across different types of lesions. We observe that a cis-syn cyclobutane thymine dimer (TT-CPD), but not a N-2-acetylaminofluorene-guanine (G-AAF) adduct, is efficiently bypassed in extracts from Rad18-deficient cells, thus demonstrating the existence of a Polη-dependent and Rad18-independent TLS pathway. In addition, by complementing Polη-deficient cells with PIP and UBZ mutants, we show that each of these domains contributes to Polη activity. The finding that the bypass of a CPD lesion in vitro does not require Ub-PCNA but nevertheless depends on the UBZ domain of Polη, reveals that this domain may play a novel role in the TLS process that is not related to the monoubiquitination status of PCNA.
INTRODUCTION
The translesion DNA synthesis (TLS) pathway is responsible for the vast majority of point mutations and is thus considered as a major process leading to genetic instability and carcinogenesis. Understanding the molecular mechanisms underlying this damage tolerance pathway and its regulation is therefore of major importance for our understanding of human pathogenesis.
In eukaryotes, TLS is carried out by specialized, low stringency, DNA polymerases belonging to the Y family (Polη, Polι, Polκ and Rev1) and the B family (Polζ). In vivo or in vitro studies have shown that these DNA polymerases have various substrate specificities and that, in many cases, TLS requires the concerted action of several TLS polymerases (1,2). Remarkably, human Polη has the unique property to replicate past cis-syn cyclobutane thymine dimers, one of the major photoproducts induced by UV irradiation, with the same efficiency and accuracy than it does on undamaged DNA (3). The loss of Polη activity underlies the high susceptibility to skin cancers of Xeroderma pigmentosum variant (XP-V) patients (4,5).
The mechanism by which TLS DNA polymerases gain access to the lesion site and take over the replicative polymerase to incorporate nucleotides opposite the damaged base is the subject of intense research. Numerous studies have highlighted the central role of replication processivity clamps (β-clamp in prokaryotes and PCNA in eucaryotes) in the fine tuning between replication and TLS. Yeast Polη and human TLS polymerases such as Polη, ι and κ functionally interact with the interconnecting loop of PCNA via their PCNA-interacting protein (PIP) motif (6–8). Mutational inactivation of the PIP domain of Polη sensitizes yeast cells to UV irradiation (9), while homologous mutations confer only moderate UV sensitivity in human cells (10,11). This suggests alternative targeting process(es) for the human polymerase. Recently, Acharya et al. (12) have identified a functional secondary PIP domain within the human Polη that may explain the above-mentioned modest sensitivity. Furthermore, treatment of yeast or human cells with agents that affect DNA replication promotes the monoubiquitination of PCNA at its K164 residue by the Rad6–Rad18 enzyme complex (13). Genetic studies in Saccharomyces cerevisiae showed an epistatic relationship between PCNA-K164R mutation (a non ubiquitinable form of PCNA) and deletion of the Polη and Polζ TLS polymerases, demonstrating that TLS in this organism is largely dependent on the monoubiquitination of the replication processivity clamp (14). Vertebrate Y-family DNA polymerases preferentially interact with the monoubiquitinated form of PCNA (15,16) via Ubiquitin (Ub) binding domains designated UBZ (Polη and Polκ) or UBM (Polι and Rev1) that are required for their relocalization in nuclear foci after UV irradiation (10,17–19). In addition, it has been observed that some mutations in the UBZ domain of human Polη have a much more drastic effect on UV cell survival than mutations in the C-terminal PIP domain (10). Consequently, it has been proposed that the binding of Y-family polymerases to the Ub moiety of PCNA is required for their access to the site of a stalled replication fork. Such a model highlighting a crucial role of PCNA ubiquitination in the regulation of TLS should however be tempered by the results on the extensive mutational analysis of the UBZ domain of Polη conducted by Acharya et al. (12,20), which suggest that the binding of the Ub moiety by Polη is not a necessary requirement for the ability of this polymerase to function in TLS of UV-induced DNA lesions.
Besides its role in facilitating the access of TLS polymerases to the lesion site, monoubiquitination of PCNA may also increase the kinetic of TLS by specialized DNA polymerases. Indeed, it has been shown that monoubiquitinated PCNA (Ub-PCNA) is more effective than the unmodified clamp in promoting yeast Polη or Rev1 TLS activity across an abasic site in vitro (21). However, such a kinetic activation by Ub-PCNA was not confirmed in another study (22). Finally, TLS in vertebrate cells appears to be only partially dependent on Rad18 activity since UV-mutagenesis is reduced only 2-fold, in Rad18KO (knock out) mouse cells (23). Furthermore, defective ubiquitination of PCNA in the chicken cell line DT40 is not epistatic to Polκ and Rev1 mutants for UV sensitivity (24,25) indicating that at least these Y-family polymerases may be recruited in a Rad18-independent manner.
Taken together these data illustrate many aspects on the regulation of TLS by the post-translational status of PCNA that remain unclear and much debated. In an effort to elucidate these mechanisms and to investigate whether a Rad18-independent TLS pathway might operate in mammalian cells, we used Rad18-deficient cells and stable XPV fibroblast cell lines (XP30RO) complemented with Polη polymerases mutated in either their PIP or UBZ motifs. Protein extracts from these cell lines were prepared in order to analyze their ability to perform TLS across different DNA lesions. We could thus monitor the activity of Polη in the presence or in the absence of Rad18 and PCNA ubiquitination, and the activity of Polη variants that are differentially affected in their binding capacity to PCNA and/or to ubiquitin. We show that while Rad18 is required for the bypass of a N-2-acetylaminofluorene-guanine (G-AAF) lesion in cell-free extracts, it is dispensable for the bypass of a TT-CPD lesion. This implies that, under the experimental conditions tested here, the Polη-dependent bypass of CPD is independent of Ub-PCNA. However, we also demonstrate that both the PIP and UBZ domains of Polη facilitate the TLS reaction across both lesions. This result indicates that, in addition to its documented role in Ub-PCNA binding, the UBZ domain of Polη may play a novel role in the TLS process that is not related to the ubiquitination status of PCNA.
MATERIAL AND METHODS
Plasmids and cell lines
Cells were grown at 37°C in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum and gentamicin (0.5 mg/ml).
The human colon carcinoma cell line HCT116-Rad18KO cell line (26) was cultured in the presence of G418 (300 µg/ml) and puromycin (0.3 µg/ml). MRC5-V1 cells are SV40-transformed normal human lung fibroblasts (27). The XP30RO cell line (SV40-transformed X. pigmentosum variant human fibroblasts) has a homozygous deletion in the Polη gene resulting in a truncated protein of only 42 amino acids (4). XP30RO cell lines expressing wild type (wt) or mutated forms of Polη were generated by transfection with a pcDNA3.1/zeo(−) plasmid harboring the corresponding Polη sequence (kindly provided by P. Kannouche); transfected cells were thereafter selected with zeocin. Mutations in the coding sequence of Polη were generated by site-directed mutagenesis. The sequences of the primers used for the generation of the D652A mutation are: 5′-CCAGAACACATGGCATATCATTTTGCA-3′ and: 5′-CATATCCCATACCGGTACCAGGGA-3′. The ΔPIP deletion, generated by introducing a stop codon at position 705, consists in a deletion of the last nine amino acids of the Polη amino acid sequence (QTLESFFKPLTH → QTL). The sequences of the primers used for mutagenesis are: 5′-GAATCATTTTTTAAGCCATTAACA-3′ and 5′-ACAATGTTTGCATGCCCTCAGGCCT-3′.
Rad18 gene silencing in MRC5 cells
siRNA design, cloning in pEBV vectors carrying a hygromycin B resistance cassette and establishment of stable knockdown or control cells were carried out as previously described (28). The RNAi sequence for Rad18 (NM_020165) stretches nucleotides 1459–1477. Control cells were obtained by stable transfection with the pBD650 vector that carries an inefficient shRNA (28). The Rad18 knockdown and control cell lines were designated as MRC5-Rad18KD and MRC5-CT, respectively. The transformed MRC5 cell lines were cultured in presence of hygromycin B (150 µg/ml, Roche).
Cell survival analysis
Cells were plated at 4000 (XP30RO) or 2500 (MRC5) cells per 10 cm diameter dish 24 h before UV irradiation (254 nm). After 7 days of culture in the presence of caffeine (1 mM), growing clones were fixed with 4% paraformaldehyde and stained with crystal violet in 10% ethanol. Clones containing more than 50 cells were counted. Experiments were performed at least four times. Colony formation was normalized as a percentage of the non-irradiated control.
Immunostaining
Cells grown on coverslips were UV-irradiated (15 J/m2) and cultured for 4 h. The cells were fixed with methanol/acetone (1/1, v/v) for 30 min at 4°C and washed three times with phosphate-buffered saline (PBS) supplemented with Tween-20 (0.1%; v/v). Cells were stained by overnight incubation (4°C) with a monoclonal anti-Rad18 antibody (Rad18 Ab57447, Abcam, 1:500 dilution) washed three times with PBS supplemented with Tween-20 (0.1%; v/v), and incubated for at least 2 h at room temperature with Alexa Fluor 488 goat anti-mouse IgG (1:2000, Molecular Probes). Observations were made with a Leica TCS4D confocal microscope equipped with an argon/krypton laser and suitable barrier filters.
Immunoblotting
Cell-free extracts (100 µg) were loaded onto SDS/polyacrylamide gels. After electrophoresis, separated proteins were transferred onto a PVDF membrane (Biorad) and probed with antibodies against Polη (C17, sc-5938), Polδ (A9, sc-17776) and PCNA (PC10, sc-56) from Santa Cruz Biotechnology. Anti-HA antibody (HA.11) was purchased from Covance.
Primer extension analysis and in vitro PCNA monoubiquitination assay
Construction of single-stranded plasmids containing either a CPD lesion (pUC-CPD.ss) or a single unique G-AAF adduct (pUC3G1-AAF.ss) has been extensively described (29). Primer extension analysis and standard ubiquitination assays were performed as previously described (30,31). Briefly, the reaction mixture (6.25 µl) containing 10 fmol of primed DNA and the whole-cell extract was incubated at 37°C in 50 mM HEPES–KOH (pH 7.8), 7 mM MgCl2, 1 mM DTT, 4 mM ATP, 500 µM of dNTPs, 40 mM creatine phosphate and 100 µg/ml creatine kinase. The reaction was stopped by adding an equal volume of proteinase K-SDS (4 mg/ml, 2%) and incubated for 30 min at 37°C. Purified replication products were further digested with EcoRI and PvuII restriction enzymes and analyzed by electrophoresis on a polyacrylamide–7 M urea denaturing gel. Quantification of the levels of TLS was determined by phospho-imager analysis.
RESULTS
Rad18 is dispensable for the in vitro bypass of a CPD lesion while it is required for the bypass of an AAF adduct
In order to investigate whether Polη functions were strictly dependent on Ub-PCNA and whether this may depend upon the nature of the DNA lesion, we compared the ability of extracts from Rad18-proficient (wild type) or Rad18-deficient (Rad18KO) HCT116 cells to synthesize DNA opposite either a CPD or an AAF lesion. HCT116-Rad18KO cells were significantly more sensitive than wild-type HCT116 to DNA-damaging agents such as cisplatin and methyl methanesulfonate but only moderately sensitive to UV (26). Strikingly, the difference between wild-type and Rad18-deficient cells was only detectable at high UV doses (Figure 1A). By using single-stranded mono-modified plasmids, we have already shown that normal cell extracts are able to bypass a blocking lesion such as a G-AAF adduct (30). In this assay, the replicative machinery that extends the radiolabeled primer (located 91 nucleotides away from the adduct site), is blocked one nucleotide before the lesion (L-1; Figure 1C). Polη carries out the incorporation of a few nucleotides opposite and beyond the lesion site, and is then probably replaced by the replicative polymerase, as judged by the comparison of the extension profiles obtained with normal and XPV cell extracts (Figure 1C), or with XPV extracts complemented with Polη (Supplementary Figure S1A). Therefore, our in vitro assay may mimic the successive steps promoting the access of TLS polymerases to damaged DNA. Indeed, we have clearly established that PCNA is required for the TLS reaction (Supplementary Figure S1B). Furthermore PCNA is monoubiquitinated during such primer extension reactions (31). This post-translational modification of PCNA depends on both relatively long tracts of DNA synthesis and upon the presence of replication hurdles such as hairpin structures or a single G-AAF adduct or a TT-CPD lesion (31 and Supplementary Figure S2). The ability of cell-free extracts to perform both PCNA monoubiquitination and DNA synthesis across a lesion gives us the unique opportunity to investigate whether the TLS activity of Polη is coupled to this modification.
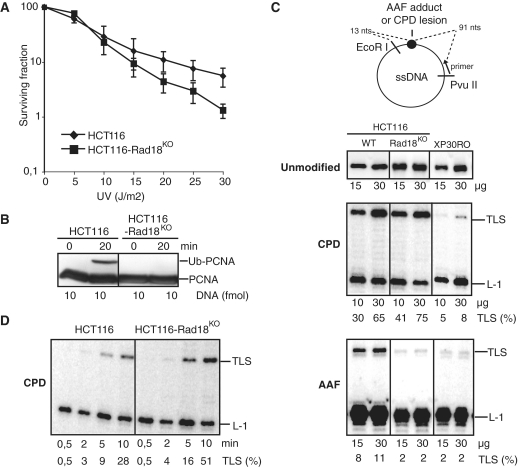
Figure 1.
Analysis of Rad18 dependence of G-AAF or TT-CPD bypass in HCT116 cells. (A) UV survival curves of HCT116 and HCT116-Rad18KO cells. (B) In vitro PCNA monoubiquitination. Cell-free extracts (30 µg) were incubated at 37°C in the presence of all four dNTPs and primed ssDNA (10 fmol) for 0 or 20 min, as indicated. Proteins were analyzed by immunoblotting with an antibody raised against PCNA. (C) Analysis of TLS catalyzed by various amounts of cell-free extracts from either HCT116, HCT116-Rad18KO or XP30RO cell lines as indicated. DNA synthesis products obtained after 20 min incubation with undamaged pUC118.ss (unmodified), pUC-CPD.ss (CPD) or pUC3G1-AAF.ss (AAF) primed templates were analyzed by electrophoresis on a denaturing gel. L-1 and TLS tags: fragment elongated up to one nucleotide before the lesion site and up to the EcoRI restriction site, respectively. (D) Kinetic analysis of the CPD bypass. The pUC-CPD.ss substrate was incubated with cell-free extracts (30 µg) at 20°C and aliquots were withdrawn and quenched at indicated time intervals.
During primer extension, Ub-PCNA formation is easily detectable in HCT116 cell-free extracts, while it is abolished in HCT116-Rad18KO cell-free extracts (Figure 1B). This demonstrates that under our conditions, the monoubiquitination reaction observed in cell-free extract is totally Rad18-dependent. As shown in Figure 1C, we observed that HCT116 cell-free extracts are able to bypass both CPD and AAF lesions, although higher efficiencies were observed for CPD. Extracts prepared from XP30RO cells which lack Polη are largely defective in TLS past either AAF or CPD lesions, indicating that Polη is involved in the bypass of both lesions. In Rad18-deficient cells, the bypass of the CPD lesion is as efficient as in wild-type cell extracts, while the bypass of the AAF adduct is totally abolished (Figure 1C). These results were further confirmed in a time course experiment showing that prolonged incubation of up to 60 min did not allow the bypass of an AAF adduct to occur in the absence of Rad18 (data not shown). Conversely, the kinetics of TLS through the CPD lesion in both HCT116 and HCT116-Rad18KO extracts are equivalent (Figure 1D). Together, these results indicate that Rad18 per se or the ubiquitination of PCNA is dispensable for the bypass of this lesion during primer extension in cell-free extracts.
We were concerned that the moderate UV sensitivity of HCT116-Rad18KO cells as compared to HCT116 cells might reflect a particular UV damage tolerance of this cell line that may interfere with the Polη pathway. In order to confirm the results obtained with HCT116-Rad18KO cells, we thus have extended our analysis on TLS efficiency through CPD and AAF lesions by using a SV40-transformed MRC5 human fibroblast cell line in which Rad18 was stably silenced by means of long-term RNA interference. To assess the efficiency of Rad18 silencing, an MRC5 cell line ectopically expressing HA-tagged Rad18 protein was transfected with pEBV coding for shRNA against Rad18. After isolation of stable transformants upon hygromycin B selection, protein extracts were analyzed by immunoblotting with an anti-HA antibody. As shown in Figure 2A, expression of HA-Rad18 was drastically reduced in three independent cell clones expressing the shRNA against Rad18, while HA-Rad18 was readily detected as two bands, in extracts from either non-transfected cells (MRC5-HARad18-NT) or cells that express a non-related shRNA sequence (MRC5-HARad18-CT). This result demonstrates the effectiveness of the Rad18 shRNA sequence for silencing Rad18 expression. Therefore, this sequence was subsequently used to generate a MRC5-Rad18KD cell line in which endogenous Rad18 depletion was monitored by immunostaining using an anti-Rad18 antibody (Figure 2B). The disappearance of the Rad18 signals in both non-irradiated and UV-irradiated MRC5-Rad18KD cells demonstrate the efficiency of Rad18 depletion in the selected clone (Figure 2B). Efficient silencing of Rad18 in this cell line was further confirmed by the absence of PCNA monoubiquitination after UV irradiation (data not shown). MRC5-Rad18KD cells also show increased sensitivity to UV as compared to MRC5 or MRC5-CT cells that express a non-related shRNA sequence (Figure 2C).
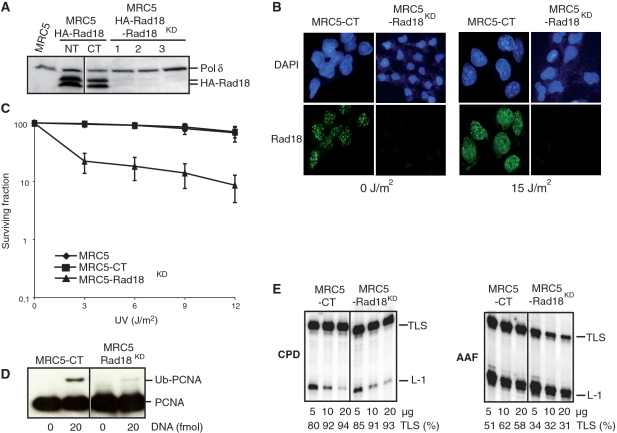
Figure 2.
Characterization of MRC5-Rad18 knockdown cells. (A) Proteins from MRC5 cells, HA-Rad18-MRC5 cells (NT), HA-Rad18-MRC5 cells expressing non related shRNA (CT) or HA-Rad18-MRC5 cells expressing shRNA against Rad18 (lanes 1, 2 and 3 correspond to three independent clones) were analyzed by immunoblotting with an antibody raised against HA. The same membrane was also blotted with antibodies against p125, the catalytic subunit of Polδ, as a loading control. (B) Representative images of MRC5-CT and MRC5-Rad18KD stained cells. Cells were fixed with methanol 4 h after mock treatment (0 J/m2) or UV irradiation (15 J/m2) and stained with a Rad18 antibody to show the efficiency of Rad18 silencing. (C) UV survival curves of MRC5, MRC5-CT and MRC5-Rad18KD cells. (D) In vitro PCNA monoubiquitination. Cell-free extracts (30 µg) were incubated for 20 min at 37°C in the presence of all four dNTPs and 0 or 20 fmol of primed ssDNA as indicated. Proteins were analyzed by immunoblotting with an antibody raised against PCNA. (E) Analysis of TLS catalyzed by cellular extracts from either MRC5-CT and MRC5-Rad18KD cells. DNA synthesis products obtained after 20 min incubation with cell-free extracts and undamaged pUC118.ss, pUC-CPD.ss or pUC3G1-AAF.ss primed templates were analyzed by electrophoresis on a denaturing gel.
We previously noticed that induction of Ub-PCNA during primer extension is much more efficient than that after UV irradiation of cultured cells (31). While Ub-PCNA is easily observable in control cell extracts, it is barely detected in MRC5-Rad18KD cell extracts (Figure 2D). As shown previously with HCT116 cells, extracts prepared from both Rad18-proficient and -deficient MRC5 cells were able to carry out TLS through a CPD lesion with equal efficiency (Figure 2E). In contrast, MRC5-Rad18KD extracts show reduced TLS capacities opposite an AAF adduct (Figure 2E). Quantitative analysis of the data shows that TLS activity through an AAF-modified substrate in the MRC5-Rad18KD cell extracts drops to about 50% of that measured in control cell extracts. We consider that the remaining PCNA monoubiquitination observed in MRC5-Rad18KD cell extracts (Figure 2D) may be responsible for the residual Polη-dependent TLS activity through an AAF adduct in these assays.
Data from both HCT116-Rad18KO and MRC5-Rad18KD cell extracts demonstrate that Rad18 activity is dispensable for the bypass of a CPD lesion in vitro while it is required for the bypass of an AAF adduct. Altogether, our observations strongly suggest the existence of both Rad18-dependent and -independent Polη-mediated TLS pathways.
UV survival of XP30RO cells expressing PIP and UBZ mutants of Polη
The Polη-deficient XP30RO cell line was used to generate several clones stably expressing different mutants of Polη. The ΔPIP mutation consists of a deletion of the last nine amino acids of the Polη sequence containing the PCNA binding site consensus sequence (6,32). The UBZ mutation consists of a D to A mutation at position 652. The solution structure of this domain shows a classical zinc-finger structure coupled with a carboxy-terminal, a helix that mediates the interaction with ubiquitin (33). The D652 residue, lying in the α-helix, is conserved in both yeast and vertebrate Polη and was shown, by co-immunoprecipitation assays and by yeast two-hybrid analysis (10; A.C., unpublished results) to be essential for Ub binding. The single D652A as well as the double mutant D652A-ΔPIP were constructed.
The UV sensitivity of stable transformants expressing mutated Polη was determined using a colony-forming assay (Figure 3). XP30RO cells expressing either wild-type Polη (WT) or the empty vector (V) were included as positive and negative controls, respectively. We found a strong positive correlation between the ability of ΔPIP Polη mutants to complement the UV sensitivity of XPV cells and the level of ectopic expression of the Polη protein in the different clones analyzed (Figure 3A and C). This result emphasizes the crucial importance of controlling the expression level of a protein in complementation assays. Indeed, while high levels of expression of the ΔPIP Polη allows an almost complete restoration of UV survival, the ΔPIP mutant clone expressing the lowest levels of Polη fails to efficiently complement the UV sensitivity of XPV cells.
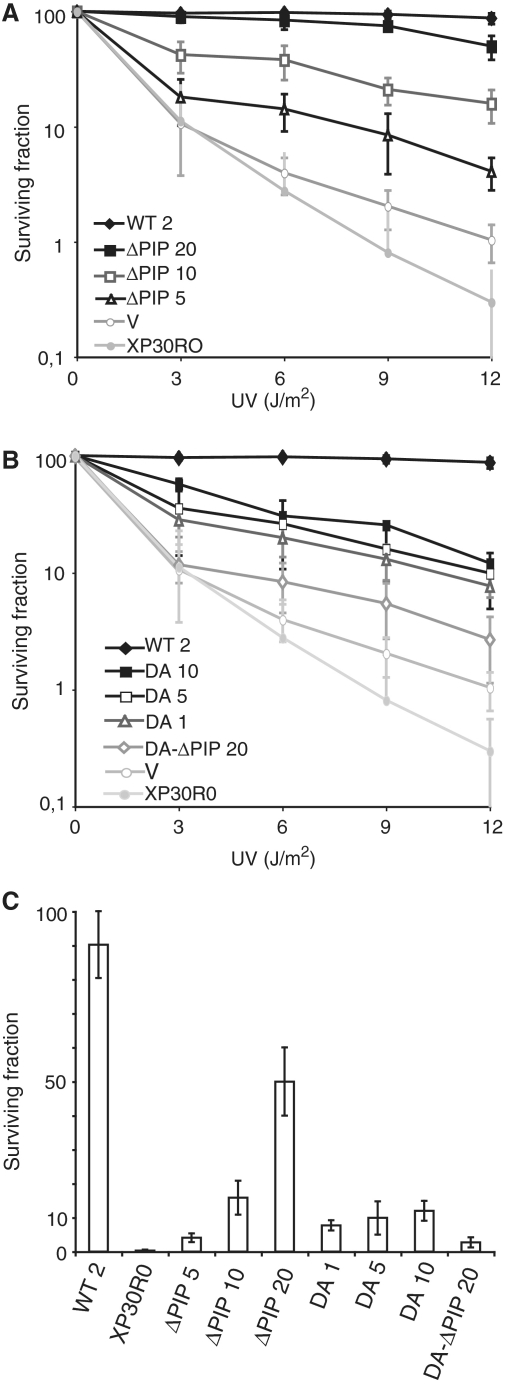
Figure 3.
Requirement of PIP and UBZ domains of Polη for its function in UV damages tolerance in vivo. UV sensitivity of XP30RO and XP30RO transformed cell lines expressing different levels of mutant Polη. (A) WT and ΔPIP mutants; V, empty vector (B) WT, D652A mutants (DA) and D652A-ΔPIP mutant (DA-ΔPIP); V, empty vector. The numbers in front of the different cell lines indicate the expression levels of ectopic Polη as compared to MRC5 cells (Figure 4B). (C) Survival of XP30RO and XP30RO transformed cell lines expressing different levels of mutant Polη following UV irradiation at 12 J/m2.
On the other hand, the UBZ domain mutant, is only partially able to complement the UV sensitivity of XPV cells, whatever its expression level (Figure 3B and C). Finally, the UV survival of the transfectant expressing a high level of the double mutant D652A-ΔPIP Polη was significantly lower than either of the single mutants (Figure 3B). Altogether, these observations are consistent with a model in which both the PIP motif and the D652 residue within UBZ domain contribute to the in vivo functions of Polη.
Both PIP and UBZ domains of Polη contribute to CPD and AAF bypass in vitro
In order to analyze the contribution of the PIP and/or UBZ domains to Polη's TLS function, extracts from cells expressing the different mutants were prepared and assayed for in vitro bypass through either CPD or AAF lesions. With extracts from cell lines expressing the mutated Polη at moderate levels (lower than 10-fold above the MRC5 basal level; Figure 4B), we observed a decrease in the amount of TLS products in comparison with the wild-type control (Figure 4A). These results suggest that mutations in either of these two domains significantly reduce the bypass efficiency through both lesions, even if the PIP truncation appears to have a more pronounced effect than the D652A mutation on the bypass efficiency (compared at identical levels of Polη expression: lanes 2 with lanes 6 and lanes 3 with lanes 7).
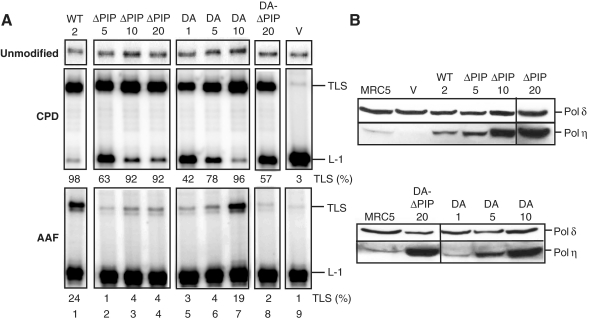
Figure 4.
Relative contribution of the PIP and UBZ domains of Polη for the in vitro bypass of CPD and AAF lesions. (A) Cell-free extracts were prepared from the various complemented XP30RO cell lines (see legend to Figure 3) and tested for their ability to perform TLS through AAF and CPD lesions, as indicated. (B) Amounts of Polη proteins in the various cell-free extracts (100 µg) were analyzed by immunoblotting with an antibody raised against Polη. As a loading control, the same membrane was blotted with antibodies against p125, the catalytic subunit of Polδ.
Finally, cell extract from the D652A-ΔPIP Polη double mutant, although containing a high level of Polη cannot restore efficient TLS through G-AAF or TT-CPD lesions. Altogether, these data show that, in accordance with the in vivo studies, both regions of Polη participate to its function. Interestingly, despite the independence of the CPD bypass with regard to Rad18, the D652 residue within the UBZ domain contributes to the bypass of a CPD lesion in vitro. In turn, this points to a specific role of the D652 residue that does not involve interaction of Polη with the ubiquitin moiety of the modified PCNA.
DISCUSSION
Bypass of TT-CPD, in contrast to G-AAF, is independent of Rad18 in cell-free extracts
Our results demonstrate that, in cell-free extracts, Rad18 is dispensable for the bypass of a CPD lesion while it is required for the bypass of an AAF adduct (Figures 1 and 2). While we cannot rule out a minor undetectable Rad18-independent monoubiquitination of PCNA (34–36), we infer from these data that CPD bypass in cell-free extracts is independent of Ub-PCNA. Our observation is consistent with a recent report showing that Polη is able to gain access to replication complexes and to catalyze TLS through CPD dimers in the presence of the non-ubiquitinable K164R PCNA mutant during replication of double-stranded DNA in HeLa cell-free extracts (37). What could be the rationale for such a difference between the CPD lesion and the AAF adduct? One obvious difference is the facility by which each of these lesions is bypassed by Polη. Steady-state kinetic analysis have shown that purified yeast and human Polη can replicate through a T–T CPD dimer with the same kinetics and fidelity as through an undamaged T–T sequence (38,39), while the bypass kinetics of a G-AAF adduct by purified human Polη is significantly lower (40). By altering the Polη–PCNA interaction, monoubiquitination of the clamp may directly stimulate Polη TLS activity through the AAF adduct, as previously shown in vitro for an abasic site (21). Ub-PCNA may also recruit additional co-factors that could facilitate the Polη-dependent bypass of the AAF adduct. Alternatively, Rad18 per se may be required for this specific reaction since it has been shown that interaction of Rad18 with Polη participates in the polymerase function in vivo (16).
Contributions of the PCNA- and ubiquitin-binding domains to the activity of Polη in vivo
We examined the ability of PIP truncation and D652A Polη mutants to complement the UV sensitivity of XP30RO cell line. Consistent with the predicted essential role of the PIP domain in PCNA binding, we found that expression of the ΔPIP Polη mutant (even fivefold above the MRC5 basal level) results in a substantial sensitization of the cells as compared to those expressing exogenous WT Polη. Surprisingly, this mutation can be compensated by high overexpression of the mutant polymerase, raising the possibility that the secondary PIP domain recently identified within the Polη sequence (12) may substitute for the C-terminal one, although with a lower efficiency. The hypothesis that the binding of the Ub moiety on PCNA via the UBZ domain may replace the Polη PIP–PCNA interaction seems unlikely since recent structural studies have shown that the PIP motif contributes more significantly to the interaction between Polη and PCNA than the UBZ domain (33,41). Alternatively, the defect in the targeting of Polη to the DNA template/primer junction by the C-terminal PIP domain might be compensated by interaction with other proteins, such as Rad18 or Rev1, that may recruit Polη to the replication stalling sites (16,42).
In agreement with our data, it has already been shown that a mutation of the D652 residue resulted in UV sensitivity in yeast and human cells (12,20,43). When overproduced about 10-fold, both D652A and ΔPIP Polη mutants partially complement the UV sensitivity of XPV cells to the same extent. While complementation by the ΔPIP Polη mutant varies notably with the expression level within a 1- to 10-fold overexpression range, complementation with the D652A Polη mutant does not change significantly with the level of overproduction (Figure 3C). We conclude from these results that the D652 residue of Polη participates in a specific role that is not exchangeable with that of the PIP domain and that is essential for full activity of Polη.
Alternative models for the role of the Polη UBZ domain in TLS through a CPD lesion
We also examined the effects of Polη PIP truncation and D652A mutation on the ability of cell-free extracts to promote TLS through a CPD lesion and an AAF adduct. We observed that whatever the lesion, both mutations impair TLS efficiency in vitro. This result together with the UV survival analysis of XP30RO cells complemented with PIP and UBZ mutants, are in agreement with a model in which both regions contribute to Polη function in TLS (10,12,20,43). The dependence of TLS on the PIP motif confirms the involvement of PCNA in the reaction since it has been shown that this motif mediates a functional interaction between Polη and PCNA (6). Interestingly, while Rad18 is not required for the bypass through a CPD lesion, we observed that the D652 residue, which is involved in the interaction with ubiquitin, contributes to this reaction. This result suggests that, for the bypass of this specific lesion, the UBZ domain of Polη may bind the ubiquitin moiety of a modified protein that is ubiquitinated in a Rad18-independent manner. Another possibility might be that the D652A mutation abrogates a novel, uncharacterized, function of the Polη UBZ domain that is distinct from ubiquitin binding. In agreement with this latter interpretation, recent genetic data provide evidences that some mutations in the C2H2 motif of the UBZ domain of Polη impair the binding to ubiquitin but have no perceptible effect on UV sensitivity and UV mutagenesis (12,20), suggesting that the binding of ubiquitin by Polη via its UBZ domain is not a necessary prerequisite for Polη to function in TLS.
CONCLUSION
Despite the recent advances in the knowledge of the TLS process, the mechanisms by which the TLS Pols gain access to the template–primer junction when the replicative machinery encounters a blocking lesion remain unclear. Several data indicate that Polη contributes not only to gap filling after the replication apparatus has moved away from the damaged site, but also helps to maintain the continuous progression of the replication fork after DNA damaging treatment. Indeed, as compared to WT cells, XPV cells accumulate shorter replication products after UV irradiation and elongation of these intermediates was markedly retarded (25,44). We propose that, depending on the nature of the lesion (and its sequence context), different mechanisms control Polη activity. Efficient bypass of lesions that do not require Rad18-dependent PCNA monoubiquitination, as a T–T CPD dimer, may occur ‘on the fly’ during processive DNA synthesis, without actual uncoupling of the replicative polymerases. If this first TLS trial is not successful, the uncoupling of leading and lagging strand synthesis provides the substrate for Rad6/Rad18-dependent PCNA monoubiquitination. Rad18 and Ub-PCNA may then facilitate the bypass reaction as exemplified here by the analysis of the AAF adduct TLS. We were concerned that our system may not be fully relevant to test this model, since the CPD lesion is located on a ssDNA template and is thus able to stimulate PCNA monoubiquitination in the absence of fork uncoupling (Supplementary Figure S2). However, as shown in Figures 1 and 2, this modification is clearly dispensable for Polη to bypass a CPD lesion.
We have established that in cell-free extracts, TLS of both an AAF adduct and a CPD lesion requires the UBZ domain. We speculate that this domain plays a dual role in the TLS process. On one hand, it may mediate the binding to Ub-PCNA during the gap filling reaction. On the other hand, the D652 residue may participate in a distinct function, such as the interaction with another protein that is necessary for the coordination of the Polη activity at the replication fork. In agreement with this model, Edmunds et al. (25) have recently shown that, in DT40 cells, PCNA monoubiquitination is dispensable for maintaining replication fork progression on damaged DNA while it is required post-replicatively. Interestingly, the data presented in this latter paper show that the UBM domains of Rev1 are required for its activity at the replication fork in the absence of Ub-PCNA. Thus, it is conceivable that the regulatory mechanism described in this study may apply to the UBD domain of Y-family polymerases in general.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
FUNDING
l’Institut National du Cancer (PL028); la Ligue Régionale contre le Cancer. Funding for open access charge: Centre National de la Recherche Scientifique.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to M. Bichara for advice and comments on the article, Dr Hubscher, U. (University of Zurich, Switzerland) for providing His-p21 and Dr Hurwitz, J. (Memorial Sloan-Kettering Cancer Center, New York) for recombinant tagged PCNA.
REFERENCES
- 1.Prakash S, Prakash L. Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 2002;16:1872–1883. doi: 10.1101/gad.1009802. [DOI] [PubMed] [Google Scholar]
- 2.Shachar S, Ziv O, Avkin S, Adar S, Wittschieben J, Reissner T, Chaney S, Friedberg EC, Wang Z, Carell T, et al. Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J. 2009;28:383–393. doi: 10.1038/emboj.2008.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCulloch SD, Kokoska RJ, Chilkova O, Welch CM, Johansson E, Burgers PM, Kunkel TA. Enzymatic switching for efficient and accurate translesion DNA replication. Nucleic Acids Res. 2004;32:4665–4675. doi: 10.1093/nar/gkh777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 5.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Poleta. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 6.Haracska L, Johnson RE, Unk I, Phillips B, Hurwitz J, Prakash L, Prakash S. Physical and functional interactions of human DNA polymerase eta with PCNA. Mol. Cell. Biol. 2001;21:7199–7206. doi: 10.1128/MCB.21.21.7199-7206.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haracska L, Johnson RE, Unk I, Phillips BB, Hurwitz J, Prakash L, Prakash S. Targeting of human DNA polymerase iota to the replication machinery via interaction with PCNA. Proc. Natl Acad. Sci. USA. 2001;98:14256–14261. doi: 10.1073/pnas.261560798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haracska L, Prakash L, Prakash S. Role of human DNA polymerase kappa as an extender in translesion synthesis. Proc. Natl Acad. Sci. USA. 2002;99:16000–16005. doi: 10.1073/pnas.252524999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haracska L, Kondratick CM, Unk I, Prakash S, Prakash L. Interaction with PCNA is essential for yeast DNA polymerase eta function. Mol. Cell. 2001;8:407–415. doi: 10.1016/s1097-2765(01)00319-7. [DOI] [PubMed] [Google Scholar]
- 10.Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 11.Gueranger Q, Stary A, Aoufouchi S, Faili A, Sarasin A, Reynaud CA, Weill JC. Role of DNA polymerases eta, iota and zeta in UV resistance and UV-induced mutagenesis in a human cell line. DNA Repair. 2008;7:1551–1562. doi: 10.1016/j.dnarep.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 12.Acharya N, Yoon JH, Gali H, Unk I, Haracska L, Johnson RE, Hurwitz J, Prakash L, Prakash S. Roles of PCNA-binding and ubiquitin-binding domains in human DNA polymerase eta in translesion DNA synthesis. Proc. Natl Acad. Sci. USA. 2008;105:17724–17729. doi: 10.1073/pnas.0809844105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 14.Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 15.Kannouche PL, Lehmann AR. Ubiquitination of PCNA and the polymerase switch in human cells. Cell Cycle. 2004;3:1011–1013. [PubMed] [Google Scholar]
- 16.Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo C, Tang TS, Bienko M, Parker JL, Bielen AB, Sonoda E, Takeda S, Ulrich HD, Dikic I, Friedberg EC. Ubiquitin-binding motifs in REV1 protein are required for its role in the tolerance of DNA damage. Mol. Cell Biol. 2006;26:8892–8900. doi: 10.1128/MCB.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plosky BS, Vidal AE, Fernandez de Henestrosa AR, McLenigan MP, McDonald JP, Mead S, Woodgate R. Controlling the subcellular localization of DNA polymerases iota and eta via interactions with ubiquitin. EMBO J. 2006;25:2847–2855. doi: 10.1038/sj.emboj.7601178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo C, Tang TS, Bienko M, Dikic I, Friedberg EC. Requirements for the interaction of mouse Polkappa with ubiquitin and its biological significance. J. Biol. Chem. 2008;283:4658–4664. doi: 10.1074/jbc.M709275200. [DOI] [PubMed] [Google Scholar]
- 20.Acharya N, Brahma A, Haracska L, Prakash L, Prakash S. Mutations in the ubiquitin binding UBZ motif of DNA polymerase eta do not impair its function in translesion synthesis during replication. Mol. Cell. Biol. 2007;27:7266–7272. doi: 10.1128/MCB.01196-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garg P, Burgers PM. Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases eta and REV1. Proc. Natl Acad. Sci. USA. 2005;102:18361–18366. doi: 10.1073/pnas.0505949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haracska L, Unk I, Prakash L, Prakash S. Ubiquitylation of yeast proliferating cell nuclear antigen and its implications for translesion DNA synthesis. Proc. Natl Acad. Sci. USA. 2006;103:6477–6482. doi: 10.1073/pnas.0510924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tateishi S, Niwa H, Miyazaki J, Fujimoto S, Inoue H, Yamaizumi M. Enhanced genomic instability and defective postreplication repair in RAD18 knockout mouse embryonic stem cells. Mol. Cell. Biol. 2003;23:474–481. doi: 10.1128/MCB.23.2.474-481.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okada T, Sonoda E, Yamashita YM, Koyoshi S, Tateishi S, Yamaizumi M, Takata M, Ogawa O, Takeda S. Involvement of vertebrate polkappa in Rad18-independent postreplication repair of UV damage. J. Biol. Chem. 2002;277:48690–48695. doi: 10.1074/jbc.M207957200. [DOI] [PubMed] [Google Scholar]
- 25.Edmunds CE, Simpson LJ, Sale JE. PCNA ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Mol. Cell. 2008;30:519–529. doi: 10.1016/j.molcel.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 26.Shiomi N, Mori M, Tsuji H, Imai T, Inoue H, Tateishi S, Yamaizumi M, Shiomi T. Human RAD18 is involved in S phase-specific single-strand break repair without PCNA monoubiquitination. Nucleic Acids Res. 2007;35:e9. doi: 10.1093/nar/gkl979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huschtscha LI, Holliday R. Limited and unlimited growth of SV40-transformed cells from human diploid MRC-5 fibroblasts. J. Cell Sci. 1983;63:77–99. doi: 10.1242/jcs.63.1.77. [DOI] [PubMed] [Google Scholar]
- 28.Biard DS, Despras E, Sarasin A, Angulo JF. Development of new EBV-based vectors for stable expression of small interfering RNA to mimick human syndromes: application to NER gene silencing. Mol. Cancer Res. 2005;3:519–529. doi: 10.1158/1541-7786.MCR-05-0044. [DOI] [PubMed] [Google Scholar]
- 29.Napolitano RL, Fuchs RP. New strategy for the construction of single-stranded plasmids with single mutagenic lesions. Chem. Res./Toxicol. 1997;10:667–671. doi: 10.1021/tx970018w. [DOI] [PubMed] [Google Scholar]
- 30.Cordonnier AM, Lehmann AR, Fuchs RP. Impaired translesion synthesis in xeroderma pigmentosum variant extracts. Mol. Cell. Biol. 1999;19:2206–2211. doi: 10.1128/mcb.19.3.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmutz V, Wagner J, Janel-Bintz R, Fuchs RP, Cordonnier AM. Requirements for PCNA monoubiquitination in human cell-free extracts. DNA Repair. 2007;6:1726–1731. doi: 10.1016/j.dnarep.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 32.Kannouche P, Broughton BC, Volker M, Hanaoka F, Mullenders LH, Lehmann AR. Domain structure, localization, and function of DNA polymerase eta, defective in xeroderma pigmentosum variant cells. Genes Dev. 2001;15:158–172. doi: 10.1101/gad.187501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bomar MG, Pai MT, Tzeng SR, Li SS, Zhou P. Structure of the ubiquitin-binding zinc finger domain of human DNA Y-polymerase eta. EMBO Rep. 2007;8:247–251. doi: 10.1038/sj.embor.7400901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simpson LJ, Ross AL, Szuts D, Alviani CA, Oestergaard VH, Patel KJ, Sale JE. RAD18-independent ubiquitination of proliferating-cell nuclear antigen in the avian cell line DT40. EMBO Rep. 2006;7:927–932. doi: 10.1038/sj.embor.7400777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang S, Chea J, Meng X, Zhou Y, Lee EY, Lee MY. PCNA is ubiquitinated by RNF8. Cell Cycle. 2008;7:3399–3404. doi: 10.4161/cc.7.21.6949. [DOI] [PubMed] [Google Scholar]
- 36.Terai K, Abbas T, Jazaeri AA, Dutta A. CRL4cdt2 E3 ubiquitin ligase monoubiquitinates PCNA to promote translesion DNA synthesis. Mol. Cell. 2010;37:143–149. doi: 10.1016/j.molcel.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nikolaishvili-Feinberg N, Jenkins GS, Nevis KR, Staus DP, Scarlett CO, Unsal-Kacmaz K, Kaufmann WK, Cordeiro-Stone M. Ubiquitylation of proliferating cell nuclear antigen and recruitment of human DNA polymerase eta. Biochemistry. 2008;47:4141–4150. doi: 10.1021/bi702329h. [DOI] [PubMed] [Google Scholar]
- 38.Johnson RE, Washington MT, Prakash S, Prakash L. Fidelity of human DNA polymerase eta. J. Biol. Chem. 2000;275:7447–7450. doi: 10.1074/jbc.275.11.7447. [DOI] [PubMed] [Google Scholar]
- 39.Washington MT, Johnson RE, Prakash S, Prakash L. Accuracy of thymine-thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase eta. Proc. Natl Acad. Sci. USA. 2000;97:3094–3099. doi: 10.1073/pnas.050491997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masutani C, Kusumoto R, Iwai S, Hanaoka F. Mechanisms of accurate translesion synthesis by human DNA polymerase eta. EMBO J. 2000;19:3100–3109. doi: 10.1093/emboj/19.12.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hishiki A, Hashimoto H, Hanafusa T, Kamei K, Ohashi E, Shimizu T, Ohmori H, Sato M. Structural basis for novel interactions between human translesion synthesis polymerases and PCNA. J. Biol. Chem. 2009 doi: 10.1074/jbc.M809745200. doi:10.1074/jbc.M809745200 [Epub ahead of print; 10 February 2009] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuasa MS, Masutani C, Hirano A, Cohn MA, Yamaizumi M, Nakatani Y, Hanaoka F. A human DNA polymerase eta complex containing Rad18, Rad6 and Rev1; proteomic analysis and targeting of the complex to the chromatin-bound fraction of cells undergoing replication fork arrest. Genes Cells. 2006;11:731–744. doi: 10.1111/j.1365-2443.2006.00974.x. [DOI] [PubMed] [Google Scholar]
- 43.Parker JL, Bielen AB, Dikic I, Ulrich HD. Contributions of ubiquitin- and PCNA-binding domains to the activity of polymerase eta in Saccharomyces cerevisiae. Nucleic Acids Res. 2007;35:881–889. doi: 10.1093/nar/gkl1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lehmann AR, Kirk-Bell S, Arlett CF, Paterson MC, Lohman PH, de Weerd-Kastelein EA, Bootsma D. Xeroderma pigmentosum cells with normal levels of excision repair have a defect in DNA synthesis after UV-irradiation. Proc. Natl Acad. Sci. USA. 1975;72:219–223. doi: 10.1073/pnas.72.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.