Abstract
Cells respond to genotoxic stress with the induction of DNA damage defence functions. Aimed at identifying novel players in this response, we analysed the genotoxic stress-induced expression of DNA repair genes in mouse fibroblasts proficient and deficient for c-Fos or c-Jun. The experiments revealed a clear up-regulation of the three prime exonuclease I (trex1) mRNA following ultraviolet (UV) light treatment. This occurred in the wild-type but not c-fos and c-jun null cells, indicating the involvement of AP-1 in trex1 induction. Trex1 up-regulation was also observed in human cells and was found on promoter, RNA and protein level. Apart from UV light, TREX1 is induced by other DNA damaging agents such as benzo(a)pyrene and hydrogen peroxide. The mouse and human trex1 promoter harbours an AP-1 binding site that is recognized by c-Fos and c-Jun, and its mutational inactivation abrogated trex1 induction. Upon genotoxic stress, TREX1 is not only up-regulated but also translocated into the nucleus. Cells deficient in TREX1 show reduced recovery from the UV and benzo(a)pyrene-induced replication inhibition and increased sensitivity towards the genotoxins compared to the isogenic control. The data revealed trex1 as a novel DNA damage-inducible repair gene that plays a protective role in the genotoxic stress response.
INTRODUCTION
The genome is perpetually endangered by endogenous and exogenous stress. Whereas endogenous stress occurs at more or less constant level, exogenous stress provoked by chemical and physical insults occurs transiently and at highly variable levels. To counteract DNA damage induced by these insults, DNA repair functions have evolved, some of which are inducible in response to genotoxic stress (1). Promoters of several DNA repair genes have been shown to be subject to modulation by genotoxins (2). Perhaps the best studied genotoxin is ultraviolet (UV) light that was shown to increase the expression of the DNA repair proteins DDB2, XPC, Pol I, Lig1 and Fen1 (3–7).
Two transcription factors play a key role in the regulation of DNA repair, p53 and AP-1. Both are induced by many types of genotoxic stress and implicated in maintaining genomic stability and cell survival. Thus, mouse embryonic fibroblasts (MEFs) deficient in p53 are more sensitive to UV light than the corresponding wild-type (wt) (8). Hypersensitivity of p53-deficient cells is ascribed to abolition of G1/S checkpoint control (9,10), impaired base excision repair (11,12) and impaired nucleotide excision repair (7,13), which leads to a high level of apoptosis (14). In contrast to the role of p53 in the UV response, the function of AP-1 is less established. AP-1 consists of different dimeric complexes containing proteins of the Jun (c-Jun, JunB and JunD), Fos (c-Fos, FosB, Fra-1, Fra2) and CREB/ATF (ATF1, ATF2) family, which exert different promoter specificities and functions (15). Dependent on the dimeric composition, AP-1 can bind to different transcription factor binding sites. Binding of Fos/Jun occurs mainly to heptameric (TGAGTCA) sites whereas Jun/ATF-2 binds to octameric CRE binding sites (TGACGTCA) (15). The different AP-1 complexes exerting different promoter affinities allow a fine-tuned stimulation of a broad spectrum of genes harboring AP-1 sites in their promoter.
In rodent cells, the c-fos gene is immediately inducible by UV light (16) and other kinds of genotoxic stress (17,18). The fact that cells lacking c-Fos are hypersensitive to genotoxins (19–21) suggests that c-Fos plays an important protective role in the cellular defence against DNA damaging agents. On the other hand, c-Fos overexpression stimulates malignant transformation (22,23), which might explain the high expression level of c-Fos in several human tumours (24,25). c-Fos overexpression also results in resistance to chemotherapy by protecting cells against the anticancer drug cisplatin (26,27).
Previously, we elucidated the mechanism leading to increased sensitivity of c-Fos-deficient cells to UV light. We showed that c-Fos is involved in the resynthesis of XPF upon DNA damage induction. Impaired XPF resynthesis in c-Fos-deficient cells leads to abrogation of repair of DNA adducts and sustained inhibition of transcription. This signals cell death pathways via down-regulation of Map kinase phosphatase 1, sustained activation of Jun kinase and subsequent induction of FasL that triggers the receptor-mediated pathway of apoptosis (28,29). Here, we further examined the role of c-Fos in the regulation of DNA repair. Comparing the expression of about 130 DNA repair genes (by means of a DNA repair microarray) in wt and c-fos knockout (fos−/−) cells after UV light exposure, we found the three prime exonuclease 1 (trex1) to be differentially expressed. This prompted us to study the regulation of trex1 in more detail. Here, we show that trex1 is induced on RNA and protein level by UV light and other genotoxic agents such as benzo(a)pyrene (B(a)P). The induction requires c-Fos/AP-1. We further show that upon UV and B(a)P treatment, TREX1 translocates from the cytoplasm into the nucleus. The data reveal trex1 as a novel DNA damage-inducible gene and suggest that TREX1 is complex regulated upon genotoxic stress, involving gene induction and nuclear translocation. We also demonstrate that cells lacking TREX1 are hypersensitive to UV light and B(a)P and respond with delayed recovery from the genotoxin-induced replication inhibition, indicating that up-regulation of TREX1 is part of the cell’s strategy to survive under genotoxic stress conditions.
EXPERIMENTAL PROCEDURES
Cell lines
MEF cell lines fos+/+1-98M (designated as wt) and fos−/−7-98M (designated as fos−/−) were described previously (8,21). Swiss albino 3T3 was purchased from the German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany). TREX1-proficient SC14+/+ and TREX1-deficient SC3−/− and SC8−/− MEFs were kindly provided by Tomas Lindahl (London). All MEF cells and human fibroblasts (GM637) were grown in Dulbecco’s minimal essential medium (DMEM) containing 10% fetal bovine serum (FBS), in 7% CO2 at 37°C.
UV light treatment and preparation of B(a)P
Growth medium was aspirated and cells were irradiated with UV-C light (320 nm) at a dose rate of 1 J/m2 per second with a radium NSE 11-270 low pressure UV lamp (Philips). Thereafter the medium was returned to the plates and cells were incubated at 37°C for the indicated time points. Activated B(a)P r-7,t-8-Dihydroxy-t-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene (anti-BPDE; CAS no. 58917-67-2) was prepared from trans-7,8-dihydroxy-7,8-dihydrobenzo[a]pyrene (30) as described (31).
Preparation of cell extracts and western blot analysis
Whole cell extracts and cytoplasmic extracts were prepared and separated by SDS-PAGE, electro-blotted onto nitrocellulose membranes and incubated with antibodies as described (32). Monoclonal antibodies against mTREX1 (611987, BD Transduction Laboratories) and PCNA (sc-56, Santa Cruz Biotechnology) and polyclonal antibodies against hTREX1 (H00011277-D01P, Abnova) were diluted 1:1000 in 5% non-fat dry milk, 0.1% Tween/PBS and incubated overnight at 4°C. Polyclonal anti-ERK2 antibody (sc-154, Santa Cruz Biotechnology) was diluted 1:3000 and incubated overnight at 4°C. The protein–antibody complexes were visualized by ECL reaction.
Preparation of RNA, RT-PCR and real-time RT-PCR
Total RNA was isolated using the RNA II Isolation Kit (Machery and Nagel). One µg RNA was transcribed into cDNA by Superscript II (Invitrogen) in a volume of 40 µl and 3 µl were subjected to RT-PCR. RT-PCR was performed by the use of specific primers (MWG Biotechnology, Germany) and Red-Taq Ready Mix (Sigma-Aldrich). The PCR program used was: 1.5 min, 94°C [(denaturation: 45 s, 94°C; annealing: 1 min, 58°C; elongation: 1 min, 72°C) 25 cycles] and 10 min 72°C. Real-time RT-PCR was performed using the LightCycler FastStart DNA Master SYBR Green I Kit (Roche Diagnostics) and the light cycler of Roche Diagnostics.
BrdU incorporation
Cells were cultured in DMEM (10% FBS) and after exposure to UVC, the thymidine analog BrdU (10 µM) was added to the medium. The incorporation was analysed 1 h later by the BrdU Incorporation Kit (Roche Diagnostics) in a microplate reader. Experiments were repeated at least three times, mean values ± SD are shown and data were statistically analysed using Student’s t-test.
Determination of apoptosis
For monitoring drug-induced apoptosis, ethanol-fixed cells were stained with propidium iodide. The Sub-G1 fraction was determined by flow cytometry. The protocol was described previously (33,34). Experiments were repeated at least three times, mean values ± SD are shown and data were statistically analysed using Student’s t-test.
Cloning and analysis of the trex1 promoter
The trex1 promoter from MEFs was cloned by RT-PCR amplification using specific primers (mTREX1-prom-up: CTGAGGGCCTGAGCATCCAGC; mTREX1-prom-low GCAGGGTCTGAGAGCCCATGC) and cloned into the pBlue-Topo vector (Invitrogen) resulting in the TREX1 reporter plasmid designated as pBlue-mTREX1-923. The Plasmids pBlue-mTREX1-730, pBlue-mTREX1-517 were generated by cloning of amplification products obtained using combination of the primer mTREX1-prom-low and two additional primers (−730: CCATGGTAACTGATCTGCCC; −517: GAGTTGTCAGTGTTGGGGCAG). The Plasmid pBlue-mTREX1-157 was generated by sub-cloning of the 157 Bp BamH1 fragment of pBlue-mTREX1prom. Vectors were used for transient transfection of MEFs (Fugene HD system, Roche Diagnostics). Two days after transfection, cells were exposed to UV, cell extracts were prepared by several freeze/thaw cycles and the β-galactosidase activity was determined by the β-Gal assay kit (Invitrogen).
Preparation of TREX1 siRNA
Down-regulation of TREX1 was obtained by the use of the Silencer siRNA Cocktail Kit from Ambion. The primers utilized for the reaction were: mTREX1-esiup: cgtaatacgactcactatagggagagTTCTCAGGGACATCCACCTC; mTREX1-esilow: cgtaatacgactcactatagggagagTGCTGAGCAGGGTTAGAACA. Transfection was performed using the Lipofectamine RNAiMAX Transfections-Kit (Invitrogen).
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) was performed as described previously (28). In brief, cellular genomic DNA and proteins were crosslinked with formaldehyde. Genomic DNA was fragmented by sonification to a fragment size of 500–1500 bp and subjected to immunoprecipitation (IP) using a c-Fos specific antibody (sc-52, Santa Cruz Biotechnology). PCR was performed using specific primers flanking the AP-1 binding site of trex1 (mTREX1-CHIP-up: GAGATGGCCCCATGGTAACTG, mTREX1-CHIP-low: CCCCCAGTCCTTGATCCAGGCC) and, as negative control, β-actin.
Preparation of nuclear extracts and electromobility shift assay
Nuclear cell extracts were prepared and subjected to electromobility shift assay (EMSA) or western blot analysis as described previously (35). The sequence of the oligonucleotides specific for the AP-1 binding site of the mouse collagenase promoter was 5′-AGTGGTGACTCATCACT-3′ and the oligonucleotide sequences specific for the AP-1 binding site of the mouse trex1 promoter were: trex1 AP-1a: 5′-GGAATTACCCTGAGTCATAGCTTTG-3′. The sequence of the oligonucleotides specific for the AP-1 binding site of the human trex1 promoter were: hTrex1-EMSA-up 5′-GGAATTGTCCTGAGTCATTGCTTTG-3′. For supershift experiments, 2 µl of antibodies specific for junD (sc74), junB (sc47) and c-jun (sc45) were pre-incubated with 2 µg protein extract for 30 min at room temperature.
Immunofluorescence
Cells were seeded on cover slips. Following genotoxin exposure, cells were fixed with 4% formaldehyde at different time points. A second fixation step was performed using methanol/aceton (3:1) (−20°C, 8 min). Cells were then blocked in 5% BSA, TBS/0.3% Triton X-100. The antibodies used were monoclonal anti-TREX1 (611987, BD Transduction Laboratories), polyclonal anti-γH2AX (#05-164, Upstate), anti-pATR (#2853S, Cell Signaling), anti-p53Bp (#4937, Cell Signaling) and as secondary antibodies Alexa Fluor 488 labelled anti-mouse (molecular probes) and Cy3 labelled anti-rabbit (Jackson Immuno Research). Between all staining steps, cells were washed three times in TBS/0.3% Triton X-100 for 5 min. Slides were mounted in anti-fade medium (Glycerol:PBS 1:1, 2.5% DABCO, pH 8.6 with HCl) and analysed using a confocal laser scanning microscope (LSM 710, Zeiss). For quantification of TREX1 localization, cells with more than 20 nuclear TREX1 foci per cell were counted in three experiments analysing each time 100 cells. Calculation of statistical significance was performed by Student’s t-test.
RESULTS
Expression of trex1 is enhanced by UV light
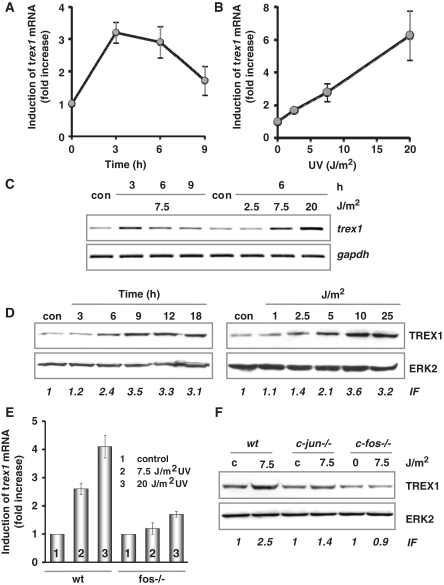
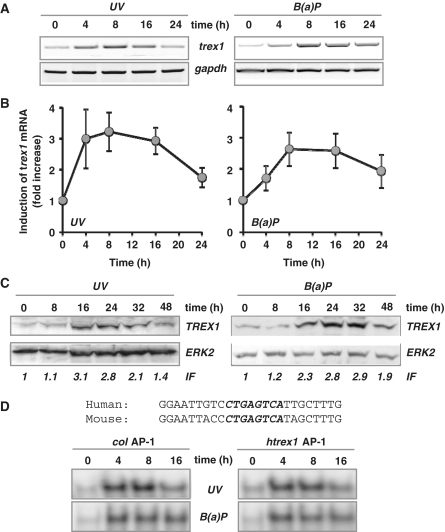
The expression of DNA repair genes was studied by microarray analysis using a self-designed DNA repair array. Comparing wt and fos−/− cells treated with UV, we observed a significant differential expression of transcripts encoding trex1 (data not shown). The findings were substantiated by quantitative real-time RT-PCR using primers located in the coding region of the gene. Treatment of wt cells with UV light transiently increased the mRNA level of trex1, reaching a maximum 3–6 h after UV exposure (Figure 1A). Accumulation of trex1 mRNA was a linear function of dose (Figure 1B). Comparable results were obtained by semi-quantitative RT-PCR, which at the same time confirmed the specificity of the amplification products (Figure 1C). To analyse whether the increased expression of trex1 mRNA leads to an increased protein level, the amount of TREX1 was studied by western blot analysis. As shown in Figure 1D, a strong time- and dose-dependent accumulation of TREX1 protein was determined in whole cell extracts. Increase in TREX1 protein followed the mRNA up-regulation, starting 6 h after UV exposure. It could be detected already at a low-dose level (≥5 J/m2).
Figure 1.
UV light-triggered induction of TREX1. (A and B) Exponentially growing wt MEFs were exposed to 7.5 J/m2 UV for 3, 6 and 9 h (A) or exposed to 2.5, 7.5 and 20 J/m2 UV for 6 h (B). Total RNA was isolated and real-time RT-PCR was performed using trex1- and gapdh-specific primers. For quantification, the expression was normalized to gapdh and the untreated control was set to 1. Data are the mean of three independent experiments +/− SD. (C) In a different set of experiments, wt MEFs were also exposed to 7.5 J/m2 UV for 3, 6 and 9 h (left panel) or exposed to 2.5, 7.5 and 20 J/m2 UV for 6 h (right panel), total RNA was isolated and semi-quantitative RT-PCR was performed using trex1 or, as loading control, gapdh-specific primers (con, non-exposed control). (D) Exponentially growing wt MEFs were exposed to 7.5 J/m2 UV for different time points or exposed to different doses of UV for 9 h. Total protein extract was isolated. Immunodetection was performed using TREX1 or, as loading control, ERK2-specific antibody. Induction factor (IF) is derived from densitometric measurement of TREX1 signal and normalized to ERK2 expression (E) Exponentially growing wt and fos−/− MEFs were exposed to 7.5 or 20 J/m2 UV for 6 h. Total RNA was isolated and real-time RT-PCR was performed using trex1- and gapdh- specific primers. For quantification, the expression was normalized to gapdh and the untreated control was set to 1. Data are the mean of three independent experiments +/− SD. (F) Exponentially growing wt, c-jun−/− and c-fos−/− MEFs were exposed to 20 J/m2 UV for 9 h. Total protein extract was isolated. Immunodetection was performed using TREX1 or, as loading control, ERK2-specific antibody.
Induction of trex1 mRNA is caused by promoter activation and depends on c-Fos
To identify the mechanism responsible for the increase in trex1 mRNA level upon UV treatment, mRNA de novo synthesis was determined. To this end, wt cells were exposed to 7.5 J/m2 UV in the presence or absence of the transcription blocking agent actinomycin D. Treatment of wt cells with the inhibitor strongly reduced the amount of trex1 mRNA, indicating its high instability (Supplementary Figure S1A). Treatment with actinomycin D completely abrogated the induction of trex1 mRNA by UV light, which demonstrates that the observed accumulation of trex1 mRNA is dependent on RNA de novo synthesis. As mentioned above, microarray analysis showed trex1 induction only in wt but not fos−/− cells. To further substantiate the role of c-Fos in the regulation of trex1 we compared the UV-mediated trex1 mRNA induction by quantitative real-time RT-PCR in MEFs wt and fos−/−. Whereas trex1 expression was increased by a factor of four in the wt, induction was marginal in fos−/− cells (Figure 1E). The abrogated induction of TREX1 in fos−/− cells was also observed on protein level (Figure 1F), supporting the view that TREX1 is regulated via c-Fos. Interestingly, also in c-Jun-deficient cells a reduced up-regulation of TREX1 was observed (Figure 1F).
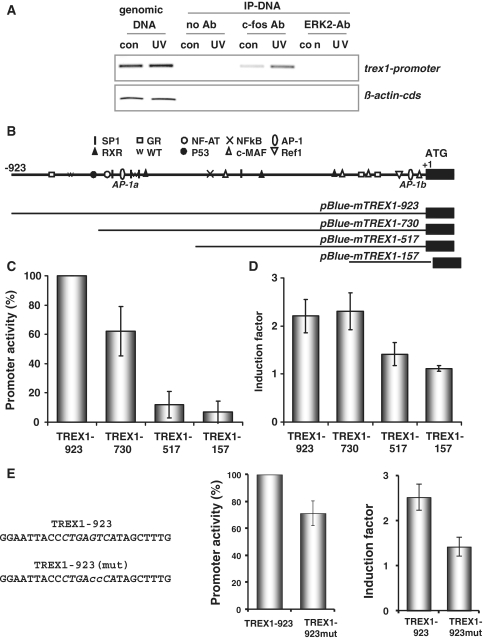
To analyse whether the trex1 promoter is recognized by c-Fos under in vivo conditions, ChIP experiments were performed using a c-Fos-specific antibody. As shown in Figure 2A, c-Fos clearly binds to the trex1 promoter. This binding was significantly enhanced in wt cells treated with UV light. As negative control, we utilized either no antibody or an unrelated one, i.e. against ERK2. In addition, we used for control the β-actin promoter that is not recognized by c-Fos.
Figure 2.
Characterization of the murine trex1 promoter. (A) ChIP analysis: exponentially growing wt cells were not exposed (con, control) or exposed to 20 J/m2 UV. Cells were harvested 6 h later and dealt with as described in ‘Material and Methods’ section. IP was performed using a c-Fos-specific antibody and (as negative control) ERK2-specific antibody. PCR was done using specific primers for the trex1 promoter and, as negative control, for the β-actin 5′-UTR. (B) Graphical visualization of several putative transcription factor binding sites identified using the program Patch 1.0 (www.gene-regulation.com) and display of generated promoter fragments. (C and D) A 923 bp genomic fragment 5′ of the ATG codon of trex1 and different trex1 promoter fragments were cloned into the pBlue-Topo vector and transiently transfected in exponentially growing Swiss albino 3T3 cells. (C) Basal promoter activity was determined by β-Gal assay and compared between untreated cells. The activity of the −923 fragment was set to 100. (D) UV-induced promoter activity was determined after exposure to 7.5 J/m2. Therefore, the promoter activity in UV-exposed cells was set in relation to the promoter activity in non-exposed cells resulting in fold induction. Data are the mean of three independent experiments +/− SD. (E) The putative AP-1 binding site (AP-1a as indicated under B) within the 923 bp genomic fragment containing pBlue-Topo vector was mutated via site-directed mutagenesis (left panel for sequences) and transiently transfected in exponentially growing Swiss albino 3T3 cells. Basal promoter activity and UV-induced promoter activity was determined after exposure to 7.5 J/m2. Data are the mean of three independent experiments +/− SD.
Identification of the minimal inducible trex1 promoter
In order to analyse whether induction of TREX1 is caused via induction of the corresponding promoter, a 923 bp genomic fragment 5′ of the ATG codon of trex1 was cloned from DNA of wt MEFs (sequence shown in Supplementary Figure S2) and its activity and UV-inducibility was studied by reporter assays (Supplementary Figure S1B). To identify potential transcription factor binding sites that were required for the induction of trex1 by UV, we performed a computer-based search for transcription factor binding sites (Patch 1.0/www.gene-regulation.com). A graphic visualization of putative binding sites is shown in Figure 2B and Supplementary Figure S2. The promoter harbours two putative AP-1 binding sites around position −676 and −33, which is in line with the requirement of c-Fos in trex1 mRNA induction. To analyse whether these binding sites are essential, several deletion constructs were cloned. As shown in Figure 2C, the region −517 to 730 was essential for basal transcription and induction of trex1. Thus, the −730 fragment showed 60% of the basal promoter activity compared to the full length fragment. In contrast, the −517 fragment showed only ∼10% promoter activity (Figure 2C). Regarding the UV provoked promoter stimulation, both the −923 and the −730 fragments were similarly inducible. In both cases, a >2-fold induction was detected, whereas neither the −517 nor the −157 fragment was inducible (Figure 2D). To further substantiate that the AP-1 binding site around position −676 is essential, the binding site was mutated via the QuickChange II XL site directed mutagenesis kit (Agilent Technology/Stratagene) at two positions (GT→CC; Figure 2E). Mutations introduced in this AP-1 site only weakly reduced basal promoter activity, but nearly completely abrogated the promoter induction following UV treatment of cells (Figure 2E, right).
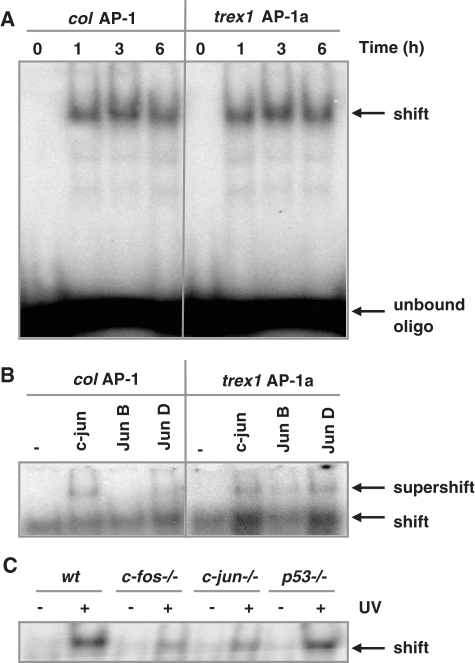
The trex1 promoter harbours a functional AP-1 site
To analyse whether the AP-1 binding site around position −676 was indeed recognized by AP-1, EMSAs were performed using radioactively labelled oligonucleotides harbouring this site (trex1 AP-1a) or, for control, the AP-1 binding site of the collagenase promoter (col AP-1). The oligonucleotides were incubated with nuclear extracts obtained from MEFs, either untreated or treated with 20 J/m2 UV light. As shown in Figure 3A, a time-dependent induction of AP-1 binding activity was observed using the collagenase AP-1 binding site (left part of the figure) or the potential AP-1 binding site (trex1 AP-1a) within the trex1 promoter. The second potential AP-1 biding site (trex1 AP-1b) derived from the computer screen presented in Figure 2B was not recognized in vitro by AP-1 (data not shown). Competition experiments with non-radioactively labelled oligonucleotides containing the AP-1 sequence of the collagenase promoter (col AP-1), but not with non-radioactively labelled oligonucleotides containing the p53 binding site of the p21 promoter (p21 p53), abrogated the recognition of the trex1 promoter by AP-1 (trex1 AP-1a), showing the specificity of the binding complex (Supplementary Figure S1C). To identify binding partners for c-Fos involved in the recognition of the trex1-specific AP-1 site, supershift experiments were performed. Whereas the collagenase AP-1 site (col AP-1) is mainly recognized by c-Jun, the trex1-specific AP-1 site (trex1 AP-1a) is recognized by both c-Jun and Jun D (Figure 3B). Binding activity of the trex1 AP-1a binding site was also compared between wt, fos−/−, c-jun−/− and p53−/− cells, showing a reduced binding activity in c-fos−/−and c-jun−/−, but not in p53−/− cells (Figure 3C). Complete abrogation of AP-1 binding activity in the null MEF cell extracts was not observed, indicating that at least in vitro it is likely that other members of the Jun and Fos family are able to recognize the trex1-specific AP-1 binding site as well.
Figure 3.
Identification of AP-1 binding sites within the trex1 promoter. (A) Binding of AP-1 to promoter fragments as determined by EMSA. Oligonucleotides containing either the AP-1 binding site of the collagenase promoter (col AP-1) or the AP-1 binding sites of the trex1 promoter (trex1 AP-1a) were incubated with nuclear extracts from wt cells exposed to 20 J/m2 UV light for 1, 3 or 6 h and subjected to EMSA. (B) EMSA supershift assay. Composition of the AP-1 factor bound to the col AP-1 and trex1 AP-1a oligonucleotide was analysed by the addition of specific antibodies to the reaction. (C) Oligonucleotides containing the AP-1 binding sites of the trex1 promoter (trex1 AP-1a) were incubated with nuclear extracts from wt cells, c-fos−/−, c-jun−/− and p53−/− cells, exposed to 20 J/m2 UV light for 6 h and subjected to EMSA.
Induction of TREX1 by different DNA damaging agents
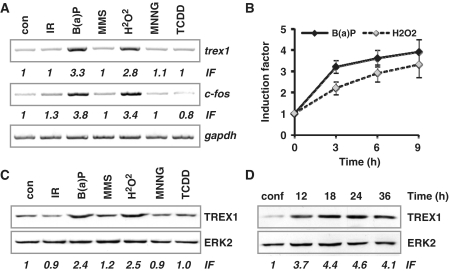
To investigate whether TREX1 can be up-regulated by genotoxic stress other than UV light, we analysed the expression of TREX1 on mRNA and protein level upon treatment of MEFs with the polycyclic aromatic hydrocarbon (B(a)P), the methylating agents MMS and MNNG, the radical forming H2O2 and ionising radiation (IR) as well as the tumour promoter and potent aryl hydrocarbone (Ah) receptor activator TCDD. As shown in Figure 4A and Supplementary Figure S1D (for semi-quantitative RT-PCR) and in Figure 4B (for quantitative real-time RT-PCR), trex1 expression was induced by B(a)P and H2O2 but not by IR, MMS, MNNG or TCDD. The same holds true for the up-regulation of the TREX1 protein that was observed after B(a)P and H2O2, but not IR, MMS, MNNG or TCDD treatment (Figure 4C, see induction factors for quantification). Parallel analysis of c-fos mRNA expression revealed that only the genotoxic agents that clearly induced c-fos mRNA, namely B(a)P and H2O2, were able to up-regulate the TREX1 protein (Figure 4A). Interestingly, the basal expression of TREX1 was dependent on cell proliferation since it was strongly reduced in confluent cells (arrested in G1) and recovered after reseeding of cells (Figure 4D). The same was true for the induced response following UV (data not shown).
Figure 4.
Induction of trex1 by genotoxins. (A) Exponentially growing wt MEFs were exposed to different genotoxins: 2.5 µM B(a)P, 1 mM H2O2, 2 Gy IR, 25 µM MNNG, 0.5 mM MMS, 2 nM TCDD for 6 h. RNA was isolated and semi-quantitative RT-PCR was performed using trex1, c-fos or, as positive control, gapdh- specific primers (con, non-exposed control). IF, induction factor. (B) Exponentially growing wt MEFs were exposed to 2.5 µM B(a)P or 0.5 mM H2O2 for 3, 6 or 9 h. Total RNA was isolated and real-time RT-PCR was performed using trex1-specific primers. For quantification, the expression was normalized with gapdh and the untreated control was set to 1. Data are the mean of three independent experiments +/− SD. (C) Exponentially growing wt MEFs were exposed to different genotoxins: 2.5 µM B(a)P, 1 mM H2O2, 2 Gy IR, 25 µM MNNG, 0.5 mM MMS, 2 nM TCDD for 9 h. Total protein extract was isolated. Immunodetection was performed using TREX1 or, as loading control, ERK2-specific antibodies. IF, induction factor. (D) Cells arrested in G1 by confluency (conf) and cells harvested at different time points after reseeding were harvested, whole cell extracts were isolated and the expression of TREX1 was measured by immunodetection using TREX1 or, as loading control, ERK2-specific antibodies.
TREX1 induction in human cells after genotoxic stress
To investigate whether induction of trex1 upon genotoxic stress occurs also in human cells, we analysed trex1 expression in the human fibroblast cell line GM637 exposed to UV light or B(a)P. As shown in Figure 5A (semi-quantitative RT-PCR), trex1 is clearly induced also in human fibroblasts after UV and B(a)P treatment. This was confirmed in GM637 cells by real-time RT-PCR (Figure 5B). To clarify whether induction of human trex1 is due to de novo synthesis and not stabilization of RNA, GM637 cells were exposed to UV light in the presence or absence of actinomycin D. As shown in Supplementary Figure S3A, treatment with the RNA synthesis inhibitor prevented induction of trex1, demonstrating that it depends on RNA de novo synthesis. Similar to MEFs, induction of human trex1 mRNA leads to increased TREX1 protein expression. As shown in Figure 5C, exposure of human GM637 cells to UV or B(a)P resulted in a time-dependent accumulation of TREX1, showing the highest induction level between 16 and 32 h.
Figure 5.
Induction of trex1 in human cells. (A) Exponentially growing human fibroblasts (cell line GM637) were exposed to 10 J/m2 UV or 2.5 µM B(a)P for different time points. Total RNA was isolated and semi-quantitative RT-PCR was performed using primers specific for trex1 or, as positive control, gapdh- specific primers (con, non-exposed control). (B) Exponentially growing GM637 cells were exposed to 10 J/m2 UV or 2.5 µM B(a)P for different time points. Total RNA was isolated and real-time RT-PCR was performed using primers specific for trex1 or, as positive control, gapdh-specific primers (con, non-exposed control). For quantification, the expression was normalized to gapdh and the untreated control was set to 1. Data are the mean of three independent experiments +/− SD. (C) Exponentially growing GM637 cells were exposed to 10 J/m2 UV or 2.5 µM B(a)P for different time points. Total protein extract was isolated. Immunodetection was performed using TREX1 or, as loading control, ERK2-specific antibodies. IF, induction factor. (D) Binding of AP-1 to promoter fragments as determined by EMSA. Oligonucleotides containing either the AP-1 binding site of the collagenase promoter (col AP-1) or the human trex1 promoter (htrex1 AP-1) were incubated with nuclear extracts from GM637 fibroblasts exposed to 10 J/m2 UV or 2.5 µM B(a)P for 4, 8 or 16 h and subjected to EMSA.
Comparing the mouse and the human trex1 promoter sequence (Supplementary Figure S4), a high similarity in the most apical region is evident. Whereas the overall identity is 61.5%, an 87 bp region (about 800 bp 5′ of the ATG) shows 82.8% identity. This region harbours the trex1-AP-1 binding site that is entirely identical to the mouse trex1-AP-1 binding site (CTGAGTCA) (Figure 5D). To demonstrate binding of AP-1 to the human promoter fragment, EMSA was performed using oligonucleotides harbouring either one of the AP-1 binding sites of the human trex1 promoter (htrex1-AP-1) or, for control, the AP-1 binding site of the collagenase promoter (col AP-1). The oligonucleotides were incubated with nuclear extracts obtained from GM637 cells, either untreated or treated with UV light or B(a)P. As shown in Figure 5D, a time-dependent induction of AP-1 binding activity was observed with the human trex1 promoter fragment containing the AP-1 binding site upon exposure to UV light and B(a)P (for presentation of the complete blot see Supplementary Figure S3B). The specificity of the binding complex was substantiated in competition experiments with non-radioactively labelled oligonucleotides (data not shown).
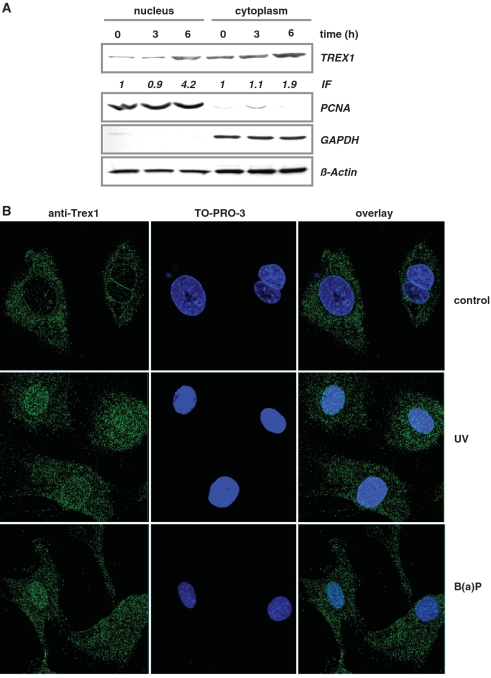
UV light and B(a)P induce nuclear translocation of TREX1
TREX1 is mostly localized in the cytoplasm. If the protein plays a role in the defence against genotoxic stress, it might be required in the nucleus. Therefore, we analysed whether nuclear translocation of TREX1 occurs following UV by determining the sub-cellular localization of TREX1. Whereas in untreated cells the major amount of TREX1 was found in the cytoplasmic fraction, upon UV exposure a strong increase in TREX1 in the nuclear fraction was observed (Figure 6A; the purity of nuclear and cytoplasmic extracts was controlled by the expression of PCNA and GAPDH). We substantiated nuclear translocation of TREX1 upon exposure of cells to UV light and B(a)P by immunofluorescence and confocal laser scanning microscopy. As shown in Figure 6B, in untreated cells TREX1 is localized in the cytoplasm. In TREX1-deficient cells (sc8−/−) TREX1 was not detectable (Supplementary Figure S5), confirming the specificity of the immunoreaction. Interestingly, we observed an accumulation of TREX1 in micronuclei (Supplementary Figure S5). Upon exposure to UV or B(a)P, a clear translocation of TREX1 from the cytoplasm into the nucleus was observed (Figure 6B). This nuclear translocation was most obvious in exponentially growing cells and strongly reduced when confluent cells were reseeded and UV irradiated in the G1 phase (Supplementary Figure S6). Analysing exponentially growing populations, cells with more than 20 nuclear TREX1 foci were observed in 3 % of non-exposed cells compared to 17 and 24 % in B(a)P- and UV-exposed cells, respectively (Figure 6B). The difference between control and UV- and B(a)P-treated cells was statistically significant (Student’s t-test: P < 0.01).
Figure 6.
Nuclear translocation of TREX1. (A) Exponentially growing wt MEFs were non-exposed or exposed to 20 J/m2 UV for 3 and 6 h. Nuclear and cytoplasmic extracts were isolated. Immunodetection was performed using TREX1, PCNA, GAPDH or, as loading control, β-Actin-specific antibody. Induction factor (IF) is derived from densitometric measurement of TREX1 signal and normalized to β-Actin expression. (B) Exponentially growing wt MEFs were not exposed (control) or exposed to 20 J/m2 UV light or 2.5 µM B(a)P for 6 h and thereafter fixed as described. TREX1 localization was visualized by the use of a specific antibody and detected by confocal laser scanning microscopy.
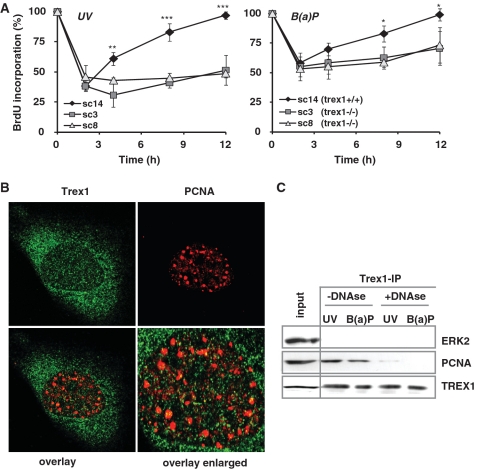
Impact of TREX1 on DNA replication
Since Trex1 foci co-localize with BrdU (36), we analysed whether TREX1 has an impact on DNA replication in cells treated with UV light or B(a)P. Hence we analyzed the genotoxin-induced replication arrest in TREX1-proficient (sc14+/+) and -deficient (sc3−/− and sc8−/−) MEFs by BrdU incorporation. As shown in Figure 7A (left panel), 2 h after treatment with 7.5 J/m2 UV DNA synthesis was reduced in sc14+/+ cells to 30% of the control level. It almost completely recovered 12 h later. In contrast, the two TREX1-deficient cell lines sc3−/− and sc8−/− displayed an incomplete recovery within the 12 h post-exposure period (Figure 7A). Similar results were obtained after B(a)P treatment, although the effect was not as pronounced as after UV treatment (Figure 7A, right panel).
Figure 7.
Impact of TREX1 on DNA replication. (A) Exponentially growing TREX1 wt cells (sc14+/+) and TREX1-deficient cells (sc3−/− and sc8−/−) were exposed to 7.5 J/m2 UV (left panel) or 2.5 µM B(a)P (right panel). Different time points later DNA replication was measured by incorporation of BrdU added to the medium 1 h before harvest. Data are the mean of three independent experiments +/− SD. *P < 0.05, **P < 0.01, ***P < 0.001. (B) Exponentially growing wt MEFs were exposed to 20 J/m2 UV light for 6 h and thereafter fixed as described. TREX1 and PCNA co-localization was analysed by the use of a specific antibody and detected by confocal laser scanning microscopy. (C) Total cell extracts were isolated from exponentially growing wt MEFs irradiated with 20 J/m2 UV or exposed to 2.5 µM B(a)P for 6 h. When indicated, RNase-free DNase I was added to the extracts at 1 U/µl for 1 h at 32°C prior to IP of TREX1 utilizing the Catch and Release® v2.0 system from Millipore. Co-immunoprecipitated PCNA was visualized as described above.
We further analysed whether TREX1 acts at the replication fork following UV by performing co-localization and co-IP experiments with PCNA. Although significant amounts of TREX1 and PCNA were found in the nucleus, TREX1 only marginally co-localized with PCNA (Figure 7B). To analyse a potential interaction of TREX1 with PCNA, protein extracts were isolated from exponentially growing wt MEFs exposed to 20 J/m2 UV or treated with 2.5 µM B(a)P for 6 h. IP occured by a specific TREX1 antibody (utilizing the Catch and Release® v2.0 system from Milipore). Co-immunoprecipitating proteins were visualized by specific antibodies against PCNA and ERK2 (negative control). Extract from 3T3 cells was included as positive control (‘input’). Co-IP of PCNA was observed following genotoxin treatment (Figure 7C). However, co-IP of PCNA and TREX1 was not based on protein–protein interaction. It was very likely due to simultaneous binding of the proteins to genomic DNA as pre-treatment with DNAse1, which degrades DNA thereby breaking the tether between DNA-bound proteins, strongly reduced the level of PCNA in the TREX1 immunoprecipitate.
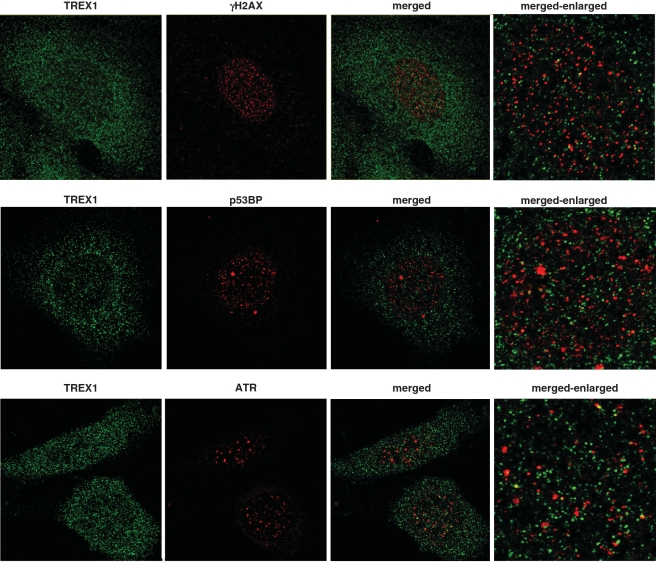
TREX1 does not co-localize with γH2AX, ATR or p53Bp
Since only marginal co-localization between TREX1 and the replication marker PCNA was observed, we addressed the question of whether upon UV exposure TREX1 localizes to the region of DNA damage, such as DNA double-strand breaks, or collapsed replication forks. Therefore, co-localization of TREX1 with either γH2AX, ATR or p53BP, all of which are implicated in the DNA damage response (DDR) [for review see (37,38)], was analyzed by immunofluorescence and confocal laser scanning microscopy. In none of the cases co-localization was observed (Figure 8).
Figure 8.
TREX1 does not co-localize with DNA damage markers. Exponentially growing wt MEFs were exposed to 20 J/m2 UV light for 6 h and thereafter fixed as described. Possible co-localization between TREX1 and γH2AX, pATR and p53BP was analyzed by the use of corresponding antibodies and detected by confocal laser scanning microscopy.
TREX1 in the defence against UV light and B(a)P
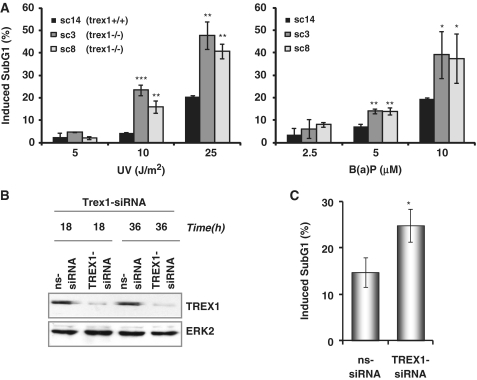
Since TREX1 is up-regulated following genotoxic stress and stimulates the recovery from the DNA replication inhibition after UV and B(a)P treatment, the question arose whether it also has impact on cell viability upon genotoxic treatments. Therefore, we determined the level of apoptosis in TREX1-proficient (sc14+/+) and -deficient (sc3−/− and sc8−/−) cells upon UV and B(a)P exposure. As shown in Figure 9A, TREX1-deficient cells were more sensitive to UV light (left) and B(a)P (right) than the isogenic wt. To substantiate the data, we analysed the sensitivity of wt cells to UV light after down-regulation of TREX1 by transfection with trex1-siRNA. The TREX1 protein was almost completely down-regulated as monitored 18 and 36 h after transfection (Figure 9B). Similar to the results with TREX1- deficient cell lines, the sensitivity to UV light was significantly enhanced under TREX1 knockdown conditions (Figure 9C). Overall, the data strongly suggest that TREX1 induction plays a significant role in protecting cells against genotoxic stress.
Figure 9.
Impact of TREX1 on sensitivity to UV and B(a)P. (A) Exponentially growing TREX1 wt cells (sc14+/+) and TREX1-deficient cells (sc3−/− and sc8−/−) were exposed to different doses of UV (left panel) or B(a)P (right panel). Cells were harvested 72 h later and the SubG1 fraction was determined. *P < 0.05, **P < 0.01, ***P < 0.001. (B) Exponentially growing wt MEFs were transiently transfected with TREX1-siRNA or a non-silencing siRNA (ns-siRNA). total protein extract was isolated 18 and 36 h later. Immunodetection was performed using TREX1 or, as loading control, ERK2-specific antibody. (C) Exponentially growing wt MEFs were transiently transfected with TREX1-siRNA or a non-silencing siRNA (ns-siRNA). Cells were exposed to UV light (20 J/m2) 18 h later. Cells were harvested 72 h later and the SubG1 fraction was determined. *P < 0.05.
DISCUSSION
Here we report on a novel player, TREX1, in the inducible genotoxic stress response. TREX1 is a 3′-5′ exonuclease that catalyses the excision of nucleoside monophosphates from the 3′-termini of DNA (39). It is active as a homodimer (40) that prefers a partial duplex DNA with multiple mispaired 3′-termini. TREX1 is also referred to as DNase III. Mutations in TREX1 are associated with the human disorders Aicardi–Goutières syndrome (41,42), autosomal dominant retinal vasculopathy with cerebral leucodystrophy (43), systemic lupus erythematosus (44) and familial chilblain lupus (45). TREX1 knockout mice develop inflammatory myocarditis, resulting in progressive cardiomyopathy leading to circulatory failure and reduced survival (46). Although mutations in TREX1 have far-reaching consequences, the biological function and regulation of TREX1 are rather unknown. It was shown that TREX1 interacts with members of the SET complex that plays a role in DNA degradation during granzyme A-mediated cell death (47). It was also shown that human TREX1 is involved in the response to IR and hydroxyurea (36), camptothecin (48) and prevents the cell-intrinsic initiation of autoimmunity (49).
We demonstrate for the first time that trex1 is a genotoxic stress-inducible gene. trex1 mRNA is induced time- and dose-dependently upon exposure of cells to UV light. The induction was not observed in MEFs derived from c-fos knockout mice indicating that c-Fos plays an essential role in the genotoxin provoked up-regulation of TREX1. Induction of TREX1 is due to promoter activation and results in a significantly enhanced level of the protein after UV treatment. To further analyse the regulation of trex1 we cloned the trex1 promoter and showed that it is inducible by UV light. The mouse promoter contains two AP-1 binding sites, one of which (676 bp upstream of the ATG start codon) is recognized by c-Fos (AP-1), as demonstrated by ChIP and EMSA. Deletion of the AP-1 binding site clearly abrogated the UV-induced activation of the trex1 promoter. In fos−/− and jun−/− cells induction of TREX1 was strongly reduced, but not completely abrogated, indicating that in addition to c-Fos and c-Jun other members of the Fos and Jun family are involved in the regulation of TREX1. This is supported by EMSA, showing the binding of JunD and, at a very low level, JunB to the trex1-specific AP-1 binding site. We should note that for the basal activity of the trex1 promoter, the AP-1 site seems to be of minor importance because deletion of this site reduces basal activity only by 25%. Also, under non-stress condition there is a weak binding activity of c-Fos/AP-1 to the promoter, and TREX1 expression in unexposed c-Fos-deficient cells is only slightly lower than in wt cells.
We observed that TREX1 is not only induced by UV light, but also by other genotoxins such as B(a)P and H2O2. Interestingly, the methylating agents MNNG and MMS as well as IR did not trigger TREX1 induction. We should note that under the treatment conditions with these agents, which elicited similar cytotoxic effects, MNNG, MMS and IR did not induce c-Fos, whereas B(a)P and H2O2 similar to UV light showed up-regulation of c-Fos. c-Fos up-regulation preceded trex1 induction (data not shown), which further supports that induction of TREX1 following UV, B(a)P and H2O2 is regulated via AP-1.
There are only few DNA repair genes that were shown to be up-regulated following genotoxic stress (2) and some of them, e.g. mgmt and fen1, were shown to be up-regulated in rodent, but not human cells upon DNA damage [(7,50), unpublished data]. Therefore, the finding attracts notice that TREX1 is induced on RNA and protein level in both mouse and human cells following genotoxic stress. A comparison of the mouse and human trex1 promoter revealed an overall identity of 61.5%. The mouse and human AP-1 binding sites in the trex1 promoter are identical (for sequence see Supplementary Figure S4); they are located within a highly conserved promoter region of 87 bp exhibiting 82.8% identity. Both the mouse and the human trex1 AP-1 binding site is recognized in vitro by AP-1. The binding is enhanced after treatment of cells with UV and B(a)P, substantiating that in mouse and human cells trex1 is regulated via AP-1. We should note that the position of the human AP-1 binding site at position −656 is nearly identical to the position of the transcription initiation site in the human trex1 promoter (position −650), which was suggested on the basis of NNPP (51).
TREX1 (DNase III) activity was first identified in nuclei of rabbit tissues (52). In human cells, DNase III is present in similar amounts in the nuclei of non-growing, e.g. adult liver, and rapidly proliferating tissues (53). In MEFs, the expression of TREX1 was related to proliferation. It was strongly reduced in confluent cells and recovered after the resumption of proliferation. Previous studies showed that murine TREX1 is mainly localized in the endoplasmatic reticulum and translocates into the nucleus upon IR and hydroxyurea (36). In human cells TREX1 was reported to translocate into the nucleus during granzyme A-mediated cell death (47). Here, we extend these observations and demonstrate that TREX1 is translocated into the nucleus upon UV light and B(a)P exposure. This nuclear translocation parallels the up-regulation of trex1 gene activity and increase in the TREX1 protein level. It therefore seems that TREX1 is complex regulated in cells exposed to genotoxins that have the ability to induce c-Fos/AP-1.
An association between TREX1 and DNA replication is further substantiated by our finding that upon UV and B(a)P treatment, TREX1-deficient cells are defective in the recovery from the genotoxin-induced block to replication. This might indicate an active role of TREX1 during replication of damaged DNA, as suggested from co-localization of TREX1 with BrdU (36) and from the fact that TREX1 knockout cells show a chronic activation of the ATM-triggered DNA damage checkpoint (36). To further examine the association of TREX1 with replication, we investigated a potential interaction with PCNA, conducting IP and co-localization experiments. Co-IP of PCNA and TREX1 was found, which was however not observed after treatment with DNAse I. Thus, the data do not support a physical interaction of TREX1 with PCNA. This was confirmed by lack of significant co-localization of TREX1 and PCNA. Overall, the data suggest that TREX1 does not directly interact with the replication machinery upon DNA damage.
Following UV irradiation, TREX1 accumulates in the nucleus and becomes visible as small foci (Figure 8). To analyse whether TREX1 foci are associated with proteins involved in the DDR we examined whether TREX1 co-localizes with well-known players of the DDR (37,38). While UV irradiation induces the formation of γH2AX, p53BP and ATR foci, a significant co-localization with TREX1 foci was not observed. The data might indicate that TREX1 is not directly involved in the DDR.
To analyse whether induction of TREX1 is required for protecting cells against genotoxins, we utilized trex1 null mouse fibroblasts and compared their cell death response with the isogenic wt following UV light and B(a)P treatment. We show that TREX1-deficient cells are more sensitive to UV and B(a)P than the wt. Cell death was executed by apoptosis. The hypersensitivity of TREX1 lacking cells was confirmed by down-regulation of TREX1 via siRNA transfection. The data suggest a functional role of TREX1 induction in the defence against DNA damaging agents. The protective function of TREX1 seems to be specific for agents inducing bulky lesions such as UV light and B(a)P since it was not observed in MEFs lacking TREX1 following IR (36). TREX1 induction was also not observed following MNNG and MMS that do not strongly induce c-Fos/AP-1 unless high toxic doses were applied (17). We should note that for IR replication is not essential for executing cell death whereas DNA replication is a prerequisite for apoptosis induction following UV and very likely also B(a)P treatment (54). This could explain the different role of TREX1 in cells exposed to IR on the one hand and UV and B(a)P on the other.
Overall, we show that the DNA repair protein TREX1 is up-regulated and becomes translocated into the nucleus in cells treated with UV light and the powerful environmental carcinogen B(a)P, both are representative of bulky lesion-inducing genotoxins. Genotoxin-induced up-regulation of TREX1 expression occured in mouse and human cells and was found to be c-Fos/AP-1-dependent. TREX1 is involved in the recovery from the genotoxin-induced inhibition of replication and protects against apoptotic cell death following UV light and B(a)P treatment. The complex regulation of TREX1 involving gene induction and nuclear translocation represents a novel mechanism in the cellular stress response that counteracts DNA damage. The data may clarify at the same time the complex phenotype of cells lacking c-Fos, which have been shown to be hypersensitive to UV light and B(a)P, but not IR (20,28). It is reasonable to posit that the observed lack of TREX1 induction in cells deficient in c-Fos contributes to the hypersensitivity of these cells. Since c-Fos is expressed at variable levels in different types of cancer (55,56), we hypothesize that TREX1 is induced during tumour therapy to different extent, which might have an impact on anticancer drug resistance. The question of whether TREX1 is induced by anticancer drugs thus contributing to tumour cell resistance will be an interesting issue of future studies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Krebshilfe 106748; Stiftung Rheinland-Pfalz; Deutsche Forschungsgemeinschaft CH 665/2-1. Funding for open access charge: Deutsche Forschungsgemeinschaft CH 665/2-1.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Birgit Rasenberger for excellent technical assistance and Dr Theodora Nikolova and Dr Wynand Roos for help with laser scanning microspcopy. We especially thank Prof. T. Lindahl for providing TREX1-deficient cells.
REFERENCES
- 1.Christmann M, Tomicic MT, Roos WP, Kaina B. Mechanisms of human DNA repair: an update. Toxicology. 2003;193:3–34. doi: 10.1016/s0300-483x(03)00287-7. [DOI] [PubMed] [Google Scholar]
- 2.Christmann M, Fritz G, Kaina B. Induction of DNA repair genes in mammalian cells in response to genotoxic stress. In: Lankenan D-H, editor. Genome Integrity, Facects and Perspectives. Berlin: Springer; 2007. pp. 383–398. [Google Scholar]
- 3.Adimoolam S, Ford JM. p53 and DNA damage-inducible expression of the xeroderma pigmentosum group C gene. Proc. Natl Acad. Sci. USA. 2002;99:12985–2990. doi: 10.1073/pnas.202485699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montecucco A, Savini E, Biamonti G, Stefanini M, Focher F, Ciarrocchi G. Late induction of human DNA ligase I after UV-C irradiation. Nucleic Acids Res. 1995;23:962–966. doi: 10.1093/nar/23.6.962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hwang BJ, Ford JM, Hanawalt PC, Chu G. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc. Natl Acad. Sci. USA. 1999;96:424–428. doi: 10.1073/pnas.96.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang J, Chen Z, Liu Y, Hickey RJ, Malkas LH. Altered DNA polymerase iota expression in breast cancer cells leads to a reduction in DNA replication fidelity and a higher rate of mutagenesis. Cancer Res. 2004;64:5597–5607. doi: 10.1158/0008-5472.CAN-04-0603. [DOI] [PubMed] [Google Scholar]
- 7.Christmann M, Tomicic MT, Origer J, Kaina B. Fen1 is induced p53 dependently and involved in the recovery from UV-light-induced replication inhibition. Oncogene. 2005;24:8304–8313. doi: 10.1038/sj.onc.1208994. [DOI] [PubMed] [Google Scholar]
- 8.Lackinger D, Eichhorn U, Kaina B. Effect of ultraviolet light, methyl methanesulfonate and ionizing radiation on the genotoxic response and apoptosis of mouse fibroblasts lacking c-Fos, p53 or both. Mutagenesis. 2001;16:233–241. doi: 10.1093/mutage/16.3.233. [DOI] [PubMed] [Google Scholar]
- 9.Hicks GG, Egan SE, Greenberg AH, Mowat M. Mutant p53 tumor suppressor alleles release ras-induced cell cycle growth arrest. Mol. Cell Biol. 1991;11:1344–1352. doi: 10.1128/mcb.11.3.1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kastan MB, Onyekwere O, Sidransky D, Vogelstein B, Craig RW. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- 11.Seo YR, Fishel ML, Amundson S, Kelley MR, Smith ML. Implication of p53 in base excision DNA repair: in vivo evidence. Oncogene. 2002;21:731–737. doi: 10.1038/sj.onc.1205129. [DOI] [PubMed] [Google Scholar]
- 12.Offer H, Milyavsky M, Erez N, Matas D, Zurer I, Harris CC, Rotter V. Structural and functional involvement of p53 in BER in vitro and in vivo. Oncogene. 2001;20:581–589. doi: 10.1038/sj.onc.1204120. [DOI] [PubMed] [Google Scholar]
- 13.Smith ML, Ford JM, Hollander MC, Bortnick RA, Amundson SA, Seo YR, Deng CX, Hanawalt PC, Fornace AJ., Jr p53-mediated DNA repair responses to UV radiation: studies of mouse cells lacking p53, p21, and/or gadd45 genes. Mol. Cell Biol. 2000;20:3705–3714. doi: 10.1128/mcb.20.10.3705-3714.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomicic MT, Christmann M, Kaina B. Apoptosis in UV-C light irradiated p53 wild-type, apaf-1 and p53 knockout mouse embryonic fibroblasts: interplay of receptor and mitochondrial pathway. Apoptosis. 2005;10:1295–1304. doi: 10.1007/s10495-005-1392-3. [DOI] [PubMed] [Google Scholar]
- 15.van Dam H, Castellazzi M. Distinct roles of Jun: Fos and Jun : ATF dimers in oncogenesis. Oncogene. 2001;20:2453–2464. doi: 10.1038/sj.onc.1204239. [DOI] [PubMed] [Google Scholar]
- 16.Buscher M, Rahmsdorf HJ, Litfin M, Karin M, Herrlich P. Activation of the c-fos gene by UV and phorbol ester: different signal transduction pathways converge to the same enhancer element. Oncogene. 1988;3:301–311. [PubMed] [Google Scholar]
- 17.Dosch J, Kaina B. Induction of c-fos, c-jun, junB and junD mRNA and AP-1 by alkylating mutagens in cells deficient and proficient for the DNA repair protein O6-methylguanine-DNA methyltransferase (MGMT) and its relationship to cell death, mutation induction and chromosomal instability. Oncogene. 1996;13:1927–1935. [PubMed] [Google Scholar]
- 18.Hollander MC, Fornace AJ., Jr Induction of fos RNA by DNA-damaging agents. Cancer Res. 1989;49:1687–1692. [PubMed] [Google Scholar]
- 19.Haas S, Kaina B. c-Fos is involved in the cellular defence against the genotoxic effect of UV radiation. Carcinogenesis. 1995;16:985–991. doi: 10.1093/carcin/16.5.985. [DOI] [PubMed] [Google Scholar]
- 20.Kaina B, Haas S, Kappes H. A general role for c-Fos in cellular protection against DNA-damaging carcinogens and cytostatic drugs. Cancer Res. 1997;57:2721–2731. [PubMed] [Google Scholar]
- 21.Lackinger D, Kaina B. Primary mouse fibroblasts deficient for c-Fos, p53 or for both proteins are hypersensitive to UV light and alkylating agent-induced chromosomal breakage and apoptosis. Mutat Res. 2000;457:113–123. doi: 10.1016/s0027-5107(00)00133-0. [DOI] [PubMed] [Google Scholar]
- 22.Miller AD, Curran T, Verma IM. c-fos protein can induce cellular transformation: a novel mechanism of activation of a cellular oncogene. Cell. 1984;36:51–60. doi: 10.1016/0092-8674(84)90073-4. [DOI] [PubMed] [Google Scholar]
- 23.Muller R, Tremblay JM, Adamson ED, Verma IM. Tissue and cell type-specific expression of two human c-onc genes. Nature. 1983;304:454–456. doi: 10.1038/304454a0. [DOI] [PubMed] [Google Scholar]
- 24.Kataki A, Sotirianakos S, Memos N, Karayiannis M, Messaris E, Leandros E, Manouras A, Androulakis G. P53 and C-FOS overexpression in patients with thyroid cancer: an immunohistochemical study. Neoplasma. 2003;50:26–30. [PubMed] [Google Scholar]
- 25.Gamberi G, Benassi MS, Bohling T, Ragazzini P, Molendini L, Sollazzo MR, Pompetti F, Merli M, Magagnoli G, Balladelli A, et al. C-myc and c-fos in human osteosarcoma: prognostic value of mRNA and protein expression. Oncology. 1998;55:556–563. doi: 10.1159/000011912. [DOI] [PubMed] [Google Scholar]
- 26.Moorehead RA, Singh G. Influence of the proto-oncogene c-fos on cisplatin sensitivity. Biochem. Pharmacol. 2000;59:337–345. doi: 10.1016/s0006-2952(99)00333-0. [DOI] [PubMed] [Google Scholar]
- 27.Funato T, Ishii T, Kanbe M, Scanlon KJ, Sasaki T. Reversal of cisplatin resistance in vivo by an anti-fos ribozyme. In Vivo. 1997;11:217–220. [PubMed] [Google Scholar]
- 28.Christmann M, Tomicic MT, Origer J, Aasland D, Kaina B. c-Fos is required for excision repair of UV-light induced DNA lesions by triggering the re-synthesis of XPF. Nucleic Acids Res. 2006;34:6530–6539. doi: 10.1093/nar/gkl895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christmann M, Tomicic MT, Aasland D, Kaina B. A role for UV-light-induced c-Fos: stimulation of nucleotide excision repair and protection against sustained JNK activation and apoptosis. Carcinogenesis. 2007;28:183–190. doi: 10.1093/carcin/bgl119. [DOI] [PubMed] [Google Scholar]
- 30.Platt K, Oesch F. Efficient synthesis of non-K-region trans-dihydro diols of polycyclic aromatic hydrocarbons from o-quinones and catechols. J. Org. Chem. 1983;48:265–268. [Google Scholar]
- 31.Yagi H, Thakker DR, Hernandez O, Koreeda M, Jerina DM. Synthesis and reactions of the highly mutagenic 7,8-diol 9,10-epoxides of the carcinogen benzo[a]pyrene. J. Am. Chem. Soc. 1977;99:1604–1611. doi: 10.1021/ja00447a053. [DOI] [PubMed] [Google Scholar]
- 32.Christmann M, Tomicic MT, Kaina B. Phosphorylation of mismatch repair proteins MSH2 and MSH6 affecting MutS{alpha} mismatch-binding activity. Nucleic Acids. Res. 2002;30:1959–1966. doi: 10.1093/nar/30.9.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomicic MT, Bey E, Wutzler P, Thust R, Kaina B. Comparative analysis of DNA breakage, chromosomal aberrations and apoptosis induced by the anti-herpes purine nucleoside analogues aciclovir, ganciclovir and penciclovir. Mutat. Res. 2002;505:1–11. doi: 10.1016/s0027-5107(02)00105-7. [DOI] [PubMed] [Google Scholar]
- 34.Tomicic MT, Friedrichs C, Christmann M, Wutzler P, Thust R, Kaina B. Apoptosis induced by (E)-5-(2-bromovinyl)-2'-deoxyuridine in varicella zoster virus thymidine kinase-expressing cells is driven by activation of c-Jun/activator protein-1 and Fas ligand/caspase-8. Mol. Pharmacol. 2003;63:439–449. doi: 10.1124/mol.63.2.439. [DOI] [PubMed] [Google Scholar]
- 35.Christmann M, Kaina B. Nuclear translocation of mismatch repair proteins MSH2 and MSH6 as a response of cells to alkylating agents. J. Biol. Chem. 2000;275:36256–36262. doi: 10.1074/jbc.M005377200. [DOI] [PubMed] [Google Scholar]
- 36.Yang YG, Lindahl T, Barnes DE. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell. 2007;131:873–886. doi: 10.1016/j.cell.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 37.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol. Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 38.Bartek J, Bartkova J, Lukas J. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene. 2007;26:7773–7779. doi: 10.1038/sj.onc.1210881. [DOI] [PubMed] [Google Scholar]
- 39.Mazur DJ, Perrino FW. Identification and expression of the TREX1 and TREX2 cDNA sequences encoding mammalian 3′–>5′ exonucleases. J. Biol. Chem. 1999;274:19655–19660. doi: 10.1074/jbc.274.28.19655. [DOI] [PubMed] [Google Scholar]
- 40.Mazur DJ, Perrino FW. Excision of 3′ termini by the Trex1 and TREX2 3′–>5′ exonucleases. Characterization of the recombinant proteins. J. Biol. Chem. 2001;276:17022–17029. doi: 10.1074/jbc.M100623200. [DOI] [PubMed] [Google Scholar]
- 41.Crow YJ, Hayward BE, Parmar R, Robins P, Leitch A, Ali M, Black DN, van Bokhoven H, Brunner HG, Hamel BC, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat. Genet. 2006;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- 42.Rice G, Newman WG, Dean J, Patrick T, Parmar R, Flintoff K, Robins P, Harvey S, Hollis T, O'Hara A, et al. Heterozygous mutations in TREX1 cause familial chilblain lupus and dominant Aicardi-Goutieres syndrome. Am. J. Hum. Genet. 2007;80:811–815. doi: 10.1086/513443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richards A, van den Maagdenberg AM, Jen JC, Kavanagh D, Bertram P, Spitzer D, Liszewski MK, Barilla-Labarca ML, Terwindt GM, Kasai Y, et al. C-terminal truncations in human 3′-5′ DNA exonuclease TREX1 cause autosomal dominant retinal vasculopathy with cerebral leukodystrophy. Nat. Genet. 2007;39:1068–1070. doi: 10.1038/ng2082. [DOI] [PubMed] [Google Scholar]
- 44.Lee-Kirsch MA, Gong M, Chowdhury D, Senenko L, Engel K, Lee YA, de Silva U, Bailey SL, Witte T, Vyse TJ, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat. Genet. 2007;39:1065–1067. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- 45.Lee-Kirsch MA, Chowdhury D, Harvey S, Gong M, Senenko L, Engel K, Pfeiffer C, Hollis T, Gahr M, Perrino FW, et al. A mutation in TREX1 that impairs susceptibility to granzyme A-mediated cell death underlies familial chilblain lupus. J. Mol. Med. 2007;85:531–537. doi: 10.1007/s00109-007-0199-9. [DOI] [PubMed] [Google Scholar]
- 46.Morita M, Stamp G, Robins P, Dulic A, Rosewell I, Hrivnak G, Daly G, Lindahl T, Barnes DE. Gene-targeted mice lacking the Trex1 (DNase III) 3′–>5′ DNA exonuclease develop inflammatory myocarditis. Mol. Cell Biol. 2004;24:6719–6727. doi: 10.1128/MCB.24.15.6719-6727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chowdhury D, Beresford PJ, Zhu P, Zhang D, Sung JS, Demple B, Perrino FW, Lieberman J. The exonuclease TREX1 is in the SET complex and acts in concert with NM23-H1 to degrade DNA during granzyme A-mediated cell death. Mol. Cell. 2006;23:133–142. doi: 10.1016/j.molcel.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 48.Wang CJ, Lam W, Bussom S, Chang HM, Cheng YC. TREX1 acts in degrading damaged DNA from drug-treated tumor cells. DNA Repair (Amst.) 2009;8:1179–1189. doi: 10.1016/j.dnarep.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fritz G, Tano K, Mitra S, Kaina B. Inducibility of the DNA repair gene encoding O6-methylguanine-DNA methyltransferase in mammalian cells by DNA-damaging treatments. Mol. Cell. Biol. 1991;11:4660–4668. doi: 10.1128/mcb.11.9.4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mazur DJ, Perrino FW. Structure and expression of the TREX1 and TREX2 3′ –> 5′ exonuclease genes. J. Biol. Chem. 2001;276:14718–14727. doi: 10.1074/jbc.M010051200. [DOI] [PubMed] [Google Scholar]
- 52.Lindahl T, Gally JA, Edelman GM. Properties of deoxyribonuclease 3 from mammalian tissues. J. Biol. Chem. 1969;244:5014–5019. [PubMed] [Google Scholar]
- 53.Hoss M, Robins P, Naven TJ, Pappin DJ, Sgouros J, Lindahl T. A human DNA editing enzyme homologous to the Escherichia coli DnaQ/MutD protein. EMBO J. 1999;18:3868–3875. doi: 10.1093/emboj/18.13.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dunkern TR, Kaina B. Cell proliferation and DNA breaks are involved in ultraviolet light-induced apoptosis in nucleotide excision repair-deficient Chinese hamster cells. Mol. Biol. Cell. 2002;13:348–361. doi: 10.1091/mbc.01-05-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Milde-Langosch K. The Fos family of transcription factors and their role in tumourigenesis. Eur. J. Cancer. 2005;41:2449–2461. doi: 10.1016/j.ejca.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 56.Durchdewald M, Angel P, Hess J. The transcription factor Fos: a Janus-type regulator in health and disease. Histol. Histopathol. 2009;24:1451–1461. doi: 10.14670/HH-24.1451. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.