Abstract
The plasmid pRN1 encodes for a multifunctional replication protein with primase, DNA polymerase and helicase activity. The minimal region required for primase activity encompasses amino-acid residues 40–370. While the N-terminal part of that minimal region (residues 47–247) folds into the prim/pol domain and bears the active site, the structure and function of the C-terminal part (residues 248–370) is unknown. Here we show that the C-terminal part of the minimal region folds into a compact domain with six helices and is stabilized by a disulfide bond. Three helices superimpose well with the C-terminal domain of the primase of the bacterial broad host range plasmid RSF1010. Structure-based site-directed mutagenesis shows that the C-terminal helix of the helix bundle domain is required for primase activity although it is distant to the active site in the crystallized conformation. Furthermore, we identified mutants of the C-terminal domain, which are defective in template binding, dinucleotide formation and conformation change prior to DNA extension.
INTRODUCTION
Primases synthesize a short oligonucleotide on single-stranded DNA. Later on this primer is extended by DNA polymerases. The action of the primase during DNA replication is required as replicative DNA polymerases are unable to initiate DNA polymerization de novo. In principle, it is not required that primases display any sequence specificity. In contrast, it could be beneficial if primer synthesis is sequence unspecific. However, many primases of the bacterial type do exhibit some sequence specificity and synthesize primers on templates that have a short (usually three bases long) recognition motif. For example the Escherichia coli DnaG protein prefers a 5′-CTG-3′ to initiate a primer (1), the T7 primase 5′-GTC-3′ (2) and SP6 primase 5′-GCA-3′ (3). In contrast, eukaryotic primases usually do not exhibit such sequence specificity.
Structures of the two major types of primase, i.e. the bacterial type primase DnaG (4,5) and its zinc-binding domain (6) and of the archaeo-eukaryotic primase (7–9) have been determined. These structures suggest that the two proteins families are not evolutionary related to each other. The bacterial type primases have a catalytic ‘toprim’ domain which is also found in topoisomerases IA and II (10). In contrast, the heterodimeric primases of the eukarya and archaea adopt a different fold that is related to the RNA recognition motif (RRM) encompassing four β-strands and two α-helices. Additionally a further β-strand (flange), which runs perpendicular to the strands of the RRM, is found in the archaeal primase structures. Despite the structural discrepancy between the bacterial and the archaeo-eukaryal primases, the architecture of the active site with catalytic aspartic residues is similar suggesting a convergent evolution of the active site and a common reaction mechanism of both types of primases.
The reaction mechanism of the primases is considered to involve two basic steps: The first step is the formation of a dinucleotide between two (cognate) nucleotides bound to the template DNA while the second step is the extension of the dinucleotide by a further nucleotide. The second step is repeated until a full-length primer is synthesized. Primases differ in the length distribution of their typical products. Although structures of some primases have been determined the structure of a primase template complex has so far escaped structure elucidation maybe reflecting the transient nature of the primase template complex.
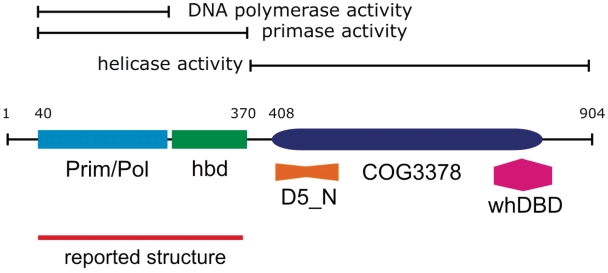
In our group we study the replication of the plasmid pRN1 of Sulfolobus islandicus. This plasmid encodes an unusual multifunctional replication protein (8). This protein termed ORF904 folds into at least three well characterized distinct domains (Figure 1). An N-terminal prim/pol domain (amino acids 47–247, pfam09250, RRM fold), a helicase domain of superfamily 3 (amino acids 408–827, COG3378) and a C-terminal winged helix DNA-binding domain (amino acids 752–844, pfam03288 and pfam02257). In addition sequence analysis suggests that the amino acids 417–522 are part of the yet functionally uncharacterized domain D5N (pfam08706). This domain structure of the protein leaves the amino acids from 248 to 407 unannotated. Nevertheless, it could be shown that this part of the protein is indispensable for primase activity (11). Although the prim/pol domain is capable to extend a primer/template substrate using dNTPs, this domain alone is unable to prime on single-stranded DNA.
Figure 1.
Domain structure of the multifunctional pRN1 replication protein. The analysis of deletion mutants allowed mapping the activities to separate regions of the 904 amino-acid long protein. The structure of the prim/pol domain (amino acids 47–247, light blue rectangle) has been previously reported. In this communication we analysed the structure of the primase domain which also encompasses the helix bundle domain (hbd, green rectangle).
The primase activity of the protein encompassing the amino acids 40–370 is particular, since the enzymes forms a mixed primer with the first base of the primer being strictly an rNTP and the remaining bases are invariable dNTPs. In the context of plasmid replication, the primer synthesis and not the DNA polymerase activity is probably the physiological function of this part of the enzyme. During our studies we discovered that in addition the primase activity of the enzyme is highly sequence specific. A primer is only synthesized on templates containing the triplet GTG (11).
In this report we determined the crystal structure of the primase including a C-terminal helix bundle domain (amino acids 256–370) whose presence is essential for the primase activity of the prim/pol domain. Guided by the structure we constructed several point mutants and a deletion mutant and discovered two amino acids which are critical for catalysis. Based on our results we suggest a model how the prim/pol domain and the helix bundle domain interact in order to synthesize a primer.
EXPERIMENTAL PROCEDURES
Materials and reagents
Oligodeoxynucleotides were purchased either HPLC-purified from biomers (Ulm, Germany) for the crystallization or from Metabion (Germany) for the activity assays. α-32P labelled deoxyadenosine 5′-triphosphate (3000 Ci/mmol) and γ-32P adenosine 5′-triphosphate (5000 Ci/mmol) were obtained from Hartmann Analytic (Germany).
Cloning, expression and purification of the proteins
The deletion mutant of ORF904 comprising the amino acids 40–370 (primase domain throughout) and the prim/pol deletion mutant from amino acids 40 to 255 was constructed and expressed with a C-terminal hexahistidine tag as described previously (11). For the crystallization the protein was concentrated to about 100 mg/ml after the purification and applied to a superose-6 gel filtration column equilibrated with 10 mM HEPES, pH 7.5, 150 mM NaCl, 5 mM DTT. After gel filtration, the fractions were pooled and again concentrated to 100 mg/ml.
The point mutations were introduced into the primase domain by an amplification of the plasmid with two overlapping mutagenic primers and a subsequent digestion of the parental DNA with DpnI. The introduced point mutations were confirmed by DNA sequencing.
The DNA fragment encoding the peptide from amino acids 256–370 was PCR amplified with the primers 5′-dCCCATATGACGGTGGTGGAGTTTGAGGA and 5′-dGCTAGTTATTGCTCAGCGG using the expression vector of the primase domain as template and cloned into the expression vector pET28c.
All proteins were expressed in E. coli Rosetta2(DE3)pLysS (Novagen) at 22°C. Cells from a 1 l culture were lysed by sonication and after centrifugation the supernatants were incubated at 65°C for 15 min. After a second centrifugation, the extracts were loaded onto 1 ml Talon gravity flow columns equilibrated with 25 mM sodium phosphate, pH 7.0, 300 mM NaCl and 6 mM imidazole. After washing the columns with 15 ml, the proteins were eluted with 150 mM imidazole in the same buffer. The protein containing fractions were pooled and concentrated by ultrafiltration and the buffer was exchanged to 25 mM sodium phosphate and 150 mM NaCl. Total yield of purified protein was about 20 mg/l culture.
Crystallization and data collection
For the crystallization experiments, the primase domain was mixed with a 1.2–1.5 molar excess of different single-stranded oligonucleotides and diluted to 50 mg/ml with gel filtration buffer. Ten millimolar ATP was included in some experiments. The crystals were grown using the hanging drop vapour diffusion technique where 1–2 µl of protein or protein/DNA solution were mixed with an equal volume of reservoir solution (0.1 M Tris–HCl pH 8.5, 30–35% PEG 4000, 200 mM lithium sulphate and 2 mM Tris-2-carboxyethyl-phosphine) and let to equilibrate against 500 µl of this reservoir solution at 20°C. Crystals grew within 1 week. For data collection the crystals were transferred to reservoir solution supplemented with 10–15% glycerol and flash-frozen in liquid nitrogen. Data were collected at 100 K using synchrotron radiation on beamline ID14-H1 at the European Synchrotron Radiation Facility (ESRF), Grenoble and processed with MOSFLM (12) and scaled with SCALA (13). Crystals belong to space group C2, with unit cell parameters a = 125.718, b = 46.556, c = 77.993, β = 124.612. Data collection statistics are summarized in Table 1.
Table 1.
Summary of the primase domain diffraction data and refinement statistics
| X-ray diffraction data | |
| Space group | C2 |
| Cell constants (Å) | |
| a | 125.718 |
| b | 46.556 |
| c | 77.993 |
| β | 124.612 |
| Resolution (Å)a | 60–1.85 (1.92–1.85) |
| Rsym (%)b | 9.1 (64.5) |
| <I/σI>c | 7.7 (1.3) |
| No. of observations | 537 575 |
| No. of unique observations | 30 362 |
| Completeness (%) | 95 (78) |
| Refinement statistics | |
| Resolution (Å) | 50–1.85 |
| R-factord | 0.2 |
| Rfreee | 0.25 |
| R.m.s.d.f bond lengths (Å) | 0.017 |
| R.m.s.d. bond angles (Å) | 0.968 |
| Overall B-factor (Å2) | 26.27 |
| φψ Angle distributiong | |
| In core region | 260 (91.2) |
| In allowed region | 25 (8.8) |
| In disallowed region | 0 (0) |
| Number of atoms | |
| Protein | 2617 |
| Ions | 6 |
| Solvent | 176 |
aHighest resolution of data set with highest resolution bin in parentheses.
bRsym = ∑h∑i|<Ih> – Ii(h)|/∑h∑iIi(h), where Ii(h) is the mean intensity of the N reflection.
cDefined as mean (I)/SD
dR-factor = ∑h |Fo – Fc|/∑h|Fo|.
eRfree is calculated from 5% of the data which were omitted during the course of the refinement.
fR.m.s.d. is the root mean square deviation from ideal geometry.
gAs defined by PROCHECK (6), the percentage distribution is given in parentheses.
Structure determination and refinement
The structure was solved by molecular replacement with the program AMoRe (14), using the previously solved residues 40–249 of the prim/pol domain of ORF904 (1RO2). Rotation and translation function calculations in the range 12.0–4.0 Å yielded two independent solutions, which had, after rigid body fitting in AMoRe, an R-factor of 0.44 and a correlation coefficient of 0.48. Rigid-body refinement as well as all the subsequent refinement was carried out in REFMAC (15). The resulting maps showed some extra density accounting for the additional primase domain and were of sufficient quality. Model building was carried out with Coot (16). After several cycles of positional refinement, B-factor refinement, TLS refinement and manual building, including placement of the zinc and sulphate ions and solvent molecules, the refinement converged to an Rcryst of 0.20 and Rfree of 0.25. The main refinement statistics are listed in Table 1. The final model encompasses residues 40-370 of ORF904, residues 250–255 were disordered and not visible in the electron density map. In none of the crystals we observed nucleic acids or ATP. Figures were generated with Chimera (17).
Primase assay
The primase assays were performed as described previously (11). Briefly, in 10 µl reactions 0.4 µM of protein was typically incubated with 4 µM oligodeoxynucleotide and 1 mM ATP in reaction buffer (25 mM Tris–HCl pH 7.5, 1 mM DTT and 10 mM MgCl2). To start the primase reaction 10 µM dNTPs supplemented with 0.6 nM [α-32P]-dATP were added and the reactions were incubated at 50°C for 10 min. The reactions were stopped by adding gel loading buffer (80% formamide, 10 mM EDTA, 0.1% xylene cyanol, 0.1% bromphenol blue) and heated to 95°C for 5 min. The reaction products were then loaded onto 20% denaturing polyacrylamide/urea gels and analysed using an InstantImager.
DNA polymerase assay
The polymerase activity of the proteins was measured using a slightly modified primer extension assay as described previously (18). A 20-mer primer (5′-dCGAACCCGTTCTCGGAGCAC) was 5′ labelled with [γ-32P]-ATP and hybridized with a 42-mer template (5′-dTTCTGCACAAAGCGGTTCTGCAGTGCTCCGAGAACGGGTTCG). In the primase reaction buffer 0.4 µM protein were incubated with 5 nM of the primer/template and 100 µM dNTPs at 50°C for 30 min. The reactions were analysed on denaturing polyacrylamide/urea gels as described above.
Fluoresence anisotropy measurements
The fluorescence anisotropy measurements to assess the DNA-binding activity of the proteins were essentially performed as previously described (11). Briefly, in a 120 μl cuvette, 10 μM of protein were mixed with 40 nM of the oligonucleotide templatefluo (Table 2) which was labelled with fluorescein at the 3′ end, in binding buffer (12,5 mM Tris–HCl, pH 8,0, 1 mM MgCl2, 10 µM ZnCl2, 50 mM KCl, 0.01% Tween). To calculate the binding constant a reverse titration was performed, where the concentration of protein was gradually reduced by replacing 30% of the measured solution with labelled oligonucleotide in binding buffer at least 10 times. The anisotropy was measured at 526 nm with an excitation wavelength of 495 nm. Each data point was measured three times and the data was fitted with a single-site binding model yielding the dissociation constant of the DNA protein equilibrium.
Table 2.
Sequences of the primase templates and the fluorescence anisotropy substrates
| template1 | 5′-TTTTTTTTGTGCACTTT |
| template2 | 5′-TTTTTTTTGTGTTT |
| templateNTG | 5′-TTTTTTTTNTGTTT |
| templateNNG | 5′-TTTTTTTTNNGTTT |
| templateNTN | 5′-TTTTTTTTNTNTTT |
| templateNNN | 5′-TTTTTTTTNNNTTT |
| templatefluo | 5′-TCTTCTGTGCACTCTTC-fluorescein |
Circular dichroism spectroscopy
Circular dichroism (CD) spectroscopy is a method to assess the secondary structure of a protein. The proteins were diluted in 25 mM sodium phosphate pH 7.0 to a concentration of 6 μM and the far-UV CD spectra were recorded on a Jasco J-600 spectropolarimeter (Jasco, Germany) at 25°C. Data was collected at a scanning speed of 10 nm/min with a bandwidth of 1 nm and a response time of 1 s. The light path of the samples was 1 mm. The buffer baseline was subtracted from the data points and they were expressed as residual ellipticity (19).
Limited proteolysis
To assess if the structure of the protein N40-C370 changes upon binding of a DNA substrate a possibly altered susceptibility to proteolytic cleavage by trypsin was investigated with limited proteolysis. The protein was diluted to 10 μM in proteolysis buffer (25 mM Tris–HCl, pH 7.5, 10 mM CaCl2, 10 mM MgCl2, 1 mM DTT). Samples were also prepared including 20 μM oligonucleotide template1 (Table 2) or oligonucleotide and 1 mM ATP. The protein was incubated with DNA and ATP for 15 min at 50°C. Next, trypsin (sequencing grade, Roche) was added at w/w ratios (trypsin: N40-C370) of 1:20 to 1:200 and incubated at 37°C for 15 min. The samples were subsequently separated on a 10% SDS gel and stained with Coomassie Blue.
RESULTS
The amino acids 256- to 370-fold into a novel helix bundle domain
Our previous results showed that the replication protein ORF904 encompassing the amino acids 40 through 370 is fully capable of primer synthesis whereas a shorter deletion mutant only up to amino acid 339, is incapable of primer synthesis. Therefore, the part of the protein from amino acids 40–370 constitutes the minimal primase active part or the primase domain (Figure 1). In previous work, we determined the structure of the prim/pol domain (amino acids 40–255) (18). Given the functional importance of the amino acids 256–370 we were interested to determine the structure of the minimal primase active part. This deletion mutant (amino acids 40–370) could be well expressed in E. coli, was purified by standard procedures and readily crystallized.
The structure of the crystallized protein was solved at 1.85 Å resolution by molecular replacement using the previously solved structure of the prim/pol domain (amino acids 40–249) as the search model (18). Of the 331 amino acids, 97% could be modelled. Only the amino-acid residues 250–255 could not be resolved, due to high flexibility and a lack of electron density.
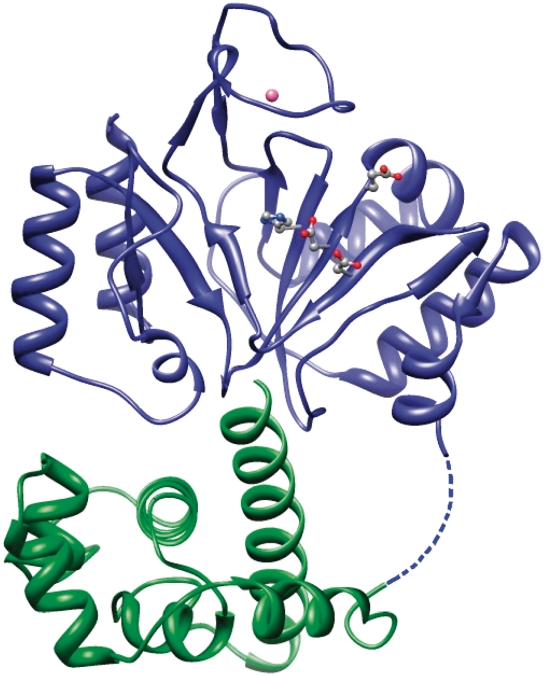
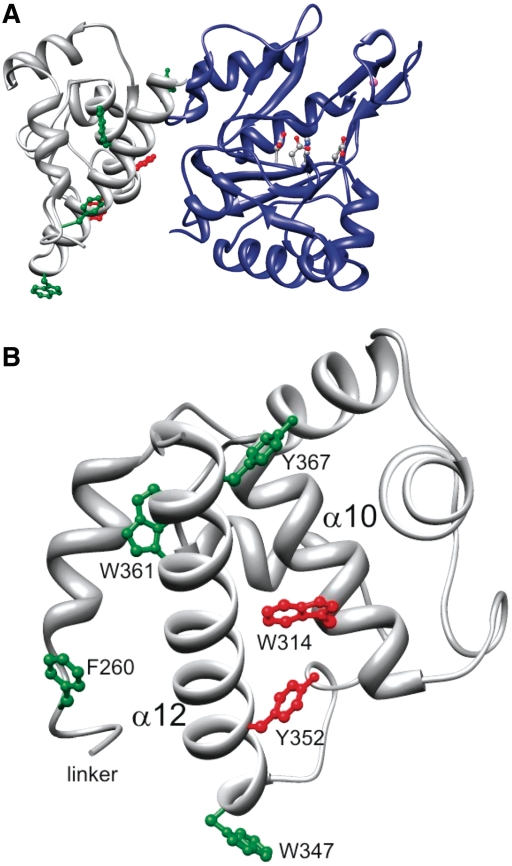
The structure of the prim-pol domain in these crystals agreed well with the structure reported previously (15). The bound zinc ion could be identified, and additionally a sulphate ion was found to be coordinated by the His141 which is also involved in the coordination of the zinc ion. In addition to the prim/pol domain, a distinct, smaller domain was observed that corresponded to a C-terminal helix bundle domain. These two domains are connected by a linker, which contains six amino acids that could not be resolved, indicating flexibility. The C-terminal domain consists of six α-helices forming a bundle with a noticeably long helix at the terminus of the protein facing the active site (Figure 2). The helix bundle domain is stabilized by a conserved disulfide bridge between C284 and C297 (Figure 3).
Figure 2.
Structure of the primase domain of the replication protein ORF904. The primase domain consists of two subdomains. The catalytic center with the amino acids D111, E113, H145 and D171 is situated in a depression of the prim/pol domain (shown in blue ribbons). The helix bundle domain (shown in green) is connected to the prim/pol domain by a flexible six amino-acid linker.
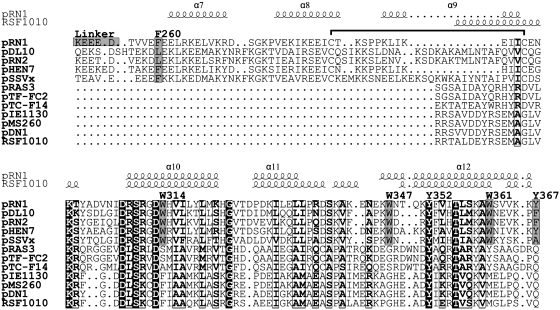
Figure 3.
Structure-based alignment of the helix bundles from the pRN1 replication protein and the RSF1010 primase. Aligned are five proteins from the pRN plasmid family (pRN1, pDL10, pRN2, pHEN7 and pSSVx) and seven primases from bacterial plasmids (pRAS3, pTF-FC2, pTC-F14, pIE1130, pMS260, pDN1 and RSF1010). Above the alignment the secondary structure elements of the helix bundle domain of ORF904 and of a part of the C-terminal domain of the RSF1010 primase are indicated. The C-terminal three helices of both structures are superimposable. With gray boxes the conserved aromatic residues which were investigated in this study are highlighted and labelled. The disulfide bridge in the pRN helix bundle domain is indicated with a horizontal bracket above the alignment.
Remarkably the helix bundle is quite distant from the active site which is situated in the prim/pol domain. Therefore, it is unlikely that the helix bundle domain could directly participate in the primase activity in this conformation. As already stated above, the C-terminal 32 amino acids of the primase domain (amino acids 339–370) are essential for the primase activity of this protein, and deletion mutants that were shortened by these 32 amino acids are not able to synthesize a primer on single-stranded plasmid DNA or oligonucleotides containing the recognition sequence (18). The crystal structure shows that 24 of these essential amino acids form a large α-helix. This suggests that this helix is either required for folding of the helix bundle domain or is directly involved in the interaction with the template DNA or the initiating ribonucleotide during the synthesis of the primer. As the prim/pol domain and the helix bundle domain are linked by a flexible linker it is possible that in the crystals the protein is in an open conformation. Possibly, upon DNA and nucleotides binding, the conformation of the domains could be altered into a closed conformation, with the C-terminal α-helix sliding towards the active site of the enzyme (see also ‘Discussion’ section and Figure 10). A similar mechanism has been proposed for the primase of the bacteriophage T7, which is also divided into two subdomains that are connected by a flexible linker (20). We wondered whether we could detect these conformational changes by limited proteolysis. We could however only see a minor increase of the proteolytic susceptibility when the primase domain was incubated with DNA but without initiating ribonucleotide ATP (data not shown).
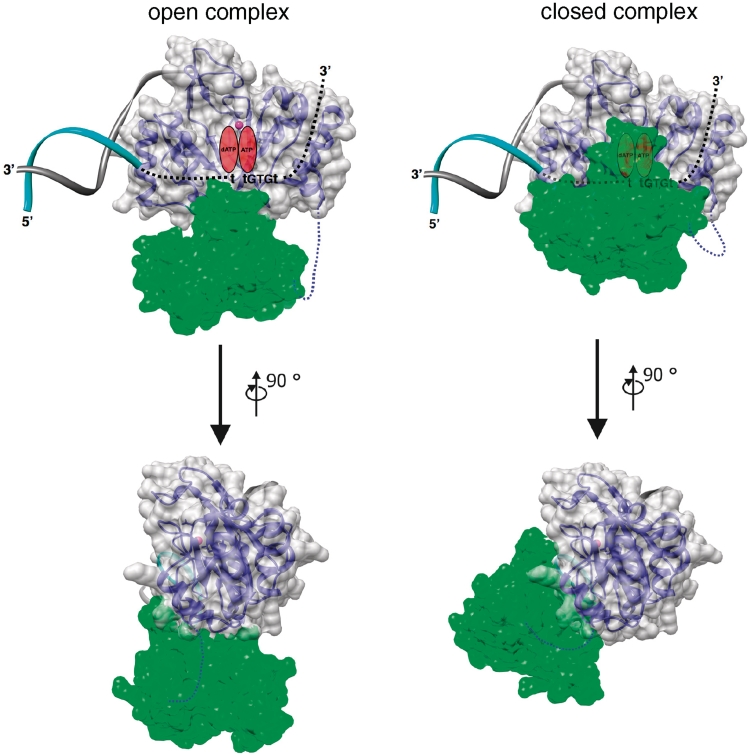
Figure 10.
Model of the pRN1 primase template complex. For modelling the prim/pol–DNA complex, the catalytic domain of prim/pol in presence of manganese (PDB ID: 1 RO2) was superimposed to the catalytic domain of primase RepB′ in complex with initiator DNA (PDB ID: 3H25). For the open complex, the primase accessory domain has been placed by superimposition of the prim/pol catalytic domain. For the closed complex, the primase helix bundle domain has been manually rotated and shifted, taking into account shape complementarity and mutational data here described. Template strand is depicted as a cyan ribbon and dashed black line while non-template strand as gray ribbon. Manganese ion is shown as a pink sphere. The prim/pol catalytic domain is represented as a blue ribbon and a gray semi-transparent molecular surface while the primase accessory domain as a yellow ribbon and a semi-transparent green molecular surface. The linker connecting the two domains, and not visible in the electron density map, is represented as a blue dashed-line.
To further analyse the interaction between both domains we expressed the catalytic domain (amino acids 40–255) and the helix bundle domain (amino acids 256–370) separately and asked whether the non-covalently linked domains would support primase activity. The helix bundle domain was expressed in high yield without any proteolytic degradation products suggesting that the peptide folded. Different molar ratios of the two proteins were used in primase assays with the C-terminal helix bundle domain in up to 100 times molar excess. In these experiments the C-terminal helix bundle domain was unable to complement the primase activity of the prim/pol domain. These observations are in contrast to the RepB′ protein of RSF1010 (see below) where the helix bundle domain could restore the activity of the separate primase domain (21).
A structure similarity search with DALI reveals that the C-terminal three helices of the helix bundle domain superimpose reasonably well (Z = 5.6) with the C-terminal helix bundle domain of the primase RepB′ from the plasmid RSF1010 which has recently been determined (21). These substructures align over about 53 residues (amino acids 308–360) with an RMSD of 2.4 Å. The primase of the bacterial broad host range plasmid RSF1010 consists of an N-terminal domain of the archeo-eukaryotic primase superfamily and a C-terminal helix bundle domain. Both domains are separated by a long helix and a coil region, which would allow conformational changes of the protein. In the crystallized conformation both domains are 29 Å apart. The catalytic domain of the primase could also be crystallized with a DNA hairpin of an initiation site. However, the crystallization with the complete initiator sites failed and the reported structure lacks 13 nucleotides including the primase recognition site.
Despite the structural similarity of both helix bundles, the sequences bear no significant similarity. To date, it cannot be said whether these similar structures arose from convergent or divergent evolution. In favour of a divergent evolution it should be mentioned that both proteins have a similar function and that their primase domains are also structurally similar.
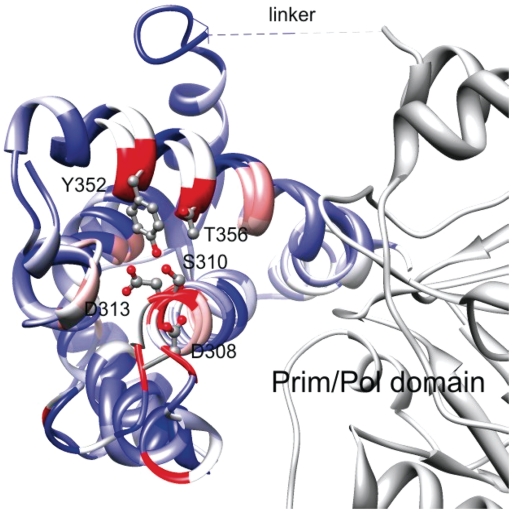
Within the three helices there are six completely conserved amino-acid positions (Figure 3). Five of the residues (D308, S310, D313, Y352 and T356 in ORF904) form a conserved patch at the surface of the helix bundle domain quite distant to the active site (Figure 4). The sixth residue G324 is located in the turn connecting helices 10 and 11. All together this analysis highlights the importance of the helices 10 and 12. Our point mutational study performed at selected position facing the prim/pol domain (see below) further substantiate that this part of the structure is critical for primase activity.
Figure 4.
Comparison of helix bundle subdomains in two primases. Superimposed is the primase domain of the pRN1 replication protein (prim/pol domain in gray, helix bundle domain in colour) and the three helices of the RSF1010 primase helical bundle. The ribbons are coloured according to the residue conservation of the underlying sequence alignment. The highly conserved residues D308, S310 and D313 (beginning of helix 10) and Y352 and T356 (center of helix 12) are shown in ball and stick representation. The view is from the ‘bottom’ of the primase when the active side (not shown) is considered to be on the ‘top’.
Is the C-terminal helix important for template recognition?
The structure of the primase domain points to the importance of the long C-terminal helix of the helix bundle domain: a deletion mutant devoid of this helix is inactive (see above) and the C-terminal helix faces the active site and could interact after a conformational change more closely with the prim/pol domain. We were therefore interested in determining the structure of the primase together in the closed conformation with template and/or nucleotides. We tested a range of templates and conditions but were unable to obtain crystals with bound DNA and or nucleotides. We therefore used the structure of the primase without ligands as a guide for mutational analysis of selected residues primarily in the C-terminal helix of the helix bundle domain. We focussed on residues which are on the surface of the subdomain and which are conserved within the pRN1 plasmid family (Figure 3). As we are specifically interested in residues which could make sequence-specific contacts with the template DNA we selected the residues W314, W347, Y352, W361 and Y367. The sequence recognized by this primase is GTG. A systematic study of RNA–protein complexes (22) pointed out that guanosine is most frequently contacted by tryptophanes and that uracil (and in analogy thymine) interacts preferentially with tyrosine.
The selected amino acids were replaced with alanine (Figure 5). As controls F260 and H141 were also changed to alanine. F260 is located at the very N-terminus of the helix bundle domain and does likely not play a role in the recognition of DNA or nucleotide, H141 is involved in the coordination of the zinc ion and replacing this residue with alanine should abolish enzymatic activity.
Figure 5.
Position of the aromatic residues investigated in this study. (A) Sideview: The prim/pol domain is shown in blue and the active site residues are in stick and ball representation. (B) View from the active site toward the helix bundle domain with the C-terminal helix 12 in front. When the aromatic residues in green F260, W347, W361 and Y367 are changed to alanine the mutant protein retains primase activity. In contrast, the amino acids W314 and Y352 (shown in red) do not support primase activity when changed to alanine.
In addition, we constructed a deletion mutant which lacked the six amino acids of the linker (Lys250–Asp255) which we suggest to be important to allow movement of the domains relative to each other.
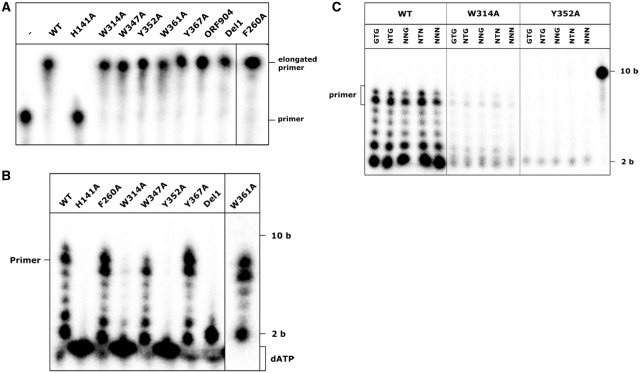
First the DNA-polymerase activity of all mutants was assessed. For the elongation of a primer bound to DNA only the amino acids 40–255 of ORF904 are necessary so this activity should not be impaired by the mutations. All proteins with the exception of H141A displayed polymerase activity (Figure 6, Table 3) indicating that the point mutations did not lead to any disruptions of the overall stability of the proteins and that the mutations in the helix bundle domain do not affect the catalysis of the elongation step.
Figure 6.
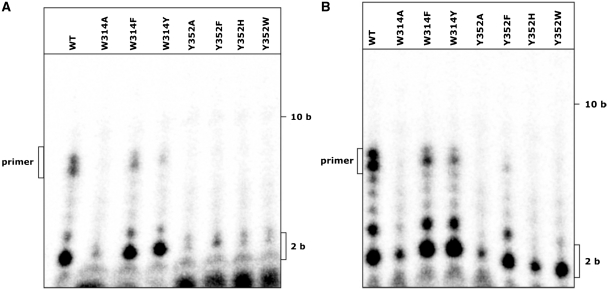
Primase activity of the point mutants with a template containing the recognition motif and with mixed templates. (A) Primer extension reaction with 0.4 µM protein and 5 nM radioactively labelled 20 base long primer hybridized to a 42 base long template in the presence of 100 µM dNTPs. –: no protein added, ORF904: full-length WT protein added. (B) Primase reactions with 0.4 µM protein and 4 µM template template2 (Table 2) were assembled in the presence of 10 µM dNTPs supplemented with [α-32P]-dATP and 1 mM ATP. The wild-type protein synthesizes mainly an 8-nt long primer, the point mutants F260A, W347A, Y367A and W361A show a comparable activity. The mutant where part of the flexible linker as been deleted (Del1) is only able to synthesize a dinucleotide. The primase activity of W314A is reduced and Y352A shows no visible primer formation. (C) As templates oligonucleotides with the sequence 5′-ttttttttXXXttt were used (Table 2). The sequence of the triplet marked with X is given above the lanes. The concentrations of the template mixes were adjusted according to the number of different triplets (GTG-2 µM, NTG-8 µM, NNG, NTN-32 µM und NNN-128 µM). The point mutant W314A shows a slight primase activity with all templates. Y352A is not able to synthesize any products with the templates.
Table 3.
Overview: activities of the wild type and the mutant primases
| DNA polymerase activitya | DNA bindingb | Dinucleotide formationb | Primase activityb | |
|---|---|---|---|---|
| WT | + | +++ | +++ | +++ |
| F260A | + | +++ | +++ | |
| W314A | + | + | (+) | (+) |
| W314F | +++ | ++ | ||
| W314Y | +++ | ++ | ||
| W347A | + | +++ | +++ | |
| Y352A | + | ++ | (+) | − |
| Y352F | ++ | + | ||
| Y352H | + | − | ||
| Y352W | ++ | − | ||
| W361A | + | +++ | +++ | |
| Y367A | +++ | +++ | ||
| H141A | − | − | − | |
| Del1 | + | +++ | − |
a+: active, –: inactive.
b+++: fully active, ++: ∼50% active, +: ∼10% active, (+): barely active, –: no activity detectable.
Next, the primase activity of the point mutants was assessed using a 17-nt oligonucleotide containing the recognition sequence GTG (template1, Table 1) as a template and deoxynucleotides supplemented with [α-32P]-dATP. This should lead to the formation of an 8-nt long primer as primer synthesis is initiated 5′ of the GTG and the full length primer is synthesized up to the 5′ end of the template. The proteins F260A, W347A, W361A and Y367A were able to synthesize a primer similar to that of the wild-type protein in this experiment indicating that these amino acids are neither involved in any specific interactions with the recognition motif (Figure 6) nor with the catalysis of the dinucleotide formation. As expected H141A did not show any primase activity since this amino-acid exchange should disrupt the zinc stem. The deletion of the flexible linker (Del1) also led to a loss of activity. Interestingly the protein still synthesizes a dinucleotide but fails to support the extension of the dinucleotide to the full-length primer, although the extension with a preformed primer template is catalysed by the mutant. This suggests that a step between dinucleotide formation and elongation is impaired in this mutant.
The point mutations W314A and Y352A appear to severely impair primer formation, for W314A a very small amount of wild-type like products is visible while the exchange of Y352 abolishes primase activity completely. Thus, these two amino acids are functionally important for faithful primer synthesis. In contrast to the mutant Del1, there is no significant synthesis of the dinucleotide suggesting that in these mutants either the initial recognition of the template or the catalysis of the dinucleotide is impaired.
The above experiments were performed under saturating template concentrations, since the Michaelis constant with ∼200 nM (8) is far below the template concentration of 4 µM. We therefore repeated these experiments at a lower template concentration in order to see more subtle changes. But even at 100 nM template concentration the mutants F260A and W367A support a primer synthesis rate as strong as the wild-type enzyme (data not shown). Only the mutant W347 shows a slightly impaired activity similar to the one seen at the higher template concentration.
Do the point mutants recognize an altered recognition sequence?
We reasoned that the strict sequence specificity of the primase activity should at least be partially contributed by the helix bundle domain. We therefore tested whether the mutant proteins now displayed a changed specificity. To identify the possibly altered recognition sequence of the mutant proteins, the point mutants W314A and Y352A were assessed in a further experiment with single oligonucleotide template mixes containing all possible triplet sequences (Table 2). Oligonucleotides were constructed based on template2 (Table 2) and the central GTG was replaced with NTG, NNG, NTN or NNN, N indicating a random base at this position. The frequency of each random base was the same and to take the concentration differences of each specific triplet in the mixes into account the concentrations of the templates used in this experiment were adjusted to 2 µM per single triplet. This should ensure that every possible base triplet was tested and could serve as a template for the synthesis of a visible primer. A primase assay was conducted using the template mixes and wild-type protein and the point mutants W314A and Y352A. The wild-type protein synthesized comparable amounts of full length primer and shorter truncated products with all template mixes indicating that the concentration of oligonucleotides containing the GTG triplet was equal in all samples (Figure 6) and that the primase domain could faithfully synthesize a primer even in the presence of a large excess of unspecific template. This is in line with our previous findings that the specificity of the pRN1 primase is extraordinarily high (11). The point mutant W314A that had shown a markedly decreased primase activity with the native recognition sequence synthesizes very low amounts of products with all mixes. The length of these products corresponds in all cases well with the wild-type products. It seems that this mutation leads to a lower affinity for the recognition sequence or possibly it also lowers the affinity for the DNA template or the growing primer without changing the recognized DNA sequence. In contrast, Y352A is not able to utilize any of the templates and does not synthesize any primers. This could indicate that the mutation of Y352 to alanine does not actually lead to a changed recognition motif but rather that the protein is unable to synthesize a primer at all.
In order to elucidate the reasons for the failure of these two mutants to synthesize a primer efficiently on all possible triplets we considered three possibilities. First, the point mutants could impede the proper folding of the helix bundle domain, secondly, the point mutants are not longer able to bind DNA with the recognition motif GTG, thirdly the mutated amino acids interfere directly with the dinucleotide formation.
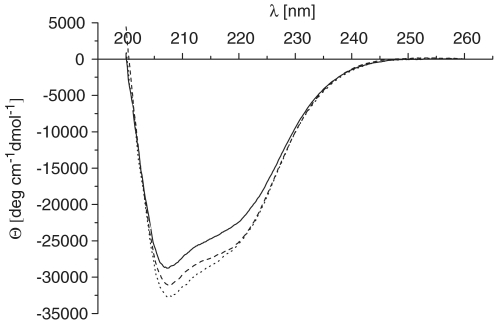
The helix bundle domain should give a strong CD signal. We therefore compared the CD spectra of the wild-type primase domain with both point mutants (Figure 7). The differences of the CD signals are rather low. All three proteins have a strong negative signal in the range of 210–220 nm which is typical for α helices. We can therefore exclude that both point mutants destroy the helical character of the domain.
Figure 7.
CD spectroscopy of the wild-type proteins and the mutants W314A and Y352A. CD spectra of the wild-type protein (solid), W314 (broken) and Y352 (points) in 25 mM sodium phosphate buffer, pH 7.0.
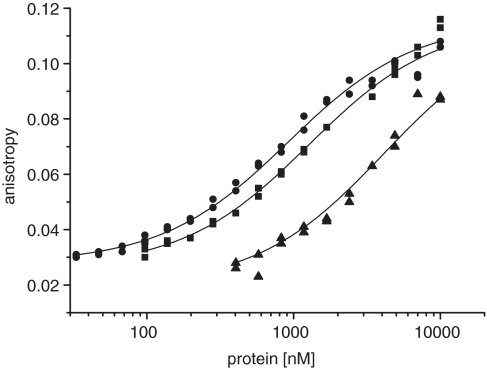
Next we determined the DNA-binding affinity of the wild-type protein and the mutants W314A and Y352A, which are impaired in primer synthesis (Figure 8). Our data shows that mutant W314A binds DNA containing the recognition sequence about 5× weaker and that Y352A binds nearly as well as the wild-type protein. The low activity of the mutant W314A can therefore be explained by its failure to bind to the specific DNA strong enough. However, the mutant Y352A binds DNA nearly as strong as the wild-type but is devoid of primase activity. This suggests that Y352 while being less important for initial template binding is crucial in the steps following, e.g. positioning of the DNA or nucleotides or even directly involved in dinucleotide formation.
Figure 8.
Fluoresence anisotropy measurements to determine the DNA-binding affinity. An amount of 40 nM of a single-stranded oligonucleotide with recognition motif (templatefluo, Table 2) was titrated with wild-type N40-C370 (circle), Y352A (square) and W314A (triangle). The data points were fitted to a single-site binding model and the affinitiy constants determined. Wild-type protein bound the DNA with a dissociation constant of 740 ± 60 nM, Y352A with 1030 ± 80 nM and W314A with 3400 ± 240 nM.
More conservative amino-acid changes at W314 and Y352 allow primer synthesis
To further characterize the function of the amino acids W314 and Y352, we constructed proteins with more conservative changes of these residues. At position Y352 mutants were constructed with tryptophane, histidine and phenylalanine in this position, at position W314 mutants with tyrosine and phenylalanine. The point mutants were first used in a primase assay with an oligonucleotide containing the native recognition sequence GTG (Table 2, template2). All amino acids showed an increased activity compared to alanine at the respective position. Replacing W314 with phenylalanine or tyrosine leads to a protein with nearly the same level of activity as the wild-type (Figure 9). When Y352 is exchanged with phenylalanine the protein retains a very low activity compared to the wild type, the products appear to have the same size but the amount synthesized is considerably smaller. Replacing it with histidine or tryptophane on the other hand reduces its activity almost completely; there is only a low amount of dinucleotide synthesized. This suggests that the interaction of Y352 with DNA or nucleotides is very specific. Only when it is replaced with the very similar phenylalanine the protein is still able to form a primer.
Figure 9.
Exchanging the aromatic residues with conservative amino acids. (A) A 4-µM template containing the recognition motif GTG was used in primase assays with 0.4 µM of protein and 10 µM dNTPs with [α-32P]dATP. Exchanging W314 with phenylalanine or tyrosine leads to a primase activity comparable to the wild-type. With Y352 only phenylalanine can be exchanged to retain a very slight primase activity, with histidine or tryptophane the protein is inactive. (B) A template mix was used (5′-ttttttttNNNttt) at 2 µM which provides all possible base triplets. Again W314F and W314Y are able to synthesize a wild-type like primer; this amino acid can apparently be replaced by similar ones.
Since exchanging the native amino acids with phenylalanine or histidine might influence the specificity of the DNA binding the point mutants were also tested with one of the templates containing a random mix of all possible base triplets (Table 2, templateNNN). With this DNA mixture the activities are similar to the template containing the GTG recognition sequence. Replacing W314 with tyrosine or phenylalanine leads to a protein that is active on this template (Figure 9). For any of the other exchanges no primase activity can be observed indicating that these exchanges do not lead to an altered recognition sequence. This suggests that the nucleotides of the recognition motif are not specifically recognized by a single amino acid of the helix bundle domain. Rather a number of amino acids possibly from the helix bundle domain and the catalytic domain provide the suitable environment for the interaction with the bases of the DNA.
DISCUSSION
To date a number of primase structures of the bacterial and archaeo-eukaryotic primase families have been solved but there is still no structure of a binary enzyme–DNA complex which would allow valuable insights into the mechanism of primer synthesis by these enzymes.
Since the structure of the primase core domain of the archaeal replication protein ORF904 had been solved (18) and the substrate requirements of the enzyme were known (11) we attempted to obtain the crystal structure of the minimal functional primase domain complexed with a DNA substrate. For this, the protein was crystallized in the presence of a variety of short single-stranded oligonucleotides. All oligonucleotides contained the recognition sequence GTG and flanking regions of different sequences and lengths (length was 6–17 nt in total). We also tried to co-crystallize the protein with a number of short primed templates. There was no DNA detectable in any of the obtained crystals even though a 17-nt single-stranded DNA template is bound with a reasonably high affinity [Kd = 225 nM (11)]. For the bacterial sliding clamp an affinity constant of 120 nM for primed DNA was reported and this complex could be crystallized (11) so it is unlikely that the affinity is the sole reason why the complex failed to crystallize. Most likely, conditions that favour crystallization disfavour DNA binding.
The X-ray structure of the pRN1 minimal primase domain was solved and showed that the protein is divided into two distinct domains. The N-terminal domain corresponded almost exactly to the previously crystallized primase core which had been identified by its proteolytic stability (18). The C-terminal domain consists entirely of α-helices and is connected to the N-terminal part of the protein via about six flexible amino acids whose positions could not be resolved. Searching for related structures of the newly resolved C-terminal domain showed highest similarity to the recently solved structure of the bacterial primase RepB′ (21). This protein is encoded by the plasmid RSF1010 and synthesizes the primers for the replication of RSF1010 on one specific initiation site on each strand (23,24) The overall organization of RepB′ resembles that of ORF904. Remarkably, the protein also consists of two clearly divided domains and the catalytic domain is separated from the helix bundle domain by a long helix and a flexible linker of 14 amino acids. The N-terminal catalytic domain shows highest similarity to other primases of the archaeo-eukaryotic primase superfamily (Pyrococcus furiosus and P. horikoshii primase), which do not have the observed bipartite organization. Superposition of the two C-terminal helix bundles shows that helices α10 and α12 (amino acids 249–361) of ORF904 and α8 and α10 of RepB′ align very well (Figure 4). This structural similarity could be due to a common ancestry of both proteins or a very specific adaption to their comparable functions as plasmid replication proteins. There is still very little known about the replication of pRN1, but the structural similarities might indicate that it is replicated with an analogous mechanism to RSF1010. For the replication of RSF1010 three proteins are necessary that are encoded by the plasmid, primase RepB′, helicase RepA and initiator protein RepC (25–27). These functions could in the case of pRN1 be performed by ORF904, which has primase and helicase activity (28) and putatively by ORF80 which is also encoded by pRN1 and has been shown to bind DNA sequence specifically (29). The replication origin of pRN1 is not known, but it is possible that ORF904 is also able to recognize a specific initiator region either through its sequence or secondary structure or both.
The here reported structure of pRN1 primase shows that the previously identified active site and the essential C-terminal helix are relatively far apart, which suggested that the protein is able to change its conformation to move these two parts closer together and allow interaction with the DNA template close to the active site.
We suggest that the template strand is bound by the pRN1 primase in the same orientation as can be expected when the hairpin in the RepB′ complex structure is extended towards the active site. This would place the 3′ end of the template DNA towards the right edge to the catalytic domain when facing the active site with the helix bundle domain below (Figure 10). In the structure of the primase of P. horikoshii (7) the nucleotide UTP is present. We propose that in the pRN1 structure the UTP occupies the position of the incoming deoxynucleotides (dATP) and is placed opposite to the second base 5′ to the GTG recognition motif. The initiating ribonucleotide (ATP) is placed right to the catalytic magnesium ion and is opposite to the first base 5′ to the GTG motif. As dinucleotide formation requires the C-terminal helix bundle, we suggest that the helix bundle domain now moves upward and makes contact with the template and the nucleotides. In the P. horikoshii structure the 2′ OH group of the UTP has no contact with the protein. It is possible that the helix bundle domain provides in the close conformation a gating mechanism and ensures that only a deoxynucleotide is accommodated as incoming nucleotide and a ribonucleotide as the first base. After dinucleotide formation, the primer/template has to be repositioned. Our data (see below) suggest that the flexibility of the connecting loop is required for this process.
This model of primer synthesis is in line with our experimental data. We showed that the C-terminal subdomain is directly involved in interaction with DNA by exchanging a number of conserved amino acids. These conserved amino acids are located in the part of ORF904 that is highly similar to the structure of RepB′. Changing these conserved amino acids to alanine showed a loss of primase activity for Y352 and a strong decrease for W314 on a template containing the recognition sequence of ORF904. This indicates that these amino acids do play a critical role in primer synthesis. We further tried to assess if these point mutants are able to recognize an altered triplet sequence using oligonucleotides with all possible triplets at equal concentrations. In this assay no primer synthesis pointing to a different recognition sequence could be observed. To rule out that the changed activities are due to conformational changes resulting from the point mutations CD spectra were recorded that showed no significant differences between the wild-type protein and the point mutants. The fact that W314A has a reduced activity to synthesize primers with all used templates could be explained by its lower affinity for DNA as measured by fluorescence anisotropy measurements. On the other hand the highly conserved Y352 seems to be involved in a step after DNA binding, e.g. this amino acid could be critical for dinucleotide formation which is greatly impaired in the mutant Y352A. In contrast, the mutant Del1 (deletion of the linker) appears to influence the step after dinucleotide formation. It is conceivable that after dinucleotide formation the dinucleotide/template pair has to move relative to the active site in order to reposition the 3′ end of the dinucleotide in the active site so that the dinucleotide can be elongated. This is not observed in the linker deletion mutant suggesting that the flexibility of the helix bundle domain relative to the catalytic prim/pol domain could be required for moving the dinucleotide/template. Therefore, it appears that the helix bundle domain could participate in three steps during full-length primer synthesis: initial binding of the template, formation of the dinucleotide and repositioning of the dinucleotide/template pair.
ACCESSION NUMBER
3M1M.
FUNDING
Deutsche Forschungsgemeinschaft (Li913/4 and Li913/6) to G.L.; Marie-Curie fellowship of the European Union to A.V.; Deutsche Forschungsgemeinschaft, SFB646, TR5, CIPSM, NIM, the Jung-Stiftung, and the Fonds der chemischen Industrie to P.C. Funding for open access charge: Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
REFERENCES
- 1.Swart JR, Griep MA. Primase from Escherichia coli primes single-stranded templates in the absence of single-stranded DNA-binding protein or other auxiliary proteins. Template sequence requirements based on the bacteriophage G4 complementary strand origin and Okazaki fragment initiation sites. J. Biol. Chem. 1993;268:12970–12976. [PubMed] [Google Scholar]
- 2.Mendelman LV, Richardson CC. Requirements for primer synthesis by bacteriophage T7 63-kDa gene 4 protein. Roles of template sequence and T7 56-kDa gene 4 protein. J. Biol. Chem. 1991;266:23240–23250. [PubMed] [Google Scholar]
- 3.Tseng TY, Frick DN, Richardson CC. Characterization of a novel DNA primase from the Salmonella typhimurium bacteriophage SP6. Biochemistry. 2000;39:1643–1654. doi: 10.1021/bi992155t. [DOI] [PubMed] [Google Scholar]
- 4.Keck JL, Roche DD, Lynch AS, Berger JM. Structure of the RNA polymerase domain of E. coli primase. Science. 2000;287:2482–2486. doi: 10.1126/science.287.5462.2482. [DOI] [PubMed] [Google Scholar]
- 5.Podobnik M, McInerney P, O'Donnell M, Kuriyan J. A TOPRIM domain in the crystal structure of the catalytic core of Escherichia coli primase confirms a structural link to DNA topoisomerases. J. Mol. Biol. 2000;300:353–362. doi: 10.1006/jmbi.2000.3844. [DOI] [PubMed] [Google Scholar]
- 6.Pan H, Wigley DB. Structure of the zinc-binding domain of Bacillus stearothermophilus DNA primase. Structure. 2000;8:231–239. doi: 10.1016/s0969-2126(00)00101-5. [DOI] [PubMed] [Google Scholar]
- 7.Ito N, Nureki O, Shirouzu M, Yokoyama S, Hanaoka F. Crystal structure of the Pyrococcus horikoshii DNA primase-UTP complex: implications for the mechanism of primer synthesis. Genes Cells. 2003;8:913–923. doi: 10.1111/j.1365-2443.2003.00693.x. [DOI] [PubMed] [Google Scholar]
- 8.Augustin MA, Huber R, Kaiser JT. Crystal structure of a DNA-dependent RNA polymerase (DNA primase) Nat. Struct. Biol. 2001;8:57–61. doi: 10.1038/83060. [DOI] [PubMed] [Google Scholar]
- 9.Lao-Sirieix SH, Nookala RK, Roversi P, Bell SD, Pellegrini L. Structure of the heterodimeric core primase. Nat. Struct. Mol. Biol. 2005;12:1137–1144. doi: 10.1038/nsmb1013. [DOI] [PubMed] [Google Scholar]
- 10.Aravind L, Leipe DD, Koonin EV. Toprim–a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 1998;26:4205–4213. doi: 10.1093/nar/26.18.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beck K, Lipps G. Properties of an unusual DNA primase from an archaeal plasmid. Nucleic Acids Res. 2007;35:5635–5645. doi: 10.1093/nar/gkm625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leslie AGW. Crystallographic Computing. Oxford: Oxford University Press; 1990. [Google Scholar]
- 13.Evans PR. Data reduction. In: Sawyer L, Issac N, Bailey S, editors. Proceedings of the CCP4 Study Weekend. Data Collection and Processing. Warrington: Daresbury Laboratory; 1993. pp. 114–122. [Google Scholar]
- 14.Navaza J. Implementation of molecular replacement in AMoRe. Acta Crystallogr. D. Biol. Crystallogr. 2001;57:1367–1372. doi: 10.1107/s0907444901012422. [DOI] [PubMed] [Google Scholar]
- 15.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 16.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 17.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera – a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 18.Lipps G, Weinzierl AO, von Scheven G, Buchen C, Cramer P. Structure of a bifunctional DNA primase-polymerase. Nat. Struct. Mol. Biol. 2004;11:157–162. doi: 10.1038/nsmb723. [DOI] [PubMed] [Google Scholar]
- 19.Schmid FX. Optical spectroscopy to characterize protein conformation and conformational changes. In: Creighton TE, editor. Protein Structure: A Practical Approach. Oxford: IRL Press; 1997. pp. 261–297. [Google Scholar]
- 20.Kato M, Ito T, Wagner G, Richardson CC, Ellenberger T. Modular architecture of the bacteriophage T7 primase couples RNA primer synthesis to DNA synthesis. Mol. Cell. 2003;11:1349–1360. doi: 10.1016/s1097-2765(03)00195-3. [DOI] [PubMed] [Google Scholar]
- 21.Geibel S, Banchenko S, Engel M, Lanka E, Saenger W. Structure and function of primase RepB' encoded by broad-host-range plasmid RSF1010 that replicates exclusively in leading-strand mode. Proc. Natl Acad. Sci. USA. 2009;106:7810–7815. doi: 10.1073/pnas.0902910106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellis JJ, Broom M, Jones S. Protein-RNA interactions: structural analysis and functional classes. Proteins. 2007;66:903–911. doi: 10.1002/prot.21211. [DOI] [PubMed] [Google Scholar]
- 23.Honda Y, Sakai H, Komano T. Two single-strand DNA initiation signals located in the oriV region of plasmid RSF1010. Gene. 1988;68:221–228. doi: 10.1016/0378-1119(88)90024-8. [DOI] [PubMed] [Google Scholar]
- 24.Honda Y, Sakai H, Komano T, Bagdasarian M. RepB' is required in trans for the two single-strand DNA initiation signals in oriV of plasmid RSF1010. Gene. 1989;80:155–159. doi: 10.1016/0378-1119(89)90261-8. [DOI] [PubMed] [Google Scholar]
- 25.Haring V, Scholz P, Scherzinger E, Frey J, Derbyshire K, Hatfull G, Willetts NS, Bagdasarian M. Protein RepC is involved in copy number control of the broad host range plasmid RSF1010. Proc. Natl Acad. Sci. USA. 1985;82:6090–6094. doi: 10.1073/pnas.82.18.6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scherzinger E, Bagdasarian MM, Scholz P, Lurz R, Ruckert B, Bagdasarian M. Replication of the broad host range plasmid RSF1010: requirement for three plasmid-encoded proteins. Proc. Natl Acad. Sci. USA. 1984;81:654–658. doi: 10.1073/pnas.81.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miao DM, Sakai H, Okamoto S, Tanaka K, Okuda M, Honda Y, Komano T, Bagdasarian M. The interaction of RepC initiator with iterons in the replication of the broad host-range plasmid RSF1010. Nucleic Acids Res. 1995;23:3295–3300. doi: 10.1093/nar/23.16.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez M, Drechsler M, Stark H, Lipps G. DNA translocation activity of the multifunctional replication protein ORF904 from the archaeal plasmid pRN1. Nucleic Acids Res. 2009;37:6831–48. doi: 10.1093/nar/gkp742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lipps G, Ibanez P, Stroessenreuther T, Hekimian K, Krauss G. The protein ORF80 from the acidophilic and thermophilic archaeon Sulfolobus islandicus binds highly site-specifically to double-stranded DNA and represents a novel type of basic leucine zipper protein. Nucleic Acids Res. 2001;29:4973–4982. doi: 10.1093/nar/29.24.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]