Abstract
The chromatin-associated enzyme PARP1 has previously been suggested to ADP-ribosylate histones, but the specific ADP-ribose acceptor sites have remained enigmatic. Here, we show that PARP1 covalently ADP-ribosylates the amino-terminal histone tails of all core histones. Using biochemical tools and novel electron transfer dissociation mass spectrometric protocols, we identify for the first time K13 of H2A, K30 of H2B, K27 and K37 of H3, as well as K16 of H4 as ADP-ribose acceptor sites. Multiple explicit water molecular dynamics simulations of the H4 tail peptide into the catalytic cleft of PARP1 indicate that two stable intermolecular salt bridges hold the peptide in an orientation that allows K16 ADP-ribosylation. Consistent with a functional cross-talk between ADP-ribosylation and other histone tail modifications, acetylation of H4K16 inhibits ADP-ribosylation by PARP1. Taken together, our computational and experimental results provide strong evidence that PARP1 modifies important regulatory lysines of the core histone tails.
INTRODUCTION
Histone proteins form the nucleosome, which is the fundamental repeating unit of chromatin (1,2). Each nucleosome contains two heterodimers of the core histones H2A and H2B, one tetramer of the core histones H3 and H4, and 146 bp of DNA (3). Dynamic chromatin structures are governed in part by post-translational modifications of the histones, modification of nucleotides, remodeling of nucleosomes, and by non-coding RNAs or non-histone DNA-binding proteins (4). The amino-terminal tails of the core histone proteins protrude from the nucleosome. They appear to be unstructured and are believed to participate in the formation of higher order chromatin structures by mediating internucleosomal interactions and to contact the linker DNA (3,5). A large number of residues within the histones are modified by post-translational modifications including acetylation, phosphorylation, methylation, ubiquitination and ADP-ribosylation, which occur in distinct patterns (6). Recent work has provided compelling evidence that these modifications influence the functional properties of chromatin.
Histones have long been known as substrates for ADP-ribosylation in vivo (7). Histones isolated from rat liver nuclei and HeLa cells incubated with radioactive NAD+ revealed that the linker histone H1 and all core histones, H2A, H2B, H3 and H4, are ADP-ribosylated, although to a variable extent (8–12). An unresolved issue regarding the mechanism of ADP-ribosylation of histones is whether this modification primarily occurs at the globular histone domains or at their unstructured amino-terminals tails. Moreover, unconfirmed ADP-ribose acceptor amino acids have previously only been proposed for H1 and H2B, but not for H2A, H3 and H4 (11,13–15).
Poly(ADP-ribose) polymerase 1 (ARTD1/PARP1) is a nuclear chromatin-associated multifunctional enzyme that is present in most eukaryotes apart from yeast (16). The enzyme is responsible for most of the cellular poly(ADP-ribose) formation. Targets of PARP1′s enzymatic activity include a variety of nuclear proteins, most prominently PARP1 itself, as well as histone proteins (17). Among the six PARP family members, PARP2 has the highest similarity with PARP1 (43% sequence identity in the catalytic domain) (18). PARP2 displays similar automodification properties as PARP1 and may account for the residual poly(ADP-ribose) synthesis observed in PARP1 knockout mice.
Recently, we showed that individual lysine residues of PARP1 and PARP2 function as acceptor sites for auto-ADP-ribosylation and not, as previously assumed, glutamic acid residues (19,20). Here, we report PARP1-mediated ADP-ribosylation of the core histone proteins. We found that PARP1, but not PARP2, ADP-ribosylates core histone proteins at their amino-terminal tails. Mass spectrometry of ADP-ribosylated histone peptides revealed that the lysine residues K13 of H2A, K30 of H2B, K27 and K37 of H3, as well as K16 of H4, are specifically ADP-ribosylated by PARP1. Acetylation of H4K16 or mutation of this residue to an alanine abrogated ADP-ribosylation. Molecular dynamics (MD) simulations of tetra- and octapeptide segments of the amino terminal tail of H4 indicated that two positively charged side chains at positions n and n + 1 in the histone sequence, which point in opposite directions, are engaged in favorable electrostatic interactions with two acidic PARP1 residues at the positions 988 and 756, buried in the catalytic cleft and on a loop at the entrance of the cleft, respectively. Taken together, our results reveal that PARP1 specifically modifies lysine residues of the core histone tails, which are known to control chromatin structure and function.
MATERIALS AND METHODS
Chemicals and antibodies
32P-NAD+ was purchased from PerkinElmer. NAD+ was obtained from Sigma-Aldrich. Anti-PAR (LP-96-10) antibody was from Becton Dickinson. Anti-PARP1 antibody was generated in this laboratory. PARP-inhibitor DAM-TIQ-A was obtained from Alexis Biochemicals, PJ34 was purchased from Enzo Life Science, 3-amino-benzamide (3AB) was from Sigma-Aldrich. Peptides were from PiProteomics. ProbondTM nickel beads were from Invitrogen. Glutathione-Sepharose 4B affinity beads were from GE-healthcare.
Plasmids
The baculovirus expression vectors pQE-TriSystem (Qiagen) and BacPak8 (Clontech) were used for the expression of recombinant proteins in Sf21 cells as described (21,22). pGEX-2T vectors (GE healthcare) were used for the cloning and expression of GST-fusion proteins. Full-length and truncated histone proteins were expressed in pET3a or pET3d vectors, as described (3). Sequencing of plasmids was performed by Microsynth (Balgach, CH).
Cloning, expression and purification of recombinant proteins
Human PARP1 and PARP2 were cloned, expressed and purified as described (19). Full-length histone proteins were generated as described (3). GST-histone tail fusion proteins were generated by PCR and cloned into pGEX-2T vector. Truncated and mutated versions of GST-fusion proteins were generated by cloning with BstBI and EcoRI restriction sites. Primers for PCR and direct cloning were obtained from Sigma-Aldrich and Microsynth. pGEX-2T plasmids were transformed into BL21(DE3) bacteria and expression was induced by addition of 1 mM IPTG into LB-medium for 3 h at 30°C. After centrifugation and resuspension in EBC-buffer (120 mM NaCl, 50 mM Tris–pH 8.0, 0.5% NP-40, 5 mM DTT, 1 mM PMSF), bacteria were lysed by the addition of lysozyme (0.5 mg/ml) and the DNA was sheared by a French Press. After centrifugation the supernatant was taken and the proteins were bound to GST-beads and washed extensively with EBC buffer. The GST-beads with the bound GST-fusion proteins were equilibrated with ADP-ribosylation buffer (50 mM Tris–HCl, pH 8.0, 4 mM MgCl2, 250 µM DTT, 20 mM NaCl, 1 µg/ml protease inhibitors pepstatin, leupeptin and bestatin) and after extensive washing eluted from the GST-beads with 10 mM reduced glutathione in ADP-ribosylation buffer.
ADP-ribosylation assays
32P-NAD+ ADP-ribosylation was performed as previously described (19). Briefly, 20 µg histone mix from calf thymus (Roche) were incubated with 10 pmol PARP1 or PARP2 in a 25 µl reaction, containing 5 pmol annealed EcoRI-linker DNA and 100 nM 32P-NAD+ in ADP-ribosylation buffer (50 mM Tris–HCl, pH 8.0, 4 mM MgCl2, 250 µM DTT, 20 mM NaCl, 1 µg/ml protease inhibitors pepstatin, leupeptin and bestatin) for 10 min at 30°C. ADP-ribosylation of full-length or truncated single histones was performed with 3 µg full-length or truncated histone proteins and 10 pmol PARP1 in a 25 µl reaction. ADP-ribosylation of GST-histone tail fusion proteins was performed with 1.5 µg purified fusion protein, together with 10 pmol of PARP1. An amount of 5 pmol EcoRI-linker DNA was always included into the reaction to activate PARP1 enzymatic activity. ADP-ribosylation assays with full-length or truncated histones were resolved by a 10–20% acrylamide gradient SDS-gel of 15 cm length (Amersham). Assays with GST-fusion proteins were resolved on standard 12% acrylamide mini-gels (Hoefer). Radiolabeled proteins were visualized by exposure to X-ray films or by quantification through the phospho-imager system (Molecular Dynamics).
For mass spectrometry, 22 nmol biotinylated H2A (aa 3–23), H2B (aa 18–37), H3 (aa 23–42), H4 (aa 1–22) or H4K16ac (aa 1–22) peptides were incubated with 10 pmol PARP1, 5 pmol annealed EcoRI-linker and 100 or 500 µM NAD+ for 15 min at 30°C in ADP-ribosylation buffer without protease inhibitors. The reaction was stopped by the addition of 3AB to a final concentration of 20 mM and subsequently 1 µg GST-ARH3 was added to the reaction and further incubated for 1 h at 30°C. The samples were acetone precipitated and the pellet was dissolved in distilled water.
GST-pulldown
Glutathione–sepharose affinity beads were incubated with a bacterial extract of the GST-fusion protein expression in EBC-buffer. After extensive washing, the GST-beads were equilibrated with ADP-ribosylation buffer containing 50 mM NaCl. After washing, 10 pmol purified PARP1 was added to the beads in a total volume of 300 µl ADP-ribosylation buffer. The protein mixture was incubated for 2 h at 4°C on rolls to allow for protein–protein interaction. The samples were washed again with ADP-ribosylation buffer and resolved on standard SDS–PAGE and subsequent western blotting with anti-PARP1 antibody.
Microvolume C-18 reversed phase purification
An amount of 22 nmol H4-peptide (aa 1–22) was incubated with 10 pmol PARP1, 5 pmol EcoRI-linker DNA and 100 nM 32P-NAD+ in 25 µl ADP-ribosylation buffer for 15 min at 30°C. The reaction was stopped by the addition of 3AB to a final concentration of 20 mM. The C18 reversed phase ZipTip (Millipore) was pre-wetted with 100% methanol and equilibrated with 3% (v/v) acetonitrile. The peptides were bound onto the column and subsequently washed extensively with 3% (v/v) acetonitrile. Bound peptides were eluted by 60% (v/v) acetonitrile directly into scintillation liquid. In the control reaction, peptides were added after the addition of 3AB to the reaction. Scintillation counts were measured and normalized to the control reaction and the relative increase in scintillation counts was calculated (cpm).
Mass spectrometry
Electron transfer dissociation (ETD) experiments were performed on a hybrid LTQ Orbitrap XL ETD mass spectrometer (Thermo Scientific, Bremen, Germany) coupled to an Eksigent nano LC system (Eksigent Technologies) and the samples were analyzed by reversed-phase liquid chromatography nanospray tandem mass spectrometry (LC–MS/MS).
Peptides were resuspended in 3% ACN and 0.2% formic acid, loaded from a cooled (10°C) Spark Holland autosampler (Emmen, Holland) and separated using an ACN/water solvent system containing 0.2% formic acid with a flow rate of 200 nl/min. Separation of the peptides was performed on a 10-cm long fused silica column (75 µm i.d.; BGB Analytik) in-house packed with 3 μm, 200 Å pore size C18 resin (Michrom BioResources, CA). Elution was achieved using a gradient of 3−48% ACN in 35 min, 48−80% ACN in 4 min and 80% ACN for 7 min.
One scan cycle was comprised of a survey full scan MS spectra from m/z 300 to 2000 acquired in the FT-Orbitrap with a resolution of R = 60 000 at m/z 400, followed by up to four sequential data-dependent ETD MS/MS scans with detection of the ETD fragment ions in the linear ion trap. AGC target values were 5 × 105 for full FTMS scans, 104 for ion trap MSn scans. Anion target value was 106 and supplementary activation was employed to enhance the fragmentation efficiency for doubly-charged precursors and charge state dependent ETD time was enabled. Data dependent decision tree was used in order to control ETD dissociation based on charge state and m/z. The ETD reaction time was set at 100 ms and the isolation width was m/z 2. For all experiments dynamic exclusion was used with one repeat count, 30-s repeat duration, and 10-s exclusion duration.
The instrument was calibrated externally according to the manufacturer's instructions. Orbitrap mass spectra were acquired using internal lock mass calibration on m/z 429.088735 and 445.120025. Spectra generated by ETD were processed using Mascot Distiller 2.2 (Matrix Science) and data was searched against a SwissProt human database using Mascot 2.2.0 to find best matching sequences. Detailed spectra analysis was done by manual evaluation.
Molecular dynamics
It is computationally prohibitive to dock the full-length histone tails to PARP1. Therefore, only the relevant tetra- and octapeptide segments of the histone tails (abbreviated H-peptides hereafter) were taken into account in the present study. A two-step procedure was used to investigate the binding of the H-peptides to PARP1 (PDB ID: 1A26). Preliminary binding modes of the flexible H-peptides into the rigid structure of the catalytic domain of PARP1 were obtained by an in-house developed docking program, which uses a combination of simulated annealing and genetic algorithm optimization of position, orientation, and rotatable bonds of the ligand (Zhao et al., unpublished). Explicit solvent MD simulations of the H-peptide/PARP1 complexes were then used to investigate the structural stability of the poses obtained by docking. In both docking and MD simulations, the N-terminus and C-terminus of the H-peptides were capped by neutral groups (acetyl and N-methlyamide, respectively) to take into account the fact that they belong to a longer polypeptide chain. To reproduce physiological pH conditions, the side chains of aspartates and glutamates were negatively charged, those of lysines and arginines were positively charged, while all other residues were considered neutral. The MD simulations were performed with the program NAMD (23) using the all-atom CHARMM PARAM27 force field (24) and the TIP3P model of water (25). The H-peptide/PARP1 complexes were inserted into a cubic water box, with a minimal distance of 12 Å between any solute atom and the boundary of the box. Chloride and sodium ions were added to neutralize the system and approximate a salt concentration of 150 mM. The water molecules overlapping with the solute atoms or the ions were removed, if the distance between the water oxygen and any atom of the complex or any ion was smaller than 2.4 Å. Periodic boundary conditions were applied to avoid finite-size effects. Electrostatic interactions were calculated within a cutoff of 12 Å, while long-range electrostatic effects were taken into account by the particle mesh Ewald summation method (26). Van der Waals interactions were treated with the use of a switch function starting at 10 Å and turning off at 12 Å, which is the default of the all-atom CHARMM force field. The temperature was kept constant at 300 K by the Langevin temperature control with a damping coefficient of 5 ps−1, while the pressure was held constant at 1 atm by applying a pressure piston. Before the production runs, water molecules and ions were subjected to energy minimization for 6000 steps, and a 1-ns equilibration with harmonic constraints applied to the positions of the C-alpha atoms. The covalent bonds involving hydrogen atoms were constrained by means of the SHAKE algorithm, and the dynamics were integrated with a time step of 2 fs. Snapshots were saved every 2 ps for trajectory analysis. Two MD runs with different initial distribution of velocities were carried out for each of the H4 peptides AKRH and AKRHRKIL for a total simulation time of 150 ns for each peptide. Analysis of the trajectories was carried out with the programs CHARMM (27) and WORDOM (28).
RESULTS
PARP1 modifies core histones
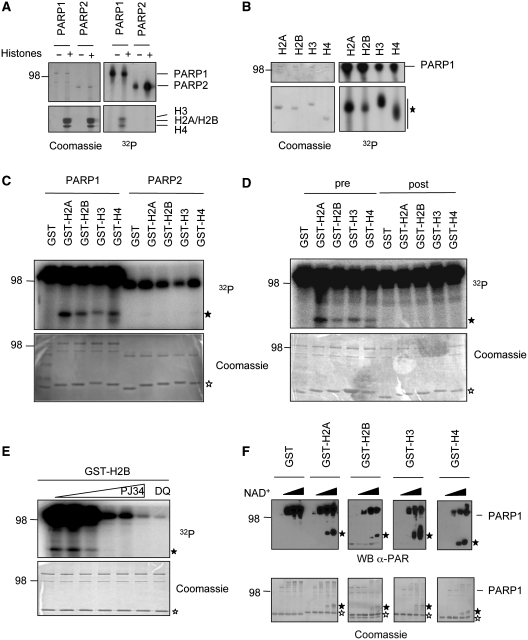
Recently, we reported the ADP-ribosylation of distinct lysine residues of PARP1 and PARP2 in cis (19,20). Since core histones were described earlier to be modified by PARP1 in vitro and in vivo [(16) and Supplementary Figure S1A and B], we set out to investigate which residues of H2A, H2B, H3 and H4 would be modified by PARP1, and possibly also by PARP2 in trans. First, human PARP1 and PARP2 were expressed and purified from insect cells using the baculovirus-system and were subsequently incubated with full-length histones isolated from calf thymus together with 100 nM radiolabeled NAD+. Although automodification of both PARP1 and PARP2 was easily detectable, only PARP1 displayed detectable trans-ADP-ribosylation of all four core histones, indicating a clear difference in the substrate specificity between PARP1 and PARP2 (Figure 1A). Similar experiments with bacterially expressed and purified single histone proteins revealed that, indeed, all four histones are modified by PARP1 (Figure 1B).
Figure 1.
PARP1 covalently modifies all four core histone tails. (A) PARP1 trans-ADP-ribosylates histones isolated from calf thymus. An amount of 10 pmol recombinant PARP1 or PARP2 were incubated for 15 min (PARP1) or 30 min (PARP2) at 30°C with 5 pmol EcoRI-linker DNA, 100 nM radiolabeled NAD+ and 20 µg extracted histones from calf thymus. The reaction was stopped by addition of SDS-lysis buffer and the proteins were resolved on a 10–20% SDS–PAGE gradient gel. The gel was stained with Coomassie-R250 and incorporated 32P was visualized by autoradiography. (B) Recombinant expressed and purified histones (3 µg) were ADP-ribosylated by PARP1 as in Figure 1A. (C) An amount of 1.5 µg of each purified GST-histone tail were used in PARP1 or PARP2 mediated trans-ADP-ribosylation reactions for 5 min at 30°C. (D) An amount of 1.5 µg of each purified GST-histone tail were used in PARP1 mediated trans-ADP-ribosylation reactions for 5 min at 30°C. Histone tails were either included during the reaction (pre) or added after the reaction had been stopped with a 100-fold excess of 3AB over radiolabeled NAD+ (post) to exclude non-covalent interaction of the histone tails with PAR. (E) Trans-ADP-ribosylation of the H2B tail is inhibited by the PARP-inhibitors PJ34 (0.01–100 µM) and DAM-TIQ-A (10 µM). (F) GST-histones were incubated with PARP1, EcoRI linker and increasing concentrations of NAD+ (0, 10, 100, 400 µM) for 10 min at 30°C. Poly(ADP-ribose) formation was assessed through western blotting with anti-PAR (LP-96-10) antibody. Unmodified GST-histone tails are marked by an empty asterisk, PARylated GST-histone tails are marked by a filled asterisk.
PARP1 mono- and poly(ADP-ribosyl)ates amino-terminal tails of core histone proteins covalently
Earlier reports suggested that histones are mainly ADP-ribosylated at their amino-terminal tails (13,29). To test whether PARP1-mediated ADP-ribosylation occurred at the amino-terminal tails or at the globular histone folds, we incubated the globular domains of histones H2B, H3 and H4 together with PARP1 and compared their ADP-ribosylation to the full-length counterpart (Supplementary Figure S2A). ADP-ribosylation of the globular domains was reduced in comparison to full-length histones, implying that indeed the amino-terminal tails are required for modification by PARP1. We then expressed the different histone tails as GST fusion proteins in bacteria and incubated them with purified PARP1 and radiolabeled NAD+. PARP1 was able to modify all four histone tails; whereas comparable PARP2-mediated ADP-ribosylation of histone tails was not detectable (Figure 1C and Supplementary Figure S2B). This finding suggests that the core histone tails are substrates specifically for PARP1 but not for PARP2 in vitro. Previous reports indicated that histones can interact with poly(ADP-ribose) non-covalently via a PAR-binding motif (30). Since the tested histone tails we analyzed did not contain this motif, it is highly unlikely that the observed labeling represented non-covalently attached PAR. To exclude this notion experimentally, the histone tail fusion proteins were added to the ADP-ribosylation reaction either together with PARP1 or after the enzymatic reaction had been stopped by addition of the PARP-inhibitor 3AB. The addition of the histone tails after the reaction did not result in their modification (Figure 1D, right panel), implying that the observed ADP-ribosylation is a covalent modification. To test whether the observed modification is an enzymatic reaction, the reaction was repeated in presence of the PARP-inhibitor PJ34. Increasing amounts of PJ34 abolished trans-ADP-ribosylation of the H2B tail, as well as automodification of PARP1 (Figure 1E). Similar results were obtained with a second PARP-inhibitor, DAM-TIQ-A (Figure 1E). Taken together these results indicate that PARP1 catalyzes the covalent ADP-ribosylation of histone tails.
PARP1 is a mono- and a poly(ADP-ribosyl) transferase (16). To determine whether the histone tails can be mono and/or poly(ADP-ribosyl)ated by PARP1, we incubated the histone tails with PARP1 in the presence of increasing amounts of NAD+. Poly(ADP-ribosyl)ation was determined by immunoblot analysis using an anti-PAR antibody, which recognizes only polymers of ADP-ribose (PAR). Both automodification of PARP1 and poly(ADP-ribosyl)ation of the histones were detected, indicating that PARP1 can attach long polymers of ADP-ribose onto histone tails (Figure 1F). Notably, the length of poly(ADP-ribose) chains of the histones increased proportionally to the amounts of NAD+, which led to retarded migration of the modified proteins due to an increased molecular weight (Figure 1F, filled asterisk).
ADP-ribosylation was earlier described to occur on glutamates of H2B and H1 (11,15). These findings were never directly attributed to PARP1 nor were they experimentally confirmed by mass spectrometry or amino-acid substitutions. The PARP1-mediated ADP-ribosylation we describe here occurred on the basic amino-terminal histone tails, most of which do not contain glutamates (Figure 2A). One exception is the tail of histone H2B that contains one single glutamate. Mutation of this residue to alanine did not affect the levels of incorporated ADP-ribose onto the H2B tail by PARP1 (Supplementary Figure S2C), suggesting that glutamates are dispensable and that additional residues can be efficiently ADP-ribosylated by PARP1 in trans. Furthermore, when we incubated poly-l-lysine or poly-l-glutamate with purified PARP1 and measured incorporated ADP-ribose, lysines but not glutamates were modified by PARP1 (Supplementary Figure S2D). These findings are consistent with our previous reports (19,20) and provided additional evidence that lysines are likely the primary target sites for PARP1-mediated ADP-ribosylation.
Figure 2.
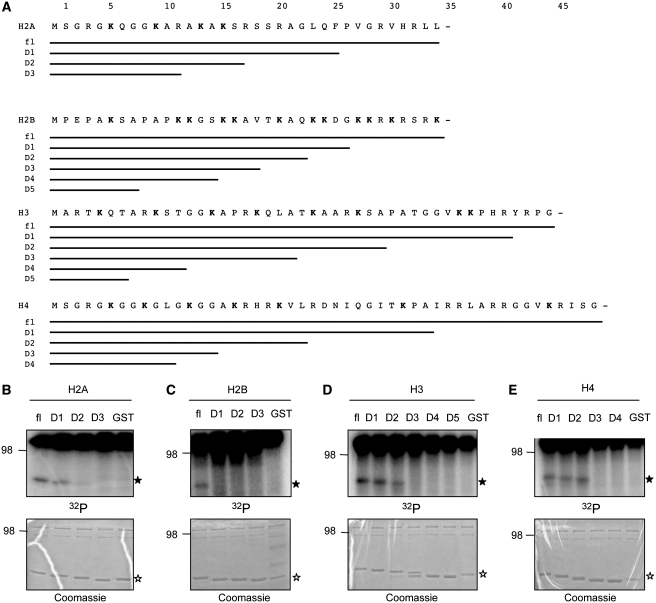
Confining the regions of putative ADP-ribose acceptor sites. (A) Schematic representation of the deletion strategy for GST-histone tails to identify the minimal ADP-ribosylated domain. (B–E) Trans-ADP-ribosylation of the indicated GST-histone deletion mutants by PARP1 with 100 nM 32P-NAD+.
Identification of ADP-ribose acceptor sites within histone tails
As the amino-terminal histone tails are frequently targeted by a variety of post-translational modifications with important physiological functions (6), we aimed at identifying the exact sites of PARP1-mediated ADP-ribosylation. To this end, we first generated a series of histone tail deletion mutants to test in ADP-ribosylation assays (Figure 2A). Successive shortening of the histone tails invariably resulted in a loss of PARP1-mediated ADP-ribosylation and defined for each histone the region comprising putative ADP-ribose acceptor sites (Figure 2B–E).
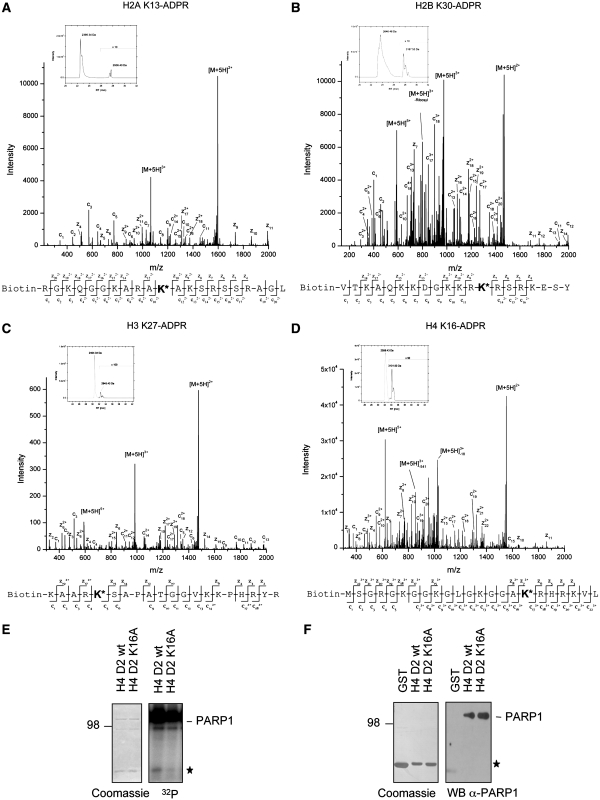
To directly identify the ADP-ribosylated amino acids within the histone tails by mass spectrometry, we used synthetic peptides covering the regions identified by our deletion strategy. We incubated these peptides with PARP1 in the presence of 100–500 µM NAD+, stopped the reactions by addition of 3AB and subsequently treated the poly(ADP-ribosyl)ated peptides with ADP-ribosylhydrolase 3 (ARH3). ARH3 is known to possess PARG-like ADP-ribose glycohydrolase activity, which hydrolyzes ester linkages between ADP-ribose units (31). Since no negatively charged amino acids (E or D), which would allow the formation of an ester linkage, were present in the polypeptides (except for H2B E35), we rationalized that treatment of the modified polypeptides with ARH3 would leave the first ADP-ribose unit bound to the peptide. The ADP-ribosylated peptides were acetone precipitated and analyzed by liquid-chromatography coupled mass-spectrometry. In the presence of PARP1 and NAD+, the attachment of a single ADP-ribose unit resulted in a mass shift of 541 Dalton (Figure 3A–D). Fragmentation of ADP-ribosylated peptides with conventional collision induced dissociation (CID) fragmentation technique completely removed the ADP-ribose moiety from the peptides, not allowing the identification of specific amino-acid acceptor sites (data not shown). In contrast, analysis of the modified peptides by electron transfer dissociation (ETD) resulted in an almost complete fragmentation of the multiply charged histone peptides, as indicated by the presence of c- and z-ions, which represent N- or C-terminal fragment ions, respectively (Figure 3A–D, Supplementary Figure S2E). Fragmentation of the mono(ADP-ribosyl)ated H2A peptide revealed specific ADP-ribosylation of K13, while H2B was mainly ADP-ribosylated at K30 (Figure 3A and B). H3 was ADP-ribosylated at K27 and K37 (Figure 3C and Supplementary Figure S2E). For H4, mass spectrometric analysis identified K16 to be ADP-ribosylated by PARP1 (Figure 3D). Control reactions performed in the presence of 500 µM NAD+ but without PARP1 or in the presence of 500µM ADP-ribose and PARP1 did not result in specific ADP-ribosylation (data not shown). The identified sites of PARP1-mediated enzymatic ADP-ribosylation represent two known sites of frequent histone modification (H3K27 and H4K16) as well as novel modification sites (H2AK13, H2BK30 and H3K37). To verify the mass spectrometric data for one histone, the H4 tail was mutated at K16 to alanine and tested for ADP-ribosylation by PARP1. The mutated H4 tail showed severely reduced ADP-ribosylation in comparison to the wild-type H4 tail (Figure 3E, filled asterisk). The reduction of ADP-ribosylation was not due to a reduced interaction of PARP1 with the mutated H4 tail, since wild-type and mutated histone tail fusion proteins were able to interact with PARP1 to comparable levels (Figure 3F).
Figure 3.
Mass spectrometric analysis of ADP-ribosylated histone peptides. (A) Extracted ion chromatogram of the biotin tagged H2A (aa 3–23) peptide incubated with 500 µM NAD+ and PARP1. The precursor masses of unmodified H2A peptide (2395.34 Da) and ADP-ribosylated H2A peptide (2936.40 Da) were plotted in a range of 10 ppm over time. ETD fragment spectrum of quintuply charged precursor ion of ADP-ribosylated H2A peptide (638.51 m/z) at K13, as indicated by the sequence plot. The spectrum shows next to the two major peaks of unfragmented charge reduced precursor ions (M+5H)n+ an almost complete series of N-terminal and C-terminal fragment ions (c-ions, z-ions, respectively). (B) Extracted ion chromatogram of the biotin tagged H2B (aa 18–37) peptide incubated with 500 µM NAD+ and PARP1 as in (A). ETD fragment spectrum of ADP-ribosylated H2B peptide (797.89 m/z) at K30, indicated by the sequence plot. (C) Extracted ion chromatogram of the biotin tagged H3 (aa 23–42) peptide incubated with PARP1 as in (A). ETD fragment spectrum of ADP-ribosylated H3 peptide (590.29 m/z) at K27, indicated by the sequence plot. (D) Extracted ion chromatogram of the biotin tagged H4 (aa 1–22) peptide incubated with 100 µM NAD+ and PARP1. ETD fragment spectrum of ADP-ribosylated H4 peptide (621.30 m/z) at K16, as indicated by the sequence plot. (E) Trans-ADP-ribosylation of the GST-H4 histone tail wild-type or K16A mutant by PARP1. (F) GST-pulldown of wild-type and mutated GST-histone H4 tails with recombinant PARP1. The GST-histone tails are marked by an asterisk.
Modeling of the histone H4 tail reveals that R17 is critical for the interaction with the catalytic domain of PARP1
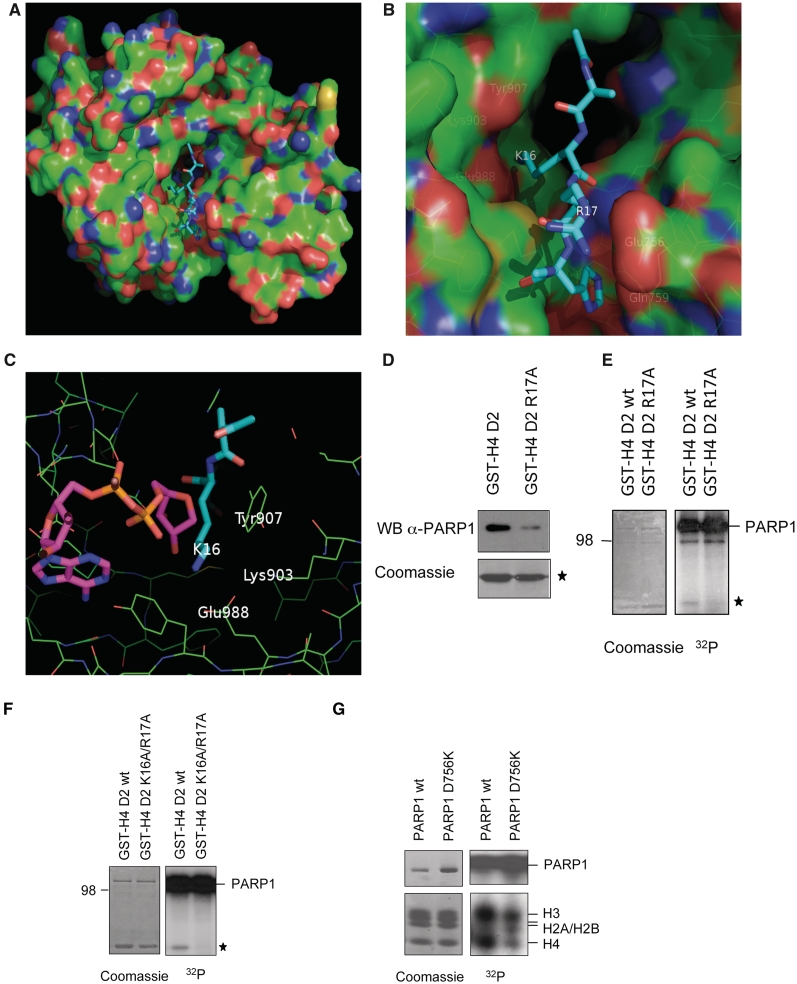
In order to test computationally, whether the tail of H4 could enter the catalytic cleft of PARP1, automatic docking followed by explicit solvent MD simulations were performed with the crystal structure from chicken PARP1 (PDB ID: 1A26), which uses the sequence numbering of human PARP1 (see ‘Materials and methods’ section). The MD simulations indicate that the H4 tetrapeptide segment AKRH (aa 15–18) binds in an extended conformation to the catalytic domain of PARP1 (Figure 4A and B). Two stable salt bridges are observed in all MD runs: between H4K16 and PARP1 Glu988 in the catalytic cleft, and between H4R17 and PARP1 Glu756 (Asp756 in human PARP1) on a loop at the entrance of the cleft. These intermolecular salt bridges lock H4 into the catalytic domain of PARP1 like two stretched arms holding two points far away from each other. In all MD runs, a single water molecule inserts between the amino group of K16 and the carboxy group of Glu988 in the first 15 ns and remains between these two charged groups until the end of the MD simulations of the tetrapeptide (Supplementary Figure S3). This water molecule occupies the same position as the water molecule that is close to Glu988 in the X-ray structure of PARP1 (water 37 in PDB code 1A26). There are two additional side chains interactions: a stacking interaction between the imidazole of H18 and the amide group of Gln759 is almost always present in all MD runs, while the hydrogen bond between the side chains of R19 and Asn906 is not very stable. In contrast to the aforementioned intermolecular salt bridges and hydrogen bonds, the K20 side chain is always exposed to solvent and very flexible. Furthermore, the backbone polar groups of the histone do not seem to be involved in hydrogen bonds with PARP1. Importantly, in all MD simulations both the N-terminal and C-terminal methyl groups point towards the solvent, which would allow the rest of the H4 polypeptide chain to position itself close to the surface of PARP1 without steric clashes. Moreover, the MD runs with the H4 tetra- and octapeptide (aa 15–22) converge towards a common extended structure of the AKRH segment with the same side chain interactions. The convergence of multiple MD simulations and the agreement with the experimental results indicate that the binding mode obtained by docking and explicit water MD is reliable. To gain insight in the putative initiation step of ADP-ribosylation, the 10 ns snapshot of the MD simulation with the H4 tetrapeptide was used for docking NAD+ into the donor site as previously published (32,33). Before docking, the nicotinamide was manually removed (Figure 4C), which mimics the NADase activity of PARP1 and creates a reactive C1-atom of the ADP-ribose that is suggested to react with the substrate amino acid (19,34,35). Interestingly, the C1-atom of the ADP-ribose is only 3.7 Å away from the ε-amino group of H4K16 and 7.8 Å from the ε-amino group of Lys903. Moreover, the distance between the catalytic active Glu988 carboxy group and the ε-amino group of H4K16 is only 3.0 Å, which would potentially allow covalent ADP-ribosylation of H4K16 (Figure 4C).
Figure 4.
Histone H4 interacts with PARP1 by salt bridges with acidic residues in the catalytic domain. (A) Representative snapshot of the binding mode saved after 10 ns MD simulation of the H4 tetrapeptide AKRH. (B) Enlarged view of the catalytic cleft for the same snapshot as in (A). Amino acids in close proximity of the H4 peptide are highlighted. PARP1 and the H4 tetrapeptide are shown in surface render and sticks, respectively. The surface is colored according to atomic elements with carbon, oxygen and nitrogen in green, red, and blue, respectively. Carbon atoms of the H4 tetrapeptide are in cyan. (C) ADP-ribose was docked into the donor-site of PARP1 catalytic domain after 10 ns MD simulation of the H4 tetrapeptide. Amino acids in close proximity of the H4 peptide are highlighted. The orientation is rotated by about 180°C with respect to (A,B). (D) GST-pulldown of wild-type and mutated GST-histone H4 tail with recombinant PARP1 and subsequent western blot with anti-PARP1 antibody. GST-histone tails are indicated by an asterisk. (E and F) Trans-ADP-ribosylation of wild-type and mutated GST-H4 tails with PARP1 and radiolabeled NAD+. Coomassie stains of the input and autoradiographies are shown. (G) Trans-ADP-ribosylation of calf-thymus extracted histones by PARP1 wild-type or PARP1 D756K mutant as in Figure 1A.
To test experimentally whether an arginine close to H4K16 is required for the interaction with PARP1, we mutated R17 (H4R17A) and analyzed the association with and the modification by PARP1. GST-pulldown experiments with recombinant PARP1 revealed that the interaction between PARP1 and the mutated H4 tail fusion protein was reduced (Figure 4D), as well as its ADP-ribosylation by PARP1 (Figure 4E). In agreement with the MD simulation and the mass spectrometry data, an H4K16A/R17A double mutant was completely defective for PARP1-mediated ADP-ribosylation (Figure 4F). Since H4R17 was suggested by molecular modeling to interact with Asp756 of PARP1, we mutated Asp756 into a lysine (which is the corresponding amino acid at this position in PARP2) and tested this mutant for its ADP-ribosylation properties. The PARP1 mutant exhibited no defect in auto(ADP-ribosyl)ation (Supplementary Figure S2F), indicating that Asp756 is not essential for automodification. This is consistent with the fact that also PARP2 is able to modify itself, although it contains a lysine at the corresponding position. Interestingly, however, the PARP1 mutant was impaired in trans-ADP-ribosylation of full-length histones (Figure 4G) and in the labeling of the H4 (1–22) peptide (Supplementary Figure S1G), confirming the MD-based prediction of the importance of this residue for stabilization of the peptide in the catalytic cleft and subsequent trans(ADP-ribosyl)ation of H4.
Acetylation of K16 inhibits ADP-ribosylation of histone H4
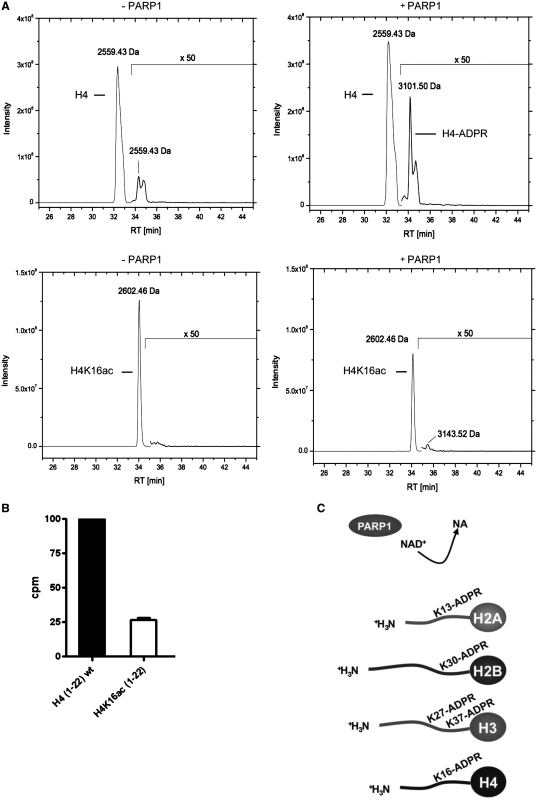
Acetylation of H4K16 occurs frequently in eukaryotic cells and has been correlated with chromatin decompaction (6). If H4K16 was indeed an acceptor site for PARP1-mediated ADP-ribosylation, acetylation at that site should impair ADP-ribosylation. In order to test this hypothesis, we employed an H4 peptide (aa 1–22) chemically acetylated at K16. LC-coupled mass spectrometry of the H4K16ac peptide revealed that PARP1 was not any longer able to induce ADP-ribosylation of the acetylated peptide above background (Figure 5A). Consistent with this result, PARP1 mediated ADP-ribosylation of both peptides (unmodified and acetylated) with radiolabeled NAD+, followed by purification over a microvolume-C18 reversed phase column and subsequent measurement of incorporated radiolabeled NAD+, provided evidence that ADP-ribosylation of the acetylated peptide was severely reduced (Figure 5B). These results confirm that H4K16 is modified by PARP1 and show that acetylation of K16 severely impairs ADP-ribosylation of the H4 peptide. Together, our results led to the identification of ADP-ribose acceptor sites within the amino-terminal tails of the four core histones (Figure 5C) and imply important cross-talks with other histone modifications such as acetylation or methylation.
Figure 5.
ADP-ribosylation of the H4 peptide is impaired by H4K16 acetylation. (A) Elution profile of biotinylated H4 peptide (aa 1–22) and biotinylated H4K16ac peptide (aa 1–22) incubated with 100 μM NAD+ for 15 min without PARP1 (–PARP1) or in the presence of PARP1 (+PARP1) and subsequent ARH3 treatment. Acetone precipitated peptides were analyzed by LC–MS/MS using a C18 reversed-phase column and subsequent detection by mass spectrometry. (B) Histone H4 (aa 1–22) and acetylated H4K16ac (1–22) peptides were ADP-ribosylated with PARP1 for 15 min at 30°C with 100 nM 32P-NAD+. The peptides were purified by microvolume-C18 reversed phase columns, eluted into scintillation liquid and counted for incorporated 32P. Relative increase of counts per minutes was calculated over background (peptides added after termination of the reaction by 3AB). (C) Overview of the identified ADP-ribose acceptor sites within the amino-terminal core histone tails.
DISCUSSION
Here, we provide evidence that PARP1 covalently modifies the tails of all four core histones. We identified H2AK13, H2BK30, H3K27, H3K37 and H4K16 as specific target sites for PARP1-mediated ADP-ribosylation. Our conclusions are based on several observations: (i) mass spectrometric analyses of PARP1-mediated ADP-ribosylated peptides, (ii) loss of function experiments by site directed mutagenesis of the putative acceptor sites in recombinant histone tail fusion proteins, (iii) cross-talk of acetylation and ADP-ribosylation at the identified acceptor site in H4 and, finally, (iv) prediction of the interaction between the histone H4 tail and PARP1 by MD and subsequent confirmation with mutated proteins.
Nearly 20 years ago, ADP-ribose acceptor sites were found in histones by biochemical approaches. Several laboratories identified glutamic acid residues in histone H1 and histone H2B to be modified when they incubated chromatin from rat liver with radioactive NAD+ (11,13–15). At that time, no other PARP family member had been identified yet, and no knockout- or knockdown-system was available. Thus, it is possible that PARP1, other PARP-family members or even unrelated NAD+ consuming enzymes were responsible for the modification at the identified glutamates. In fact, we could show here by mutational analyses, that E2 of H2B is dispensable for PARP1-mediated ADP-ribosylation and that additional amino-acid residues are acceptors for ADP-ribose moieties. Consistent with this, we identify lysines in the amino terminal histone tails and in particular lysine 13 of H2A, lysine 30 of H2B, lysines 27 and 37 of histone H3, as well as lysine 16 of histone H4 as target sites for enzymatic ADP-ribosylation by PARP1, both by mass spectrometry and amino-acid substitution. Therefore, we propose the ADP-ribosylation of the ε-amino group of lysines by PARP1 as a new canonical histone tail modification.
Remarkably, explicit solvent MD revealed that the tetra- and octapeptides of H4 interact strongly with specific amino-acid side chains of the catalytic cleft of PARP1, suggesting that specific binding of a relatively short segment of H4 is sufficient to allow poly(ADP-ribosyl)ation of the histone tail. The positively charged amino acid at the +1 position of the ADP-ribosylated residue formed a salt bridge with Glu756 of chicken PARP1, which corresponds to Asp756 in human PARP1. In contrast, the corresponding amino acid in PARP2 is a lysine at position 312. Although the catalytic domains of PARP1 and PARP2 are highly similar (18), the substrate specificity of those enzymes might be regulated by subtle differences in the catalytic cleft. This could explain why PARP2 does not modify histones to a detectable extent at least in vitro (Figure 1A, C and Supplementary Figure S2B).
Application of novel mass spectrometry techniques allowed us to identify distinct amino acids as acceptors of ADP-ribose. We took advantage of the ETD technique (36), which allows the fragmentation of highly charged peptides, leaving most post-translational modifications intact. Recent publications describe the technical basis for the fragmentation of chemically ADP-ribosylated peptides by electron capture dissociation (ECD) (37) and the closely related ETD (38). It is noteworthy, that, in contrast to other reports, we observed partial fragmentation of the ADP-ribose at the phosphodiester bond by application of ETD, as revealed by the presence of a m/z 348 ion. However, conventional CID mass spectrometry of ADP-ribosylated H4 peptide did not reveal any ADP-ribose acceptor sites, since the ADP-ribose was cleaved off from the peptide during fragmentation. The commonly used CID, instead of ETD, might thus explain why numerous efforts to identify ADP-ribosylated residues failed in the past. Consequently, we would strongly recommend ETD as standard technique to detect ADP-ribosylated peptides. Of note, a previous study employing CID failed to detect ADP-ribosylated lysine residues in the catalytic PARP1 mutant E988Q (39). In summary, ETD can be expected to facilitate future investigations on ADP-ribosylated peptides, opening new opportunities to screen for ADP-ribosylated residues in a systems-biology setup.
Since poly(ADP-ribosyl)ated peptides cannot be detected by MS and PARP1 mainly attaches poly(ADP-ribose) to target proteins, we removed the poly(ADP-ribose) with ARH3, which degrades poly(ADP-ribose). However, this treatment was rather inefficient (data not shown), which could partly explain the observed low mono(ADP-ribosyl)ation of the histone peptides. Moreover, we cannot exclude that PARP1 modifies additional residues, which were not identified by these mass spectrometric analyses.
Our data provide strong evidence that PARP1-mediated ADP-ribosylation of histones occurs as post-translational modification at distinct lysine residues within the amino-terminal basic tails. From a chemical perspective, modification of a lysine residue by ADP-ribose results in an unstable Schiff base, which can undergo an Amadori rearrangement to form a stable ketoamine (19,40). It will be interesting to investigate whether histone lysine ADP-ribosylation can be reversed by a previously identified but still poorly characterized ADP-ribosyl protein lyase (41). Since the attachment of ADP-ribose not only neutralizes the positive charge of the amino-acid side chain, but instead reverses it into a negative charge, the functional consequences of lysine ADP-ribosylation can be assumed to be even more drastic than those of other modifications, such as acetylation. Therefore, the possible effects on chromatin architecture, histone dynamics, histone degradation, and histone variant incorporation may be dramatic. Possibly, ADP-ribosylation of histones interferes with other post-translational modifications of the histone tails. For example, H3K27 is methylated by EZH2 (enhancer of zeste homolog 2), which is in the polycomb group complex that is involved in maintenance of the inactive X-chromosome (42). Interestingly, PARP1 was demonstrated to participate in the maintenance of X-chromosome silencing as well (43). On the other hand, the amino-terminal tail of histone H4 was shown to be required for chromatin fiber formation, since the positively charged stretch between K16 and K20 makes internucleosomal contacts to two acidic patches on the carboxy-terminal α-helices of histone H2A (44). In addition to its function for chromatin topology, the stretch between K16 and K20 is required for the interaction with various non-histone modulators. For example, the ISWI-containing ATP dependent chromatin remodeler ACF solely engages histone H4, but is repelled, if H4K16 is acetylated (45,46). Furthermore, the chromatin remodeler Alc1 was shown to require the K16 to K20 stretch of H4 for its activity (47). Interestingly, recent reports provide evidence that the ATPase activity of Alc1 is highly stimulated by binding to poly(ADP-ribosyl)ated PARP1 (47,48). Whether ADP-ribosylated H4 would activate Alc1, remains to be investigated. Another intriguing possibility of how histone tail ADP-ribosylation could affect chromatin function is implied by a recent study showing that macrodomain-containing histone variants specifically bind to poly(ADP-ribose) generated after DNA damage (49). Using biochemical, crystallographic and state-of-the-art imaging techniques, it was shown that macroH2A1.1 senses PARP1 activation and directly binds poly(ADP-ribose) to cause a transient compaction of the chromatin. It will be interesting to investigate whether macrodomains preferentially bind to automodified PARP1 or also function as ‘readers’ of poly(ADP-ribosyl)ated histone tails.
Taken together, the work presented here sheds new light on a well known but neglected histone modification and builds the basis for future investigations exploring the function of histone lysine ADP-ribosylation in chromatin dynamics, transcription, DNA repair signaling and other nuclear processes influenced by histone modifications.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Swiss National Science Foundation (Grant 31-122421 to S.M. and M.A.); The Kanton of Zurich (to M.O.H.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Timothy Richmond (ETH Zurich, Switzerland) and Achim Leutz (Max-Delbrück-Centrum for Molecular Medicine, Berlin, Germany) for the generous provision of reagents and sequences. Brighton Samatanga (University of Zurich, Switzerland) is acknowledged for his technical support. The MD stimulations were carried out on the Schroedinger computer cluster at the University of Zurich. The authors thank Raffaella Santoro (University of Zurich, Institute of Veterinary Biochemistry and Molecular Biology) for critical and constructive comments.
REFERENCES
- 1.Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 2.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 3.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 4.Campos E, Reinberg D. Histones: annotating chromatin. Annu. Rev. Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 5.Angelov D, Vitolo JM, Mutskov V, Dimitrov S, Hayes JJ. Preferential interaction of the core histone tail domains with linker DNA. Proc. Natl Acad. Sci. USA. 2001;98:6599–6604. doi: 10.1073/pnas.121171498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Ueda K, Omachi A, Kawaichi M, Hayaishi O. Natural occurrence of poly(ADP-ribosyl) histones in rat liver. Proc. Natl Acad. Sci. USA. 1975;72:205–209. doi: 10.1073/pnas.72.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Otake H, Miwa M, Fujimura S, Sugimura T. Binding of ADP-ribose polymer with histone. J. Biochem. 1969;65:145–146. [PubMed] [Google Scholar]
- 9.Adamietz P, Bredehorst R, Hilz H. ADP-ribosylated histone H1 from HeLa cultures. Fundamental differences to (ADP-ribose)n-histone H1 conjugates formed into vitro. Eur. J. Biochem. 1978;91:317–326. doi: 10.1111/j.1432-1033.1978.tb12682.x. [DOI] [PubMed] [Google Scholar]
- 10.Minaga T, Romaschin AD, Kirsten E, Kun E. The in vivo distribution of immunoreactive larger than tetrameric polyadenosine diphosphoribose in histone and non-histone protein fractions of rat liver. J. Biol. Chem. 1979;254:9663–9668. [PubMed] [Google Scholar]
- 11.Ogata N, Ueda K, Hayaishi O. ADP-ribosylation of histone H2B. Identification of glutamic acid residue 2 as the modification site. J. Biol. Chem. 1980;255:7610–7615. [PubMed] [Google Scholar]
- 12.Boulikas T. At least 60 ADP-ribosylated variant histones are present in nuclei from dimethylsulfate-treated and untreated cells. EMBO J. 1988;7:57–67. doi: 10.1002/j.1460-2075.1988.tb02783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burzio LO, Riquelme PT, Koide SS. ADP ribosylation of rat liver nucleosomal core histones. J. Biol. Chem. 1979;254:3029–3037. [PubMed] [Google Scholar]
- 14.Riquelme PT, Burzio LO, Koide SS. ADP ribosylation of rat liver lysine-rich histone in vitro. J. Biol. Chem. 1979;254:3018–3028. [PubMed] [Google Scholar]
- 15.Ogata N, Ueda K, Kagamiyama H, Hayaishi O. ADP-ribosylation of histone H1. Identification of glutamic acid residues 2, 14, and the COOH-terminal lysine residue as modification sites. J. Biol. Chem. 1980;255:7616–7620. [PubMed] [Google Scholar]
- 16.Hassa PO, Haenni SS, Elser M, Hottiger MO. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol. Mol. Biol. Rev. 2006;70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quénet D, El Ramy R, Schreiber V, Dantzer F. The role of poly(ADP-ribosyl)ation in epigenetic events. Int. J. Biochem. Cell Biol. 2009;41:60–65. doi: 10.1016/j.biocel.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 18.Amé JC, Rolli V, Schreiber V, Niedergang C, Apiou F, Decker P, Muller S, Höger T, Ménissier-de Murcia J, de Murcia G. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem. 1999;274:17860–17868. doi: 10.1074/jbc.274.25.17860. [DOI] [PubMed] [Google Scholar]
- 19.Altmeyer M, Messner S, Hassa PO, Fey M, Hottiger MO. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 2009;37:3723–3738. doi: 10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haenni SS, Hassa PO, Altmeyer M, Fey M, Imhof R, Hottiger MO. Identification of lysines 36 and 37 of PARP-2 as targets for acetylation and auto-ADP-ribosylation. Int. J. Biochem. Cell Biol. 2008;40:2274–2283. doi: 10.1016/j.biocel.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 21.Hassa PO, Haenni SS, Buerki C, Meier NI, Lane WS, Owen H, Gersbach M, Imhof R, Hottiger MO. Acetylation of poly(ADP-ribose) polymerase-1 by p300/CREB-binding protein regulates coactivation of NF-kappaB-dependent transcription. J. Biol. Chem. 2005;280:40450–40464. doi: 10.1074/jbc.M507553200. [DOI] [PubMed] [Google Scholar]
- 22.Hassa PO, Buerki C, Lombardi C, Imhof R, Hottiger MO. Transcriptional coactivation of nuclear factor-kappaB-dependent gene expression by p300 is regulated by poly(ADP)-ribose polymerase-1. J. Biol. Chem. 2003;278:45145–45153. doi: 10.1074/jbc.M307957200. [DOI] [PubMed] [Google Scholar]
- 23.Kale L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, Phillips J, Shinozaki A, Vraradarajan K, Schulten K. NAMD2: greater scalability for parallel molecular dynamics. J. Comput. Phys. 1999;151:283–312. [Google Scholar]
- 24.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J . Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 25.Jorgensen WL, Chandrasekhar J, Madura JD. Comparison of simple potential functions for simulating liquid water. J . Phys. Chem. B. 1983;79:926–936. [Google Scholar]
- 26.Darden T, York D, Pedersen L. Particle mesh Ewald: An N.log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 27.Brooks BR, Brooks CL, Mackerell AD, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, et al. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seeber M, Cecchini M, Rao F, Settanni G, Caflisch A. Wordom: a program for efficient analysis of molecular dynamics simulations. Bioinformatics. 2007;23:2625–2627. doi: 10.1093/bioinformatics/btm378. [DOI] [PubMed] [Google Scholar]
- 29.Jump DB, Sudhakar S, Tew KD, Smulson M. Probes to study the effect of methyl nitrosourea on ADP-ribosylation and chromatin structure at the subunit level. Chem. Biol. Interact. 1980;30:35–51. doi: 10.1016/0009-2797(80)90112-x. [DOI] [PubMed] [Google Scholar]
- 30.Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 2000;275:40974–40980. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- 31.Oka S, Kato J, Moss J. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J. Biol. Chem. 2006;281:705–713. doi: 10.1074/jbc.M510290200. [DOI] [PubMed] [Google Scholar]
- 32.Ruf A, Rolli V, de Murcia G, Schulz GE. The mechanism of the elongation and branching reaction of poly(ADP-ribose) polymerase as derived from crystal structures and mutagenesis. J. Mol. Biol. 1998;278:57–65. doi: 10.1006/jmbi.1998.1673. [DOI] [PubMed] [Google Scholar]
- 33.Ruf A, de Murcia G, Schulz GE. Inhibitor and NAD+ binding to poly(ADP-ribose) polymerase as derived from crystal structures and homology modeling. Biochemistry. 1998;37:3893–3900. doi: 10.1021/bi972383s. [DOI] [PubMed] [Google Scholar]
- 34.Alvarez-Gonzalez R, Pacheco-Rodriguez G, Mendoza-Alvarez H. Enzymology of ADP-ribose polymer synthesis. Mol. Cell Biochem. 1994;138:33–37. doi: 10.1007/BF00928440. [DOI] [PubMed] [Google Scholar]
- 35.Bürkle A. Poly(ADP-ribose). The most elaborate metabolite of NAD+ FEBS J. 2005;272:4576–4589. doi: 10.1111/j.1742-4658.2005.04864.x. [DOI] [PubMed] [Google Scholar]
- 36.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl Acad. Sci. USA. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hengel SM, Shaffer SA, Nunn BL, Goodlett DR. Tandem mass spectrometry investigation of ADP-ribosylated kemptide. J. Am. Soc. Mass Spectrom. 2009;20:477–483. doi: 10.1016/j.jasms.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zee BM, Garcia BA. Electron transfer dissociation facilitates sequencing of adenosine diphosphate-ribosylated peptides. Anal. Chem. 2010;82:28–31. doi: 10.1021/ac902134y. [DOI] [PubMed] [Google Scholar]
- 39.Tao Z, Gao P, Liu H. Identification of the ADP-ribosylation sites in the PARP-1 automodification domain: analysis and implications. J. Am. Chem. Soc. 2009;131:14258–14260. doi: 10.1021/ja906135d. [DOI] [PubMed] [Google Scholar]
- 40.Cervantes-Laurean D, Jacobson EL, Jacobson MK. Glycation and glycoxidation of histones by ADP-ribose. J. Biol. Chem. 1996;271:10461–10469. doi: 10.1074/jbc.271.18.10461. [DOI] [PubMed] [Google Scholar]
- 41.Oka J, Ueda K, Hayaishi O, Komura H, Nakanishi K. ADP-ribosyl protein lyase. Purification, properties, and identification of the product. J. Biol. Chem. 1984;259:986–995. [PubMed] [Google Scholar]
- 42.Heard E. Delving into the diversity of facultative heterochromatin: the epigenetics of the inactive X chromosome. Curr. Opin. Genet. Dev. 2005;15:482–489. doi: 10.1016/j.gde.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 43.Nusinow DA, Hernández-Muñoz I, Fazzio TG, Shah GM, Kraus WL, Panning B. Poly(ADP-ribose) polymerase 1 is inhibited by a histone H2A variant, MacroH2A, and contributes to silencing of the inactive X chromosome. J. Biol. Chem. 2007;282:12851–12859. doi: 10.1074/jbc.M610502200. [DOI] [PubMed] [Google Scholar]
- 44.Zhou J, Fan JY, Rangasamy D, Tremethick DJ. The nucleosome surface regulates chromatin compaction and couples it with transcriptional repression. Nat. Struct. Mol. Biol. 2007;14:1070–1076. doi: 10.1038/nsmb1323. [DOI] [PubMed] [Google Scholar]
- 45.Clapier CR, Längst G, Corona DF, Becker PB, Nightingale KP. Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI. Mol. Cell. Biol. 2001;21:875–883. doi: 10.1128/MCB.21.3.875-883.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shogren-Knaak M, Ishii H, Sun J-M, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 47.Ahel D, Horejsi Z, Wiechens N, Polo S, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West S, Jackson S, et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gottschalk A, Timinszky G, Kong S, Jin J, Cai Y, Swanson S, Washburn M, Florens L, Ladurner A, Conaway J, et al. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc. Natl Acad. Sci. USA. 2009;106:13770–13774. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Timinszky G, Till S, Hassa P, Hothorn M, Kustatscher G, Nijmeijer B, Colombelli J, Altmeyer M, Stelzer E, Scheffzek K, et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat. Struct. Mol. Biol. 2009;16:923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.