Abstract
Several different approaches exist to generate expressed RNA interference (RNAi) precursors for multiple target inhibition, a strategy referred to as combinatorial (co)RNAi. One such approach makes use of RNA Pol III-expressed long hairpin RNAs (lhRNAs), which are processed by Dicer to generate multiple unique short interfering siRNA effectors. However, because of inefficient intracellular Dicer processing, lhRNA duplexes have been limited to generating two independent effective siRNA species. In this study, we describe a novel strategy whereby four separate anti-HIV siRNAs were generated from a single RNA Pol III-expressed transcript. Two optimized lhRNAs, each comprising two active anti-HIV siRNAs, were placed in tandem to form a double long hairpin (dlhRNA) expression cassette, which encodes four unique and effective siRNA sequences. Processing of the 3′ position lhRNA was more variable but effective multiple processing was possible by manipulating the order of the siRNA-encoding sequences. Importantly, unlike shRNAs, Pol III-expressed dlhRNAs did not compete with endogenous and exogenous microRNAs to disrupt the RNAi pathway. The versatility of expressed lhRNAs is greatly expanded and we provide a mechanism for generating transcripts with modular lhRNAs motifs that contribute to improved coRNAi.
INTRODUCTION
Exogenous RNA intermediates of the RNA interference (RNAi) pathway have become powerful tools for the development of reverse genetics approaches and novel therapeutics against a wide variety of diseases (1). Specifically, expressed short hairpin RNAs (shRNAs), which mimic precursor microRNA (pre-miRNA), are Dicer substrates of the mammalian miRNA pathway and have been extensively exploited for the production of effectors of the RNAi pathway (2). However, to prevent emergence of resistant viral mutants from highly mutable targets, such as those of human immunodeficiency virus (HIV) and hepatitis C virus (HCV), a combination of multiple short interfering RNA (siRNA) effectors is required (3,4). Therefore, an effective combinatorial RNAi (coRNAi) system aimed at generating active siRNA products, and which is capable of targeting multiple sites simultaneously, remains an important objective.
Harnessing the RNAi pathway to suppress multiple gene targets concurrently has been attempted using several different coRNAi strategies with varying success. The most common approach is to place shRNA expression cassettes adjacent to each other to achieve combined expression of multiple shRNAs (5–9). However, there are several limitations to combining multiple shRNA cassettes. Typically shRNAs are expressed from powerful constitutively active promoters (e.g. U6, H1 and U1 promoters). A coRNAi approach, with several shRNA expression cassettes in tandem, risks overwhelming the endogenous RNAi pathway. High levels of specific shRNAs may have serious and potentially fatal consequences, which are important for application to gene therapy (10). Another concern relates to the use of lentiviral vectors, which are often employed to deliver therapeutic anti-HIV RNAi sequences to T cells. These vectors are prone to recombining or deleting repeat sequences (11,12). Although this problem may be alleviated by simultaneous use of different promoters (8), such an arrangement of expressed silencing sequences would require careful empirical assessment to optimize expression of each shRNA. Other coRNAi approaches have made use of a combination of primary microRNA (pri-miRNA) shuttles, where siRNA guide sequences are placed within mimics of endogenous polycistronic pri-miRNAs (13–18). Although polycistronic pri-miRNA shuttles represent a promising coRNAi approach, efficient processing of multiple siRNAs from these pri-miRNA precursors is variable (13,15,18) and development of new methods of achieving coRNAi remains important.
Exploiting the action of endogenous Dicer to process long dsRNA templates and form multiple siRNA species is potentially a useful approach for achieving coRNAi. The most well-studied method is to employ Pol III promoters to generate transcripts with defined 5′- and 3′-termini that fold into long hairpins with duplex regions of between 30 and 100 bp (19–27). These lhRNAs, which have 2 nt 3′-OH overhangs have been shown to be intracellular Dicer substrates and produce multiple siRNA species (19–23). We and others have shown that siRNAs are processed by Dicer in a gradient of decreasing efficiency, which starts from the base of the dsRNA hairpin duplex and moves towards the apical loop (19,20,22,26).
Here we describe a novel lhRNA-based strategy whereby four independent siRNAs were produced in effective doses from a single RNA Pol III-expressed transcript. Two optimized dual-targeting lhRNAs were placed in tandem within a single transcript to form a double long hairpin RNA (dlhRNA) template. We show that the dlhRNA is processed into two lhRNAs, which in turn produce four active anti-HIV siRNAs. Moreover, there was no evidence of disruption of the endogenous miRNA pathway by dlhRNAs. This work describes a novel and safe dlhRNA approach to coRNAi and expands the versatility of expressed lhRNAs for applications requiring simultaneous silencing of multiple targets.
MATERIALS AND METHODS
Hairpin expression plasmids
The procedure for generating hairpin RNAs is a modification of the PCR-based method described by Castanotto et al. (28) and later adapted by ourselves to generate lhRNAs (20). A panel of twelve U6-driven lhRNAs encoding two putative siRNA sequences targeted to the HIV-1 tat and nef or int and LTR sequences was constructed using two rounds of PCR. Selection of target sites was based on previously reported effective knockdown of HIV-1 tat (29), nef (23), int (30) and LTR (20) sequences by shRNAs. Oligonucleotide sequences used in each of the amplification reactions are provided in Supplementary Table 1. During the first round of PCR a U6 promoter-containing plasmid DNA, pTZ-U6+1, was used as template (31). A universal U6 forward primer complementary to the 5′-end of the U6 promoter was used for both rounds of PCR. The reverse primers for the first round of PCR were complementary to 18–21 nt of the 3′-end of the U6 promoter and included sequences encoding the sense strand and loop of the lhRNA. The first round of PCR product was used as template for the second round of PCR. The round two reverse primer sequences were designed to hybridize to the loop-encoding region at the 3′ extremity of the first round PCR amplicon. These primers included the sequence encoding the antisense strand of the lhRNA as well as a RNA Pol III termination signal. Double-lhRNA expression cassettes were similarly generated using the two-round PCR method described above, but previously generated lhRNAs were used as templates. For dlhRNAs, round one reverse primers were complementary to the last 18 nt of the lhRNA template that would be positioned at the 5′-side. These primers included a spacer of two adenosine residues and a sequence encoding the sense strand of the 3′-lhRNA. To complete synthesis of the dlhRNA cassettes, round two reverse primers were employed that were the same as those used during the second round of PCR to generate lhRNA cassettes. To propagate the expression cassettes, the final PCR products were ligated directly to the TA cloning vector pTZ57R/T (Fermentas, WI, USA), and sequences were confirmed using standard procedures. Twelve lhRNA plasmids were generated: plhLTR-int +1, plhLTR-int +2, plhLTR-int +3, plhint-LTR +1, plhint-LTR +2, plhint-LTR +3, plhtat-nef +1, plhtat-nef +2, plhtat-nef +3, plhnef-tat +1, plhnef-tat +2 and plhnef-tat +3. Two dlhRNA plasmids were propagated: plhLI-TN and plhTN-LI. U6-driven shRNAs encoding siRNAs corresponding to those included in the lhRNA expression constructs were generated using a single round of PCR. The U6 promoter-containing plasmid DNA was used as template. Amplification with the universal U6 forward primer and specific reverse primers enabled formation of the shRNA-encoding sequences and these were inserted into pTZ57R/T (Fermentas) to generate: pshLTR, pshint, pshtat and pshnef. pH1-lhtat-nef +1 and p7SK-lhLTR-int +1 are plasmids expressing two lhRNAs from H1 and 7SK promoters, respectively. These transcriptional regulatory elements were incorporated using a similar procedure to that described above, but genomic DNA from HEK293 cells was used as a template. Forward and reverse primers used during the first round of PCR are listed in Supplementary Table 1. Second round reverse primers were the same as those used to generate U6-driven lhRNAs.
Target plasmids
Dual luciferase sense and antisense target plasmids were constructed by inserting annealed oligonucleotides encoding the HIV-1 targets into the 3′-UTR of the Renilla Luciferase cassette of the psiCheck-2 plasmid (Promega, WI, USA). After treatment with polynucleotide kinase (Promega) sticky end-containing duplex oligonucleotides were ligated to XhoI–NotI sites (sense targets) or XhoI–SpeI sites (antisense targets). Sequences of these oligonucleotides are provided in Supplementary Table 2. Identification of correct clones was facilitated by the introduction of an EcoRV site within each insert.
Cell culture and transfections
HEK293 and Huh7 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; BioWhittaker, MD, USA) supplemented with 10% heat inactivated fetal calf serum (FCS; Delta Bioproducts, Johannesburg, South Africa) at 37°C and 5% CO2. U87.CD4.CCR5 cells were maintained in DMEM supplemented with 15% FCS, 1 µg/ml puromycin, 300 µg/ml G418, glutamine, penicillin and streptomycin. Transfections were carried out using a ratio of 1 µl Lipofectamine2000 (Invitrogen, CA, USA) to 1 µg total DNA according to the manufacturer’s instructions. Medium was changed 24 h after transfection and analyses were carried out a further 24 h thereafter. Co-transfecting a plasmid that constitutively produces enhanced green fluorescent protein (pCI-eGFP) followed by fluorescence microscopy was used to verify equivalent transfection efficiencies (32).
To evaluate the effects of the lhRNA- and shRNA-encoding plasmids on a reporter target, 120 000 HEK293 cells were seeded 24 h prior to transfection in each well of a 24-well culture dish. Cells were transfected with 150 ng target plasmid, 750 ng of lhRNA or shRNA encoding plasmid and 100 ng of pCI-eGFP, unless otherwise stated. For northern blot analysis HEK293 cells were seeded at 80% confluency in 10-cm culture dishes 24 h prior to transfection. Cells were transfected with 18 µg of hairpin-encoding plasmid and 1 µg pCI-eGFP. To determine the induction of interferon (IFN) response-related genes, HEK293 cells were seeded as described above and transfected with 900 ng of lhRNA or shRNA encoding plasmid and 100 ng pCI-eGFP per well. Control double-stranded RNA, Poly (I:C; Sigma, MO, USA), was transfected at equivalent amounts to the hairpin encoding plasmids. Measurement of IFN response gene mRNA concentrations were then determined using quantitative PCR according to previously described methods (33).
To assess saturation of the endogenous miRNA pathway caused by the lhRNA constructs, Huh7 cells were co-transfected with 80 ng psiCheck-miR-16T × 7 (18), 750 ng hairpin expression plasmid or pTZ-U6-miR-16S × 7 sponge and 150 ng pCI-eGFP. psiCheck-miR-16T × 7 is a psiCheck plasmid-containing 7 miR-16 target sites downstream of the Renilla luciferase ORF and pTZ-U6-miR-16S × 7 sponge includes a Pol III expression cassette that transcribes RNA containing seven copies of a miR-16 target site. To determine the effects of the lhRNA constructs on the silencing efficacy of an exogenous miRNA, HEK293 cells were cotransfected with 100 ng pCMV miR-31 HBx, 100 ng of psiCheck-HBx together with the hairpin constructs (18,34). pCMV miR-31 HBx encodes a miRNA shuttle that has a guide cognate in the X gene (HBx) of hepatitis B virus, and psiCheck-HBx is the reporter target vector.
Dual luciferase reporter assay
Firefly and Renilla luciferase activity was determined using a Veritas dual-injection luminometer (Turner Biosystems, C A, USA) according to the instructions of the manufacturer of the Dual-Luciferase Reporter kit (Promega). Target-specific Renilla luciferase expression was normalized to background Firefly luciferase expression. Average expression ratios for a control plasmid were set to 100%, and relative expression for other samples was calculated accordingly. Two independent experiments in triplicate were performed and the data were expressed as the mean ± SD.
Inhibition of gene targets in the full-length HIV luciferase reporter molecular clone
Hairpin expression cassettes were assessed for efficacy against a full-length HIV-1 target by determining knockdown in a HIV-1 molecular reporter pNL4-3.Luc.R-E-, as has been previously described (35,36). This molecular clone has a Firefly luciferase gene inserted into the nef gene and is capable of only a single round of replication. Suppression of viral gene expression was measured by determining Firefly luciferase activity. HEK293 cells (120 000) were co-transfected with 1:1 (150 ng:150 ng) ratio of hairpin expression plasmid to pNL4-3.Luc.R-E-. Approximately 50 ng of phRL-CMV (Promega), which encodes Renila luciferase, was used to control for transfection efficiency. Dual luciferase reporter assays were carried out as described above.
Northern blot analysis
Total RNA, extracted from HEK293 cells 48 h after transfection, was prepared using TriReagentTM (Sigma) according the manufacturer’s instructions. Thirty micrograms of RNA was resolved on urea denaturing 15% polyacrylamide gels and blotted onto nylon membranes. DecadeTM Marker (Ambion, TX, USA) was prepared according the manufacturer’s instructions and run alongside the cellular RNA. Blots were hybridized to DNA oligonucleotides to detect products of hairpin processing. These probes were complementary to regions spanning the target sense or antisense sequences of the hairpins. Probes were labelled at their 5′-ends using [γ-32P] ATP with T4 Polynucleotide kinase then purified using standard procedures. Northern blot hybridization and washing were carried out according to previously described methods (26). After analysis using a FLA-7000 phosphorimager (Fujifilm, Japan), blots were stripped and reprobed. An oligonucleotide probe that was complementary to U6 small nuclear RNA was used as a control to verify equal loading of cellular RNA. Sense probe oligonucleotide sequences were the following: tat(S) probe: 5′-GCG GAG ACA GCG ACG AAG AGC-3′, nef(S) probe: 5′-GTG CCT GGC TAG AAG CAC AAG -3′, LTR(S) probe: 5′-GTA ACT AGA GAC CTC TCA GAC-3′, int(S) probe: 5′-GCC GGA GAG CAA TGG CTA GTC-3′. Antisense probe oligonucleotide sequences were: tat (AS) probe: 5′-GCT CTT CGT CGC TGT CTC CGC-3′; nef (AS) probe: 5′-CTT GTG CTT CTA GCC AGG CAC-3′; LTR (AS) probe: 5′-GTC TGA GAG GTC TCT AGT TAC-3′; and int (AS) probe: 5′-GAC TAG CCA TTG CTC TCC GGC-3′.
RESULTS
Optimal design of Pol III-driven single lhRNA cassettes encoding two unique siRNA sequences
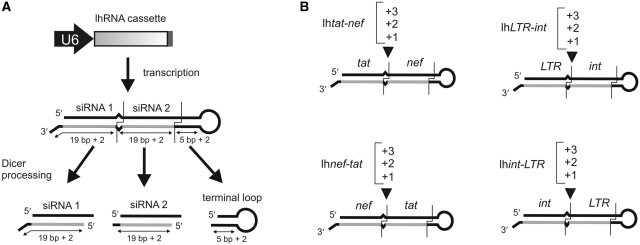
We and others have previously established that Pol III-expressed lhRNA sequences, which include a 2–3 nt uridine 3′-end overhang, can generate independent effective siRNAs in cells (19,21). However, Dicer cleavage is initiated at the base of the hairpin duplex, and proceeds towards the loop with decreasing cleavage efficiency. Previously, Liu et al. (21) have shown that two rounds of Dicer cleavage can yield two effective siRNAs when lhRNAs are generated from a H1 Pol III promoter. The arrangement of each siRNA along a hairpin duplex was optimal for 44 and 45 bp stems (21), where siRNA precursors of 19 bp are separated by 3–4 bp. To avoid second Dicer cleavage occurring within the terminal loop, we generated lhRNAs with a longer 48–50 bp complementary duplex sequence and a 9 nt loop (Figure 1A). We designed several sets of U6 lhRNA expression plasmids that each encode two siRNA sequences. The intended siRNAs targeted four unique HIV-1 sites within tat (29), nef (23), LTR (20) and int (30), which have previously been shown to be susceptible to RNAi-mediated silencing (Figure 1A and B). In total, four different series of lhRNAs were generated: two sets of lhRNAs targeted against tat and nef and two sets targeted against int and the LTR, allowing for each siRNA-encoding sequence to be situated in the stem base or loop side positions of the hairpin duplex (Figure 1B). The position of these putative 19 bp +2 nt siRNAs were adjusted within each of the four lhRNA series by inserting 1, 2 or 3 symmetrical paired mismatched bases at the junction between first and second siRNAs (Figure 1B). In addition, four U6-expressing shRNA controls were generated for each target. All lhRNAs and shRNAs included G:U pairings that were introduced at regular intervals along the sense strand of the duplex. The antisense sequence was retained to ensure complete hybridization to each intended target RNA. The addition of G:U wobbles facilitated propagation of lhRNA expressing plasmids in Escherichia coli (24,25).
Figure 1.
Design of dual targeting lhRNAs. (A) Schematic representation of an lhRNA expression cassette showing upstream U6 promoter and the predicted structures of the transcribed lhRNAs. Dual targeting lhRNAs encode two 19 bp +2 nt siRNAs and have a 5 bp +2 nt spacer before the loop sequence. (B) Four different series of 48–50 bp lhRNAs were generated that target tat and nef as well as int and LTR. Each 19 bp +2 nt siRNA-encoding sequence is positioned either in the stem or loopside of the lhRNA duplex and separated by 1, 2 or 3 mismatched paired bases.
The processing and silencing efficacy of two siRNAs derived from a single U6-driven lhRNA in cell culture
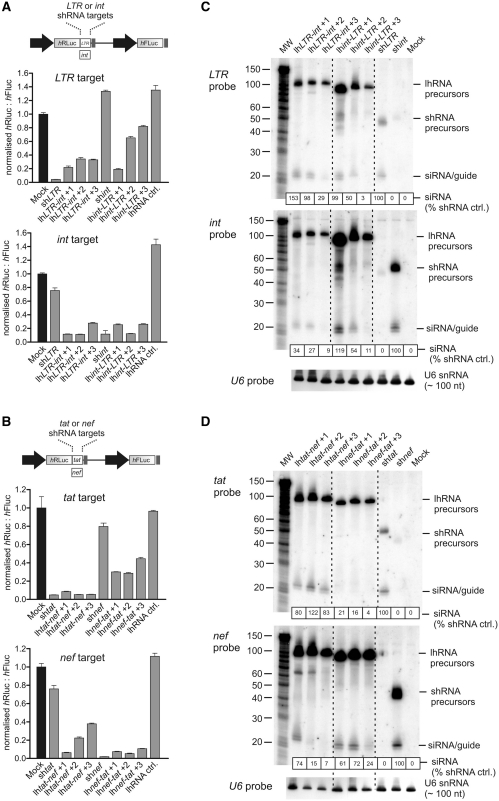
To assess the ability of variable length dual targeting lhRNAs to inhibit their targets in cultured cells, HEK293 cells were transfected with lhRNAs (Figure 1B) or with individual shRNA expression plasmids. These hairpin constructs were transiently co-transfected with a psiCheck dual luciferase target reporter plasmid containing the tat, nef, LTR or int target sequence within the 3′-UTR of the human Renilla luciferase (hRLuc) transcript (top panel Figure 2A and B). A lhRNA targeted against the HBx gene of HBV (26) was used as a negative lhRNA control. Knockdown was determined according to the ratios of Renilla to Firefly luciferase activities and values were normalized relative to that obtained after co-transfection with the empty U6 vector pTZU6+1 (Mock). Regardless of the spacing at the siRNA junctions, or the relative arrangement of the siRNA sequence, siRNAs in the first position of the hairpin (at the base of the duplex) were consistently capable of suppressing their respective targets by ∼80%, which is comparable to the knockdown achieved by individual control shRNAs. Target inhibition from the siRNA derived from the loopside position of the hairpin duplex was most efficient when only one mismatched base pair was present between siRNA encoding regions (+1 configuration). A gradual decrease in efficacy was observed with increased spacing at the junction of the first and second siRNAs (Figure 2A and B).
Figure 2.
Knockdown efficacy and processing of dual targeting lhRNAs. Dual luciferase reporter assays showing knockdown of the LTR and int targets (A) and tat and nef targets (B) when the target sequence was inserted downstream of the Renilla luciferase (hRLuc) open reading frame. Values represented are mean ratios of Renilla to Firefly luciferase (n = 3, SEM) and are normalized to cells transfected with a plasmid containing a U6 promoter only with no RNAi effector sequence (mock). Small RNA (PAGE) northern blot analysis was carried out on total RNA extracted from cells transfected with the indicated transcripts. Labelled probes complementary to the guide strand of LTR and int (C) or tat and nef (D) were hybridized to immobilized RNA and exposed to a phosphorimaging plate. lhRNA and shRNA precursor RNA as well as processed siRNAs are indicated. The amount of processed guide strand is shown and normalized for each blot relative to the shRNA (set at 100). Decade MarkerTM indicates fragment size and a probe complementary to small nuclear U6 RNA was used to detect U6 snRNA as a loading control.
siRNA and hairpin-intermediates derived from long processed hairpin precursors were analysed by using small RNA northern blot hybridization carried out on total RNA extracted from transfected HEK293 cells. Figure 2C and D show representative blots using four probes complementary to each of the intended siRNA guide strands. The signal for siRNAs derived from the first position of the lhRNAs with a +1 configuration was similar to that of the corresponding shRNA cassette. With the exception of the lhtat-nef series, the guide signal of the first position siRNA decreased with an increase in the mismatched paired bases inserted between the putative siRNAs of the hairpin duplex. For example, in the case of the LTR and int probes, very weak guide hybridization signals were detected when three mismatched bases were inserted between the siRNA-encoding regions. With the exception of lhnef-tat, processed products from the second position of the hairpin were easily detected after transfection of plasmids encoding lhRNAs with a +1 configuration. The tat siRNA from the lhnef-tat set was not detected for all spacing arrangements and correlates with poor knockdown that was observed with tat reporter targets (Figure 2A and B). The decrease in siRNA production from the second position was observed in a gradient fashion; an increase in mismatched paired bases at the junction between siRNA encoding sequences resulted in a decreased detection of processed second-position guide strand. This result is in accordance with the gradient in inhibition observed for the psiCheck reporter gene targets (Figure 2A and B) and previously reported for longer hairpins (19). When only one pair of mismatched bases was inserted at the junction, siRNAs in the second position were detected at similar levels to those of siRNAs at the base of the stem. It should be noted that 2–3 bands, differing in size by 1 nt, are often visible for single siRNA guide strands. This indicates that Dicer does not consistently cleave the duplex at the same position and therefore generates guide strands ranging in size from 19–22 nt. While the potency of individual shRNAs differ (in the order: shtat > shnef >> shint > shLTR), reporter gene inhibition was effective for both siRNAs along the duplex, even when transfecting with decreasing concentrations of hairpin-expressing plasmid (Supplementary Figure 1), using different Pol III promoters (H1 and 7SK), and when combining two different hairpin expression cassettes in a single vector (Supplementary Figure 2A). Thus lhRNAs lhtat-nef +1 and lhLTR-int +1 are optimally designed to allow for efficient processing along the entire hairpin duplex to produce a strong dual siRNA response.
The generation of four independent and effective siRNAs from a single U6-driven double lhRNA (dlhRNA) expression cassette
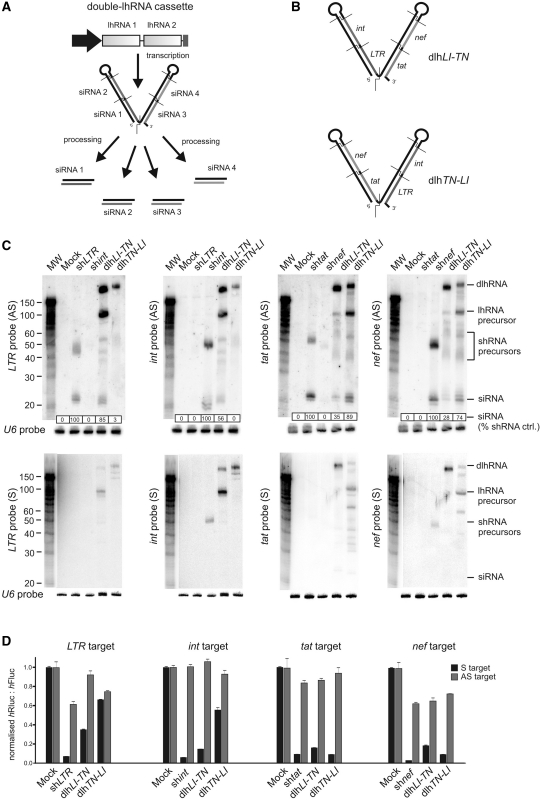
Placement of two shRNAs together within the same transcript has been shown to produce siRNAs targeted to two independent sites (37,38). Although Pol III-expressed shRNAs and lhRNAs consist of defined 5′- and 3′-termini that facilitate Dicer recognition and cleavage, shRNAs expressed from Pol II promoters produce longer 5′-leader and 3′-trailer sequences which often results in variable production of active siRNAs (39–41). To develop a system for generating four separate siRNAs from a single transcript, we combined the two most effective lhRNAs described here. lhtat-nef +1 and lhLTR-int +1 were placed in tandem to generate two dlhRNA expression constructs (Figure 3A). A 2-nt UU bridge was included between each lhRNA to mimic the 3′-overhang generated for a single Pol III-generated lhRNA. dlhLI-TN and dlhTN-LI included lhRNAs lhLTR-int +1 and lhtat-nef +1 in either the 5′-position or 3′-position of the dlhRNA transcripts, respectively (Figure 3B).
Figure 3.
Generation of four effective individual siRNAs from a single dlhRNA transcript containing two adjacent lhRNAs. (A) Schematic representation of a dlhRNA expression cassette driven by a single promoter showing the predicted structure and derivation of four siRNAs. (B) Effective dual targeting long hairpin RNAs lhtat-nef +1 and lhLTR-int +1 were both combined in 5′ or 3′ positions within the dlhRNA transcript to generate lhLI-TN and lhTN-LI. (C) Low molecular weight northern blot analysis was carried out on total RNA extracted from cells transfected with the dlhRNA expression cassettes with individual targeting shRNAs used as positive controls. Labelled probes complementary to the guide and antisense strand of LTR, int, tat and nef were hybridized to immobilized RNA and exposed to a phosphorimaging plate. Precursor hairpin RNAs as well as processed siRNAs are indicated. The amount of processed guide strand is shown and normalized for each blot relative to the shRNA (set at 100). Decade MarkerTM indicates fragment size and a probe complementary to small nuclear U6 RNA was used to detect U6 snRNA as a loading control. (D) Dual luciferase reporter assays showing knockdown of the sense (S) and antisense (AS) targets of LTR, int, tat and nef when the target sequence was inserted downstream of the Renilla luciferase open reading frame. Values represented are mean ratios of Renilla to Firefly luciferase (n = 3, ± SEM) and are normalized to cells transfected with a plasmid containing a U6 promoter only with no RNAi effector sequence (mock).
To characterize the processing of dlhRNAs, small RNA northern blot analysis was performed as before. Figure 3C shows the signals obtained following hybridization with antisense (AS) and sense (S) probes to detect siRNA guide and passenger strands. All probes detected the full-length dlhRNA transcripts from both dlhRNA constructs. However, only dlhLI-TN generated detectable processed precursors representing both lhRNAs. For dlhTN-LI, strong signals were detected for the 5′-position lhRNA precursor, lhtat-nef +1, with S and AS probes to tat and nef. No hybridization signal was detected by either S or AS probes to LTR and int for the 3′-position lhLTR-int +1, suggesting that this lhRNA precursor is rapidly degraded following initial Dicer cleavage of the dlhRNA transcript. For dlhLI-TN, the 5′-position lhRNA, lhLTR-int +1, was detected by S and AS probes targeted to LTR and int. The lhRNA in the 3′-position (lhtat-nef +1) was detected by S and AS tat and nef probes, but at a lower concentration than that of the 5′-position lhRNA precursor. Again this suggests that processing of the dlhRNA renders lhRNA at the 3′-position less stable. Nevertheless, all four siRNAs guide sequences were detected from dlhLI-TN demonstrating that a single dlhRNA construct is capable of successfully generating four independent siRNA guide strands.
To determine whether the guide strands were capable of effecting knockdown of defined HIV targets, a dual-luciferase reporter gene assay was performed as before. Double long hairpin RNA dlhLI-TN inhibited all four targets by 70–80% (Figure 3D), indicating that four effective siRNAs were generated from this dlhRNA and that a decreased amount of tat and nef guide strand derived from the second lhRNA did not affect knockdown under these conditions of transient transfection. As expected, dlhTN-LI, only inhibited tat and nef reporter targets, confirming that compromised processing of the second position lhRNA affects downstream guide strand production and subsequent target knockdown. To determine whether the passenger strand of each putative processed siRNA was active, dual-luciferase reporter constructs were generated that included AS targets. The passenger strands were largely ineffective, and guide strand formation was according to the intended bias.
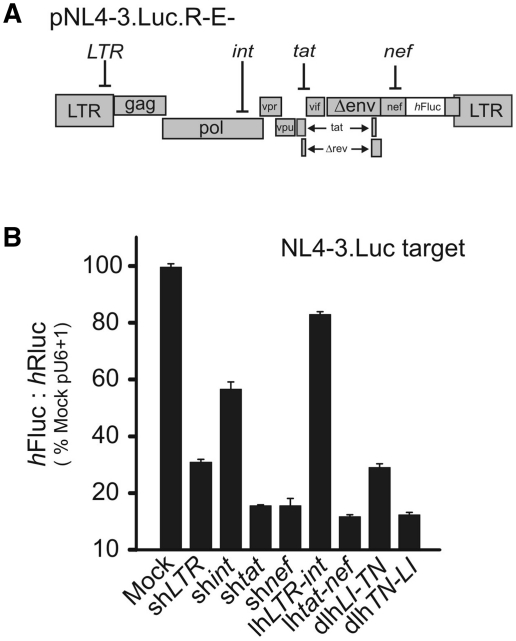
Ability of the dlhRNAs to inhibit cognate targets within the context of a full-length HIV-1 sequence was also determined (Figure 4A and B). To quantify the anti-viral effects of these hairpins, we co-transfected the shRNA-, lhRNA- and dlhRNA-expressing plasmids with the HIV-1 molecular clone pNL4-3.Luc.E-R-. This reporter plasmid lacks functional env and the nef reading frame is substituted with a Firefly luciferase ORF (Figure 4A). Knockdown was measured by determining Firefly luciferase activity, which was normalized to activity of Renilla luciferase that was consitutively expressed from a co-transfected plasmid. Luciferase ratios were determined relative to that of a mock control vector (Figure 4B). The individual shRNAs targeted to the tat and nef sequences showed potent luciferase inhibition (>80%), whilst shRNAs targeted to LTR and int were less effective (70% and 40% knockdown, respectively). This may reflect a lower potency of individual LTR and int siRNAs (Supplementary Figure 1). As expected, inhibition of the HIV-1 reporter by lhRNAtat-nef was similar to that achieved by each of the tat or nef shRNAs. However, the dual targeting hairpin lhLTR-int was only capable of inhibiting the HIV-1 reporter by ∼20%. This confirms previous observations that LTR and int targets were silenced less effectively when produced from lhLTR-int. Importantly, both double lhRNA expression cassettes achieved good silencing of the reporter gene. However, dlhTN-LI achieved slightly better silencing than dlhLI-TN. This may be a result of better knockdown achieved by the more efficiently processed nef- and tat-targeting guides (Figure 3C). Collectively these data indicate that although there may be variability in the silencing efficacy of individual siRNAs, compensatory effects may result in good overall silencing which is a desirable property of coRNAi applications.
Figure 4.
Inhibition of full-length HIV-1 molecular clone pNL4-3.Luc.R-E-. (A) The separate regions of the NL4-3.Luc sequence targeted by the four generated siRNAs are shown schematically. (B) HEK293 cells were transfected in a 1:1 ratio of pNL4-3.Luc.R-E- together with indicated hairpin constructs and trace amounts of Renilla luciferase plasmid phRL-CMV. The four siRNA-targeted regions are indicated above in the modified HIV genome of pNL4-3.Luc.R-E-. Values represented are mean ratios of Firefly luciferase normalized to Renilla luciferase (n = 3, ±SEM) and expressed as a percentage of mock (pU6 + 1) transfected cells (set at 100%).
lhRNA cassettes do not disrupt independent RNAi-mediated gene silencing or stimulate the innate IFN response
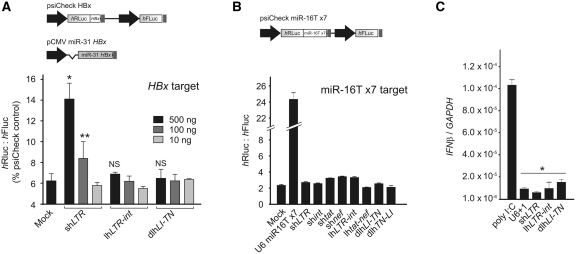
An important safety concern of potentially therapeutic lhRNAs is the possible disruptive effect that they may have on the endogenous cellular miRNA pathway. Both exogenous siRNAs and shRNAs can compete with each other or with endogenous miRNAs for access to components of the RNAi machinery (42,43). However, RNAi activators that are present in lower concentrations, are less likely to saturate the RNAi pathway (34,43,44). To assess effects of lhRNA cassettes on independent RNAi, two assays were performed to determine whether lhRNA expression cassettes have any derepression effect on silencing caused by an endogenous or an exogenous miRNA. In the first assay, HEK293 cells were co-transfected in a 1:1 ratio with an exogenous miRNA shuttle, pCMV miR-31 HBx (targeted to a unique site within the HBV genome), and its cognate dual-luciferase reporter target plasmid, psiCheck HBx (34). Diminishing amounts of plasmids expressing shRNA, lhRNA or dlhRNAs relative to pCMV miR-31 HBx were co-transfected in ratios of 5:1, 1:1 and 0.1:1, respectively. U6 promoter-driven shLTR showed significant derepression of miR-31 HBx mediated knockdown at ratios of 5:1 and 1:1 but not at a ratio of 0.1:1. However, equivalent ratios of U6 promoter-driven lhLTR-int or dlhLI-TN had no effect on miR-31 HBx knockdown efficacy (Figure 5A). In the second assay, Huh-7 cells were co-transfected with a panel of hairpin-expressing plasmids together with a dual-luciferase reporter plasmid containing seven copies of a natural miR-16 target site (18). A miRNA ‘sponge’ construct expressing seven copies of an imperfectly matched miR-16 target, pU6 miR16Tx7, significantly derepressed miR-16 silencing of its reporter cognate (Figure 5B). However, at a concentration of 5:1 (sh/lhRNA expression cassette:target reporter plasmid), none of the hairpin RNAs had any detectable disruptive effect on endogenous miR-16 silencing of its reporter cognate. Together, these results suggest that lhRNAs and dlhRNAs, although expressed from a potent U6 promoter, are less likely than shRNAs to compete with RNAi pathway components necessary for either exogenous or endogenous miRNA function. Although lhRNA and shRNA expression cassettes are expressed from U6 Pol III promoters, intracellular precursors and siRNA guide strands from lhRNA cassettes are present at lower concentrations than those derived from shRNA expression cassettes (Figure 3C). Lastly, to exclude the possibility of non-specific effects caused by the induction of an interferon response, IFN-β mRNA concentrations were measured in transfected cells. As previously shown for other Pol-III generated lhRNAs (21,22,26), quantitative qRT–PCR demonstrated that none of the hairpin cassettes induced expression of IFN-β (Figure 5C).
Figure 5.
Assessment of non-specific effects mediated by long hairpin RNAs. (A) Analysis of effects of a shRNA, lhRNA and dlhRNA expression cassette on the repression of HBx target reporter sequence by an exogenously introduced miR-31 HBx shuttle using a dual luciferase assay. Co-transfection of reporter plasmid, containing an HBx target sequence downstream of the Renilla luciferase ORF, was carried out together with three different concentrations of RNAi expression cassettes and empty backbone plasmid (mock). Mean ratios of Renilla to Firefly luciferase (as a percentage of psiCheck2 empty backbone vector) were used to determine derepression of miR-31 HBx. Statistical significance was determined using a one-way ANOVA relative to mock transfected control (pU6 +1, *P < 0.05 and **P < 0.001). (B) The effect of hairpin expression cassettes on the function of endogenous miR-16 was analysed following co-transfection of a dual luciferase reporter plasmid containing seven copies of the miR-16 target downstream of the hRluc ORF together with the indicated hairpin-expressing plasmids or miR-16 sponge plasmid expressing seven copies of an imperfectly matched miR-16 target. Mean ratios of Renilla to Firefly luciferase were used to determine derepression of miR-16. (C) The induction of the IFN response was assessed by measuring IFN-β mRNA concentration in total RNA extracted from cells transfected with the indicated shRNA, lhRNA and multi-lhRNA expression cassettes. Poly I:C served as a positive control. Mean normalized ratios of IFN-β:GAPDH (n = 3, ± SEM) determined by using quantitative RT–PCR are indicated. Statistical significance was determined using a one-way ANOVA relative to mock transfected control (pU6 +1, *P < 0.001).
DISCUSSION
Despite extensive optimization of hairpin stem length, siRNA sequence, and the spatial arrangement of unique siRNAs along a lhRNA duplex, it seemed unlikely that expressed lhRNAs can be designed to produce more than two, possibly three, separate effective siRNAs (19,21,27). Deriving high concentrations of more than two independent functional siRNAs from an lhRNA scaffold remains difficult and requires new ways of exploiting the processing of dsRNA substrates by Dicer. To develop effective Pol III-driven lhRNAs expressing two functional siRNAs, we tested the effects of combining two 19 bp +2 nt siRNAs within a single 48–50 bp lhRNA duplex, and included up to three mismatched paired bases between each siRNA-encoding sequence. Liu et al. (21) showed that a single mismatched paired base between two effective siRNAs at the centre of the lhRNA stem is well tolerated and results in the same level of processing or siRNA activity within the context of a 43 bp lhRNA. In total, we established a panel of four unique anti-HIV siRNAs was used to generate 12 dual-targeting lhRNA structures. When tested against respective HIV targets, we observed an inverse correlation between siRNA silencing potency and increased spacing between each siRNA along the duplex. Optimal siRNA processing from lhRNAs occurred when only one mismatched paired base was placed between each 19 bp + 2 nt siRNA, and this was irrespective of siRNA position or sequence. These data are in accordance with previously published data reporting on dual-targeting lhRNAs (21), and is in accordance with predicted Dicer cleavage intervals of ∼22 nt (20 bp +2 nt) in human cells (45). With some dual-acting lhRNAs, processing of the siRNA at the first position diminished when more mismatches were inserted in the lhRNA duplex at the junction of each the siRNAs. This is likely to be caused by an inhibitory effect on processing which is caused by bulges occurring at the Dicer cleavage sites (46). However, the +1 configuration did not affect Dicer processing through the lhRNA duplex. Importantly, we were able to identify two effective and optimized dual-targeting anti-HIV lhRNAs, lhtat-nef +1 and lhLTR-int +1, which together produce high levels of four siRNAs that inhibit their cognate targets. These lhRNAs were ideally optimized for inclusion into a single combinatorial dlhRNA expression cassette.
Based on HIV reverse-transcriptase error rates, it has been determined that a minimum of four separate HIV target sites should be targeted simultaneously to prevent the emergence of RNAi-resistant viral species (47,48). Therefore, it was encouraging that the dlhRNA design, with two highly effective lhRNAs joined together within a single expressed transcript, enabled accurate processing into four active anti-HIV siRNAs. Apart from polycistronic miRNA mimics (13–17), this is the first example of four active guide strands being derived from a single Pol III expression cassette, and provides a useful framework for generating effective coRNAi strategies against highly evolving viruses or multiple rogue genetic elements. Nevertheless, the mechanism by which these dlhRNA structures are processed is unclear. Although previous attempts to generate binary or dual shRNA-containing transcripts suggest that these duplexes can be processed into siRNA-sized products, the mechanism for their intracellular cleavage remains unexplained (37,38). Although we do not exclude involvement of other RNases in the processing of dlhRNA precursors, it seems likely that Dicer is responsible for the initial cleavage to form lhRNA species. This is supported by evidence that Dicer is capable of cleaving hairpins with either 5′- and 3′-extensions, albeit less efficiently (39–41). The fact that the 3′-position lhRNA is present in reduced amounts (or degraded completely) suggests that, once cleaved, the 5′-position lhRNAs is initially protected by Dicer before being processed into shorter hairpin products. Since the intrinsic stability of the two lhRNAs is the same within the dlhRNA system, it remains odd that the 3′-position lhRNA in dlhLI-TN is processed but is degraded in the context of dlhTN-LI. The only difference appears to be the sequence presented for second round Dicer cleavage. Moreover, the 5′-terminus of the lhLTR-int within dlhLI-TN has a triphosphate moiety generated by Pol III transcription, a feature which is lacking for lhLTR-int in the context of dlhTN-LI, and which may add to the stability of lhLTR-int. However a clearer picture is likely to emerge with the study of more dlhRNA combinations comprising different lhRNAs.
Although coRNAi aims to induce strong silencing from multiple guide strands, high levels of shRNA produced from Pol III promoters are known to be associated with unwanted cellular toxicities. High levels of expressed shRNAs in the liver may cause fatality in mice as a result of saturation of the endogenous RNAi machinery (10). In addition, McBride and colleagues have observed toxicities caused by shRNA-based vectors in brain, which may have been caused by a buildup of guide strand RNA (49,50). Replacement of shRNAs with miRNA shuttles reduced neurotoxicities, suggesting that natural RNAi pathway precursors are less likely to interfere with endogenous miRNA functions. Although not fully understood, it is likely that highly expressed shRNAs abrogate the function of natural and exogenous miRNAs (10,34,43,44,51,52). Here, we have shown that both lhRNAs and dlhRNAs do not induce the same disruptive effects on endogenous miRNA that were observed after transfection of a U6-generated shRNA. This augurs well for the safety and potential therapeutic application of these constructs. Moreover, although there have been concerns about induction of the IFN response by long (>30 bp) duplex RNA, we did not detect any IFN-β mRNA activation for any of the hairpins tested. This confirms previously reported results from analysis of expressed lhRNAs (24,25,53), and suggests that intracellular transcription of dsRNA hairpins is less likely than exogenous synthetic RNA to activate the type 1 IFN response. This is explained by the fact that expressed lhRNAs, unlike transfected synthetic duplex RNA, do not traverse the endosome which contain Toll-like receptors that are typically activated by introduced siRNAs (54,55).
In conclusion, we show that RNA Pol III-expressed dlhRNA transcripts may be processed to generate four independent siRNAs that can effect significant knockdown of non-contiguous siRNA-susceptible regions of HIV-1. Although there is some variation in the processing efficiency of the 3′-lhRNA, effective coRNAi can be achieved. Importantly, the dlhRNA constructs described here do not appear to disrupt the natural miRNA pathway, which represents an important objective for their implementation as potential therapeutic agents. This design of dlhRNA cassettes improves on the limited versatility of expressed lhRNAs and provides a useful approach for generating transcripts with modular lhRNAs motifs that achieve effective coRNAi in mammalian cells.
FUNDING
South African National Research Foundation (NRF); South African Medical Research Council (MRC); Poliomyelitis Research Foundation (PRF); Sheena Saayman is a recipient of a Stella and Paul Loewenstein Studentship.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rossi JJ. Expression strategies for short hairpin RNA interference triggers. Hum. Gene Ther. 2008;19:313–317. doi: 10.1089/hum.2008.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henry SD, van der Wegen P, Metselaar HJ, Tilanus HW, Scholte BJ, van der Laan LJ. Simultaneous targeting of HCV replication and viral binding with a single lentiviral vector containing multiple RNA interference expression cassettes. Mol. Ther. 2006;14:485–493. doi: 10.1016/j.ymthe.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 4.ter Brake O, Konstantinova P, Ceylan M, Berkhout B. Silencing of HIV-1 with RNA interference: a multiple shRNA approach. Mol. Ther. 2006;14:883–892. doi: 10.1016/j.ymthe.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 5.McIntyre GJ, Groneman JL, Tran A, Applegate TL. An infinitely expandable cloning strategy plus repeat-proof PCR for working with multiple shRNA. PLoS ONE. 2008;3:e3827. doi: 10.1371/journal.pone.0003827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng TL, Teng CF, Tsai WH, Yeh CW, Wu MP, Hsu HC, Hung CF, Chang WT. Multitarget therapy of malignant cancers by the head-to-tail tandem array multiple shRNAs expression system. Cancer Gene Ther. 2009;16:516–531. doi: 10.1038/cgt.2008.102. [DOI] [PubMed] [Google Scholar]
- 7.Song J, Giang A, Lu Y, Pang S, Chiu R. Multiple shRNA expressing vector enhances efficiency of gene silencing. BMB Rep. 2008;41:358–362. doi: 10.5483/bmbrep.2008.41.5.358. [DOI] [PubMed] [Google Scholar]
- 8.ter Brake O, te Hooft K, Liu YP, Centlivre M, von Eije KJ, Berkhout B. Lentiviral vector design for multiple shRNA expression and durable HIV-1 inhibition. Mol. Ther. 2008;16:557–564. doi: 10.1038/sj.mt.6300382. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez S, Castanotto D, Li H, Olivares S, Jensen MC, Forman SJ, Rossi JJ, Cooper LJ. Amplification of RNAi–targeting HLA mRNAs. Mol. Ther. 2005;11:811–818. doi: 10.1016/j.ymthe.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 10.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 11.Jetzt AE, Yu H, Klarmann GJ, Ron Y, Preston BD, Dougherty JP. High rate of recombination throughout the human immunodeficiency virus type 1 genome. J. Virol. 2000;74:1234–1240. doi: 10.1128/jvi.74.3.1234-1240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.An W, Telesnitsky A. Frequency of direct repeat deletion in a human immunodeficiency virus type 1 vector during reverse transcription in human cells. Virology. 2001;286:475–482. doi: 10.1006/viro.2001.1025. [DOI] [PubMed] [Google Scholar]
- 13.Aagaard LA, Zhang J, von Eije KJ, Li H, Saetrom P, Amarzguioui M, Rossi JJ. Engineering and optimization of the miR-106b cluster for ectopic expression of multiplexed anti-HIV RNAs. Gene Ther. 2008;15:1536–1549. doi: 10.1038/gt.2008.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung KH, Hart CC, Al-Bassam S, Avery A, Taylor J, Patel PD, Vojtek AB, Turner DL. Polycistronic RNA polymerase II expression vectors for RNA interference based on BIC/miR-155. Nucleic Acids Res. 2006;34:e53. doi: 10.1093/nar/gkl143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu YP, Haasnoot J, ter Brake O, Berkhout B, Konstantinova P. Inhibition of HIV-1 by multiple siRNAs expressed from a single microRNA polycistron. Nucleic Acids Res. 2008;36:2811–2824. doi: 10.1093/nar/gkn109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun D, Melegari M, Sridhar S, Rogler CE, Zhu L. Multi-miRNA hairpin method that improves gene knockdown efficiency and provides linked multi-gene knockdown. Biotechniques. 2006;41:59–63. doi: 10.2144/000112203. [DOI] [PubMed] [Google Scholar]
- 17.Zhou H, Huang C, Xia XG. A tightly regulated Pol III promoter for synthesis of miRNA genes in tandem. Biochim. Biophys. Acta. 2008;1779:773–779. doi: 10.1016/j.bbagrm.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ely A, Naidoo T, Arbuthnot P. Efficient silencing of gene expression with modular trimeric Pol II expression cassettes comprising microRNA shuttles. Nucleic Acids Res. 2009;37:e91. doi: 10.1093/nar/gkp446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saayman S, Barichievy S, Capovilla A, Morris KV, Arbuthnot P, Weinberg MS. The efficacy of generating three independent anti-HIV-1 siRNAs from a single U6 RNA Pol III-expressed long hairpin RNA. PLoS ONE. 2008;3:e2602. doi: 10.1371/journal.pone.0002602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barichievy S, Saayman S, von Eije KJ, Morris KV, Arbuthnot P, Weinberg MS. The inhibitory efficacy of RNA POL III-expressed long hairpin RNAs targeted to untranslated regions of the HIV-1 5′ long terminal repeat. Oligonucleotides. 2007;17:419–431. doi: 10.1089/oli.2007.0095. [DOI] [PubMed] [Google Scholar]
- 21.Liu YP, Haasnoot J, Berkhout B. Design of extended short hairpin RNAs for HIV-1 inhibition. Nucleic Acids Res. 2007;35:5683–5693. doi: 10.1093/nar/gkm596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sano M, Li H, Nakanishi M, Rossi JJ. Expression of long anti-HIV-1 hairpin RNAs for the generation of multiple siRNAs: advantages and limitations. Mol. Ther. 2008;16:170–177. doi: 10.1038/sj.mt.6300298. [DOI] [PubMed] [Google Scholar]
- 23.Nishitsuji H, Kohara M, Kannagi M, Masuda T. Effective suppression of human immunodeficiency virus type 1 through a combination of short- or long-hairpin RNAs targeting essential sequences for retroviral integration. J. Virol. 2006;80:7658–7666. doi: 10.1128/JVI.00078-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akashi H, Miyagishi M, Yokota T, Watanabe T, Hino T, Nishina K, Kohara M, Taira K. Escape from the interferon response associated with RNA interference using vectors that encode long modified hairpin-RNA. Mol. Biosyst. 2005;1:382–390. doi: 10.1039/b510159j. [DOI] [PubMed] [Google Scholar]
- 25.Watanabe T, Sudoh M, Miyagishi M, Akashi H, Arai M, Inoue K, Taira K, Yoshiba M, Kohara M. Intracellular-diced dsRNA has enhanced efficacy for silencing HCV RNA and overcomes variation in the viral genotype. Gene Ther. 2006;13:883–892. doi: 10.1038/sj.gt.3302734. [DOI] [PubMed] [Google Scholar]
- 26.Weinberg MS, Ely A, Barichievy S, Crowther C, Mufamadi S, Carmona S, Arbuthnot P. Specific inhibition of HBV replication in vitro and in vivo with expressed long hairpin RNA. Mol. Ther. 2007;15:534–541. doi: 10.1038/sj.mt.6300077. [DOI] [PubMed] [Google Scholar]
- 27.Liu YP, von Eije KJ, Schopman NC, Westerink JT, Brake O, Haasnoot J, Berkhout B. Combinatorial RNAi against HIV-1 using extended short hairpin RNAs. Mol. Ther. 2009;17:1712–1723. doi: 10.1038/mt.2009.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castanotto D, Li H, Rossi JJ. Functional siRNA expression from transfected PCR products. RNA. 2002;8:1454–1460. doi: 10.1017/s1355838202021362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee NS, Dohjima T, Bauer G, Li H, Li MJ, Ehsani A, Salvaterra P, Rossi J. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 2002;20:500–505. doi: 10.1038/nbt0502-500. [DOI] [PubMed] [Google Scholar]
- 30.Westerhout EM, Vink M, Haasnoot PC, Das AT, Berkhout B. A conditionally replicating HIV-based vector that stably expresses an antiviral shRNA against HIV-1 replication. Mol. Ther. 2006;14:268–275. doi: 10.1016/j.ymthe.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 31.Bertrand E, Castanotto D, Zhou C, Carbonnelle C, Lee NS, Good P, Chatterjee S, Grange T, Pictet R, Kohn D, et al. The expression cassette determines the functional activity of ribozymes in mammalian cells by controlling their intracellular localization. RNA. 1997;3:75–88. [PMC free article] [PubMed] [Google Scholar]
- 32.Passman M, Weinberg M, Kew M, Arbuthnot P. In situ demonstration of inhibitory effects of hammerhead ribozymes that are targeted to the hepatitis Bx sequence in cultured cells. Biochem. Biophys. Res. Commun. 2000;268:728–733. doi: 10.1006/bbrc.2000.2209. [DOI] [PubMed] [Google Scholar]
- 33.Carmona S, Ely A, Crowther C, Moolla N, Salazar FH, Marion PL, Ferry N, Weinberg MS, Arbuthnot P. Effective inhibition of HBV replication in vivo by anti-HBx short hairpin RNAs. Mol. Ther. 2006;13:411–421. doi: 10.1016/j.ymthe.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 34.Ely A, Naidoo T, Mufamadi S, Crowther C, Arbuthnot P. Expressed anti-HBV primary microRNA shuttles inhibit viral replication efficiently in vitro and in vivo. Mol. Ther. 2008;16:1105–1112. doi: 10.1038/mt.2008.82. [DOI] [PubMed] [Google Scholar]
- 35.Connor RI, Chen BK, Choe S, Landau NR. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 36.He J, Choe S, Walker R, Di Marzio P, Morgan DO, Landau NR. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J. Virol. 1995;69:6705–6711. doi: 10.1128/jvi.69.11.6705-6711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson J, Banerjea A, Akkina R. Bispecific short hairpin siRNA constructs targeted to CD4, CXCR4, and CCR5 confer HIV-1 resistance. Oligonucleotides. 2003;13:303–312. doi: 10.1089/154545703322616989. [DOI] [PubMed] [Google Scholar]
- 38.Leirdal M, Sioud M. Gene silencing in mammalian cells by preformed small RNA duplexes. Biochem. Biophys. Res. Commun. 2002;295:744–748. doi: 10.1016/s0006-291x(02)00736-2. [DOI] [PubMed] [Google Scholar]
- 39.Giering JC, Grimm D, Storm TA, Kay MA. Expression of shRNA from a tissue-specific pol II promoter is an effective and safe RNAi therapeutic. Mol. Ther. 2008;16:1630–1636. doi: 10.1038/mt.2008.144. [DOI] [PubMed] [Google Scholar]
- 40.Xia H, Mao Q, Paulson HL, Davidson BL. siRNA-mediated gene silencing in vitro and in vivo. Nat. Biotechnol. 2002;20:1006–1010. doi: 10.1038/nbt739. [DOI] [PubMed] [Google Scholar]
- 41.Denti MA, Rosa A, Sthandier O, De Angelis FG, Bozzoni I. A new vector, based on the PolII promoter of the U1 snRNA gene, for the expression of siRNAs in mammalian cells. Mol. Ther. 2004;10:191–199. doi: 10.1016/j.ymthe.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 42.Koller E, Propp S, Murray H, Lima W, Bhat B, Prakash TP, Allerson CR, Swayze EE, Marcusson EG, Dean NM. Competition for RISC binding predicts in vitro potency of siRNA. Nucleic Acids Res. 2006;34:4467–4476. doi: 10.1093/nar/gkl589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keck K, Volper EM, Spengler RM, Long DD, Chan CY, Ding Y, McCaffrey AP. Rational design leads to more potent RNA Interference against hepatitis B virus: factors effecting silencing efficiency. Mol. Ther. 2009;17:538–547. doi: 10.1038/mt.2008.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Castanotto D, Sakurai K, Lingeman R, Li H, Shively L, Aagaard L, Soifer H, Gatignol A, Riggs A, Rossi JJ. Combinatorial delivery of small interfering RNAs reduces RNAi efficacy by selective incorporation into RISC. Nucleic Acids Res. 2007;35:5154–5164. doi: 10.1093/nar/gkm543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siolas D, Lerner C, Burchard J, Ge W, Linsley PS, Paddison PJ, Hannon GJ, Cleary MA. Synthetic shRNAs as potent RNAi triggers. Nat. Biotechnol. 2005;23:227–231. doi: 10.1038/nbt1052. [DOI] [PubMed] [Google Scholar]
- 46.Soifer HS, Sano M, Sakurai K, Chomchan P, Saetrom P, Sherman MA, Collingwood MA, Behlke MA, Rossi JJ. A role for the Dicer helicase domain in the processing of thermodynamically unstable hairpin RNAs. Nucleic Acids Res. 2008;36:6511–6522. doi: 10.1093/nar/gkn687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leonard JN, Schaffer DV. Computational design of antiviral RNA interference strategies that resist human immunodeficiency virus escape. J. Virol. 2005;79:1645–1654. doi: 10.1128/JVI.79.3.1645-1654.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.ter Brake O, Berkhout B. Lentiviral vectors that carry anti-HIV shRNAs: problems and solutions. J. Gene Med. 2007;9:743–750. doi: 10.1002/jgm.1078. [DOI] [PubMed] [Google Scholar]
- 49.Boudreau RL, Martins I, Davidson BL. Artificial microRNAs as siRNA shuttles: improved safety as compared to shRNAs in vitro and in vivo. Mol. Ther. 2009;17:169–175. doi: 10.1038/mt.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McBride JL, Boudreau RL, Harper SQ, Staber PD, Monteys AM, Martins I, Gilmore BL, Burstein H, Peluso RW, Polisky B, et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc. Natl Acad. Sci. USA. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.An DS, Qin FX, Auyeung VC, Mao SH, Kung SK, Baltimore D, Chen IS. Optimization and functional effects of stable short hairpin RNA expression in primary human lymphocytes via lentiviral vectors. Mol. Ther. 2006;14:494–504. doi: 10.1016/j.ymthe.2006.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ely A, Naidoo T, Arbuthnot P. Efficient silencing of gene expression with modular trimeric Pol II expression cassettes comprising microRNA shuttles. Nucleic Acids Res. 2009;37:e91. doi: 10.1093/nar/gkp446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weinberg MS, Ely A, Barichievy S, Mufamadi S, Carmona S, Arbuthnot P. Specific inhibition of HBV replication in vitro and in vivo with expressed long hairpin RNA. Mol. Ther. 2007;15:534–541. doi: 10.1038/sj.mt.6300077. [DOI] [PubMed] [Google Scholar]
- 54.Marques JT, Devosse T, Wang D, Zamanian-Daryoush M, Serbinowski P, Hartmann R, Fujita T, Behlke MA, Williams BRG. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat. Biotechnol. 2006;24:559–565. doi: 10.1038/nbt1205. [DOI] [PubMed] [Google Scholar]
- 55.Robbins MA, Li M, Leung I, Li H, Boyer DV, Song Y, Behlke MA, Rossi JJ. Stable expression of shRNAs in human CD34+ progenitor cells can avoid induction of interferon responses to siRNAs in vitro. Nat. Biotechnol. 2006;24:566–571. doi: 10.1038/nbt1206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.