Abstract
Using an experimental approach, we investigated the RNome of the pathogen Staphylococcus aureus to identify 30 small RNAs (sRNAs) including 14 that are newly confirmed. Among the latter, 10 are encoded in intergenic regions, three are generated by premature transcription termination associated with riboswitch activities, and one is expressed from the complementary strand of a transposase gene. The expression of four sRNAs increases during the transition from exponential to stationary phase. We focused our study on RsaE, an sRNA that is highly conserved in the bacillales order and is deleterious when over-expressed. We show that RsaE interacts in vitro with the 5′ region of opp3A mRNA, encoding an ABC transporter component, to prevent formation of the ribosomal initiation complex. A previous report showed that RsaE targets opp3B which is co-transcribed with opp3A. Thus, our results identify an unusual case of riboregulation where the same sRNA controls an operon mRNA by targeting two of its cistrons. A combination of biocomputational and transcriptional analyses revealed a remarkably coordinated RsaE-dependent downregulation of numerous metabolic enzymes involved in the citrate cycle and the folate-dependent one-carbon metabolism. As we observed that RsaE accumulates transiently in late exponential growth, we propose that RsaE functions to ensure a coordinate downregulation of the central metabolism when carbon sources become scarce.
INTRODUCTION
Regulatory RNAs in prokaryotes are frequently referred to as small RNA (sRNAs). The sRNAs are typically between 50 and 300 nts in length. Most sRNAs with identified regulatory functions pair with mRNAs to modulate their translation, stability or transcription (1). The sRNAs encoded opposite to mRNA genes are referred to as ‘cis-encoded sRNAs’, since they often regulate the associated mRNA expressed from the complementary coding strand by extended perfect base-pairing (2). In contrast, sRNAs expressed from intergenic regions (IGRs) often act on target mRNAs transcribed from genetically unlinked regions. Base-pairing between these trans-encoded sRNAs and mRNA targets is usually imperfect and limited to short sequences. In many bacteria, including the Gram-negative bacterium Escherichia coli, the activity of trans-encoded sRNAs requires the RNA-chaperone Hfq (3). However, the Gram-positive bacterium Staphylococcus aureus might not require Hfq for sRNA activity since an hfq deletion mutant of this organism has no discernable phenotype (4). A third class of sRNAs can exert a regulatory function by directly interacting with proteins (5–7). Lastly, some mRNAs possess structured 5′ untranslated regions (UTR) that, under specific growth conditions, can adopt alternative conformations to affect gene expression of the downstream genes in cis. Binding of molecules such as metabolites or tRNAs to these so-called riboswitches induces RNA structure changes of the transcribed UTR, causing premature transcriptional termination or translational inhibition (8). Some riboswitches are known to produce sRNAs (9), which might independently function on trans-encoded target mRNAs by base-pairing mechanisms (10).
Several methods have been used to identify sRNAs, including computational predictions, direct labeling, DNA microarrays and shotgun sequencing (11,12). Escherichia coli and Salmonella each encode an estimated 100 sRNAs (11). These respond to diverse environmental factors such as glucose availability, iron limitation, envelope stress, or transition to anaerobiosis, to facilitate adjustments, respectively, in carbon metabolism (13,14), of levels of iron-requiring enzymes (15–17), amounts of metabolic transporters (18,19) or quantities of enzymes required for anaerobic growth (20,21). Other regulatory functions of sRNAs include quorum sensing and virulence (22–26).
Staphylococcus aureus is a human commensal of skin and nares and an opportunistic pathogen responsible for a wide variety of human infections including superficial skin and wound infections to deep abscesses (endocarditis and meningitis), septicemia or toxin-associated syndromes (27). It is a leading cause of nosocomial and community acquired diseases and many strains acquire antibiotic resistance (28). Staphylococcus aureus virulence gene expression is regulated by the coordinated action of numerous regulators. One of the best characterized regulators is an sRNA called RNAIII (514 nt), which modulates virulence gene expression (29). Its 3′ domains can mediate translational repression of S. aureus virulence genes directly by interacting with spa mRNA, which encodes the surface-anchored Protein A, or indirectly, by interacting with rot mRNA, which encodes Rot, a transcriptional regulator of several exoproteins (30–32).
Staphylococcus aureus has recently emerged as a model organism for the study of bacterial sRNAs (33). A pioneering study based on a computational analysis of IGRs led to the identification of 12 sRNAs that were experimentally confirmed, among which seven were expressed from pathogenicity islands (34). Thirty-four new sRNAs, including some that likely originate from putative riboswitches were recently identified (35–37). Finally, one deep sequencing and two global transcriptome studies on S. aureus, performed using different growth conditions suggested the existence of numerous non-coding small stable RNAs; however, most of them were not confirmed by alternative methods (37–39).
As sRNAs are potent regulators that can potentially affect viability, adaptability to different growth conditions, or virulence of this deadly pathogen, we undertook a search for new sRNAs in S. aureus. The characterization of one of these new sRNAs indicates that it coordinately downregulates multiple enzymes of two major metabolic pathways, likely to help S. aureus prepare for the entry into stationary phase.
MATERIALS AND METHODS
Bacterial strains, plasmids and growth conditions
The RNomics approach was performed on strain N315 (27). Transcriptome analysis was performed on strain RN6390 (40), a laboratory derivative of the NCTC8325 clinical strain (41) that was shown to be virulent in different animal models (42,43). Note that for simplicity, we only used the N315 gene name nomenclature. Engineered plasmids were constructed in E. coli TG1, and transferred to RN4220 (rsbU, agr), which is transformable with exogenous DNA (44), and subsequently to RN6390.
Staphylococcus aureus strains were routinely grown in BHI medium with aeration at 37°C. Escherichia coli TG1 used for cloning experiments was grown in LB broth at 37°C. Chloramphenicol was added to media as needed at the following concentrations: 5 µg/ml for S. aureus and 10 µg/ml for E. coli. To determine the effect of sRNA induction, staphylococcal cultures were prepared as follows: 10-fold serial dilutions of exponential cultures were spotted on BHI plates containing or not 1 µM anhydrotetracycline (aTc) and incubated at 37°C for 24 h.
pAT12 contains the repA, repC, tetR genes and the xyl/tetO promoter (45). The pAT12 derivative containing rsaA, rsaH, rsaE or expressing RNA sequences antisense of RsaA, RsaH and RsaE (named pAT12-RsaA, pAT12-RsaH, pAT12-RsaE, pAT12-ASRsaA, pAT12-ASRsaH and pAT12-ASRsaE, respectively) were constructed as follows: DNA sequences of interest were amplified from N315 genomic DNA by PCR, using primers indicated in Supplementary Data S1. The resulting products were digested by EcoRI and NotI, ligated to EcoRI-NotI-treated pAT12. Expression of the cloned sequences on constructed plasmids was induced by adding anhydrotetracycline (aTc) to the media, and confirmed by northern blot analysis.
DNA preparation
Lysis of S. aureus cell suspensions was achieved by treatment with 100 µg/ml of lysostaphin (Sigma-Aldrich, St. Louis, MO, USA) for 1 h at 37°C, followed by standard methods for DNA preparation. DNA transformation of E. coli and for S. aureus was performed as described (46,47).
RNA manipulation
For northern blot analysis and 454 pyrosequencing, overnight cultures of staphylococcal strains grown in BHI medium were diluted 1000-fold in the same medium and grown at 37°C; samples were withdrawn at mid-or post-exponential or stationary phase cultures. To study the effect of stress conditions, cultures at OD600 = 0.2 were either shifted to 45°C, or supplemented with NaCl 1.5 M or H2O2 10 mM for 2 h. Total RNAs were extracted as described (48) or using NucleoSpin RNAII (Macherey Nagel, Düren, Germany).
Northern blots were performed as described (35,46,49). Samples were separated by either PAGE or agarose gels and probed with γ32-P 5′ labeled oligonucleotides (described in Supplementary Data S1).
Rapid amplification of cDNA ends (RACE) mapping was performed as described (50,51).
Synthetic RNAs were generated using the T7 MEGAshortscript kit (Ambion, Foster City, CA, USA) according to manufacturer’s instructions.
RNA gel mobility shift assays were performed as described (52). [α32-P]UTP was added to the transcription mix to produce radioactive transcripts. The transcripts were denatured in 50 mM HEPES (pH 6.9), 50 mM NaCl, 5 mM KCl and 1 mM MgCl2 for 3 min at 85°C, followed by refolding for 10 min at 30°C. Labeled RsaE (0.04 pmol) was incubated with increasing amounts of unlabeled synthetic RNAs or yeast tRNAs in 10 mM Tris–HCl (pH 7.5), 6 mM NaCl, 10 mM EDTA and 5 mM dithiothreitol for 10 min at 30°C. Samples were supplemented with 10% glycerol (final concentration) and loaded on a native 5% polyacrylamide gel containing 5% glycerol.
Toeprint experiments were performed as described (53,54) with the following modifications. Annealing mixtures contained 0.2 pmol unlabeled opp3A mRNA and 1 pmol labeled SA0849 mRNA (primer 404) in a buffer containing 10 mM Tris–acetate (pH 7.5), 60 mM NH4Cl and 1 mM DTT. The effect of RsaE was tested by adding various quantity of RsaE (25–300 pmol) to the annealing mixtures prior to the addition of purified E. coli 70S ribosomes. Ribosomes were activated for 15 min at 37°C and diluted in the reaction buffer in the presence of 1 mM MgCl2. For each sample, 4 pmol of 70S were added, followed by 5 min incubation and adjustment of MgCl2 to 10 mM. Ten picomoles of uncharged tRNAfMet were then added and samples were incubated 15 min. cDNA was synthesized using two units of AMV RT (New England Biolabs, Ipswich, MA, USA) for 15 min at 37°C. Reactions were stopped by addition of 10 µl loading buffer II (Ambion, Foster City, CA, USA). The cDNAs were loaded and separated by Urea 8% PAGE. The toeprint position on opp3A mRNA was determined by DNA sequencing.
cDNA library from sRNAs, 454 pyrosequencing and analysis of data
RNA preparations (cf. above) were treated twice with DNase (DNA-free, Ambion, Foster City, CA, USA) according to manufacturer’s instructions. For cDNA library construction, S. aureus RNAs from six different growth conditions (see ‘Results’ section) were mixed in a equimolar ratio. In addition, we used a new treatment protocol to enrich for primary bacterial transcripts (most mRNAs and sRNAs) (73). For this purpose, the pooled RNA was treated with Terminator 5′-phosphate dependent exonuclease (Epicentre, Madison, WI, USA). From this RNA, a cDNA library was prepared and analyzed on a Roche GS20 sequencer (F. Hoffmann-La Roche, Basel, Switzerland) as described (55). The cDNA library was constructed by Vertis Biotechnology AG (http://www.vertis-biotech.com/). In brief, the Terminator exonuclease-treated RNA was first treated with tobacco acid pyrophosphatase (TAP). The RNA samples were subsequently poly(A)-tailed using poly(A) polymerase followed by ligation of an RNA adapter to the 5′-monophosphate of the sRNAs. First-strand cDNA synthesis was then performed using an oligo(dT)-adapter primer and MMLV H- reverse transcriptase. The resulting cDNAs were then PCR-amplified and the specific barcode sequence CAGC was attached to the 5′ ends of the cDNAs.
DNA sequences obtained from a multiplex 454 run on a Roche GS20 sequencer that contained the specific barcode tag CAGC were extracted. The 5′ end linker and polyA-tails were clipped, yielding 38 052 insert sequences (reads). 543 reads with lengths <14 nt were excluded from the study. The remaining 37 507 reads (≥14 nt) were blasted with WU-blast (http://blast.wustl.edu/, chosen parameters were E = 0.0001, m = 1, n = –3, Q = 3, R = 3) against the S. aureus N315 genome sequence (NC_002745.fna), and 36 021 reads that could be mapped were kept for further analysis. Those with blast hits that corresponded to rRNA, tRNA, tmRNA, RNaseP, SRP RNA and known mRNA were removed. 202 reads (Supplementary Data S2) corresponded to sequences putatively issued from IGRs, and were assembled with cd-hit-est (similarity threshold c = 0.92) into 160 clusters (56). Further blast analysis of these 160 remaining clusters revealed additional tRNA or mRNA sequences that were excluded. The assembled sequences were manually analyzed to identify the sRNAs presented in this study. For example, at this last step, reads issued from sequences adjacent to open reading frames were considered as likely mRNA UTRs and discarded.
Transcriptome analysis
Overnight cultures grown in BHI supplemented with chloramphenicol were diluted 1000-fold in the same medium at 37°C. At OD600 = 0.6, aTc (1 µM) was added to the medium and 5 min later, the cultures were sampled. Total RNAs were extracted and treated twice with DNase I. RNA quality was controlled with a 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA). Two conditions were tested, RN6390 containing pAT12-RsaE and pAT12-AS-RsaE. Trancriptome experiments were performed using S. aureus Affymetrix DNA chips (Santa Clara, CA, USA) by Cogenics (Newton, MA, USA) according to the standard manufacturer’s instructions.
Quantitative real-time PCR (qRT–PCR)
Overnight cultures grown in BHI supplemented with chloramphenicol were diluted 1000-fold in the same medium at 37°C. At OD600 = 0.3, aTc (1 µM) was added to the medium and cultures were sampled at 0, 5, 10, 20 and 30 min. RNAs were prepared as described (48). Genes for normalization were selected, qRT–PCR experiments were performed and data were analyzed as described (57). The geometric mean of 3 genes (recA, gyrA and ftsZ) chosen among 13 was used to normalize the samples.
RESULTS
Screening for new S. aureus sRNAs
As compared to enteric bacteria, comparatively few sRNAs have been experimentally confirmed to date in S. aureus (34–37,58). Our goal was to identify new sRNAs without any a priori restrictions in the selection; we therefore used an RNomics approach based on pyrosequencing of cDNA libraries (12). The study was performed on N315, a methicillin-resistant S. aureus strain that was isolated in a case of a hospital-acquired infection in 1982 (27). As sRNAs are often expressed in response to specific signals, total RNA was extracted from six different conditions. N315 was inoculated in rich medium (BHI) under aeration at 37°C and samples were withdrawn in (i) exponential phase, (ii) post-exponential phase, (iii) stationary phase and after 2 h of exposure to (iv) osmotic, (v) thermal or (vi) oxidative stress (cf. experimental procedures). For cDNA library preparation the RNAs extracted from these six growth conditions were pooled in an equimolar ratio, treated with Terminator 5′-phosphate dependent exonuclease to deplete processed sRNAs, thus enriching primary transcripts, and converted to cDNA libraries to be analyzed by 454 pyrosequencing as described (55,59,73).
A total of 36 021 cDNA sequences (referred to as reads) > 14 nt matched to the N315 S. aureus reference DNA sequence. The vast majority of these reads corresponded to rRNAs (33 094, mainly 5S) or tRNAs (2548), and were not analyzed further. Likewise, 173 reads matching small mRNAs or probable mRNA degradation products were discarded. A total of 126 reads that did not correspond to the above-described RNAs were retained for further analysis (Supplementary Data S2). Of these, 50 were likely issued from mRNA UTR regions, rather than sRNAs per se, and 73 were putatively derived from IGRs. In addition, seven sequences mapped to the complementary strand of coding sequences (Supplementary Data S2), suggesting the expression of cis-encoded RNAs. The 71 sequences of interest were clustered in 32 IGRs. None of the sequences matched RNAIII, as expected since the N315 strain expresses little if any RNAIII. 4.5S RNA, tmRNA, RNAse P RNA and 6S RNA were found in S. aureus via a computational screening (34); we identified all of these (corresponding to 54 reads, Table 1). In addition, six of the seven previously reported sRNAs of S. aureus pathogenicity islands, named SprA through SprG (34), were recovered by our RNomics approach (Table 1); the exception was SprE, which has turned out to be an rRNA fragment and not an sRNA (S.C. and B.F., unpublished results). In addition to already known sRNAs, our global RNomics search suggested the existence of new trans-encoded sRNAs [of which two were suggested, but not confirmed, by previous studies (34,38,39)]. We confirmed 16 of these putative sRNAs by northern blot (two other tested regions could not be confirmed). We named them RsaOH to RsaOW [for RNA S. aureus Orsay (35)] and the corresponding genes rsaOH to rsaOW (Table 1). While the present work was being completed, the sRNAs RsaOJ, RsaOK and RsaON were independently reported; we will refer to them using the recently published names, RsaA, RsaH and RsaE, respectively (36).
Table 1.
sRNAs identified in this RNomics study
| RNA namea | IGR | Flanking genes | Strand Orientation | Nb of readsb | Alternative names | Conservationc |
|---|---|---|---|---|---|---|
| RsaOH | 72446/73508 | SA0065/kdpE | < > < | 1 | IGR59 (34) | S. aureus, S. epidermidis, S. haemolyticus |
| 4.5S RNA (34) | 501358/502001 | SA0434/SA0435 | > > > | 23 | SRP RNA | Conserved |
| RsaOI | 626797/627130 | proP/SA0532 | > < > | 4 | Sau-6477 (37) | S. aureus |
| RsaA (36) | 636758/637419 | SA0543/SA0544 | < > < | 2 | RsaOJ (this work), Sau-64 (37) | S. aureus, S. epidermidis |
| RsaC (36) | 679620/680738 | SA0586/SA0587 | < < < | 1 | S. aureus | |
| RsaH (36) | 829363/830064 | SA0724/SA0725 | < > > | 2 | RsaOK (this work) | Staphylococcal strains |
| tmRNA (34) | 843706/844543 | smpB/SA0737 | > > > | 27 | SsrA | Conserved |
| RsaOL | 856992/857482 | cspC/SA0748 | > < < | 1 | Sau-07 (37) | S. aureus, S. epidermidis |
| RsaOM | 875349/875597 | SAS023/SA0769 | > > > | 1 | Predicted SAM-I riboswitch | Conserved |
| RsaE (36) | 975283/975742 | SA0859/SA0860 | > > < | 3 | RsaON (this work) Sau-20 (37) | Bacillales order |
| Sau-02 (37) | 1006365/1007020 | SAS028/SAS029 | > < > | 1 | S. aureus, S. epidermidis, phages, plasmids | |
| RsaOO | 1012811/1013050 | SA0891/SA0892 | > < < | 1 | S. aureus | |
| RsaOP | 1089149/1089566 | SA0961/SA0962 | > > > | 1 | Staphylococcal strains | |
| RsaOQ | 1372074/1372564 | SA1198/SA1199 | > > > | 1 | Predicted T-box | Conserved |
| RNaseP RNA (34) | 1483578–1484012 | SA1277/SA1278 | < < < | 2 | S. aureus, phages | |
| 6S RNA (34) | 1660651–1660795 | SAS050/aspS | < < < | 2 | Srr80 (38,39) | Conserved |
| SprA (34) | 1856223/1856978 | SA1623/tnp IS232 | < > < | 1 | Staphylococcal strains | |
| SprB (34) | 1866662–1867133 | SA1633/SA1634 | > < < | 3 | S. aureus | |
| SprC (34) | 1871168–1872530 | luckE/SA1639 | < < > | 1 | S. aureus, S. epidermidis, S. haemolyticus | |
| SprD (34) | 2006879–2007560 | SA1754/SA1755 | < < > | 3 | S. aureus | |
| RsaOR | 2008572–2009085 | SA1756–57/sak | < < < | 1 | Srr6 (38,39, SprX) | S. aureus, phages |
| SprF3 | 2010790–2011000 | SA1956/SAS069 | > > > | 1 | RsaOS (this work) | Staphylococcal strains |
| Sau-30 (37) | 2294189/2294725 | cbiO/rplQ | < < < | 2 | Staphylococcal strains | |
| RsaOG (35) | 2367825/2368208 | SA2104/SA2105 | < < > | 2 | RsaI (36) | Staphylococcal strains |
| SprA2 (34) | 2490680–2491385 | SA2217/SA2218 | < < > | 1 | WAN014FZW (38,39) | Staphylococcal strains |
| RsaOT | 2544336–2544594 | SA2267/SA2268 | > < < | 7 | Srr43 (38,39) | S. aureus |
| RsaOU | 2611619/2611918 | ptsG/SA2327 | < < < | 2 | Rat sequence | |
| RsaOV | 2632123/2632361 | SA2343/copA | > > > | 1 | Sau-40 (37) | S. aureus |
| RsaOW | Eight possible positions | Tnp for IS1181 | < > – | 4 | ||
| RsaOX | SA0062 | Cis-encoded | 1 |
aGene names corresponding to sRNAs confirmed by northern blot previously or in this study (cf. Figure 2 and Supplementary Data S4).
bNumber of reads obtained by 454 sequencing matching the corresponding indicated IGR in given orientations (cf. Supplementary Data S2).
cConservation determined by Blast analysis (default parameters) using the identified or predicted sRNA gene sequence. Conserved indicates that the sequence can be found in phylogenetically distantly related bacteria.
Expression of S. aureus sRNA genes encoded in IGRs
Expression of the 15 identified RNAs was followed as a function of the growth phase. An overnight culture of N315 was diluted into a fresh rich medium and grown aerobically at 37°C; samples were withdrawn at different stages of growth and sRNA amounts were evaluated by northern blot analysis (Figure 1). We also examined the expression of six sRNAs in the six growth and stress conditions used for the RNomics approach (Supplementary Data S4). The studied sRNAs presented below are ordered according to their position on the N315 genomic map. Conservation of the identified sRNAs in other species is indicated in Table 1.
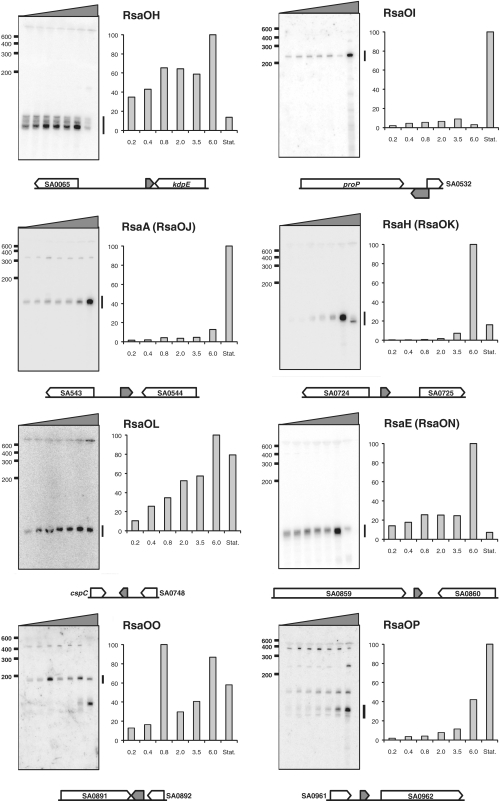
Figure 1.
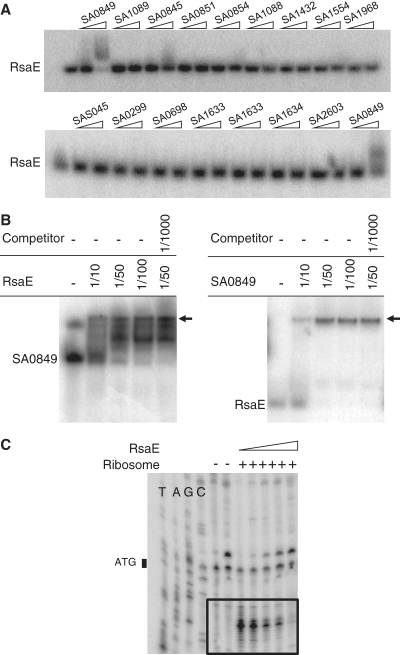
Identification and characterization of new N315 S. aureus sRNAs derived from the RNomics approach. Northern blots showing the expression of various sRNA candidates in nutrient-rich medium are presented. Total RNAs were prepared from cultures harvested at OD600: 0.2, 0.4, 0.8, 2, 3.5, 6 and 10. Black wedges indicate the increasing OD600 values from left to right. All blots presented were performed using the same RNA extracts. Marker sizes are shown to the left of each blot. RNAs were detected using γ32P-labeled oligonucleotides designed to pair with the predicted RNAs (cf. Supplementary Data S1). 5S rRNA was used as a loading control. Histograms indicate the relative expression of each sRNA in different growth phases (the intensity of each band was divided by the intensity of the 5S band from the same RNA extraction; and the maximum value for each histogram was set to 100). A bar on the right side of the blot indicates the band(s) selected for quantification. Flanking or overlapping genes (open arrows) of each sRNA (grey arrow) is presented to indicate genetic localization. Arrows indicate the direction of transcription and are proportional to known or expected transcript sizes.
RsaOH is about 100-nt long. Its gene, rsaOH, is located between SA0065 and kdpE genes encoding a putative C4-dicarboxylate transporter and a putative transcriptional regulator belonging to the OmpR family of two-component response regulators, respectively. The rsaOH detected sequence is adjacent to the 3′-end of kdpE (Supplementary Data S3). One may speculate that RsaOH interacts with the 3′-end of the kdpD-kdpE mRNA and therefore could regulate its stability. RsaOH increases with the growth phase but strongly decreases in the stationary phase (Figure 1).
RsaOI is about 240-nt long; it is the longest sRNA that we identified, and contains no expected ORF. Its gene, rsaOI, is located between proP, encoding a putative transporter and partially (38 nt) overlaps in antisense orientation with the 5′ end of SA0532 encoding a Staphylococcus-specific hypothetical protein (Supplementary Data S3). It is therefore possible that RsaOI affects transcription or translation of SA0532. In contrast to RsaOH, RsaOI expression drastically increases in stationary phase (Figure 1).
RsaA (RsaOJ) is 142-nt long (36). Its gene, rsaA, is located between SA0543 encoding a conserved putative membrane protein and SA0544 encoding a putative heme peroxidase. Like RsaOI, RsaA, is detected in all growth phases and drastically increases in stationary phase (Figure 1). RsaA did not accumulate upon thermal, salt or oxidative stress (Supplementary Data S4).
RsaH (RsaOK) is 128-nt long (36). Its gene is located between SA0724, encoding a hypothetical protein similar to a cell-division inhibitor, and SA0725, encoding a protein of unknown function. RsaH could encode a putative 23 amino-acid polypeptide (MCKTLHDTNEGKNLTPFSSGPVR). In the corresponding IGR of S. aureus strain RF122 the SAB0725 ORF of a putative 52 amino-acid protein with no significant similarity to proteins from other organisms has been annotated. However, in the majority of S. aureus strains, including N315, the coding sequence is interrupted by a stop codon leading to the putative 23 amino-acid peptide. RsaH was previously reported to be induced in stationary phase (36). We found that in N315, RsaH is barely detectable in early exponential phase, but accumulates during pre-stationary phase (Figure 1), and then strongly decreases in stationary phase.
RsaOL is estimated to be about 100-nt long, according to our northern blot probing. It is transcribed from a gene located in the IGR between cspC (encoding a cold shock protein from the CspA family) and SA0748 (encoding a poorly conserved putative protein of unknown function). RsaOL is expressed in all growth phases tested. Its quantity increases with the growth phase, except in stationary phase, where levels are slightly decreased (Figure 1).
RsaE (RsaON) is 93-nt long (36). The rsaE gene is located between SA0859, encoding a predicted dithiol-disulfide isomerase involved in polyketide biosynthesis, and SA0860 encoding an oligoendopeptidase. The RsaE sequence is present in the bacillales order and the synteny is conserved within the Firmicute phylum with the noticeable exception of the Listeriaceae family. The rsaE gene is identical in all S. aureus strains sequenced to date. RsaE is thus likely under a strong selective pressure for the conservation of its primary sequence: one possibility is that RsaE interacts with a specific protein requiring a defined nucleotide sequence for interaction and subsequent function. However, experiments based on the incubation of synthetic RsaE with protein extracts did not identify an RsaE-specific binding protein (data not shown). RsaE was also reported as induced in stationary phase (36). However, a detailed analysis reveals that in N315, although RsaE is detectable during all growth phases, it accumulates specifically during pre-stationary phase (OD600 = 6) and almost disappears in stationary phase (Figure 1). Its expression profile is similar to that of RsaH. In several strains, RNAIII is also induced in late exponential phase. Transition to stationary phase and possibly virulence activate the expression of several sRNAs, which make them candidates for being key regulators of transition adaptation processes. Furthermore, RsaE is reduced under heat-shock (Supplementary Figure S4).
RsaOO is about 180-nt long. Its gene, rsaOO, is located between SA0891 and SA0892, both of which encode membrane proteins. A second product, about 140-nt long, accumulates in stationary phase, possibly issued from the processing of RsaOO (Figure 1).
The rsaOP gene is located between SA0961 and SA0962, which encode a protein of unknown function and an integral membrane protein (FtsW) involved in cell division, respectively. Several products were detected when probing for RsaOP; the major one is about 100-nt long and accumulates in stationary phase (Figure 1).
RsaOR (alias SprX) is about 160-nt long with a possible longer form. Its gene is located between SA1757, encoding a putative truncated amidase, and sak, encoding staphylokinase. The sequence comprising this region, including adjacent gene SA1757 is also found in several phages (tp310-3, 52A, phi12, tp310-2, 80apha, phiNM4, etc.). RsaOR is thus likely acquired via horizontal transfer and expressed by phages. RsaOR is expressed throughout growth, but amounts are reduced in stationary phase (Figure 1). It also accumulates upon salt stress (Supplementary Data S4). N315 RsaOR may encode a putative polypeptide of 40 amino acids (MHQLFTSLFT QACHWVFFLM IESIVFILLP RSIYDFSIPV). However, the initiation codon is absent in most S. aureus sequenced genomes and there is no obvious ribosome binding site sequence. Therefore, if expressed, this peptide would be expressed only in few S. aureus strains. We therefore favor the hypothesis that RsaOR is a regulatory sRNA.
The intergenic SA1760/SAS059 region contains sprF and on its complementary strand sprG. In N315, the corresponding RNAs SprF and SprG were proposed to have paralogs expressed from two other separated regions located on pathogenicity islands (34). A read obtained by 454 (Supplementary Data S3) matched with the SA1956/SAS069 IGR containing the putative sprF3 (rsaOS) gene. Northern blot experiments show that the expression of sprF3 increases gradually with the growth phase (Figure 1). 5′ RACE mapping experiments (Supplementary Data S3) indicate that the sequence of the first 47 nt of sprF3 does not match sprF and sprF2, suggesting that targets of SprF3 could differ from those of SprF and SprF2.
RsaOT is about 180-nt long. It is poorly expressed and we could not identify its extremities. rsaOT expression is at a maximum during pre-stationary phase (Figure 1) and is also stimulated by the presence of H2O2 in the medium (Supplementary Data S4). As one of the RsaOT cDNA sequences obtained via our RNomic study is only at 6 nt downstream from the SA2268 stop codon, it is possible that the sRNA detected by northern blot is issued from 3′ end processing of SA2268.
RsaOV is about 100-nt long. Its gene, rsaOV, is located between SA2343 and copA, which encode a putative poorly conserved protein and a copper translocating ATPase, respectively. RsaOV accumulates in stationary phase (Figure 1).
The rsaOW gene is present in eight copies (rsaOW1 to rsa0W8) on the N315 genome; it is located immediately upstream of the ORF encoding the IS1181 transposase (Supplementary Data S3). RsaOW is therefore perfectly complementary to the transposase 5′ UTR region (including its ribosome binding site). As RsaOW is detected in all growth conditions (Figure 1), one may hypothesize that its role is to control, likely lowering, the expression of the IS1181 transposase. RsaOW would therefore affect the S. aureus genome evolution.
Our results confirm the existence of several trans-encoded S. aureus sRNAs and show that their expression is, for the most part, dependent on growth conditions.
Expression of cis-encoded S. aureus sRNA genes
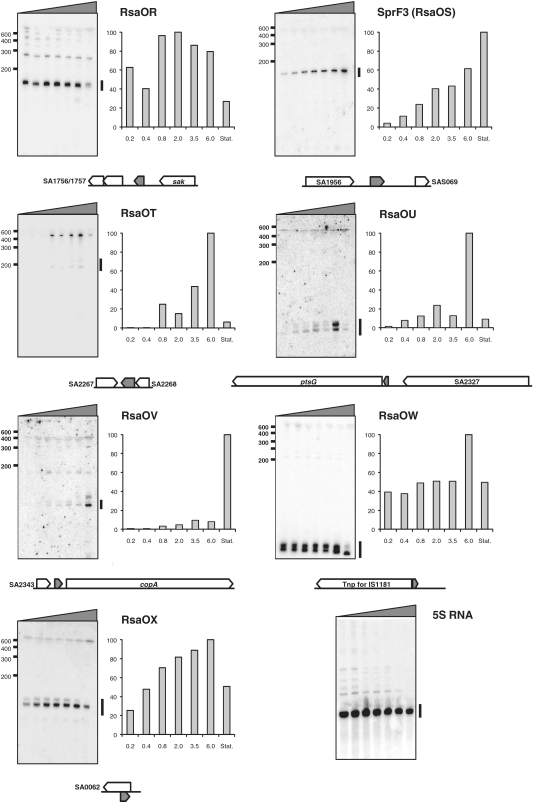
Our RNomics approach predicted seven candidates of cis-encoded sRNAs; we were able to detect one of them, RsaOX, by northern blot analysis (Figure 1). The others gave either no hybridization signal or signals corresponding to high-molecular weight RNAs (data not shown). RsaOX is about 120-nt long and is transcribed from the complementary strand of SA0062; it was expressed in all of the tested growth phases. As SA0062 encodes a putative transposase, the existence of RsaOX is reminiscent of the system described for the E. coli IS10 transposase, whose mRNA (called RNAin) is controlled by RNAout, its cis-encoded sRNA (60,61). SA0062 mRNA was recently proposed to be the target of MazFSA, a sequence-specific endoribonuclease (62). One can postulate that in addition to MazFSA, RsaOX could prevent the synthesis of the likely deleterious SA0062-encoded transposase and therefore might limit transpositions.
Identification and expression of sRNA for putative S. aureus riboswitches
The RNomics analysis allowed us to identify transcripts that likely result from premature transcriptional termination associated with riboswitch activities. RsaOM corresponds to the sequence upstream of SA0769 encoding a d-methionine transport system ATP-binding protein. Probing for this region revealed several small transcripts more expressed in the high salt stress condition (Supplementary Data S4). The SAS023-SA0769 IGR is predicted to contain a SAM riboswitch [also known as S-box leader or SAM-I riboswitch; (63)], which is found upstream of genes involved in methionine or cysteine biosynthesis (64). The effector, S-adenosyl-methionine, specifically binds to the nascent S-box RNA leading to a premature transcription arrest (65,66), which would explain the observed short transcripts (Supplementary Data S4). Surprisingly, the N315 predicted SAM riboswitch may encode a putative protein of 60 amino acids conserved in S. aureus genomes. However, this ORF does not exist in other species, suggesting that if expressed in S. aureus, its activity might not be related to the SAM riboswitch control.
RsaOQ corresponds to the upstream sequence of SA1199, which encodes component I of the anthranilate synthase involved in phenylalanine, tyrosine and tryptophan biosynthesis. Probing for this sequence revealed several bands with a different profile in stationary phase as compared to the exponential phase or stress conditions (Supplementary Figure S4). The SA1198-SA1199 IGR is predicted to contain two tandem T-boxes (63,67). These leader elements sense uncharged tRNA to regulate the expression of aminoacyl–tRNA synthetase genes and amino-acid biosynthetic genes such as anthranilate synthase (68).
RsaOU is a tiny sRNA (estimated length is <70 nt) originating immediately upstream of ptsG, which encodes a component of the glucose-specific phosphotransferase system. RsaOU is highly expressed in pre-stationary phase (Figure 1). It is likely that rsaOU acts as a cis-element on ptsG expression. Indeed, the ptsG operon of Staphylococcus carnosus is regulated by a termination/antitermination mechanism associated with a sequence called RAT for ribonucleic antiterminator (69,70). HPr-dependent phosphorylation of GlcT facilitates antitermination (69,70). A similar regulation is likely to exist in S. aureus; in addition to ptsG, glcA, which encodes another enzyme from the PTS system, is preceded by a RAT sequence that generates an sRNA (36).
RsaE downregulates the one carbon pool by folate and central metabolic pathways
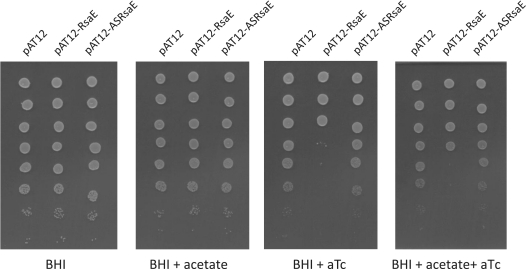
The rsaA (rsaOJ), rsaE (rsaON) and rsaH (rsaOK) genes (corresponding to RNAs highly expressed during specific growth phases) were cloned under the regulatory control of the Pxyl/tetO promoter of pAT12, giving rise to pAT12-RsaA, pAT12-RsaE and pAT12-RsaH, respectively. These plasmids express the Tet repressor that binds to the tetO operator; sRNA expression was induced by addition of anhydrotetracycline (aTc) to the media. We also constructed derivatives, pAT12-ASRsaA, pAT12-ASRsaH and pAT12-ASRsaE expressing antisense RNAs of RsaA, RsaH and RsaE, respectively. RN6390 containing pAT12-RsaE, but not the other above described plasmids, was sensitive to aTc (100 nM) in a solid rich media at 30, 37 or 42°C (Figure 2 and data not shown). Accumulation of RsaE may cause toxicity by affecting essential functions; we therefore focused our interest on the functional characterization of RsaE.
Figure 2.
Accumulation of RsaE induces a growth defect that is partly alleviated by acetate. Overnight cultures were prepared in BHI from the strains listed below. Five-microliter spots of ten-fold serial dilutions of cultures were deposited from top (most concentrated) to bottom on BHI plates supplemented or not with aTc and acetate (100 mM) as indicated, and incubated at 37°C. Strains are RN6390 derivatives containing the indicated plasmids pAT12, pAT12-RsaE and pAT12-ASRsaE. pAT12-RsaE and pAT12-ASRsaE contain the rsaE gene and its antisense, respectively, both of which are under the transcriptional control of the Ptet promoter (inducible by aTc). Experiments were also performed at 30 and 42°C, and in LB medium, and gave similar results (data not shown).
Many sRNAs act via base-pairing to target mRNAs to affect their stability and/or translation. We hypothesized that RNA targets of RsaE would vary upon RsaE accumulation, and could thus be identified by transcriptome analysis. We therefore performed an mRNA analysis of strains expressing a plasmid-encoded RsaE (pAT12-RsaE) or RsaE-antisense (pAT12-ASRsaE), sampling RNA 5 min after induction of RsaE or its antisense sequence; this approach was designed to identify those mRNAs with which RsaE directly interacts, and is similar to the pulse-expression approaches that facilitated sRNA target discovery in E. coli and Salmonella (16,19). Preliminary northern blot analysis showed that 5 min of induction was sufficient for strong accumulation of either RsaE or its antisense RNA (data not shown). Induction was performed at OD600 = 0.6, when the expression of the chromosomally encoded RsaE is low (Figure 1). RsaE accumulation led to the downregulation of 25 genes; surprisingly, it also upregulated 39 genes at least 2-fold (Table 2 and Supplementary Data S5). To consolidate our results, the expression of 16-selected genes was studied via a kinetic of RsaE induction at time 0, 5, 10, 20 and 30 min by qRT–PCR (Table 2 and Supplementary Data S6). With the exception of one gene (opp3A, cf. Table 2 legend), the qRT–PCR results fully confirmed the transcriptome data. The mRNAs modulated either directly or indirectly by RsaE mainly encode membrane proteins or metabolic enzymes.
Table 2.
Variation of gene expressions upon 5 min of RsaE induction
| Gene namea | Affim.b | qRT–PCRc | Δgd | Function |
|---|---|---|---|---|
| SA0845 (opp3B) | 1.10 | 0.79 | −25.47 | Oligopeptide transport system |
| SA0849 (opp3A)e | 2.18 | 0.76 | −24.31 | Oligopeptide transport system |
| SA0850 (opp4A) | 4.38 | 6.06 | Peptide/nickel transport system | |
| SA0851 (opp4D) | 3.15 | 4.59 | −14.70 | Peptide/nickel transport system |
| SA0873 | 0.48f | −21.13 | Hypothetical protein | |
| SA1088 (sucC) | 0.54f | −18.24 | Succinyl–CoA synthetase subunit beta | |
| SA1089 (sucD) | 0.47f | −15.51 | Succinyl–CoA synthetase subunit alpha | |
| SA1184 (citB) | 0.47 | −15.61 | Aconitate hydratase | |
| SA1365 (gcvPB) | 0.48 | Glycine dehydrogenase subunit 2 | ||
| SA1366 (gcvPA) | 0.36 | 0.48 | −21.88 | Glycine dehydrogenase subunit 1 |
| SA1367 (gcvT) | 0.32 | 0.36 | Aminomethyltransferase | |
| SA1432 | 0.43 | 0.46 | −18.04 | Hypothetical Mn2+ and Fe2+ transporter |
| SA1434 | 0.43 | Acetyl–CoA carboxylase | ||
| SA1435 | 0.45 | Acetyl–CoA carboxylase | ||
| SA1517 (citC) | 0.40 | Isocitrate dehydrogenase | ||
| SA1518 (citZ) | 0.37 | 0.48 | −14.80 | Methylcitrate synthase |
| SA1553 (fhs) | 0.30f | −14.08 | Formate–tetrahydrofolate ligase | |
| SA1858 (ilvD) | 3.45 | 1.90 | Dihydroxy-acid dehydratase | |
| SA1859 (ilvB) | 2.75 | Acetolactate synthase large subunit | ||
| SA1860 (ilvH) | 3.12 | Acetolactate synthase 1 regulatory subunit | ||
| SA1861 (ilvC) | 2.17 | Ketol-acid reductoisomerase | ||
| SA1862 (leuA) | 2.77 | 2-Isopropylmalate synthase | ||
| SA1863 (leuB) | 2.56 | 3-Isopropylmalate dehydrogenase | ||
| SA1865 (leuD) | 3.11 | Isopropylmalate isomerase small subunit | ||
| SA1968 (arg) | 0.35 | 0.44 | −18.81 | Arginase |
RN6390 strains containing either pAT12-RsaE or p12-ASRsaE were grown at 37°C in BHI to OD600: 0.6. aTc was added to the medium and after 5 min, samples were removed and total RNA were extracted.
aSA0845 to SA849, SA1088-SA1089, SA1367-SA1365, SA1435 to SA1432, SA1518-SA1517, SA1858 to SA1865, likely organized in six distinct operons, respectively (genes for the same operon are expected to have a RsaE-dependent coordinated regulation).
bGene expression ratios between strains containing pAT12-RsaE versus p12-ASRsaE obtained by Affimetrix microarray analysis (see also Supplementary Data S6).
cGene expression ratios between strains containing pAT12-RsaE versus p12-ASRsaE obtained by qRT–PCR analysis. Experiments were also performed with samples taken at 0, 10, 20 and 30 min after the aTc addition to the media (Supplementary Data S7).
dFree energy of pairing between RsaE and the 5′ region of corresponding mRNAs as calculated by RNAup software (71).
eDiscrepancy was observed between transcriptome and qRT–PCR experiments in the case of opp3A mRNA is possibly due to cross-hybridization of opp4A cDNA with the transcriptome opp3A probes.
fResults supported by published transcriptome results (36).
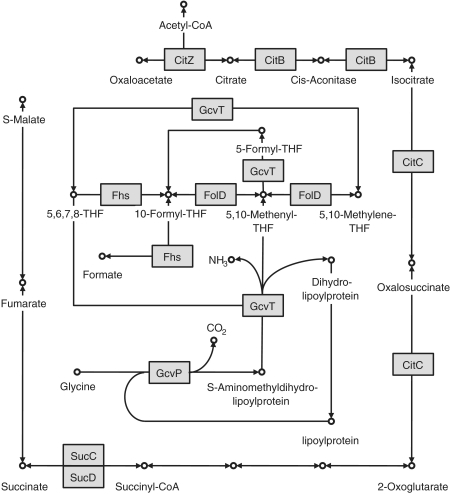
Strikingly, mRNAs that are downregulated by RsaE can be clustered around two essential pathways (Figure 3): (i) SA1366-SA1367 (gcvP-gcvT) and SA1553 (fhs) mRNAs are associated with production of methenyl tetrahydrofolate (THF) required for the one-carbon shuffle, and (ii) SA1088-SA1089 (sucC-sucD), SA1184 (citB) and SA1517-SA1518 (citC-citZ) mRNA, which produce enzymes of the Krebs cycle. The growth inhibitory effect of RsaE overproduction is consistent with the observed downregulation of these genes; furthermore, addition of acetate to the media, which should allow acetyl–CoA formation for partial bypass of the TCA cycle, partly suppressed the toxicity (Figure 2).
Figure 3.
RsaE accumulation downregulates specific metabolic pathways. The metabolic charts are adapted from the Kegg database (90). Enzymes corresponding to downregulated genes by RsaE are indicated by square boxes highlighted in grey [FolD [SA0915] indicated on the figure was not found in this study, but was previously reported (36)]; metabolites are shown as open circles; biochemical reactions that are demonstrated or predicted in N315 by arrows, respectively. The outer pathway is the TCA cycle. The inner pathway is the THF biosynthesis pathway. In certain organisms, these two pathways are connected via the glyoxylate shunt, which leads to a modified version of the TCA cycle that bypasses certain steps. However, connecting enzymes in N315 are not yet known.
Genes upregulated by RsaE include the operon for valine, leucine and isoleucine biosynthesis. In addition, several genes, likely encoding membrane proteins involved in peptide transport, are upregulated. Among them, two encode subunits of an oligopeptide permease (Opp) [SA0850 (opp4A), SA0851 (opp4D)].
The RsaE accumulation appears to correlate with an adaptation response aimed at reducing central metabolic pathways and increasing the amino acid pool.
RsaE binds the opp3A mRNA ribosome-binding site
To identify the primary RsaE targets among the putative candidates revealed by the transcriptome analysis, we performed a computational screen using RNAup. This software determines pairing energies between sRNAs and putative mRNA targets (71). Pairing energies between RsaE and the 5′ region (−150/+20) of all S. aureus CDSs were evaluated; one of the best scores was obtained with SA0849 (ΔG:−21 kcal/M) encoding Opp3A. The transcript of the two immediate opp3A-downstream genes strongly accumulates upon RsaE induction (Table 2 and Supplementary Data S5 and S6). These opp genes encode components of ABC transporters possibly involved in peptide/nickel transport (72). The combination of computational and transcriptome data indicates that RsaE could affect opp regulation by pairing with the SA0849 mRNA.
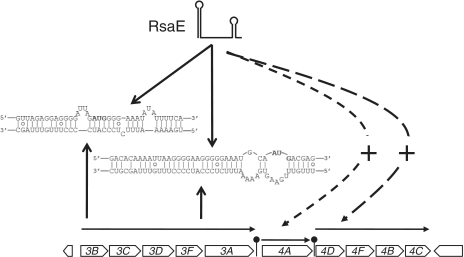
The interaction of RsaE with different mRNAs predicted to be RsaE targets (i.e. by trancriptome results and computational analyses using RNAup) was tested by gel retardation assays. The candidates correspond to (i) opp3A (SA0849) and opp4D (SA0851) encoding components of oligopeptide transport systems (72), (ii) SA1432 and SA0645 encoding putative manganese and anion transporters, respectively, (iii) sucC (SA1088) and sucD (SA1089) encoding the succinyl–CoA synthetase alpha and beta subunits, respectively, (iv) gcvP (SA1366) and gcvT (SA1367), encoding enzymes from the glycine cleavage pathway, (v) acsA (SA1554) encoding the acetyl–CoA synthetase and (vi) arg (SA1968) encoding arginase. (Additional candidates based solely on bio-computing data were also tested: opp3B [SA0845], opp4C [SA0854], SAS045, SA0308, SA0785 and SA2118.) A synthetic full-length RsaE and truncated mRNAs (∼180 nt long; for oligonucleotides used, see Supplementary Data S1) were produced from PCR-generated genes placed under the control of the T7 RNA polymerase using an in vitro transcription assay. The putative RsaE targets tested were about 140-nt long with about 80-nt corresponding to the Shine-Dalgarno (SD) upstream sequences. Among these, only one mRNA, corresponding to opp3A (SA0849), formed a complex with RsaE in vitro (Figure 4A and B). Complex formation between RsaE and SA0849 is specific since a thousand fold excess of tRNA did not disrupt the sRNA–mRNA duplex (Figure 4B).
Figure 4.
RsaE interacts with the 5′ end region of opp3A (SA0849) mRNA. (A) Gel retardation assay of labeled RsaE (0.4 pmol) incubated with synthetic RNAs (4 and 40 pmol), corresponding to different 5′ gene ends of the indicated genes (see Supplementary Data S1 for the sequences). The RsaE/SA0849 mRNA interaction is shown by a mobility shift of labeled RsaE in the presence of SA0849 mRNA (RsaE mobility was not affected by the other tested RNAs). (B) Left: Gel retardation assay of synthetic labeled SA0849 mRNA 5′ end fragment (0.4 pmol) incubated with increasing concentrations (molar ratio is indicated) of synthetic RsaE. Right: Gel retardation assay of labeled RsaE (0.4 pmol) incubated with increasing concentrations (molar ratio is indicated) of synthetic SA0849 mRNA 5′ end fragment. Addition of a large excess of unlabelled tRNAs did not alter the RsaE/SA0849 mRNA interactions in either assay. (C) RsaE prevents ribosome loading and translation initiation on opp3A SA0849 mRNA. Ribosome toeprints on SA0849 mRNA. ‘–/+’ indicates the absence or presence of purified ribosomes. The wedge indicates increasing amounts of RsaE. The experimentally determined toeprints are boxed. T, A, G and C refer to the SA0849 mRNA sequencing ladders. The position of the SA0849 mRNA initiation codon is indicated (ATG).
Predicted pairings between RsaE and the SA0849 mRNA (Figure 6) suggest that RsaE interacts with the opp3A SD sequence, as well as part of the AUG initiation codon and therefore could prevent translation initiation. To test this, toeprint assays were performed on ternary initiation complexes including purified 70S ribosomes, initiator tRNAfMet and the SA0849 mRNA. Two ribosome toeprints were detected on the mRNA, at 14 and 15-nt downstream from the initiation codon respectively (Figure 4C), supporting the location of the SA0849 mRNA start codon as drawn on Figure 6. RsaE reduced ribosome loading onto the SA0849 mRNA in a concentration-dependent manner (Figure 4C). This demonstrates the formation of a translational initiation complex on the SA0849 mRNA. We conclude that RsaE inhibits SA0849 mRNA translation by antisense pairing, thus preventing ribosome binding. The fact that RsaE (i) is predicted to efficiently pair with the opp3A mRNA at the SD site, (ii) forms a specific complex in vitro with the opp3A mRNA transcript and (iii) prevents in vitro formation of an opp3A mRNA translation initiation complex, together suggest that RsaE targets interacts with the oppA3 mRNA and prevents its translation.
Figure 6.
RsaE targets two SD sequences within the same mRNA. The transcriptional organization of the S. aureus N315 opp-3 and opp-4 operons is presented [based on (72)]. RsaE prevents ribosome loading onto opp-3 mRNA in front of opp3B (36) and opp3A ORF (this work). Predicted base pairing between RsaE and the opp-3 operon is indicated. RsaE positively regulates opp-3 downstream genes. Dashed lines indicate RsaE-mediated activation, as deduced from microarray and qRT–PCR results.
DISCUSSION
We used an RNomics approach based on deep sequencing of RNA enriched for primary transcripts (73) to search for novel staphylococcal sRNAs. Our analysis of 36 000 reads identified 30 known or putative sRNAs encoded within IGRs (Table 1 and Figure 5). Fourteen sRNAs confirmed by northern blots were categorized as: (i) sRNAs that could target genetically unlinked mRNAs (eight new instances) or (ii) sRNAs that pair with the UTRs of mRNAs expressed from the adjacent gene (two new instances). We also detected small transcripts likely associated with three riboswitch activities. Seven sRNAs were expressed from the opposite strand of ORFs; one is a bona fide cis-encoded RNA. We recently predicted 50 regions as encoding sRNAs; out of 24 tested, seven were confirmed to express sRNAs (35). Additionally, five of the newly reported sRNAs (RsaE, RsaOM, RsaOP, RsaOQ and RsaOW) are among the untested previously predicted regions. This observation brings further support for this computational screen based on phylogenetic profiling, which is particularly efficient at detecting novel bacterial sRNAs (35).
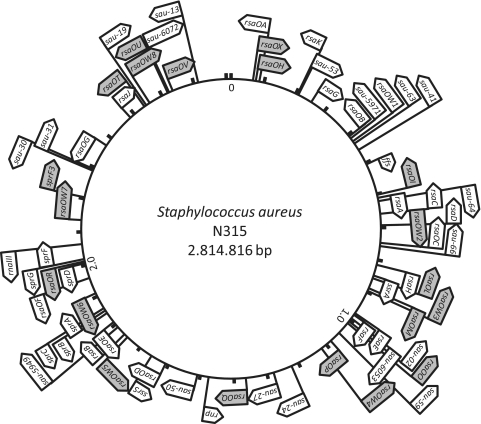
Figure 5.
Genomic map showing the distribution of S. aureus sRNAs. Flags indicate the names of sRNAs that were predicted and confirmed by northern blot analysis; sRNAs uncovered by the present work are highlighted in grey (34–37,58).
Most S. aureus RNAs are differentially expressed in a growth phase-dependent fashion (34,36,58). The detailed analysis of sRNA expression performed in this study reveals that some sRNAs accumulate in exponential (RsaOH, RsaOR), pre-stationary (RsaH, RsaE, RsaOT, RsaOU) or stationary (RsaOI, RsaA, RsaOP, RsaOV) phases. We observed abrupt changes in the expression levels of several sRNAs between OD600 = 6.0 and stationary phase (OD600 = 10). It is striking that several sRNAs such as RsaH and RsaE accumulate specifically in pre-stationary phase, and disappear in stationary phase, suggesting a role in adaptation to growth phase changes. Numerous staphylococcal regulatory functions are coordinated with environmental conditions or growth phase. Virulence factor expression is correlated with growth phase: surface adhesins are generally expressed in exponential phase, whereas exoproteins are expressed in stationary phase. The newly identified sRNAs conceivably play role(s) in this regulation, in addition to RNAIII (29). We tested the effect of several regulators on the expression of rsaE. RsaE quantities were compared in isogenic 8325-4 strains deficient for regulators SigB (74), SarA (75), Agr (76), PerR (77), Fur (78) or Spa (79), or proficient for RsbU (80); no significant differences were observed in any of those genetic contexts, suggesting that RsaE expression is probably regulated independently of these regulators (data not shown). A σA-consensus binding site is located just upstream of the rsaE start site, indicating expression driven by the S. aureus vegetative σ factor.
The primary RsaE nucleotide sequence is surprisingly well conserved among the sequenced bacillales, and RsaE over-expression is toxic. Altogether, these observations suggest important functional role(s) for RsaE. A variation in the quantity of RsaE induced a significant change in the transcriptional pattern. Among the up or downregulated genes, many express mRNAs that are predicted to pair with RsaE. Structure probing (36) and computational analyses suggest that RsaE folds as two stem loops separated by a 17-nt-long RNA single strand. RsaE has a duplicated, CCCCUUUGUUU accessible sequence with one copy in the loop of the first hairpin and a second within the central single stranded domain. The predicted pairings between RsaE and the targeted mRNAs involve the duplicated sequence and the mRNA SD sequences (Supplementary Data S7). Repetition of that interacting sequence on the sRNA suggests several pairing combinations with its mRNA substrates, involving either the RsaE apical loop 1 or its unstructured central domain.
The opp operons encode ABC transporters involved in the transport of small peptides. Staphylococcus aureus has several putative opp operons, three of which (opp-1, opp-2 and opp-3) are conserved in all strains (72). The opp-3 operon is dedicated to nitrogen nutrition by peptide import and is also involved in the regulation of extracellular proteases (SspA and Aur) via an unknown mechanism (81). In several Enterobacteriales, expression of dipeptide, oligopeptide and microcin ABC transport systems are partly controlled by sRNAs such as GcvB and RydC (52,82–84). RsaE targets the opp operons and is thus a possible GcvB/RydC functional analog. However, we detected no sequence similarities between RsaE and these sRNAs. In N315, the opp-3 operon comprises opp3B, opp3C, opp3D, opp3F and opp3A; it is followed by a second opp operon, opp-4, comprising opp4A and opp4D (72). The function of the opp-4 operon is at present unknown. Toeprint experiments show that RsaE interacts with the opp-3 mRNA (Figure 4). Surprisingly, RsaE induction had no significant effects on the amounts of opp-3 mRNAs, but strongly upregulated the downstream operon, opp-4 (quantified by qRT–PCR, cf. Table 2). Several interpretations can be considered for these results: (i) the observed RsaE mediated inhibition of ribosome loading on opp3A mRNA (Figure 4) could in turn, upregulate the opp-4 operon (i.e. Opp proteins could mediate this regulation) (ii) interactions between RsaE and opp-3 mRNA generate an anti-termination mechanism that leads to increased opp-4 operon transcription or (iii) RsaE interacts directly with the opp-4 mRNA preventing its degradation; however, we did not observe RsaE gel retardation by the opp4D 5′ mRNA. Recently, RsaE was reported to target the SD sequence of the opp3B mRNA, the first gene of the opp-3 operon (36). Here, we provide evidence that RsaE targets opp3A, the last gene of the opp3 operon. These observations suggest that RsaE operates simultaneously on two genes of the same mRNA, revealing an unusual mode of sRNA action (Figure 6).
Our results point to a regulatory role of RsaE in primary metabolism. It is astonishing that four enzymes converging directly to the 5, 10 methenyl-THF have their mRNA coordinately downregulated by the same sRNA, RsaE (Figure 3). They correspond to glycine decarboxylase (GcvP) and aminomethyltransferase (GcvT) which belong to the glycine cleavage pathway that provides the one-carbon units to methenyl-THF, and the formate-tetrahydrofolate ligase (Fhs) and methylene-THF enzyme (FolD) (36) which convert THF into methenyl-THF. This latter compound is the supplier of one-carbon units for the synthesis of purines and the formyl group of N-formylmethionyl-tRNA(f). It is also striking that RsaE downregulates four key enzymes of the tricarboxylic acid (TCA) cycle: the succinyl–CoA synthetase (SucC-SucD) [also in ref (36)], the aconitase (CitB), the isocitrate dehydrogenase (CitC) and the citrate synthase (CitZ) (Table 2, Figure 3). The TCA cycle is involved in the synthesis of usable energy. General downregulation of these different pathways might induce the observed toxicity of rsaE over-expression via an ectopic copy. The partial toxicity alleviation by acetate could be due to the utilization of glyoxylate pathway that shunts some RsaE-repressed steps of the TCA cycle. As endogenous RsaE is induced in a pre-stationary phase, an expected role for this sRNA would be to coordinate downregulation of energy metabolism (TCA cycle), and of cofactors, vitamins, translation, and purine biosynthesis (folate-dependent one-carbon metabolism) to facilitate adaptation to the entry into stationary phase.
CcpA is a master regulator for carbon catabolite regulation in numerous Gram-positive bacteria. Many genes that are also modulated by RsaE (e.g. sucC, sucD, arg, citC, citZ and ilvB) show CcpA-dependent regulation (85–87). This suggested that RsaE could affect a CcpA. However, accumulation of RsaE (by means of pAT12-RsaE) did not affect the quantity of the ccpA mRNA (Supplementary Data S5 and S6). In addition, we did not observe any band shift of the ccpA UTR mRNA sequence by RsaE (data not shown) suggesting that the RsaE effect is likely not via CcpA.
Computer predictions indicate the possibility that RsaE-dependent downregulation is mediated via pairing of RsaE with mRNA 5′ UTRs. However, in vitro experiments performed between RsaE and synthetic transcripts of the candidate UTR sequences displayed gel retardation with only one mRNA sequence. It is possible that the sRNA/RNA target binding in S. aureus is more labile in vitro than with sRNAs from other species such as E. coli (possibly because sequences are A/U-rich). For example, the sucD mRNA was previously identified and validated as an RsaE target using in vitro toeprinting (36); however, the RsaE-sucD interaction seemed too labile to be detected in our gel retardation assays (Figure 3A). In E. coli, the activity of trans-encoded RNA usually requires the RNA chaperone protein Hfq, but it is not the case for known sRNA–mRNA interactions in S. aureus (4,30,32,36); thus an unknown factor might be required to promote such regulatory interactions in S. aureus. Further evidence for RsaE-dependent gene downregulation via a direct interaction of RsaE with 5′ mRNA UTRs was obtained using a statistical method: Indeed, the probability that the seven of the 22 most downregulated genes (among a total of 2790 S. aureus genes) are found by random among the 139 best RsaE/mRNA matching scores (determined using the RNAup software) is 0.000054 (data from Table 2 and Supplementary Table S5). This result rules out the possibility of the random appearance of so many genes matching RsaE among the RsaE-dependent downregulated genes. Strikingly, among these 22 genes that are the most downregulated, 11 are directly involved in Krebs/one carbon by folate metabolism, providing further indication that a pairing with RsaE is required for the observed downregulation of metabolic pathways.
Since RsaE is induced in late exponential phase when the growth medium is depleted, we speculate that RsaE would contribute to downregulating bacterial metabolism (one carbon by folate and TCA cycle), and at the same time activate Opp transporters, as a last-ditch effort to adapt to limited nutrient conditions as cells approach stationary phase.
sRNAs are often expressed during specific growth conditions and have numerous targets. As such, they constitute key regulators of an intricate network of metabolic pathways and nutrient uptake systems. So far, most of the evidence for these roles comes from studies on enteric bacteria, while little is known concerning Gram-positive bacteria (36,88,89); results on RsaE make it obvious that the concept of coordinated control of metabolic enzymes by sRNAs should be extended to the Gram-positive pathogen, S. aureus.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grant ‘Microbiologie-Immunologie-Maladies Emergentes’ from the ‘Agence Nationale pour la Recherche’ (ANR-06-MIME-016-01); ‘Centre National de la Recherche Scientifique (CNRS)/Conseil Général de l′Essonne’ and ‘Agence Nationale pour la Recherche’ fellowship (to C.R.); ‘DIM Malinf’ fellowship from ‘Région Ile de France’ (to P.S.). Funding for open access charge: CNRS.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Pascale Romby and her colleagues for sharing results prior publication, and Armel Guyonvarch, Michelle David and Stella Artois for helpful discussions, technical assistance and warm support. The authors thank Richard Reinhardt and Bernd Timmermann for pyrosequencing and Vertis Biotechnology AG for cDNA construction. The authors thank Sandy Gruss for critical reading of the manuscript and helpful discussions.
REFERENCES
- 1.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brantl S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr. Opin. Microbiol. 2007;10:102–109. doi: 10.1016/j.mib.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 3.Brennan RG, Link TM. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 2007;10:125–133. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 4.Bohn C, Rigoulay C, Bouloc P. No detectable effect of RNA-binding protein Hfq absence in Staphylococcus aureus. BMC Microbiol. 2007;7:10. doi: 10.1186/1471-2180-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Babitzke P, Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr. Opin. Microbiol. 2007;10:156–163. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 6.Wassarman KM. 6S RNA: a small RNA regulator of transcription. Curr. Opin. Microbiol. 2007;10:164–168. doi: 10.1016/j.mib.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 7.Pichon C, Felden B. Proteins that interact with bacterial small RNA regulators. FEMS Microbiol. Rev. 2007;31:614–625. doi: 10.1111/j.1574-6976.2007.00079.x. [DOI] [PubMed] [Google Scholar]
- 8.Henkin TM. Riboswitch RNAs: using RNA to sense cellular metabolism. Genes Dev. 2008;22:3383–3390. doi: 10.1101/gad.1747308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jager JG, Huttenhofer A, Wagner EG. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 2003;31:6435–6443. doi: 10.1093/nar/gkg867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, Repoila F, Buchrieser C, Cossart P, Johansson J. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell. 2009;139:770–779. doi: 10.1016/j.cell.2009.08.046. [DOI] [PubMed] [Google Scholar]
- 11.Altuvia S. Identification of bacterial small non-coding RNAs: experimental approaches. Curr. Opin. Microbiol. 2007;10:257–261. doi: 10.1016/j.mib.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Sharma CM, Vogel J. Experimental approaches for the discovery and characterization of regulatory small RNA. Curr. Opin. Microbiol. 2009;12:536–546. doi: 10.1016/j.mib.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 13.Gorke B, Vogel J. Noncoding RNA control of the making and breaking of sugars. Genes Dev. 2008;22:2914–2925. doi: 10.1101/gad.1717808. [DOI] [PubMed] [Google Scholar]
- 14.Horler RS, Vanderpool CK. Homologs of the small RNA SgrS are broadly distributed in enteric bacteria but have diverged in size and sequence. Nucleic Acids Res. 2009;37:5465–5476. doi: 10.1093/nar/gkp501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masse E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl Acad. Sci. USA. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masse E, Vanderpool CK, Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 2005;187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacques JF, Jang S, Prevost K, Desnoyers G, Desmarais M, Imlay J, Masse E. RyhB small RNA modulates the free intracellular iron pool and is essential for normal growth during iron limitation in Escherichia coli. Mol. Microbiol. 2006;62:1181–1190. doi: 10.1111/j.1365-2958.2006.05439.x. [DOI] [PubMed] [Google Scholar]
- 18.Guillier M, Gottesman S, Storz G. Modulating the outer membrane with small RNAs. Genes Dev. 2006;20:2338–2348. doi: 10.1101/gad.1457506. [DOI] [PubMed] [Google Scholar]
- 19.Papenfort K, Pfeiffer V, Mika F, Lucchini S, Hinton JC, Vogel J. SigmaE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol. 2006;62:1674–1688. doi: 10.1111/j.1365-2958.2006.05524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boysen A, Moller-Jensen J, Kallipolitis BH, Valentin-Hansen P, Overgaard M. Translational regulation of gene expression by an anaerobically induced small non-coding RNA in Escherichia coli. J. Biol. Chem. 2010;285:10690–10702. doi: 10.1074/jbc.M109.089755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Durand S, Storz G. Reprogramming of anaerobic metabolism by the FnrS small RNA. Mol. Microbiol. 2010;75:1215–1231. doi: 10.1111/j.1365-2958.2010.07044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 23.Christiansen JK, Nielsen JS, Ebersbach T, Valentin-Hansen P, Sogaard-Andersen L, Kallipolitis BH. Identification of small Hfq-binding RNAs in Listeria monocytogenes. RNA. 2006;12:1383–1396. doi: 10.1261/rna.49706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sousa SA, Ramos CG, Moreira LM, Leitao JH. The hfq gene is required for stress resistance and full virulence of Burkholderia cepacia to the nematode Caenorhabditis elegans. Microbiology. 2010;156:896–908. doi: 10.1099/mic.0.035139-0. [DOI] [PubMed] [Google Scholar]
- 25.Chao Y, Vogel J. The role of Hfq in bacterial pathogens. Curr. Opin. Microbiol. 2010;13:24–33. doi: 10.1016/j.mib.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 26.Toledo-Arana A, Repoila F, Cossart P. Small noncoding RNAs controlling pathogenesis. Curr. Opin. Microbiol. 2007;10:182–188. doi: 10.1016/j.mib.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 27.Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai YF. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet. 2001;357:1225–1240. doi: 10.1016/s0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 28.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 29.Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 2003;48:1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 30.Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007;21:1353–1366. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huntzinger E, Boisset S, Saveanu C, Benito Y, Geissmann T, Namane A, Lina G, Etienne J, Ehresmann B, Ehresmann C, et al. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 2005;24:824–835. doi: 10.1038/sj.emboj.7600572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geisinger E, Adhikari RP, Jin R, Ross HF, Novick RP. Inhibition of rot translation by RNAIII, a key feature of agr function. Mol. Microbiol. 2006;61:1038–1048. doi: 10.1111/j.1365-2958.2006.05292.x. [DOI] [PubMed] [Google Scholar]
- 33.Anderson KL, Dunman PM. Messenger RNA turnover processes in Escherichia coli, Bacillus subtilis, and emerging studies in Staphylococcus aureus. Int. J. Microbiol. 2009;2009:525491. doi: 10.1155/2009/525491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pichon C, Felden B. Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc. Natl Acad. Sci. USA. 2005;102:14249–14254. doi: 10.1073/pnas.0503838102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marchais A, Naville M, Bohn C, Bouloc P, Gautheret D. Single-pass classification of all noncoding sequences in a bacterial genome using phylogenetic profiles. Genome Res. 2009;19:1084–1092. doi: 10.1101/gr.089714.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geissmann T, Chevalier C, Cros MJ, Boisset S, Fechter P, Noirot C, Schrenzel J, Francois P, Vandenesch F, Gaspin C, et al. A search for small noncoding RNAs in Staphylococcus aureus reveals a conserved sequence motif for regulation. Nucleic Acids Res. 2009;37:7239–7257. doi: 10.1093/nar/gkp668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abu-Qatouseh LF, Chinni SV, Seggewiss J, Proctor RA, Brosius J, Rozhdestvensky TS, Peters G, von Eiff C, Becker K. Identification of differentially expressed small non-protein-coding RNAs in Staphylococcus aureus displaying both the normal and the small-colony variant phenotype. J. Mol. Med. 2010;88:565–75. doi: 10.1007/s00109-010-0597-2. [DOI] [PubMed] [Google Scholar]
- 38.Anderson KL, Roberts C, Disz T, Vonstein V, Hwang K, Overbeek R, Olson PD, Projan SJ, Dunman PM. Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J. Bacteriol. 2006;188:6739–6756. doi: 10.1128/JB.00609-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts C, Anderson KL, Murphy E, Projan SJ, Mounts W, Hurlburt B, Smeltzer M, Overbeek R, Disz T, Dunman PM. Characterizing the effect of the Staphylococcus aureus virulence factor regulator, SarA, on log-phase mRNA half-lives. J. Bacteriol. 2006;188:2593–2603. doi: 10.1128/JB.188.7.2593-2603.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng HL, Novick RP, Kreiswirth B, Kornblum J, Schlievert P. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 1988;170:4365–4372. doi: 10.1128/jb.170.9.4365-4372.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iordanescu S, Surdeanu M. Two restriction and modification systems in Staphylococcus aureus NCTC8325. J. Gen. Microbiol. 1976;96:277–281. doi: 10.1099/00221287-96-2-277. [DOI] [PubMed] [Google Scholar]
- 42.Cheung AL, Eberhardt KJ, Chung E, Yeaman MR, Sullam PM, Ramos M, Bayer AS. Diminished virulence of a sar-/agr- mutant of Staphylococcus aureus in the rabbit model of endocarditis. J. Clin. Invest. 1994;94:1815–1822. doi: 10.1172/JCI117530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Booth MC, Cheung AL, Hatter KL, Jett BD, Callegan MC, Gilmore MS. Staphylococcal accessory regulator (sar) in conjunction with agr contributes to Staphylococcus aureus virulence in endophthalmitis. Infect. Immun. 1997;65:1550–1556. doi: 10.1128/iai.65.4.1550-1556.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kreiswirth BN, Lofdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 45.Bateman BT, Donegan NP, Jarry TM, Palma M, Cheung AL. Evaluation of a tetracycline-inducible promoter in Staphylococcus aureus in vitro and in vivo and its application in demonstrating the role of sigB in microcolony formation. Infect. Immun. 2001;69:7851–7857. doi: 10.1128/IAI.69.12.7851-7857.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 47.Rigoulay C, Entenza JM, Halpern D, Widmer E, Moreillon P, Poquet I, Gruss A. Comparative analysis of the roles of HtrA-like surface proteases in two virulent Staphylococcus aureus strains. Infect. Immun. 2005;73:563–572. doi: 10.1128/IAI.73.1.563-572.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oh ET, So JS. A rapid method for RNA preparation from Gram-positive bacteria. J. Microbiol. Methods. 2003;52:395–398. doi: 10.1016/s0167-7012(02)00218-x. [DOI] [PubMed] [Google Scholar]
- 49.Douchin V, Bohn C, Bouloc P. Down-regulation of porins by a small RNA bypasses the essentiality of the regulated intramembrane proteolysis protease RseP in Escherichia coli. J. Biol. Chem. 2006;281:12253–12259. doi: 10.1074/jbc.M600819200. [DOI] [PubMed] [Google Scholar]
- 50.Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 51.Bensing BA, Meyer BJ, Dunny GM. Sensitive detection of bacterial transcription initiation sites and differentiation from RNA processing sites in the pheromone-induced plasmid transfer system of Enterococcus faecalis. Proc. Natl Acad. Sci. USA. 1996;93:7794–7799. doi: 10.1073/pnas.93.15.7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Antal M, Bordeau V, Douchin V, Felden B. A small bacterial RNA regulates a putative ABC transporter. J. Biol. Chem. 2005;280:7901–7908. doi: 10.1074/jbc.M413071200. [DOI] [PubMed] [Google Scholar]
- 53.Benito Y, Kolb FA, Romby P, Lina G, Etienne J, Vandenesch F. Probing the structure of RNAIII, the Staphylococcus aureus agr regulatory RNA, and identification of the RNA domain involved in repression of protein A expression. RNA. 2000;6:668–679. doi: 10.1017/s1355838200992550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hartz D, McPheeters DS, Traut R, Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- 55.Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JC, Vogel J. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 57.Bury-Mone S, Nomane Y, Reymond N, Barbet R, Jacquet E, Imbeaud S, Jacq A, Bouloc P. Global analysis of extracytoplasmic stress signaling in Escherichia coli. PLoS Genet. 2009;5:e1000651. doi: 10.1371/journal.pgen.1000651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Berghoff BA, Glaeser J, Sharma CM, Vogel J, Klug G. Photooxidative stress-induced and abundant small RNAs in Rhodobacter sphaeroides. Mol. Microbiol. 2009;74:1497–1512. doi: 10.1111/j.1365-2958.2009.06949.x. [DOI] [PubMed] [Google Scholar]
- 60.Case CC, Simons EL, Simons RW. The IS10 transposase mRNA is destabilized during antisense RNA control. EMBO J. 1990;9:1259–1266. doi: 10.1002/j.1460-2075.1990.tb08234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma C, Simons RW. The IS10 antisense RNA blocks ribosome binding at the transposase translation initiation site. EMBO J. 1990;9:1267–1274. doi: 10.1002/j.1460-2075.1990.tb08235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu L, Inoue K, Yoshizumi S, Kobayashi H, Zhang Y, Ouyang M, Kato F, Sugai M, Inouye M. Staphylococcus aureus MazF specifically cleaves a pentad sequence, UACAU, which is unusually abundant in the mRNA for pathogenic adhesive factor SraP. J. Bacteriol. 2009;191:3248–3255. doi: 10.1128/JB.01815-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, et al. Rfam: updates to the RNA families database. Nucleic Acids Res. 2009;37:D136–D140. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grundy FJ, Henkin TM. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol. Microbiol. 1998;30:737–749. doi: 10.1046/j.1365-2958.1998.01105.x. [DOI] [PubMed] [Google Scholar]
- 65.Epshtein V, Mironov AS, Nudler E. The riboswitch-mediated control of sulfur metabolism in bacteria. Proc. Natl Acad. Sci. USA. 2003;100:5052–5056. doi: 10.1073/pnas.0531307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat. Struct. Biol. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- 67.Naville M, Gautheret D. Transcription attenuation in bacteria: theme and variations. Brief. Funct. Genomic Proteomic. 2009;8:482–492. doi: 10.1093/bfgp/elp025. [DOI] [PubMed] [Google Scholar]
- 68.Grundy FJ, Rollins SM, Henkin TM. Interaction between the acceptor end of tRNA and the T box stimulates antitermination in the Bacillus subtilis tyrS gene: a new role for the discriminator base. J. Bacteriol. 1994;176:4518–4526. doi: 10.1128/jb.176.15.4518-4526.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Langbein I, Bachem S, Stulke J. Specific interaction of the RNA-binding domain of the bacillus subtilis transcriptional antiterminator GlcT with its RNA target, RAT. J. Mol. Biol. 1999;293:795–805. doi: 10.1006/jmbi.1999.3176. [DOI] [PubMed] [Google Scholar]
- 70.Knezevic I, Bachem S, Sickmann A, Meyer HE, Stulke J, Hengstenberg W. Regulation of the glucose-specific phosphotransferase system (PTS) of Staphylococcus carnosus by the antiterminator protein GlcT. Microbiology. 2000;146(Pt 9):2333–2342. doi: 10.1099/00221287-146-9-2333. [DOI] [PubMed] [Google Scholar]
- 71.Muckstein U, Tafer H, Hackermuller J, Bernhart SH, Stadler PF, Hofacker IL. Thermodynamics of RNA-RNA binding. Bioinformatics. 2006;22:1177–1182. doi: 10.1093/bioinformatics/btl024. [DOI] [PubMed] [Google Scholar]
- 72.Hiron A, Borezee-Durant E, Piard JC, Juillard V. Only one of four oligopeptide transport systems mediates nitrogen nutrition in Staphylococcus aureus. J. Bacteriol. 2007;189:5119–5129. doi: 10.1128/JB.00274-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermuller J, Reinhardt R, et al. The primary transcriptome of the major human pathogen. Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 74.Chan PF, Foster SJ, Ingham E, Clements MO. The Staphylococcus aureus alternative sigma factor sigmaB controls the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model. J. Bacteriol. 1998;180:6082–6089. doi: 10.1128/jb.180.23.6082-6089.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chan PF, Foster SJ. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J. Bacteriol. 1998;180:6232–6241. doi: 10.1128/jb.180.23.6232-6241.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chan PF, Foster SJ. The role of environmental factors in the regulation of virulence-determinant expression in Staphylococcus aureus 8325-4. Microbiology. 1998;144(Pt 9):2469–2479. doi: 10.1099/00221287-144-9-2469. [DOI] [PubMed] [Google Scholar]
- 77.Horsburgh MJ, Clements MO, Crossley H, Ingham E, Foster SJ. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect. Immun. 2001;69:3744–3754. doi: 10.1128/IAI.69.6.3744-3754.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Horsburgh MJ, Ingham E, Foster SJ. In Staphylococcus aureus, Fur is an interactive regulator with PerR, contributes to virulence, and Is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J. Bacteriol. 2001;183:468–475. doi: 10.1128/JB.183.2.468-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McDevitt D, Francois P, Vaudaux P, Foster TJ. Identification of the ligand-binding domain of the surface-located fibrinogen receptor (clumping factor) of Staphylococcus aureus. Mol. Microbiol. 1995;16:895–907. doi: 10.1111/j.1365-2958.1995.tb02316.x. [DOI] [PubMed] [Google Scholar]
- 80.Horsburgh MJ, Aish JL, White IJ, Shaw L, Lithgow JK, Foster SJ. sigmaB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 2002;184:5457–5467. doi: 10.1128/JB.184.19.5457-5467.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Borezee-Durant E, Hiron A, Piard JC, Juillard V. Dual role of the oligopeptide permease Opp3 during growth of Staphylococcus aureus in milk. Appl. Environ. Microbiol. 2009;75:3355–3357. doi: 10.1128/AEM.02819-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Urbanowski ML, Stauffer LT, Stauffer GV. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol. Microbiol. 2000;37:856–868. doi: 10.1046/j.1365-2958.2000.02051.x. [DOI] [PubMed] [Google Scholar]
- 83.Sharma CM, Darfeuille F, Plantinga TH, Vogel J. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev. 2007;21:2804–2817. doi: 10.1101/gad.447207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McArthur SD, Pulvermacher SC, Stauffer GV. The Yersinia pestis gcvB gene encodes two small regulatory RNA molecules. BMC Microbiol. 2006;6:52. doi: 10.1186/1471-2180-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Moreno MS, Schneider BL, Maile RR, Weyler W, Saier MH., Jr Catabolite repression mediated by the CcpA protein in Bacillus subtilis: novel modes of regulation revealed by whole-genome analyses. Mol. Microbiol. 2001;39:1366–1381. doi: 10.1111/j.1365-2958.2001.02328.x. [DOI] [PubMed] [Google Scholar]
- 86.Seidl K, Goerke C, Wolz C, Mack D, Berger-Bachi B, Bischoff M. Staphylococcus aureus CcpA affects biofilm formation. Infect. Immun. 2008;76:2044–2050. doi: 10.1128/IAI.00035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Seidl K, Muller S, Francois P, Kriebitzsch C, Schrenzel J, Engelmann S, Bischoff M, Berger-Bachi B. Effect of a glucose impulse on the CcpA regulon in Staphylococcus aureus. BMC Microbiol. 2009;9:95. doi: 10.1186/1471-2180-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Heidrich N, Moll I, Brantl S. In vitro analysis of the interaction between the small RNA SR1 and its primary target ahrC mRNA. Nucleic Acids Res. 2007;35:4331–4346. doi: 10.1093/nar/gkm439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Preis H, Eckart RA, Gudipati RK, Heidrich N, Brantl S. CodY activates transcription of a small RNA in Bacillus subtilis. J. Bacteriol. 2009;191:5446–5457. doi: 10.1128/JB.00602-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.