Abstract
Two archaeal tRNA methyltransferases belonging to the SPOUT superfamily and displaying unexpected activities are identified. These enzymes are orthologous to the yeast Trm10p methyltransferase, which catalyses the formation of 1-methylguanosine at position 9 of tRNA. In contrast, the Trm10p orthologue from the crenarchaeon Sulfolobus acidocaldarius forms 1-methyladenosine at the same position. Even more surprisingly, the Trm10p orthologue from the euryarchaeon Thermococcus kodakaraensis methylates the N1-atom of either adenosine or guanosine at position 9 in different tRNAs. This is to our knowledge the first example of a tRNA methyltransferase with a broadened nucleoside recognition capability. The evolution of tRNA methyltransferases methylating the N1 atom of a purine residue is discussed.
INTRODUCTION
Cellular RNAs possess numerous chemically modified nucleosides, but the largest number and the greatest variety are found in transfer RNA (tRNA). These modifications are introduced by many different enzymes during the complex process of RNA maturation. The functions of these modified nucleosides are not well known, but it seems that modifications in the anticodon region play a direct role in increasing translational efficiency and fidelity, while modifications outside the anticodon region are typically involved in the maintenance of the structural integrity of tRNA. Among naturally occurring nucleoside modifications, base and ribose methylations are by far the most frequently encountered (1,2). These methylations are catalysed by tRNA methyltransferases (MTases) that use S-adenosyl-l-methionine (AdoMet) as the methyl donor, with a single exception of a recently identified enzyme, which uses a folate as the methyl donor (3).
AdoMet-dependent MTases belong to at least seven evolutionarily and structurally unrelated classes/superfamilies (4–6). Most of the known RNA MTases belong to class I. They possess a fold similar to the Rossmann fold, and are therefore called Rossmann fold MTases (RFM). The structure comprises a seven-stranded β sheet with a central topological switch-point and a characteristic reversed β hairpin at the carboxyl end of the sheet (6↑7↓5↑4↑1↑2↑3↑). This β sheet is flanked by α helices to form an αβα sandwich (4). RFM enzymes are typically monomeric although di-tri- or tetrameric structures have been reported. Class IV MTases, also named the SPOUT class MTases, a nomenclature coming from the first two members of this class, SpoU and TrmD, act also on RNA, but are structurally different from the class I MTases (7). They possess a five-stranded β sheet core (5↑3↑4↑1↑2↑) flanked by seven α helices. The most characteristic feature of these enzymes is the presence of a deep topological knot in the C-terminal part of the sheet. This knot is responsible for AdoMet binding. All known SPOUT MTases are dimers, with the catalytic site formed at the interface of two monomers (8).
There are only a few archaeal tRNA specific MTases functionally characterised to date. Those MTases can be very different in terms of structure, of target or in their mechanism. Some act as base MTases, while others are ribose MTases. Among the MTases acting on ribose, there exist two kinds of methylation pathways. In the first one, called ‘all-protein enzyme’, the MTase recognizes on its own the target nucleoside and catalyses the methyl transfer on the ribose. The second mechanism involves an MTase taking part of a ribonucleoprotein (RNP) complex, in which the recognition of the target nucleoside is carried out by a small guide RNA. In different organisms, the same modification can be accomplished by the two different mechanisms. This is the case of the Cm56 ribose methylation of tRNA in Archaea. In Pyrobaculum aerophilum, this modification is catalysed by a RNP complex, where the small RNA sR35 targets the C56 nucleoside for modification by a protein catalytic subunit, a member of the RFM superfamily (9). This case is an exception in Archaea, because all other archaeal genomes sequenced to date possess a SPOUT MTase homologous to aTrm56 from Pyrococcus abyssi, which catalyses non-guided methylation of C56 (9,10).
The six archaeal RFM MTases characterised to date act on the base of nucleosides. Trm-G10 from P. abyssi methylates or dimethylates guanosine 10 from tRNA to form m2G10 or 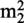 G10 (11,12). The same modification can be found at position 26 of tRNA and is catalysed by Trm-G26 (also known as Trm1) (13,14). The MTase encoded by the open reading frame (ORF) PAB1947 from P. abyssi acts on several cytosines in tRNA to form m5C, with a preference for C49. The specificity of this MTase for C49 is increased by an archaease (15). Another important tRNA MTase identified in Archaea is Trm5, which methylates G37 in tRNA (16). Guanosine in position 37 is commonly methylated to form m1G37 in tRNA from organisms belonging to the three domains of life, and this modification prevents frame-shifting by assuring correct codon–anticodon pairing (17). The tRNA MTase TrmU54 catalyses the methylation of atom C5 in uridine to form ribothymidine in tRNA from P. abyssi (18). This modification is invariably found at position 54 in the TΨC loop of tRNAs of most organisms. And finally, the MTase TrmI from P. abyssi catalyses the methylation of position N1 of adenosine to form m1A. This enzyme displays region specificity, methylating A58 and A57 from TΨC loop of tRNA, m1A57 being the obligatory intermediate in the biosynthesis of 1-methylinosine (m1I) (19).
G10 (11,12). The same modification can be found at position 26 of tRNA and is catalysed by Trm-G26 (also known as Trm1) (13,14). The MTase encoded by the open reading frame (ORF) PAB1947 from P. abyssi acts on several cytosines in tRNA to form m5C, with a preference for C49. The specificity of this MTase for C49 is increased by an archaease (15). Another important tRNA MTase identified in Archaea is Trm5, which methylates G37 in tRNA (16). Guanosine in position 37 is commonly methylated to form m1G37 in tRNA from organisms belonging to the three domains of life, and this modification prevents frame-shifting by assuring correct codon–anticodon pairing (17). The tRNA MTase TrmU54 catalyses the methylation of atom C5 in uridine to form ribothymidine in tRNA from P. abyssi (18). This modification is invariably found at position 54 in the TΨC loop of tRNAs of most organisms. And finally, the MTase TrmI from P. abyssi catalyses the methylation of position N1 of adenosine to form m1A. This enzyme displays region specificity, methylating A58 and A57 from TΨC loop of tRNA, m1A57 being the obligatory intermediate in the biosynthesis of 1-methylinosine (m1I) (19).
Until now, only one tRNA from hyperthermophilic archaea has been fully sequenced, the initiator methionine tRNA from the crenarchaeon Sulfolobus acidocaldarius (20). This tRNA contains 10 modified nucleosides, 9 of them bearing a methylation either on the base or on the ribose, or even both on base and ribose. However, the nature of the modified nucleoside at position 9 is unknown. In yeast, some tRNAs with a guanosine at this position are methylated by the Trm10p MTase, to form m1G9 (21). As a protein distantly related to the yeast enzyme is encoded by the Saci_1677 gene of S. acidocaldarius, it could be a potential candidate for the modification at position 9 of S. acidocaldarius tRN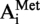 .
.
In this article, we show that the S. acidocaldarius Saci_1677p enzyme indeed acts at position 9 of tRNA, catalysing m1A formation. Furthermore, in Euryarchaeota, the homologous protein from Thermococcus kodakaraensis also acts at position 9 of tRNA, but catalyses both m1A and m1G formation. To our knowledge, this is the first MTase found to methylate the two purine bases at the same position.
MATERIALS AND METHODS
Strains, media, growth conditions and general procedures
Pwo DNA polymerase, T4 DNA ligase, T7 RNA polymerase, and T4 polynucleotide kinase were purchased from Roche. Ribonuclease A was from Fermentas. Genomic DNA from T. kodakaraensis was a gift from H. Grosjean (CNRS, France) and T.J. Santangelo (Ohio State University, USA). Genomic DNA from S. acidocaldarius was a gift from D. Charlier (VUB, Belgium). The S. cerevisiae Trm10-GST clone plasmid (pYCG_YOL93w) and S. cerevisiae Y16243 strain (BY4742; MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; yol093w::kanMX4) were purchased from Euroscarf.
Cloning of the T. kodakaraensis TK0422 ORF, S. acidocaldarius Saci_1677 ORF and of S. cerevisiae TRM10 ORF
The TK0422 ORF was amplified from T. kodakaraensis genomic DNA using Pwo polymerase (Roche) and the primers TKF (5′-CTAGCATATGAAGACCCTCGCAGATG-3′) and TKR (5′-CTAGCTCGAGTCAGCAGTTGTAGCAGAGC-3′) containing the NdeI and XhoI restriction sites, respectively. After cloning the PCR product in pCR-Blunt vector (Zero Blunt®, Invitrogen), the NdeI/XhoI fragment was extracted and cloned in pET-28b expression vector (Novagen), generating the pTK1 plasmid, allowing expression of an N-terminal His-tagged T. kodakaraensis protein in Escherichia coli. The Saci_1677 ORF was amplified from S. acidocaldarius genomic DNA using Pwo polymerase (Roche) and the primers SAF (5′-CTAGCATATGACACTTGCAAAGGTTTTTTCGC-3′) and SAR (5′-CTAGCTCGAGTCAATTTTTTCCCAGTCTAC-3′) containing the NdeI and XhoI restriction sites, respectively. After cloning the PCR product in pJET1.2/blunt cloning vector (CloneJETTM Fermentas), the NdeI/XhoI fragment was extracted and cloned in pET-28b expression vector, generating the pSA1 plasmid, allowing expression of an N-terminal His-tagged S. acidocaldarius protein in E. coli. The TRM10 ORF was amplified from the Trm10-GST clone using Pwo polymerase (Roche) and the primers SCF (5′-CTACATATGTCCAATGATGAGATAAACC-3′) and SCR (5′-CTACTCGAGTGTGTCCTTTGGAGCTGG-3′) containing the NdeI and XhoI restriction sites, respectively. After cloning the pCR product in pJET1.2/blunt cloning vector, the NdeI/XhoI fragment was extracted and cloned in pET-28b expression vector, generating the pSC1 plasmid, allowing expression of an N-terminal His-tagged S. cerevisiae protein in E. coli. The sequences of all clones were checked.
Expression and purification of the recombinant T. kodakaraensis TK0422p, S. acidocaldarius Saci_1677p and S. cerevisiae Trm10p
The His-tagged TK0422p, Saci_1677p and Trm10p recombinant proteins were expressed in E. coli strain Rosetta (DE3) (Novagen) carrying extra copies of tRNA genes (argU, argW, ileX, glyT, leuW, proL, metT, thrT, tyrU and thrU) specific for rare E. coli codons, to aid this expression. Freshly transformed cells were grown to an OD660 of 0.5–0.6 at 37°C in 1 l of Luria broth with kanamycin (30 µg/ml). Isopropyl-β-d-thiogalactopyranoside (IPTG) (Roche Diagnostics) was then added to a final concentration of 1 mM to induce recombinant protein expression. Cells were harvested after 3 h incubation at 37°C and resuspended in 100 ml of buffer A (Tris–HCl 50 mM pH 8, KCl 500 mM) complemented with protease inhibitors (Complete, EDTA-free protease inhibitor; Roche Diagnostics) prior to cell disruption by sonication. The lysate was cleared by centrifugation (20 000g for 30 min), and was applied to a Chelating-Sepharose fast flow column (GE Healthcare) charged with Ni2+ and equilibrated with buffer A. The column was washed with the same buffer, and the adsorbed material was eluted with a linear gradient (210 ml, from 0 to 1.0 M) of imidazole in buffer A. The fractions containing TK0422p, Saci_1677p and Trm10p were separately pooled. The purified proteins were then submitted to a gel filtration chromatography (Superdex G200; GE Healthcare), leading to almost completely pure TK0422p, Saci_1677p and Trm10p.
T7 in vitro transcription of tRNA genes
The general procedure for generating in vitro transcripts of tRNA genes is based on the method described previously (22). The sequence of the DNA product obtained after amplification of S. acidocaldarius genomic DNA with oligonucleotides MK1 (5′-TCTGCGTAATACGACTC ACTATAGGCGGCGTAGGGAAGCCTGGTATCCC-3′) and MK2 (5′-TCTGCGCTGCAGTGGTGGCGGCGCCTGGATTTGAACCAGGGACCTCAGGGTTA-3′) together with the sequence of this region of the genome in the database revealed differences with that of the 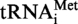 sequence (20). The amplification product for in vitro transcription was therefore corrected in respect to the published tRNA sequence (D-loop in the published sequence was ACUGGGAGUA and the corrected sequence is AGCCUGGUA). The sequence coding for S. acidocaldarius
sequence (20). The amplification product for in vitro transcription was therefore corrected in respect to the published tRNA sequence (D-loop in the published sequence was ACUGGGAGUA and the corrected sequence is AGCCUGGUA). The sequence coding for S. acidocaldarius
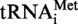 was thus PCR amplified using the oligonucleotides MK1 (5′-TCTGCGTAATACGACTCACTATAGGCGGCGTAGGGAAGCCTGGTATCCC-3′) and MK2 (5′-TCTGCGCTGCAGTGGTGGCGGCGCCTGGATTTGAACCAGGGACCTCAGGGTTA-3′) as primers and oligonucleotides MK3 (5′-GGAAGCCTGGTATCCCGCAGGGCTCATAACCCTGAGGTCCCTGGTTC-3′) and MK4 (5′-GAACCAGGGACCTCAGGGTTATGAGCCCTGCGGGATACCAGGCTTCC-3′) as template DNA. The
was thus PCR amplified using the oligonucleotides MK1 (5′-TCTGCGTAATACGACTCACTATAGGCGGCGTAGGGAAGCCTGGTATCCC-3′) and MK2 (5′-TCTGCGCTGCAGTGGTGGCGGCGCCTGGATTTGAACCAGGGACCTCAGGGTTA-3′) as primers and oligonucleotides MK3 (5′-GGAAGCCTGGTATCCCGCAGGGCTCATAACCCTGAGGTCCCTGGTTC-3′) and MK4 (5′-GAACCAGGGACCTCAGGGTTATGAGCCCTGCGGGATACCAGGCTTCC-3′) as template DNA. The 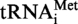 (A9G) was amplified as described for S. acidocaldarius
(A9G) was amplified as described for S. acidocaldarius
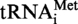 but with oligonucleotide MK1′ (same as MK1 but introducing the A9G mutation) instead of MK1. The sequence coding for T. kodakaraensis tRNAAla was PCR amplified using oligonucleotides MK5 (5′-TCTGGAATTCTAATACG ACTCACTATAGGGCCGGTAGCTCAGCCTGGTAT G-3′) and MK6 (5′-TCTGGAATTCCTGCAGTGGTGGACCGGCCGGGATTTGAACCC-3′) as primers and oligonucleotides MK7 (5′-CAGCCTGGTATGAGCGCCGCCTTGGCAAGGCGGAGGCCCCGGGTTCAAATCC-3′) and MK8 (5′-GGATTTGAACCCGGGGCCTCCGCCTTGCCAAGGCGGCGCTCATACCAGGCTG-3′) as template DNA. T. kodakaraensis tRNAAsp was PCR amplified using oligonucleotides MK9 (5′-TCTGGAATTCTAATACGACTCACTATAGCCCGGGTGGTGTAGCCCGGCCCATC-3′) and MK10 (5′-TCTGGAATTCCCTGGCGCCCGGGCCGGGATTTGAACCCGG-3′) as primers and oligonucleotides MK11 (5′-GTAGCCCGGCCCATCATACGGGACTGTCACTCCCGTGACCCGGGTTCAAATC-3′) and MK12 (5′-GATTTGAACCCGGGTCACGGGAGTGACAGTCCCGTATGATGGGCCGGGCTAC-3′) as template DNA. The DNA sequences encoding
but with oligonucleotide MK1′ (same as MK1 but introducing the A9G mutation) instead of MK1. The sequence coding for T. kodakaraensis tRNAAla was PCR amplified using oligonucleotides MK5 (5′-TCTGGAATTCTAATACG ACTCACTATAGGGCCGGTAGCTCAGCCTGGTAT G-3′) and MK6 (5′-TCTGGAATTCCTGCAGTGGTGGACCGGCCGGGATTTGAACCC-3′) as primers and oligonucleotides MK7 (5′-CAGCCTGGTATGAGCGCCGCCTTGGCAAGGCGGAGGCCCCGGGTTCAAATCC-3′) and MK8 (5′-GGATTTGAACCCGGGGCCTCCGCCTTGCCAAGGCGGCGCTCATACCAGGCTG-3′) as template DNA. T. kodakaraensis tRNAAsp was PCR amplified using oligonucleotides MK9 (5′-TCTGGAATTCTAATACGACTCACTATAGCCCGGGTGGTGTAGCCCGGCCCATC-3′) and MK10 (5′-TCTGGAATTCCCTGGCGCCCGGGCCGGGATTTGAACCCGG-3′) as primers and oligonucleotides MK11 (5′-GTAGCCCGGCCCATCATACGGGACTGTCACTCCCGTGACCCGGGTTCAAATC-3′) and MK12 (5′-GATTTGAACCCGGGTCACGGGAGTGACAGTCCCGTATGATGGGCCGGGCTAC-3′) as template DNA. The DNA sequences encoding 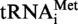 and
and 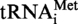 A9G were cloned in the SmaI site of the pUC18 vector, giving plasmids pUC18-
A9G were cloned in the SmaI site of the pUC18 vector, giving plasmids pUC18-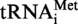 and pUC18-
and pUC18- A9G. The DNA sequences encoding tRNAAla and tRNAAsp were cloned in the EcoRI site of pUC18, giving plasmids pUC18-tRNAAla and pUC18-tRNAAsp. The sequences of all the clones were checked. These plasmids allow T7 transcription of S. acidocaldarius
A9G. The DNA sequences encoding tRNAAla and tRNAAsp were cloned in the EcoRI site of pUC18, giving plasmids pUC18-tRNAAla and pUC18-tRNAAsp. The sequences of all the clones were checked. These plasmids allow T7 transcription of S. acidocaldarius
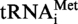 and
and 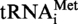 (A9G) and T. kodakaraensis tRNAAla and tRNAAsp, respectively. Radioactive (32P) in vitro transcripts were obtained using PstI (for S. acidocaldarius tRNA) or MvaI-digested plasmids (for T. kodakaraensis tRNA). [α32P]ATP and [α32P]GTP were purchased from Perkin Elmer. Radioactive transcripts were purified by 10% polyacrylamide gel electrophoresis.
(A9G) and T. kodakaraensis tRNAAla and tRNAAsp, respectively. Radioactive (32P) in vitro transcripts were obtained using PstI (for S. acidocaldarius tRNA) or MvaI-digested plasmids (for T. kodakaraensis tRNA). [α32P]ATP and [α32P]GTP were purchased from Perkin Elmer. Radioactive transcripts were purified by 10% polyacrylamide gel electrophoresis.
tRNA MTase assays
The two types of tRNA MTase assays used in this work were described in (23). The first method consisted of measuring the amount of 14C transferred to total yeast tRNA (Y16243 strain), or total E. coli tRNA using [methyl-14C]AdoMet as the methyl donor. The reaction mixture (300 µl) consisted of 50 mM Tris–HCl pH 8, 10 mM MgCl2, 100 µg total tRNA, 25 nCi [methyl-14C]AdoMet (50 mCi/mmol; GE Healthcare) and enzyme. The effect of TK0422p concentration on the MTase reaction was carried out using 80 µg total E. coli tRNA, and variable amount of TK0422p (5, 2.5, 1, 0.5 and 0.01 µg). The effect of pH on MTase reaction was measured using 50 µg E. coli tRNA as substrate and 5 µg enzyme. The buffers were: acetate buffer pH 4.8, MES buffer pH 5.5, phosphate buffer pH 7, HEPES buffer pH 7, CHES buffer pH 9 and CAPS buffer pH 9.75. All buffers concentrations were 50 mM. The second type of tRNA MTase assay involved in vitro transcribed, 32P-labelled tRNA as substrates. Modified nucleotides were analysed by 2D thin layer chromatography (2D–TLC) on cellulose plates (Merck). First dimension was with solvent A (isobutyric acid/concentrated NH4OH/water; 66/1/33; v/v/v); second dimension was with solvent B [0.1 M sodium phosphate pH 6.8/solid (NH4)2SO4/n-propanol; 100/60/2; v/w/v)] . The nucleotides were identified using a reference map (24).
Analysis of the presence of m1A or m1G in E. coli tRNA in vivo
Prior to tRNA extraction, the E. coli strain overexpressing TK0422p and the control strain (no expression of TK0422p) were incubated for 2 h at 50°C. Then, total tRNA was extracted. An amount of 200 µg of totally hydrolysed tRNA was injected on a Supelco Discovery C18 (250 × 4.6) mm HPLC column equilibrated with ammonium acetate 0.25 M, pH 6.5. The column was eluted with a linear gradient of acetonitrile/water (40/60; v/v) at a flow rate of 1.2 ml/min. The nucleosides were detected by measuring UV absorbance at 254 nm. The standard HPLC curve was obtained by injecting 20 µl of 1 mM canonical nucleosides together with m1A and m1G modified nucleosides.
Localization of m1A and m1G in S. acidocaldarius
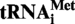
The transcript of 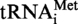 or
or 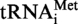 (A9G) from S. acidocaldarius was used as substrate for the MTases TK0422p or Saci_1677p. An amount of 2 µg of tRNA was incubated in presence of 4 µg of enzyme and 0.3 mM AdoMet (Sigma) for 1 h at 50°C. As a control, transcripts were incubated in the MTase assay conditions, but without enzyme. Then the reaction was stopped by phenol extraction, transcripts were ethanol precipitated and the pellet was resuspended in 30 µl of RNase A buffer (Tris–Cl 10 mM pH 7.5, 15 mM NaCl). The resuspended solution was heated at 85°C for 5 min and then slowly cooled down to 37°C. Then 10 µg of RNAse A was added and the digestion was carried out for 1 h at 70°C. Half of the RNase A digestion was 5′-end radiolabelled with 50 µCi of [γ32P] ATP and 20 units of T4 polynucleotide kinase. The radiolabelled fragments were then separated by 20 or 30% polyacrylamide gel electrophoresis and revealed by autoradiography. The 8-nt long fragments (A9 or G9 at 5′-end), and 5-nt long fragments (control) were identified by their position and their intensities on the autoradiogram, excised from the gel and eluted overnight at 45°C in 200 µl water. Subsequently, they were digested by P1 nuclease and mononucleotides were separated by 2D–TLC as described in the MTase assay. The radiolabelled nucleotides present at the 5′-end of the fragments were revealed by autoradiography.
(A9G) from S. acidocaldarius was used as substrate for the MTases TK0422p or Saci_1677p. An amount of 2 µg of tRNA was incubated in presence of 4 µg of enzyme and 0.3 mM AdoMet (Sigma) for 1 h at 50°C. As a control, transcripts were incubated in the MTase assay conditions, but without enzyme. Then the reaction was stopped by phenol extraction, transcripts were ethanol precipitated and the pellet was resuspended in 30 µl of RNase A buffer (Tris–Cl 10 mM pH 7.5, 15 mM NaCl). The resuspended solution was heated at 85°C for 5 min and then slowly cooled down to 37°C. Then 10 µg of RNAse A was added and the digestion was carried out for 1 h at 70°C. Half of the RNase A digestion was 5′-end radiolabelled with 50 µCi of [γ32P] ATP and 20 units of T4 polynucleotide kinase. The radiolabelled fragments were then separated by 20 or 30% polyacrylamide gel electrophoresis and revealed by autoradiography. The 8-nt long fragments (A9 or G9 at 5′-end), and 5-nt long fragments (control) were identified by their position and their intensities on the autoradiogram, excised from the gel and eluted overnight at 45°C in 200 µl water. Subsequently, they were digested by P1 nuclease and mononucleotides were separated by 2D–TLC as described in the MTase assay. The radiolabelled nucleotides present at the 5′-end of the fragments were revealed by autoradiography.
Sequence analyses
Searches of the current version of non-redundant sequence database (nr) were carried out using a local version of PSI-BLAST (25) with E-value threshold of 1e – 5, until convergence. This threshold ensured identification of members of the Trm10 family, without paralogous members of other, more distantly related families of the SPOUT superfamily. All sequences were extracted and a multiple sequence alignment was calculated using MUSCLE (26). Structure prediction was carried out via the GeneSilico metaserver (27).
RESULTS
Sulfolobus acidocaldarius Saci_1677p and T. kodakaraensis TK0422p are distantly related to Trm10p from S. cerevisiae
The 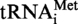 from S. acidocaldarius is the only tRNA from an hyperthermophilic archaeon sequenced to date (20). It bears an unknown modification at position 9. In yeast, the enzyme Trm10p catalyses m1G formation at this position (21). By iterative Psi-Blast analysis, the authors identified proteins from S. solfataricus and P. furiosus which are distantly related to S. cerevisiae Trm10p. We used these two archaeal sequences as query for a Psi-Blast analysis of archaeal proteomes, and identified the proteins Saci_1677p from S. acidocaldarius (phylum Crenarchaeota) and TK0422p from T. kodakaraensis (phylum Euryarchaeota). These two enzymes are orthologues of S. cerevisiae Trm10p.
from S. acidocaldarius is the only tRNA from an hyperthermophilic archaeon sequenced to date (20). It bears an unknown modification at position 9. In yeast, the enzyme Trm10p catalyses m1G formation at this position (21). By iterative Psi-Blast analysis, the authors identified proteins from S. solfataricus and P. furiosus which are distantly related to S. cerevisiae Trm10p. We used these two archaeal sequences as query for a Psi-Blast analysis of archaeal proteomes, and identified the proteins Saci_1677p from S. acidocaldarius (phylum Crenarchaeota) and TK0422p from T. kodakaraensis (phylum Euryarchaeota). These two enzymes are orthologues of S. cerevisiae Trm10p.
The proteins Saci_1677p from S. acidocaldarius and TK0422p from T. kodakaraensis show different tRNA MTase activities in vitro
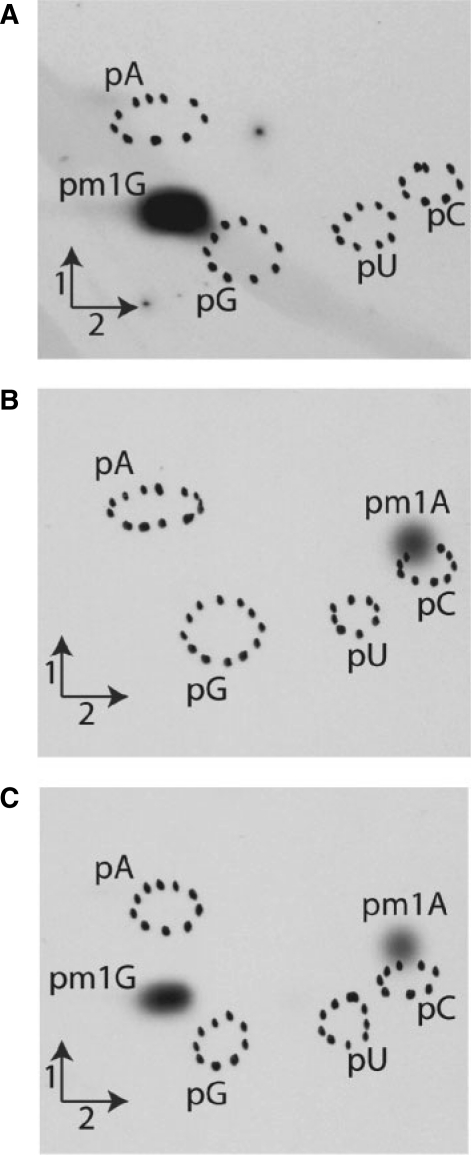
Since Saci_1677p and TK0422p are distantly related to S. cerevisiae Trm10p, the activities of these enzymes were tested in vitro using unfractionated (bulk) tRNA from the S. cerevisiae Y16243 strain (in which the TRM10 gene is inactivated) as substrate. The purified recombinant proteins (see ‘Materials and Methods’ section) were incubated at 30°C for the yeast enzyme, and at 50°C for the archaeal enzymes for an hour with [methyl-14C]AdoMet and tRNA. The choice of a reaction temperature of 50°C was based on a compromise between hyperthermophilic enzyme activity and mesophilic tRNA stability. After incubation, the tRNA was recovered by phenol extraction and ethanol precipitation and further completely hydrolysed into 5′-phosphate nucleosides by nuclease P1. The resulting hydrolysates were then analysed by 2D–TLC followed by autoradiography. The results presented in Figure 1 confirm that S. cerevisiae Trm10p catalyses formation of one single radioactive compound with migration characteristics identical to 1-methylguanosine (m1G) 5′-phosphate according to reference maps (24). Interestingly, the S. acidocaldarius enzyme catalyses the formation of one single radioactive compound, but it corresponds to 1-methyladenosine (m1A) 5′-phosphate. Surprisingly, the T. kodakaraensis TK0422p enzyme catalyses formation of two radioactive compounds, m1A and m1G in tRNA. The enzyme TK0422p forms approximately the same amount of m1A and m1G when the tRNA of the yeast strain Y16243 is used as substrate. Given that occurrence of A9 and G9 in this tRNA population is almost equal (∼50% each) this result indicates that the enzyme TK0422p does not show any preference for one of these two nucleosides.
Figure 1.
Analysis of MTase activities of S. cerevisiae Trm10p, S. acidocaldarius Saci_1677p and T. kodakaraensis TK0422p on total (bulk) tRNA from the S. cerevisiae Y16243 strain, in which the TRM10 gene is inactivated. tRNA was incubated in presence of [methyl-14C]AdoMet and purified Trm10p (A), Saci_1677p (B) and TK0422p (C) as described in ‘Materials and Methods’ section. After incubation, tRNA was recovered and digested by nuclease P1. The resulting nucleotides were analysed by 2D–TLC and autoradiography. Circles in dotted lines show the migration of the four canonical nucleotides used as UV markers.
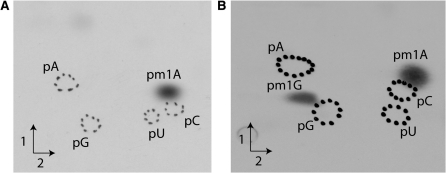
To further confirm the activities of the archaeal enzymes, the MTase activity assay was carried out as described above on unfractionated E. coli tRNA. The rationale behind using bulk E. coli tRNA arises from the fact that position 9 of E. coli tRNA is never modified. The results shown in Figure 2 confirm those obtained using tRNA from the yeast Y16243 (trm10::kan) strain. Indeed, the Saci_1677p from S. acidocaldarius catalyses the formation of m1A, while the TK0422p from T. kodakaraensis catalyses the formation of two radioactive compounds, m1A and m1G. The ratio m1A/m1G is higher than what we observed using tRNA from the yeast Y16243 strain. This is consistent with the fact that there are about two times more tRNAs with A9 than with G9 in E. coli. Again, this confirms the absence of preference of TK0422p for one of these two nucleosides. To our knowledge this is the first description of a tRNA MTase with a broadened substrate recognition capability.
Figure 2.
Analysis of MTase activities of S. acidocaldarius Saci_1677p and T. kodakaraensis TK0422p on total E. coli tRNA. E. coli tRNA was incubated in presence of 5 µg of purified Saci_1677p (A) or TK0422p (B) and [methyl-14C]AdoMet (see ‘Materials and Methods’ section). tRNA was recovered and digested by nuclease P1. The resulting 5′-phosphate mononucleosides were analysed by 2D–TLC followed by autoradiography.
Saci_1677p catalyses m1A formation, while TK0422p catalyses m1A and m1G formation on in vitro transcripts of archaeal tRNAs
The activity of Saci_1677p from S. acidocaldarius was confirmed under identical experimental conditions as above, but using [α32P]ATP-labelled tRNA substrates obtained after in vitro transcription by T7 RNA polymerase of synthetic S. acidocaldarius
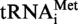 gene or
gene or 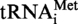 A9G gene. After incubation, the formation of N1-methyladenosine was analysed as above, and the 32P phosphate is only present in AMP (5′P) and AMP (5′P) derivatives. Figure 3 demonstrates unambiguously the presence of m1A in the WT
A9G gene. After incubation, the formation of N1-methyladenosine was analysed as above, and the 32P phosphate is only present in AMP (5′P) and AMP (5′P) derivatives. Figure 3 demonstrates unambiguously the presence of m1A in the WT 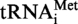 from S. acidocaldarius, showing that the enzyme is indeed a tRNA m1A MTase. Interestingly, no m1A was formed in a mutant of the
from S. acidocaldarius, showing that the enzyme is indeed a tRNA m1A MTase. Interestingly, no m1A was formed in a mutant of the 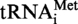 where position 9 is occupied by a guanosine (
where position 9 is occupied by a guanosine ( A9G), suggesting that S. acidocaldarius Saci_1677p acts at position 9 of tRNA, as its yeast orthologue does. Quantification of the relative amount of 32P in the different radioactive spots on TLC plates revealed that about 1 mole of m1A was formed per mole of tRNA after 1 h incubation at 50°C.
A9G), suggesting that S. acidocaldarius Saci_1677p acts at position 9 of tRNA, as its yeast orthologue does. Quantification of the relative amount of 32P in the different radioactive spots on TLC plates revealed that about 1 mole of m1A was formed per mole of tRNA after 1 h incubation at 50°C.
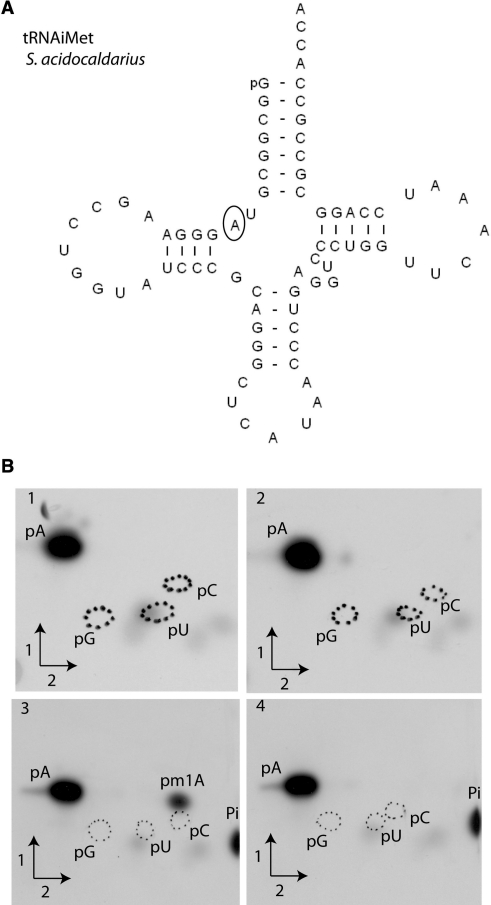
Figure 3.
Saci_1677p from S. acidocaldarius methylates atom N1 of adenosine of in vitro transcribed S. acidocaldarius
 . (A) Nucleotide sequence of the in vitro transcript of S. acidocaldarius
. (A) Nucleotide sequence of the in vitro transcript of S. acidocaldarius
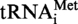 . Position 9 is circled. (B) Autoradiograms of 2D–TLC chromatograms of P1 hydrolysates of [α32P]ATP-labelled S. acidocaldarius
. Position 9 is circled. (B) Autoradiograms of 2D–TLC chromatograms of P1 hydrolysates of [α32P]ATP-labelled S. acidocaldarius
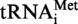 transcripts (B1 and B3) and S. acidocaldarius
transcripts (B1 and B3) and S. acidocaldarius
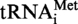 A9G transcripts (B2 and B4) incubated in presence of Saci_1677p for 1 h at 50°C (B3 and B4) or in absence of enzyme (B1 and B2). Circles of dotted lines show the migration of pG, pC and pU nucleotides used as UV markers.
A9G transcripts (B2 and B4) incubated in presence of Saci_1677p for 1 h at 50°C (B3 and B4) or in absence of enzyme (B1 and B2). Circles of dotted lines show the migration of pG, pC and pU nucleotides used as UV markers.
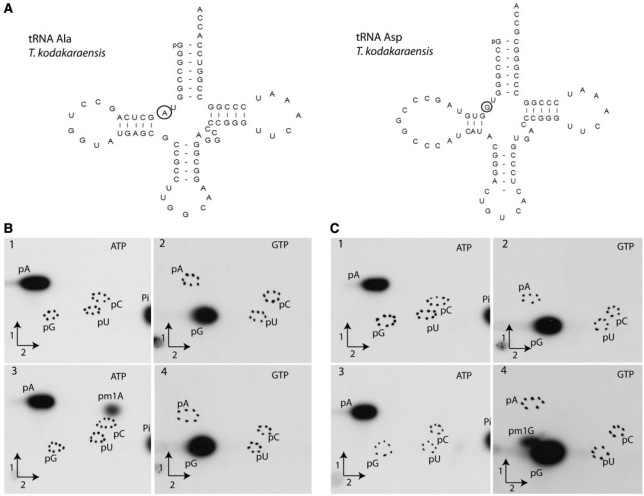
Similarly, the activity of T. kodakaraensis TK0422p was also confirmed using in vitro [α32P]ATP- and [α32P]GTP-labelled transcripts of T. kodakaraensis tRNAAla and tRNAAsp genes. The tRNAAla contains an adenosine at position 9, while tRNAAsp contains a guanosine at this position. Figure 4 shows that TK0422p catalyses formation of m1A in the [α32P]ATP-tRNAAla (A in position 9), but not in the [α32P]ATP-tRNAAsp that possesses G in position 9. Similarly, there was only formation of m1G in the GTP-labelled tRNAAsp (G9), but not in the tRNAAla (A9). Quantification of the relative amount of 32P in the different radioactive spots on TLC plates revealed that about 1 mol of m1A or 1 mol of m1G was formed per mole of tRNA after 1 h incubation at 50°C. Taken together, these results confirm that TK0422p is capable of methylating the two purine bases, and they suggest that the T. kodakaraensis enzyme acts also at position 9 of tRNA.
Figure 4.
TK0422p from T. kodakaraensis methylates atom N1 of both adenosine and guanosine of in vitro transcripts of T. kodakaraensis tRNAAla and tRNAAsp. (A) Nucleotide sequence of in vitro transcripts of T. kodakaraensis tRNAAla and tRNAAsp. Position 9 is circled. (B) Autoradiograms of 2D–TLC chromatograms of P1 hydrolysates of [α32P]ATP- or [α32P]GTP labelled T. kodakaraensis tRNAAla transcripts and (C) of T. kodakaraensis tRNAAsp transcripts. Transcripts were incubated in presence of TK0422p for 1 h at 50°C (B3, B4, C3 and C4) or in the absence of enzyme (B1, B2, C1 and C2). Circles of dotted lines show the migration of the four canonical nucleotides used as UV markers. The radioactive nucleotide used to label the transcripts is indicated on the autoradiograms.
The archaeal enzymes act at position 9 of tRNA
The MTase Trm10p from S. cerevisiae modifies guanosine at position 9 of tRNA (21). The results described above suggest that the archaeal enzymes also target this position. To precisely identify the localisation of nucleosides methylated by the archaeal enzymes, modified and unmodified S. acidocaldarius
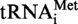 or S. acidocaldarius
or S. acidocaldarius
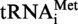 A9G were digested by RNase A and the products of this reaction were 32P-labelled at the 5′ extremity (see ‘Materials and Methods’ section). The resulting 8-nt long fragments bearing A9 or G9, and 5-nt long fragments (controls) were hydrolysed by nuclease P1 and the 5′-phosphate mononucleosides were separated by 2D–TLC. The nature of the nucleotide present at the 5′-end of each fragment was revealed by autoradiography. Figure 5 shows that m1A or m1G is only present at the 5′-end of the 8-nt long fragment from the modified transcript, confirming that both archaeal enzymes act at position 9 of tRNA. Furthermore, it underlines the dual tRNA m1A9-m1G9 specificity of the T. kodakaraensis TK0422p.
A9G were digested by RNase A and the products of this reaction were 32P-labelled at the 5′ extremity (see ‘Materials and Methods’ section). The resulting 8-nt long fragments bearing A9 or G9, and 5-nt long fragments (controls) were hydrolysed by nuclease P1 and the 5′-phosphate mononucleosides were separated by 2D–TLC. The nature of the nucleotide present at the 5′-end of each fragment was revealed by autoradiography. Figure 5 shows that m1A or m1G is only present at the 5′-end of the 8-nt long fragment from the modified transcript, confirming that both archaeal enzymes act at position 9 of tRNA. Furthermore, it underlines the dual tRNA m1A9-m1G9 specificity of the T. kodakaraensis TK0422p.
Figure 5.
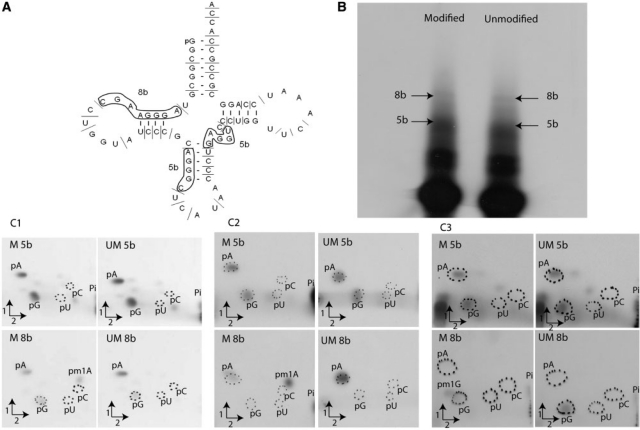
Localisation of m1A and m1G modifications in S. acidocaldarius
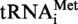 and
and 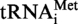 A9G. (A) RNase A digestion profile of S. acidocaldarius
A9G. (A) RNase A digestion profile of S. acidocaldarius
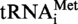 . The 8-nt long fragments (8b) and the 5-nt long fragments (5b) are boxed. (B) Autoradiogram of the polyacrylamide gel used to separate the RNase A fragments of the modified and unmodified
. The 8-nt long fragments (8b) and the 5-nt long fragments (5b) are boxed. (B) Autoradiogram of the polyacrylamide gel used to separate the RNase A fragments of the modified and unmodified 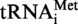 transcripts. The 8-nt long fragments (8b) and the 5-nt long fragments (5b) are shown by arrows. (C) The 8-nt long fragments and the 5-nt long fragments obtained from transcripts of
transcripts. The 8-nt long fragments (8b) and the 5-nt long fragments (5b) are shown by arrows. (C) The 8-nt long fragments and the 5-nt long fragments obtained from transcripts of 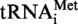 (panels C1 and C2) or
(panels C1 and C2) or 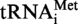 A9G (panel C3) incubated in the presence or absence of Saci_1677p (panel C1) or TK0422p (panels C2 and C3) were hydrolysed by nuclease P1. The nucleotides in the hydrolysates were separated by 2D–TLC. The nature of the nucleotide at the 5′end of the fragments was revealed by autoradiography. M is for the transcripts enzymatically modified and UM for those left unmodified.
A9G (panel C3) incubated in the presence or absence of Saci_1677p (panel C1) or TK0422p (panels C2 and C3) were hydrolysed by nuclease P1. The nucleotides in the hydrolysates were separated by 2D–TLC. The nature of the nucleotide at the 5′end of the fragments was revealed by autoradiography. M is for the transcripts enzymatically modified and UM for those left unmodified.
Further characterisation of the unique m1A-m1G MTase activity of T. kodakaraensis TK0422p
To determine whether the formation of m1A and m1G resulted from the use of a too high enzyme/tRNA ratio, various concentrations of the enzyme were incubated with the same amount of E. coli tRNA and [methyl-14C]AdoMet (see ‘Materials and Methods’ section). Both m1A and m1G are formed in E. coli tRNA incubated in presence of TK0422p, and the intensity of the spots are related to the concentration of TK0422p. Furthermore, the ratio between m1A and m1G is conserved throughout the dilutions (results not shown).
Interestingly, T. kodakaraensis TK0422p methylates atom N1 of both adenosine and guanosine, whose protonation states are different at physiological pH. Indeed, position N1 of adenosine is unprotonated at physiological pH, whereas position N1 of guanosine is protonated. To get some insight into the catalysis by TK0422p, influence of the pH on the methylation reaction was analysed (Table 1). At pH 4.8, the T. kodakaraensis TK0422p is not active. The MTase is active in a pH range going from pH 5.5 to 9.75, but the intensity of m1A and m1G spots vary greatly as a function of the pH. At pH 5.5, m1A MTase activity of TK0422p is predominant over m1G. At pH 7 or higher, both m1A and m1G were detected, m1G intensity growing with increasing pH.
Table 1.
pH dependence of m1A and m1G formation by TK0422p from T. kodakaraensis
| pH | m1G/m1A ratio |
|---|---|
| 4.8 | 0 |
| 5.5 | 0.06 |
| 7 | 0.42 |
| 9 | 0.18 |
| 9.75 | 1.43 |
The MTase activity was measured using total (bulk) E. coli tRNA as substrate. tRNA was incubated in presence of [methyl-14C]AdoMet and purified TK0422p protein. The different buffers are described in ‘Materials and Methods’ section. After incubation, tRNA was recovered and the formation of m1G and m1A was measured as described in ‘Materials and Methods’ section.
The T. kodakaraensis MTase TK0422p catalyses m1A and m1G formation in E. coli tRNA in vivo
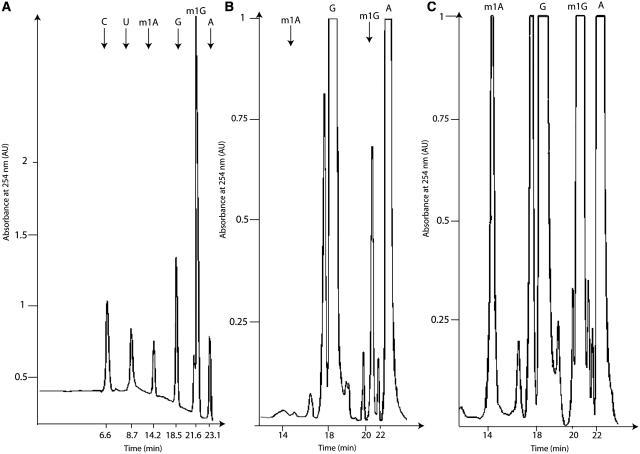
To get some insight into the in vivo activity of T. kodakaraensis TK0422p, total tRNA was extracted from the E. coli strains expressing or not TK0422p. The tRNA preparations were then hydrolysed by P1 nuclease, snake venom phosphodiesterase and alkaline phosphatase and the resulting mononucleosides were analysed by HPLC (Figure 6). As E. coli does not naturally possess any tRNA (m1A) MTase, no m1A could be detected in tRNA from the control strain. On the other hand, a small amount of m1G could be detected, consistent with the presence of a tRNA m1G37 activity in E. coli. When we then analysed the HPLC profile of the tRNA from the recombinant strain overexpressing TK0422p, m1A could clearly be identified (Figure 6), and more m1G was present in this tRNA preparation compared to the control strain. This result shows that TK0422p can act as a tRNA m1A and m1G MTase in E. coli.
Figure 6.
Thermococcus kodakaraensis TK0422p MTase displays tRNA (m1A and m1G) MTase activity in E. coli in vivo. tRNA from the E. coli strain expressing (C) or not (B) TK0422p were extracted and hydrolysed by nuclease P1, snake venom phosphodiesterase and alkaline phosphatase, and then analysed by HPLC. The peaks corresponding to m1A and m1G are indicated by arrows. A standard curve with the reference nucleosides is given in (A).
DISCUSSION
In this work, we have identified and characterised two archaeal orthologues of Trm10p from S. cerevisiae. We have shown that the protein Saci_1677p from the crenarchaeon S. acidocaldarius catalyses N1 methylation of an adenosine to form m1A at position 9 of tRNA. Our data also show that Saci_1677p is specific for A9 of tRNA substrates. Interestingly, we have shown that the orthologous protein TK0422p from the euryarchaeon T. kodakaraensis catalyses both m1A and m1G formation in tRNA. We have demonstrated that both m1A and m1G are formed at position 9 of tRNA. To our knowledge, this is the first description of an MTase with a broadened nucleoside recognition capability. It is interesting to point out that the protonation state of position N1 of both adenosine and guanosine differs at physiological pH. Indeed, the N1 atom of adenosine is not protonated at this pH, while the same position of guanosine bears a proton. This difference of protonation state raises questions about the enzymatic mechanism of TK0422p. In this article, we have shown that the pH of the methylation reaction has an influence on the efficacy of m1A or m1G formation. Indeed, the relative efficiency of m1G formation by TK0422p increases with increasing pH, especially at pH values above 9.5 which corresponds to the pKa of N1 of G (28). This could reflect the need of this enzyme to deprotonate G to be able to catalyse the methyl transfer, although other possibilities are not excluded, such as an influence of pH on tRNA or enzyme structure.
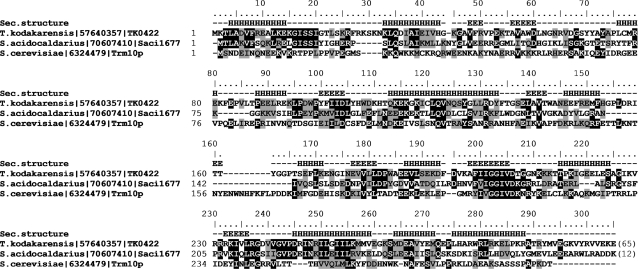
Figure 7 illustrates sequence conservation between Trm10p from yeast and the two archaeal proteins analysed in this work. According to structure prediction methods, the catalytic core of the SPOUT domain spans residues 100–260 of TK0422p (and the corresponding positions in the homologous sequences), and includes five β-strands and six α-helices. The N-terminus is conserved only between the archaeal proteins, but not with the eukaryotic protein. Due to uncertain alignments with known SPOUT structures we were unable to generate a confident 3D model of the catalytic domain, nonetheless the mapping of clusters of conserved residues on predicted secondary structures suggests that the active site is composed of nearly invariant (on the level of the whole family) residues D104, Q122 and D206 (numbering for TK0422p). Interestingly, there appears to be no equivalent catalytic residues between the Trm10p family and the bacterial TrmD family of m1G MTases, also members of the SPOUT superfamily that act on the position G37 in tRNA (29). This suggests that the N1 purine methylation activity has evolved independently at least two times in the SPOUT superfamily. Independent origin of the N1 purine methylation activity has been also postulated for MTases of the RFM superfamily, i.e. tRNA:m1A58 MTase TrmI, tRNA:m1A22 MTase TrmK, and 16S rRNA:m1A1408 MTase KamB, which also present different active sites despite the common fold of the catalytic domain (30,31). This variety of protein folds and active sites available to carry out the same type of reaction suggests that N1 purine methylation is probably relatively easy for an enzyme to evolve (e.g. from an MTase of a different specificity).
Figure 7.
Alignment of three experimentally characterised members of the Trm10p family, including the eukaryotic founding member of this family and two archaeal proteins analysed in this work. Residues that are conserved and physicochemically similar in >50% of the sequences are indicated by black and grey shading, respectively. Secondary structures predicted for TK0422p are indicated as E (extended) and H (helix).
Unfortunately, we were unable to identify a negatively charged residue conserved between TK0422p (m1G and m1A MTase) and Trm10p (m1G MTase) but absent from Saci_1677p (m1A MTase) that could explain the inability of the latter protein to deprotonate and methylate guanosine. Therefore, the molecular basis of the different specificities in the Trm10p family remains a mystery. This problem may be approached in the future by experimental determination of a high-resolution structure for one of the family members, characterisation of specificities of numerous additional members of the family, or by mutagenesis of individual members. In particular the availability of a crystal structure for a Trm10p family member would enable its comparison with the known structures of evolutionarily related TrmD MTases and would help to evaluate the hypothesis of convergent evolution of their active sites for an analogous reaction, upon homologous protein scaffold.
Interestingly, the distribution of Trm10p orthologues in Archaea is restricted to hyperthermophiles. We found single representatives of this family in Crenarchaeota (S. acidocaldarius, S. tokodaii, S. solfataricus, S. islandicus, Hyperthermus butylicus, Ignicoccus hospitalis, Aeropyrum pernix, Pyrobaculum aerophilum, P. arsenaticum, P. calidifontis, P. islandicum, Thermoproteus neutrophilus, Caldivirga maquilingensis, Metallosphaera sedula) and in Euryarchaeota (T. kodakaraensis, T. gammatolerans, T. onnurineus, T. barophilus, T. sibiricus, T. sp. AM4, Archaeoglobus fulgidus, Methanopyrus kandleri, Pyrococcus furiosus, P. abyssi and P. horikoshii), as well as in Nanoarchaeum equitans. On the other hand, Trm10p is absent from the mesophilic archaeon Halobacterium volcanii, which is known to lack m1A or m1G at position 9 (32). This suggests that m1G or m1A modification may contribute to the thermostability of archaeal tRNAs.
A stabilizing function of base methylation has been recently identified for the m7G46 modification in a thermophilic bacterium T. thermophilus (33). In tRNA crystal structures the base A9 is stacked between bases 45 and 46. Modelling of the S. acidocaldarius tRNA with and without modifications (data not shown) suggests that the methyl group of m7G46 would be located only about 5–6 Å from the methyl group of m1A9. However, G46 is not methylated in Archaea due to the absence of the TrmB family MTase responsible for this activity in Bacteria and Eukaryota (34). The methyl group on residue A9 may fill a cavity between bases 45 and 46 and the backbone of residue 22. This could increase the surface of interaction between this residue and G46, which may contribute to the increased stability of the tRNA molecule. Furthermore, the positive charge on m1A may have a stabilisatory function due to electrostatic interactions with the neighbouring phosphate groups of residues 22 and 23. Thus, it is tempting to speculate that m1A9 may fulfil a similar stabilisatory role in thermophilic Archaea as m7G46 does in thermophilic bacteria.
FUNDING
Fonds pour le Recherche Fondamentale Collective [grant number 2.4.520.05F] and by the Fonds D. et A. Van Buuren; Fonds pour la Formation à la Recherche dans l’Industrie et dans l’Agriculture fellowship (to M.K.); EURASNET Network of Excellence grant in the 6th Framework Program of the European Commission and by the Polish Ministry of Science and Higher Education [grant number 301 2396 33] (to J.M.B.); Polish Ministry of Science and Higher Education [grant number 301 105 32/3599] and by the START fellowship from the Foundation for Polish Science (to K.L.T.). Funding for open access charge: Fonds pour la Recherche Fondamentale Collective.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors warmly acknowledge D. Gigot (ULB, Belgium) for help and advice during protein purifications and HPLC analyses. We are grateful to D. Charlier (VUB, Belgium), H. Grosjean (CNRS, France) and T.J. Santangelo (Ohio State University, USA) for the gift of archaeal genomic DNAs. J.M.B. thanks Tomasz Puton (Adam Mickiewicz University, Poznan, Poland) and Irina Tuszynska (IIMCB, Warsaw, Poland) for their help with tRNA modelling and structural analysis.
REFERENCES
- 1.Jühling F, Mörl M, Hartmann RK, Sprinzl M, Stadler PF, Pütz J. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–D162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Czerwoniec A, Dunin-Horkawicz S, Purta E, Kaminska KH, Kasprzak JM, Bujnicki JM, Grosjean H, Rother K. MODOMICS: a database of RNA modification pathways 2008 update. Nucleic Acids Res. 2009;37:D118–D121. doi: 10.1093/nar/gkn710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Urbonavicius J, Skouloubris S, Myllykallio H, Grosjean H. Identification of a novel gene encoding a flavin-dependent tRNA:m5U methyltransferase in bacteria-evolutionary implications. Nucleic Acids Res. 2005;33:3955–3964. doi: 10.1093/nar/gki703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schubert HL, Blumenthal RM, Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem. Sci. 2003;28:329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kozbial PZ, Mushegian AR. Natural history of S-adenosylmethionine-binding proteins. BMC Struct. Biol. 2005;5:19. doi: 10.1186/1472-6807-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaminska KH, Purta E, Hansen LH, Bujnicki JM, Vester B, Long KS. Insights into the structure, function and evolution of the radical-SAM 23S rRNA methyltransferase Cfr that confers antibiotic resistance in bacteria. Nucleic Acids Res. 2009;38:1652–1663. doi: 10.1093/nar/gkp1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anantharaman V, Koonin EV, Aravind L. SPOUT: a class of methyltransferases that includes SpoU and TrmD RNA methylase superfamilies novel superfamilies of predicted prokaryotic RNA methylases. J. Mol. Microbiol. Biotechnol. 2002;4:71–75. [PubMed] [Google Scholar]
- 8.Tkaczuk KL, Dunin-Horkawicz S, Purta E, Bujnicki JM. Structural and evolutionary bioinformatics of the SPOUT superfamily of methyltransferases. BMC Bioinformatics. 2007;8:73–103. doi: 10.1186/1471-2105-8-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Renalier MH, Joseph N, Gaspin C, Thebault P, Mougin A. The Cm56 tRNA modification in Archaea is catalyzed either by a specific 2′-O-methylase, or a C/D sRNP. RNA. 2005;11:1051–1063. doi: 10.1261/rna.2110805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuratani M, Bessho Y, Nishimoto M, Grosjean H, Yokoyama S. Crystal structure and mutational study of a unique SpoU family archaeal methylase that forms 2′-O-methylcytidine at position 56 of tRNA. J. Mol. Biol. 2008;375:1064–1075. doi: 10.1016/j.jmb.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 11.Armengaud J, Urbonavicius J, Fernandez B, Chaussinand G, Bujnicki JM, Grosjean H. N2-methylation of guanosine at position 10 in tRNA is catalyzed by a THUMP domain-containing, S-adenosylmethionine-dependent methyltransferase, conserved in Archaea and Eukaryota. J. Biol. Chem. 2004;279:37142–37152. doi: 10.1074/jbc.M403845200. [DOI] [PubMed] [Google Scholar]
- 12.Urbonavicius J, Armengaud J, Grosjean H. Identity elements required for enzymatic formation of N2, N2-dimethylguanosine from N2-monomethylated derivative and its possible role in avoiding alternative conformations in archaeal tRNA. J. Mol. Biol. 2006;357:387–399. doi: 10.1016/j.jmb.2005.12.087. [DOI] [PubMed] [Google Scholar]
- 13.Constantinesco F, Motorin Y, Grosjean H. Characterization and enzymatic properties of tRNA (guanine 26, N2,N2)-dimethyltransferase (Trm1p) from Pyrococcus furiosus. J. Mol. Biol. 1999;291:375–392. doi: 10.1006/jmbi.1999.2976. [DOI] [PubMed] [Google Scholar]
- 14.Ihsanawati, Nishimoto M, Higashijima K, Shirouzu M, Grosjean H, Bessho Y, Yokoyama S. Crystal structure of tRNA N2,N2-guanosine dimethyltransferase Trm1 from Pyrococcus horikoshii. J. Mol. Biol. 2008;383:871–884. doi: 10.1016/j.jmb.2008.08.068. [DOI] [PubMed] [Google Scholar]
- 15.Auxilien S, El Khadali F, Rasmussen A, Douthwaite S, Grosjean H. Archaease from Pyrococcus abyssi improves substrate specificity and solubility of a tRNA m5C methyltransferase. J. Biol. Chem. 2007;282:18711–18721. doi: 10.1074/jbc.M607459200. [DOI] [PubMed] [Google Scholar]
- 16.Christian T, Evilia C, Williams S, Hou Y-M. Distinct origins of tRNA(m1G37) methyltransferase. J. Mol. Biol. 2004;339:707–719. doi: 10.1016/j.jmb.2004.04.025. [DOI] [PubMed] [Google Scholar]
- 17.Björk GR, Jacobsson K, Nilsson K, Johansson MJ, Byström AS, Persson OP. A primordial tRNA modification required for the evolution of life? EMBO J. 2001;20:231–239. doi: 10.1093/emboj/20.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walbott H, Leulliot N, Grosjean H, Golinelli-Pimpaneau B. The crystal structure of Pyrococcus abyssi tRNA (uracil-54, C5)-methyltransferase provides insights into its tRNA specificity. Nucleic Acids Res. 2008;36:4929–4940. doi: 10.1093/nar/gkn437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roovers M, Wouters J, Bujnicki J, Tricot C, Stalon V, Grosjean H, Droogmans L. A primordial RNA modification enzyme: the case of tRNA (m1A) methyltransferase. Nucleic Acids Res. 2004;32:465–476. doi: 10.1093/nar/gkh191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuchino Y, Ihara M, Yabusaki Y, Nishimura S. Initiator tRNAs from archaebacteria show common unique sequence characteristics. Nature. 1982;298:684–685. doi: 10.1038/298684a0. [DOI] [PubMed] [Google Scholar]
- 21.Jackman JE, Montange RK, Malik HS, Phizicky EM. Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. RNA. 2003;9:574–585. doi: 10.1261/rna.5070303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reyes VM, Abelson J. A synthetic substrate for tRNA splicing. Anal. Biochem. 1987;166:90–106. doi: 10.1016/0003-2697(87)90551-3. [DOI] [PubMed] [Google Scholar]
- 23.Droogmans L, Roovers M, Bujnicki JM, Tricot C, Hartsch T, Stalon V, Grosjean H. Cloning and characterization of tRNA (m1A58) methyltransferase (TrmI) from Thermus thermophilus HB27, a protein required for cell growth at extreme temperatures. Nucleic Acids Res. 2003;31:2148–2156. doi: 10.1093/nar/gkg314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grosjean H, Keith G, Droogmans L. Detection and quantification of modified nucleotides in RNA using thin-layer chromatography. Methods Mol. Biol. 2004;265:357–391. doi: 10.1385/1-59259-775-0:357. [DOI] [PubMed] [Google Scholar]
- 25.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurowski MA, Bujnicki JM. GeneSilico protein structure prediction meta-server. Nucleic Acids Res. 2003;31:3305–3307. doi: 10.1093/nar/gkg557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rogstad K, Jang Y, Sowers L, Goddard W. First principles calculations of the pKa values and tautomers of isoguanine and xanthine. Chem. Res. Toxicol. 2003;16:1455–1462. doi: 10.1021/tx034068e. [DOI] [PubMed] [Google Scholar]
- 29.Elkins PA, Watts JM, Zalacain M, van Thiel A, Vitazka PR, Redlak M, Andraos-Selim C, Rastinejad F, Holmes WM. Insights into catalysis by a knotted TrmD tRNA methyltransferase. J. Mol. Biol. 2003;333:931–949. doi: 10.1016/j.jmb.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 30.Roovers M, Kaminska KH, Tkaczuk KL, Gigot D, Droogmans L, Bujnicki JM. The YqfN protein of Bacillus subtilis is the tRNA:m1A22 methyltransferase (TrmK) Nucleic Acids Res. 2008;36:3252–3262. doi: 10.1093/nar/gkn169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koscinski L, Feder M, Bujnicki JM. Identification of a missing sequence and functionally important residues of 16S rRNA:m1A1408 methyltransferase KamB that causes bacterial resistance to aminoglycoside antibiotics. Cell Cycle. 2007;6:1268–1271. doi: 10.4161/cc.6.10.4231. [DOI] [PubMed] [Google Scholar]
- 32.Gupta R. Halobacterium volcanii tRNAs. Identification of 41 tRNAs covering all amino acids, and the sequences of 33 class I tRNAs. J. Biol. Chem. 1984;259:9461–9471. [PubMed] [Google Scholar]
- 33.Tomikawa C, Yokogawa T, Kanai T, Hori H. N7-methylguanine at position 46 (m7G46) in tRNA from Thermus thermophilus is required for cell viability at high temperatures through a tRNA modification network. Nucleic Acids Res. 2010;38:942–957. doi: 10.1093/nar/gkp1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Purta E, van Vliet F, Tricot C, De Bie LG, Feder M, Skowronek K, Droogmans L, Bujnicki JM. Sequence-structure-function relationships of a tRNA (m7G46) methyltransferase studied by homology modeling and site-directed mutagenesis. Proteins. 2005;59:482–488. doi: 10.1002/prot.20454. [DOI] [PubMed] [Google Scholar]