Abstract
The dynamic exchange of histone lysine methylation status by histone methyltransferases and demethylases has been previously implicated as an important factor in chromatin structure and transcriptional regulation. Using immunoaffinity TAP analysis, we purified the WHISTLE-interacting protein complexes, which include the heat shock protein HSP90α and the jumonji C-domain harboring the histone demethylase JMJD1C. In this study, we demonstrate that JMJD1C specifically demethylates histone H3K9 mono- and di-methylation, and mediates transcriptional activation. We also provide evidence suggesting that both WHISTLE and JMJD1C performs functions in the development of mouse testes by regulating the expression of the steroidogenesis marker, p450c17, via SF-1-mediated transcription. Furthermore, we demonstrate that WHISTLE is recruited to the p450c17 promoter via SF-1 and represses the transcription of prepubertal stages of steroidogenesis, after which JMJD1C replaces WHISTLE and activates the expression of target genes via SF-1-mediated interactions. Our results demonstrate that the histone methylation balance mediated by HMTase WHISTLE and demethylase JMJD1C perform a transcriptional regulatory function in mouse testis development.
INTRODUCTION
The basic building block of chromatin structure, the nucleosome, is composed of 146 bp of DNA wrapped around a histone octamer (1). A growing body of evidence suggests that chromatin structure is modulated by a variety of histone modifications that perform pivotal regulatory functions in the control of gene expression. Histone methylation has been studied extensively, and recognized as an essential marker for a variety of biological processes, including transcriptional regulation. Histone methylation is known to be facilitated by target lysines and arginine residues of histones; in particular, the methylation of a specific lysine residue results in transcriptional activation or in the repression of target genes.
Histone lysine demethylation is carried out by two families of enzymes: the amine oxidases, such as LSD1, and the hydroxylases of a family of proteins that harbor the jumonji C (JmjC)-domain (2,3). Considering the pivotal biological functions that histone methylation has been shown to perform, histone demethylation is expected to be involved in the same processes, and with similar effects (4). Therefore, both histone lysine methylation and demethylation can be employed to demonstrate the dynamic status of methylation marks, in addition to their epigenetic control of transcriptional regulation (5).
Recently, an increasing number of researchers are suggesting that the status of histone methylation is tightly regulated by HMTases and demethylases in a specific target gene, in a tissue-dependent and more importantly, mutually non-exclusive manner in transcriptional time and localization. Both transcriptional activation and repression methylation markers have been detected in pluripotent cells via genome-wide profiling (6). Physical interactions occurring between the demethylase UTX and MLL complexes in the transcriptional regulation of HOX genes have also been theorized to occur (7,8). The recruitment of G9a and GLP to the cyclin D1 promoter via the expression of Jarid2 reflects the formation of a protein complex between the histone-modifying enzymes (9). Recently, coordinate regulations of histone methylation and demethylation for the fine control of target gene transcription have been demonstrated (10,11).
Steroidogenesis begins with the transfer of cholesterol from the cytoplasm into the inner membranes of mitochondria by steroidogenic acute regulatory protein (StAR). A series of sequential conversions of cholesterol into testosterone involves different cleavage enzymes, including p450scc and p450c17. Steroidogenic factor 1 (SF-1) primarily regulates steroidogenesis in the testes via the regulation of steroid-synthesizing enzymes (12). In the regulation of postnatal testicular steroidogenesis, luteinizing hormone (LH) functions as a primary regulator and stimulates Leydig cells to generate testosterone (13).
We demonstrated previously that an HMTase WHISTLE evidences both histone H3K4 and H3K27 methylating activity and transcriptional repression activity via HDAC recruitment (14). We further demonstrated that WHISTLE induces caspase-dependent apoptotic cell death in a SET domain-dependent manner (15). In this study, we immunoaffinity-purified the WHISTLE-interacting proteins, including HSP90α and JMJD1C, and subsequently biochemically characterized their specific HMTase cofactor and H3K9 demethylase activities, respectively. We further present both in vitro and in vivo evidence suggesting the coordinate regulatory role of WHISTLE and JMJD1C in steroidogenesis in the mouse testes. Our findings demonstrate that the differential time-scale occupancy of the steroidogenic marker p450c17 promoter by WHISTLE and JMJD1C regulates transcription via interaction with SF-1. We suggest that a possible transcriptional regulatory mechanism in the development of the mouse testes occurs via the coordinated regulation of histone methylation and demethylation.
MATERIALS AND METHODS
Plasmids
The pGEX-4T1-WHISTLE plasmid was described previously (14). For the purification of TAP, WHISTLE was cloned into the pNTAP vector (Stratagene) in-frame with the SBP and CBP affinity tags located at the 5′ end of the fusion gene. pCMV-sport6-JMJD1C (BC068318) and pCMV-HSP90α (BC023006) were purchased from Openbiosystems and KUGI, respectively. The partial regions (amino acids 1836–2487) of the JMJD1C cDNA were inserted into the pGEX-4T1 bacterial expression vector (Amersham Biosciences) to construct the GST-JMJD1C fusion proteins. JMJD1C (H2326A) point mutants were constructed by Cosmo Genetech. In order to construct the mammalian expression vectors, we employed modified pcDNA6-HA-myc-his (Invitrogen) and used pFLAG-JMJD1C to create the HA, myc and his-tagged JMJD1C (H2326A) point mutant and flag-tagged JMJD1C proteins. The shRNA against mouse WHISTLE (M-060989-01-0010) was purchased from Openbiosystems. The siRNAs against mouse JMJD1C (L-062872-00-0010) and SF-1 (sc-37902) were purchased from Dharmacon and Santa Cruz Biotechnology, respectively.
Antibodies
Antibodies against WHISTLE (sc-50151, Santa Cruz Biotechnology), calmodulin-binding epitope (07-482, Milipore), HSP90α (ab59459, Abcam), JMJD1C (ab31215, Abcam), histone H3 (06-755, Milipore) and β-actin (sc-47778, Santa Cruz Biotechnology) were employed for immunoblot analysis, immunoprecipitation or immunohistochemistry; FLAG (f3165, Sigma), myc (sc-40, Santa Cruz Biotechnology) for immunocytochemistry, histone H3K4-me2 (07-030, Milipore), H3K9-me1 (07-450, Milipore)/-me2 (07-441, Milipore)/-me3 (17-625, Milipore), H3K27-me2 (07-452, Milipore) and H3K36-me2 (07-274, Milipore) were used for immunoblot analysis or immunocytochemistry; and SF-1 (sc-28740, Santa Cruz Biotechnology) was used for chromatin immunoprecipitation (ChIP) analysis.
Cell lines and stable cell-line establishment
Mouse embryonic fibroblast NIH3T3 cells and mouse testis Leydig TM3 cells were maintained in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum and 0.5% penicillin–streptomycin. In order to establish the cell-line stably expressing TAP-WHISTLE, the NIH3T3 cells were transfected with pNTAP-WHISTLE using Lipofectamine 2000 (Invitrogen), after which the stably transfected clones were selected in media containing 2.5 mg/ml of G418 (Sigma).
Tandem affinity purification and LC–MS/MS analysis
Tandem affinity purification (TAP) purifications were conducted as described previously (16) with several modifications using an InterPlay mammalian TAP system (Stratagene). After the two-step purification, the protein samples were subjected to protease digestion, followed by LC–MS/MS analysis.
HMTase assay
In vitro and in vivo HMTase assays were conducted as previously described (15). For in vivo analysis, NIH3T3 cells were cotransfected with pcDNA6-WHISTLE, pCMV-HSP90α using Lipofectamine 2000 (Invitrogen), and the transfected cells were lysed with 1× RSB lysis buffer for 1 h at 4°C. The cell lysates were fractionated in 14% SDS–PAGE, transferred, and each lysine specificity of histone H3 methylation was detected using rabbit α-H3K4-me2, H3K9-me1/-me2/-me3, H3K27-me2 and H3K36-me2 antibodies.
Demethylase assay and mass spectrometry
Demethylase assays were conducted as described previously (17). H3K9-me1 peptide [TARK(me1)STG], H3K9-me2 peptide [TARK(me2)STG] and H3K9-me3 peptide [TARK(me3)STG] were synthesized on the basis of the N-terminal amino-acid sequences of histone H3 (Peptron). Synthetic peptides (0.1 µg) were employed as substrates in the demethylase assay.
Immunocytochemistry
NIH3T3 cells were grown on chamber slides and transfected with pFLAG-JMJD1C expression plasmids using Lipofectamine 2000. After 48 h of transfection, the cells were fixed, incubated with each antibody, washed, stained with SYTO61 and mounted.
Immunohistochemistry
The testis specimens were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 12–14 h at 4°C, then washed, dehydrated and paraffin-embedded. The 6 µM thick serial sections were obtained with a rotary microtome. The sections were stained immunohistochemically using an LSAB2 system-HRP kit (Dako), in accordance with the manufacturer’s instructions. In brief, the sections were deparaffinized, hydrated and placed in peroxidase block solution for 10 min. After three washings in PBS, the sections were incubated with α-WHISTLE and α-JMJD1C antibodies for 2 h, respectively, and treated for 20 min with biotinylated link solution, after which streptavidin–HRP solution was applied for an additional 20 min. As a negative control, primary antiserum was replaced with PBS. The sections were subsequently treated with DAB substrate–chromogen solution for 10 min and counterstained with Harris hematoxylin. Images were photographed on a light microscope (Olympus).
RT–PCR and quantitative PCR
Total RNA samples from each of the construct-transfected TM3 cells and experimental mice at different developmental stages were extracted using Trizol reagent (Invitrogen), in accordance with the manufacturer’s recommendations. One microgram of total RNA was used to synthesize the cDNA. The cDNA synthesis was primed using oligo-dT primer (Fermentas) and the quantified cDNA was applied for p450c17, LHR, WHISTLE and JMJD1C mRNA expression pattern analysis. The primer sequences were as follows: p450c17: sense 5′-CAGCTGGCCATCTGCCTACAC-3′ and antisense 5′-ATCTGGGGCCGACCAGAGAAT-3′; LHR: sense 5′-ATGGGGCGGCGGGTCCCGGCT-3′ and antisense 5′-CTGTGCATCTTCTCCAGGTAG-3′; WHISTLE: sense 5′-AAGCTGGCAAGAAACTGCAT-3′ and antisense 5′-CAAAGCCTCCTTGCTTTCAC-3′; JMJD1C: sense 5′-GAGGACTTCAAGGCC-3′ and antisense 5′-AATTAGGTGTCTTCC-3′. The amplification reaction was performed under the following conditions: 35 cycles of denaturation at 94°C, annealing at 58°C and extension at 72°C. Dissociation curves were generated after each PCR run to ensure that a single product of the appropriate length was amplified. The mean threshold cycle (Ct) and standard error were calculated from individual Ct values obtained from three replicates per stage. The normalized mean Ct was estimated as ΔCt by subtracting the mean Ct of β-actin from that of p450c17. The value ΔΔCt was calculated as the difference between control ΔCt and values obtained at each sample. The n-fold change in gene expression, relative to untreated control, was calculated as 2−ΔΔCt.
Transcriptional activity assay
In the transcriptional activity assay (2.5 × 104 cells/well/48-well plate at their 50–60% confluence stage), TM3 cells were cotransfected with 100–300 ng of the expression plasmid and 200 ng of the SV40-driven reporter or SF-1-RE-luc reporter plasmid using Lipofectamine 2000 reagents, again in accordance with the manufacturer’s recommendations. After 48 h, the cells were harvested and subjected to the luciferase assay (Promega). The amount of DNA in each transfection was maintained at constant levels via the addition of fixed amounts of pcDNA3.0 vector.
In vitro transcription and translation reactions
For in vitro transcription and translation, the [35S]-labeled JMJD1C proteins were constructed using a coupled transcription and translation (TNT) system (Promega). In brief, 1 µg of DNA was added directly to TNT rabbit reticulocyte lysates, then permitted to react for 2 h at 30°C. The reaction mixtures were then employed in a GST-pull down assay, using GST-WHISTLE, GST and TM3 cell lysates. The precipitated proteins were then separated via 8% SDS–PAGE and autoradiographically visualized.
Immunoprecipitation
For the interaction assays, the transfected cells were lysed in RIPA lysis buffer and immunoprecipitated with α-myc or α-SF-1 antibodies (Santa Cruz Biotechnology) and protein A agarose beads (GenDEPOT). The bound proteins were analyzed in immunoblots with α-JMJD1C, α-WHISTLE and α-SF-1antibodies.
ChIP assay and quantitative PCR
The TM3 cells were transfected, harvested for 48 h and then cross-linked with 1% formaldehyde in the medium for 10 min at room temperature, followed by the addition of 125 mM glycine for 5 min at RT. The samples were sonicated and immunoprecipitated using the indicated antibodies. The immunoprecipitates were eluted and reverse cross-linked, after which the DNA fragments were purified for PCR amplification. Primer pairs were selected for experimental studies of the potential site of SF-1 in the 541-bp region (–837 to −296: sense 5′-TTTCAGGGGCCAGAAGGTG-3′ and antisense 5′-TCCTCCCAGAGGCAAATGC-3′) of mouse p450c17 promoter. For p450c17 promoter exon region analysis, the primer sets comprised the 454-bp region (+1 to +454: sense 5′-CAGGCTTGGAGACACTCCAGG A-3′ and antisense 5′-CATCTGGGGCCGACCAGAGAATT-3′). The primer concentration employed for real-time PCR was 0.2 µM/25 µl. The thermal cycler conditions were as follows: 15 min of holding at 95°C, followed by three steps of PCR for 50 cycles at 94°C for 15 s, 60°C for 30 s and 72°C for 30 s (Corbett Research). The percentage (Bound/Input) is percentage recruited to the target promoter compare to that of input [calculated via the 2−ΔCt method].
Radioimmunoassay
Testosterone concentrations were assayed via a RIA technique, as described previously (18). In brief, TM3 cells seeded onto 60-mm culture dishes were cultured for 24 h in DMEM medium supplemented with 10% FBS. Culture medium was collected for radioimmunoassay (RIA) at the appropriate time points after exchanging with fresh medium containing 200 ng/ml of LH, and the culture media were then directly assayed with no additional purification. Each treatment group included duplicate cultures, and each experiment was repeated at least twice. The assay procedures were followed as described previously using labeled testosterone ([1,2,6,7-3H]testosterone; 96 Ci/mol) and progesterone ([1,2,6,7-3H] progesterone; 96 Ci/mol) obtained from NEN.
Statistical analysis
The data were expressed as the means ± SD of three or more independent experiments. Statistical significance effects (P < 0.05) were evaluated with Microsoft EXCEL software. Differences between groups were evaluated via one-way analysis of variance (ANOVA), followed by Student’s t-tests or Bonferroni’s tests, as appropriate.
RESULTS
Purification of TAP-WHISTLE and interacting proteins
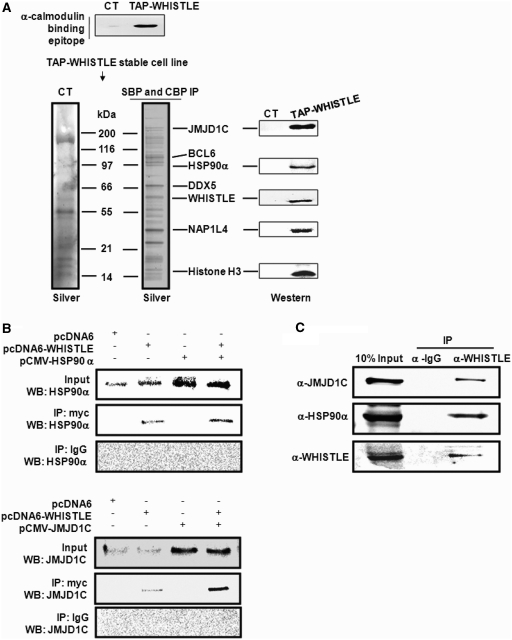
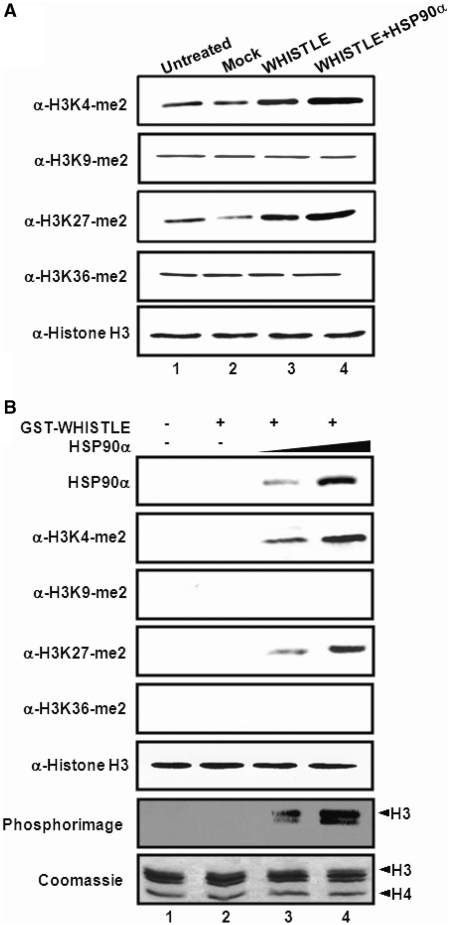
In an effort to obtain greater insight into the in vivo functions of WHISTLE, we developed NIH3T3 cell lines that stably expressed TAP-tagged WHISTLE, and then purified the WHISTLE-interacting complexes using a TAP system. In an effort to identify the interacting complexes, the affinity eluates were analyzed via SDS–PAGE followed by silver staining, and subsequently subjected to mass spectrometric analysis (Figure 1A). Mass analysis revealed a sizeable collection of the WHISTLE-interacting complexes, including HSP90α and histone demethylase JMJD1C (Figure 1A and Supplementary Table S1). Parts of the identified proteins were immunoblotted using specific antibodies. Immunoprecipitation (IP) assays were conducted to verify the interactions between WHISTLE and the identified proteins, HSP90α and JMJD1C, using NIH3T3 cell extracts (Figure 1B). In a previous study, we reported that WHISTLE was abundantly expressed in mouse testes (14). Co-IPs have been conducted in mouse testis Leydig TM3 cells using α-WHISTLE antibodies, and pull-downs of endogenous JMJD1C and HSP90α proteins were performed (Figure 1C). Considering the results of our previous studies, we hypothesized that WHISTLE-interacting HSP90α might operate as a cofactor with WHISTLE to achieve maximal HMTase activity (15). The transient transfection of pcDNA6-WHISTLE elicited methylations of both H3K4-me2 and H3K27-me2 as compared to what was observed in mock vector-transfected cells (Figure 2A and Supplementary Figure S1A). When pCMV-HSP90α was cotransfected with WHISTLE, further increases in the methylation of H3K4 and H3K27 were noted (Figure 2A and Supplementary Figure S1A). No changes in the methylation levels of H3K9 and H3K36 were noted throughout the processes. The expressions of the WHISTLE and HSP90α constructs employed in transfection were confirmed by the results of immunoblotting (Supplementary Figure S1B). The effect of HSP90α on the HMTase activity of WHISTLE was corroborated further by the results of an in vitro HMTase assay. The incubation of cold-SAM, histones and recombinant WHISTLE yielded almost no enzyme activity. The histone H3K4-me2 and H3K27-me2 methylating activities of WHISTLE were shown to be increased upon the addition of increasing quantities of purified HSP90α (Figure 2B). Consistent results were observed when [14C]-SAM was employed as a methyl donor (Figure 2B, phosphorimage). These results demonstrate that HSP90α functions as a cofactor for the full HMTase activity of WHISTLE.
Figure 1.
WHISTLE interacts with HSP90α and JMJD1C. (A) Schematic representation of TAP-WHISTLE purification. Upper panel: verification of CT (TAP-empty vector) and TAP-WHISTLE stably expressing NIH3T3 cells by α-calmodulin antibody. Lower panel: silver staining of the final purified sample on 9–16% SDS gradient gel and immunoblot analysis of identified proteins using each of the indicated antibodies. (B) Interaction between WHISTLE and HSP90α, JMJD1C. Verification of JMJD1C and HSP90α interaction with WHISTLE via co-immunoprecipitation in NIH3T3 cells. Co-IPs were conducted using α-myc (WHISTLE) antibodies, after which immunoblots were performed using α-HSP90α and JMJD1C antibodies. (C) Endogenous interaction between WHISTLE and JMJD1C and HSP90α, respectively. HSP90α and JMJD1C with TM3 cells were immunoprecipitated using α-WHISTLE and α-IgG, then immunoblotted against α-JMJD1C and α-HSP90α antibodies.
Figure 2.
HSP90α functions as a cofactor for the full HMTase activity of WHISTLE. (A) In vivo HMTase activity. pcDNA6-WHISTLE and pCMV-HSP90α were transfected into NIH3T3 cells, and altered histone methylation patterns were detected on immunoblot analyses using specific antibodies. Equal amount of analyzed samples were verified by immunoblot against α-histone antibodies. (B) In vitro HMTase assay with increasing concentration of HSP90α proteins. The amounts of HSP90α proteins, histone H3 and histone methylation status were detected via immunoblots using the indicated antibodies. The phosphorimage shows methylated histone H3 via the same in vitro HMTase with methyl donor isotope. Core histones used for assays were shown by Coomassie Blue staining.
Histone H3K9 mono- and di-methylation-specific demethylase activity of JMJD1C
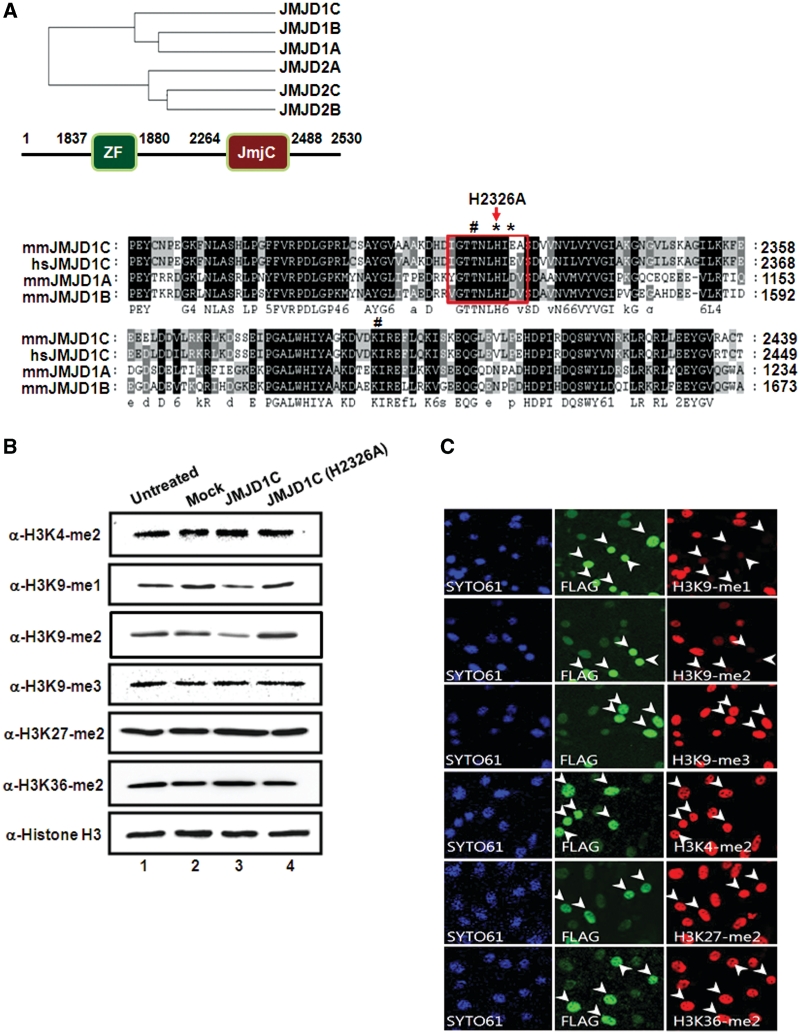
The phylogenic analysis of human JmjC-containing proteins classified JMJD1C among the subfamily of demethylases that includes JMJD1A (JHDM2A) and JMJD1B (JHDM2B) (Figure 3A, upper panel). One of the distinct domain characteristics of the JMJD1 family in different species is that they all harbor a zinc-finger (ZF)-like domain, as well as a JmjC domain. Both Fe(II) and α-ketoglutarate (α-KG)-binding sites are conserved among JMJD1A orthologs, which indicates that these proteins may exhibit H3K9-specific demethylase activity (19). Sequence alignment of the mammalian JmjC domain-harboring proteins from the JMJD1 family showed that the position of the histidine residue is highly conserved (Figure 3A, lower panel, boxed). In a recent study, it was determined that JMJD1A effects the demethylation of H3K9-me1 and H3K9-me2 in embryonic stem (ES) cells (20). JMJD1A evidences demethylase activity on H3K9-me1 and H3K9-me2, and is also sometimes referred to as testis-specific gene A (TSGA) (21). TSGA has been implicated in mouse spermatogenesis and androgen receptor-mediated gene activation (17,22).
Figure 3.
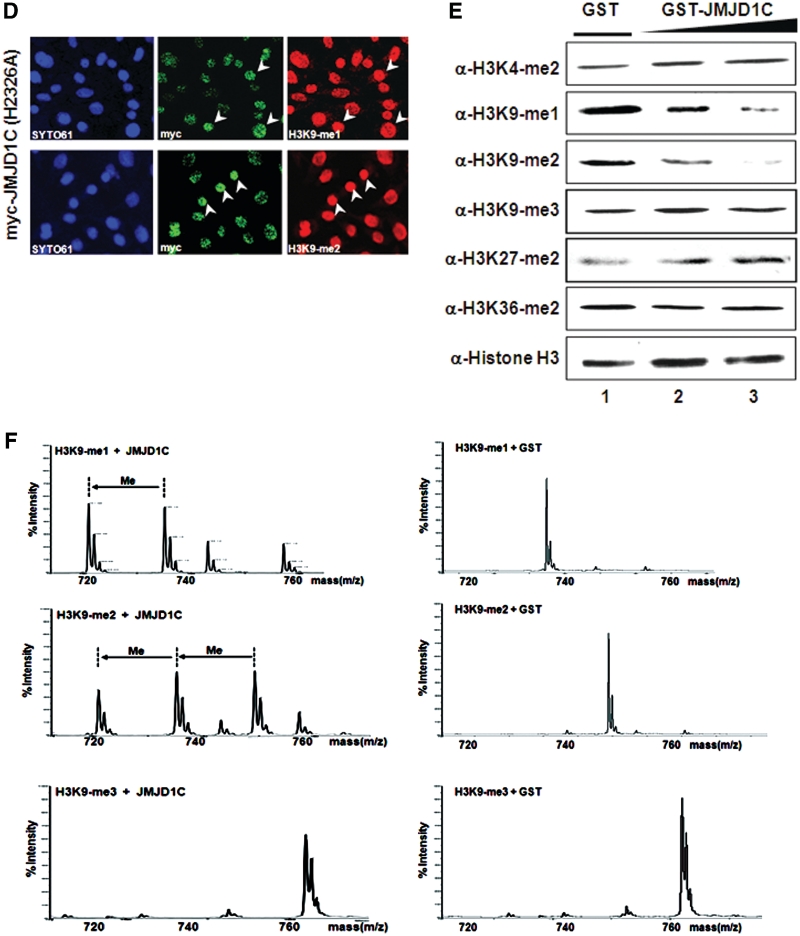
Histone H3K9 specific demethylase activity of JMJD1C. (A) Upper panel: phylogenetic analysis based on protein sequence data of the JMJD1 and JMJD2 families. Schematic representation of the JMJD1C with functional domains is shown. Lower panel: alignment of parts of the JmjC domain of the human and mouse JMJD1 family members. The position of catalytically inactive point mutation changing an amino acid 2326 from histidine to alanine in the JmjC domain of demethylase JMJD1C is shown. Conserved amino-acid residues are shown in black and similar residues are shaded in grey. Indicated asterisk and hash marks represent Fe(II) and α-ketoglutarate (α-KG) binding site, respectively. (B) JMJD1C and JMJD1C (H2326A) were transfected in TM3 cells, and demethylation activities were analyzed via immunoblot analysis with indicated antibodies. (C) TM3 cells transfected with flag-JMJD1C were immunostained with antibodies against flag-JMJD1C and the indicated histone H3 lysine residues with different methylation marks. SYTO61 staining represents nucleosomes. Arrowheads indicate transfected cells. (D) TM3 cells were transfected with myc-JMJD1C (H2326A) and stained with α-myc, α-H3K9-me1 and α-H3K9-me2. Arrowheads indicate the transfected cells. (E) In vitro demethylase assays of GST-purified JMJD1C were conducted with core histones using α-KG, ascorbate and Fe(II) as cofactors, then immunoblotted against the indicated antibodies. (F) H3K9-me1/-me2/-me3 peptides were incubated with GST-JMJD1C or GST and the modified peptides were analyzed via MALDI-TOF mass spectrometry.
In order to evaluate the lysine specificity of demethylase JMJD1C, TM3 cells were transiently transfected and the nucleosomes were immunoblotted against specific antibodies for H3K4-me2, H3K9-me1/-me2/-me3, H3K27-me2 and H3K36-me2. We initially detected a reduction of H3K9-me2 as the consequence of JMJD1C treatment; further immunoblot analysis revealed that the levels of H3K9-me1 were also reduced, whereas the level of H3K9-me3 remained unaltered (Figure 3B and Supplementary Figure S2A). No demethylase activities of JMJD1C to H3K9-me1/H3K9-me2 were noted with a JMJD1C point mutant (H2326A), which illustrates the importance of the conserved residue in the JmjC domain in regard to demethylase activity (Figure 3B). Deletions of the JmjC domain (JMJD1C-N) or ZF-like motif (JMJD1C-C) failed to reduce the levels of H3K9-me1/-me2 in both immunoblot analyses (Supplementary Figure S3A–C). The lysine specificity of JMJD1C was also evaluated via immunocytochemistry in this study. Ectopic JMJD1C expression induced a loss of H3K9-me1/-me2, whereas no such reductions in the levels of H3K9-me3, H3K4-me2, H3K27-me2 and H3K36-me2 were observed (Figure 3C). The overexpression of the enzymatically defective mutant JMJD1C (H2326A) did not influence the levels of H3K9-me1/-me2 (Figure 3D). Histones were subjected to in vitro demethylase assays using purified GST-JMJD1C. The results of immunoblot analysis demonstrated that H3K9-me1/-me2 underwent a significant reduction in methylation levels as compared to the H3K9-me3, H3K27-me2 and H3K36-me2 samples (Figure 3E and Supplementary Figure S2B). In an effort to further evaluate the H3K9-me1 and H3K9-me2 specificity of JMJD1C, H3K9-me1/-me2/-me3 peptides were analyzed via demethylase assays. Mass spectrometry analysis demonstrated that H3K9-me1/-me2 peptides were converted to mono- and unmethylated peptides after incubation with JMJD1C (Figure 3F). When the H3K9-me3 peptides were incubated with JMJD1C, no demethylation was detected.
Expression profile of WHISTLE and JMJD1C during mouse testis development
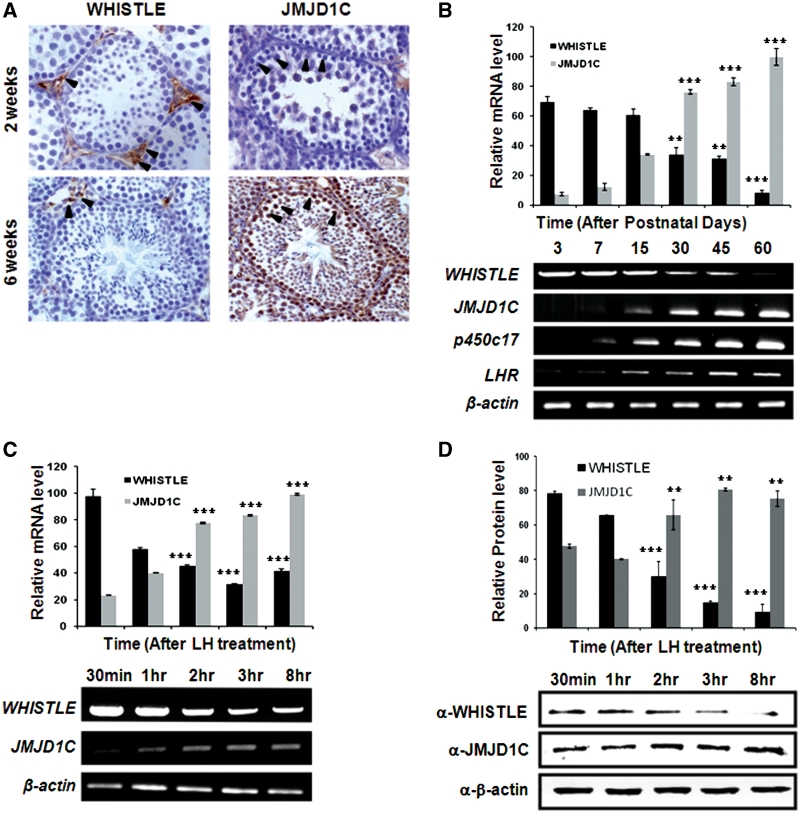
To evaluate the localization and expression patterns of WHISTLE and JMJD1C in the testes of mice, we probed sections of the testicular tissues of 2 and 6 weeks’ postnatal mice with an antiserum against JMJD1C and WHISTLE. The postnatal 2- and 6-week-old mice were selected because both genes were expressed at sufficient levels around these periods. Immunohistochemical studies of mouse testis have demonstrated that WHISTLE was abundant in the interstitial cells of Leydig and vascular endothelial cells at 2 weeks of age staining (Figure 4A, brown color), and WHISTLE-positive cells were less abundantly detected in the 6-week-old sections. As compared to WHISTLE, specific immunoreactions of JMJD1C evidenced much stronger signals in the spermatogonia and primary spermatocyte within the seminiferous tubules and the interstitum in the 6-week-old sections (Figure 4A). It is interesting to note that these two proteins evidence partial cell-type specificity within the testicular tissues, even though they are also expressed in the same types of cells.
Figure 4.
WHISTLE and JMJD1C are involved in mouse testis development. (A) Immunohistochemical analysis of WHISTLE and JMJD1C expression in 2- and 6-week-old mouse testes. Arrowheads indicate the expression of WHISTLE and JMJD1C in different stages of mouse testis tissue, as indicated. (B) The expression levels of WHISTLE, JMJD1C, p450c17 and LHR in indicated postnatal mouse testes are detected via RT–PCR. The quantified RT–PCR results are shown in the upper panel. (C) Expression pattern of WHISTLE and JMJD1C in different time points after LH (200 ng/ml) treatment in TM3 cell. Upper panel indicates quantitated RT–PCR results and the lower panels indicate RT–PCR analysis. (D) Western blot analysis of WHISTLE and JMJD1C protein expression levels in LH-treated TM3 cells. The quantification of western blots is shown in the upper panel. All data are shown as means ± SD; n = 3 at each time point. **P < 0.01 and ***P < 0.001.
To further evaluate the physiological functions of WHISTLE and JMJD1C in the development of mouse testes, we evaluated their expression patterns in postnatal developmental stages of the mouse testes. The results of RT–PCR analysis demonstrated that WHISTLE mRNA was expressed abundantly in the very early stages of development, and was reduced gradually for up to 60 days after birth (Figure 4B). By way of contrast, JMJD1C mRNA was detected at very low levels during early developmental stages, and then concentrations began to increase at 7 days, with high expression levels being detected after 30 days (Figure 4B). Because the cholesterol side chain cleavage enzyme, p450c17 and the pituitary gonadotropin LH receptor, LHR, are important regulators and differentiation markers of steroidogenesis, the expression patterns of these proteins were assessed as controls. As LH treatment induces testosterone secretion in TM3 cells during steroidogenesis, we treated the TM3 cells with LH and then evaluated the expression patterns of WHISTLE and JMJD1C. Abundant WHISTLE expression was maintained until 1 h after treatment, followed by a significant decline after 2 h (Figure 4C). By way of contrast, the expression of JMJD1C mRNA evidenced a steady increase beginning 1 h after treatment. Similar protein expression patterns of WHISTLE and JMJD1C were observed on immunoblot analysis (Figure 4D). Collectively, these data indicate that WHISTLE and JMJD1C may perform distinct roles in testis steroidogenesis, via the regulation of target gene transcriptions via specific modifications of histone methylation status.
Regulatory role of WHISTLE and JMJD1C in SF-1-mediated mouse steroidogenesis
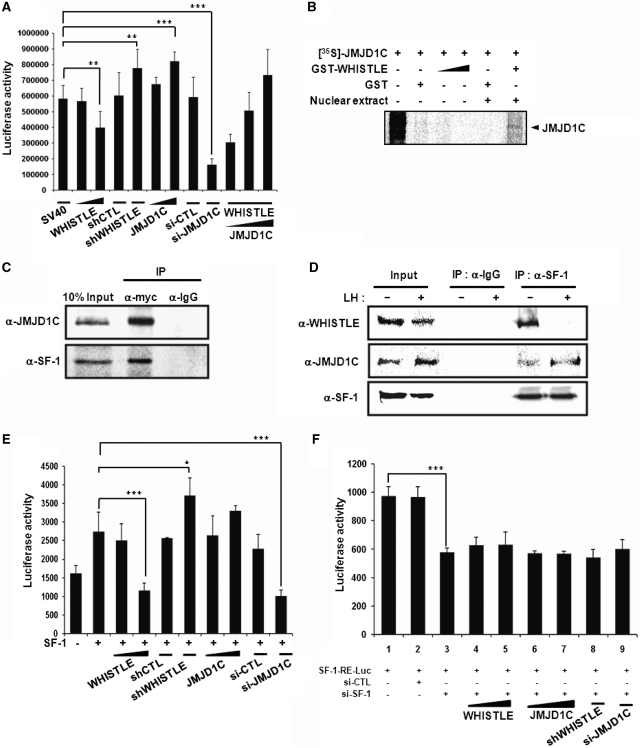
In an effort to elucidate more thoroughly the effects of JMJD1C on WHISTLE-mediated transcriptional repression, we conducted a transient transfection assay using the pCMX-Gal4-SV40 reporter system. As was previously reported, transfection with WHISTLE repressed luciferase activity and the overexpression of shWHISTLE ameliorated WHISTLE-mediated repression and further activated the transcription of SV40 in TM3 cells (Figure 5A). We then attempted to determine whether WHISTLE-interacting JMJD1C could reverse the observed WHISTLE-mediated transcriptional repression, by demethylating H3K9 methylation. When transfected alone, JMJD1C moderately induced SV40 transcription to levels similar to those of shWHISTLE (Figure 5A). The significant reduction in transcription noted with si-JMJD1C treatment confirmed the role of JMJD1C in SV40 transactivation. As anticipated, increasing quantities of JMJD1C overexpression significantly reversed WHISTLE-mediated transcriptional repression in a dose-dependent manner (Figure 5A). These results demonstrate that JMJD1C may function as a substitute for WHISTLE-mediated transcriptional repression activity for activation, via the demethylation of H3K9 methylation.
Figure 5.
WHISTLE and JMJD1C exert different effects on transcriptional activity of SF-1. (A) TM3 cells were transfected with pCMX-Gal4-SV40 and indicated DNA constructs and siRNA, and their cell extracts were assayed for luciferase activity. The results are representative of at least five independent experiments (±SD). **P < 0.01 and ***P < 0.001, compared with untreated control. (B) In vitro translated JMJD1C proteins were permitted to interact with GST-WHISTLE proteins and TM3 cell nuclear extract was added, followed by the analysis of the proteins on 10% SDS–PAGE gels. (C) In vivo interaction analysis between WHISTLE and JMJD1C and SF-1 via co-IP in TM3 cells. TM3 cells were transfected with myc-WHISTLE, JMJD1C and SF-1. IP was performed using α-myc antibodies, after which immunoblot analysis was conducted using α-JMJD1C and SF-1 antibodies. (D) Co-IP between WHISTLE and JMJD1C with SF-1 prior to and after LH treatment in TM3 cells. (E and F) Regulatory effects of WHISTLE and JMJD1C through SF-1-mediated transcription. TM3 cells were transfected with SF-1 (E) or treated with si-SF-1 (F) and a three copy SF-1-RE-luc, and indicated DNA constructs and siRNAs. All data are expressed as means ± SD; n = 5. *P < 0.05 and ***P < 0.001.
As an orphan nuclear receptor, steroidogenic factor 1 (SF-1) has been shown to regulate the expression of many of the genes involved in steroidogenesis, including CYP variants and StAR (23). These genes perform important roles in the function and development of reproductive tissues including the adrenals and gonads (12). The differential expression patterns of WHISTLE and JMJD1C in the mouse testes during developmental stages, as well as the observed induction of steroidogenesis, compelled us to determine with more specificity whether or not WHISTLE and JMJD1C are involved in steroidogenesis via interactions with SF-1. In order to characterize in detail the functions of WHISTLE and JMJD1C in SF-1-mediated steroidogenesis, we attempted to determine the manner in which these proteins interact with one another. Interaction assays using in vitro transcription-translated JMJD1C and immunoprecipitates against WHISTLE were conducted, and we determined that WHISTLE and JMJD1C did not interact directly with one another; rather, they appear to associate with one another in a protein complex in vivo (Figure 5B and C). Co-IP assays were conducted to confirm complex formation among WHISTLE, JMJD1C and SF-1 (Figure 5C). We further analyzed the mechanisms underlying SF-1-mediated WHISTLE and JMJD1C interaction prior to and after LH treatment. The IP assays showed that SF-1 interacted strongly with WHISTLE prior to LH treatment and then switched to JMJD1C after the induction of steroidogenesis by LH (Figure 5D).
The complex formation and differential interactions between SF-1 and WHISTLE/JMJD1C indicate that WHISTLE and JMJD1C are involved in steroidogenesis via the regulation of SF-1 transcription. We conducted a transient transfection assay using the SF-1-RE luciferase reporter system. As had been anticipated, the transfection of SF-1 into TM3 cells resulted in the activation of luciferase activity. Consistent with its transcriptional repression activity, WHISTLE was shown to repress the transcriptional activity of SF-1 (Figure 5E). The transcriptional repressive activity of WHISTLE was clearly verified by two independent RNAi-mediated knock-downs, shWHISTLE and si-WHISTLE treatment, by virtue of the recovery of SF-1-mediated transcriptional activity (Figure 5E and Supplementary Figure S4). Interestingly, the transcriptional activation of SF-1 was further enhanced when JMJD1C was cotransfected with SF-1 (Figure 5E). Treatment with si-JMJD1C downregulated the JMJD1C-induced enhancements of transcriptional activation. Identical results were noted when different 293T cell lines were used, although the overall luciferase activities were lower than those of the TM3 cell lines (data not shown). The knockdown of the SF-1 by si-SF-1 definitively abolished the transcriptional activation induced by endogenous SF-1, and none of the tested reagents (WHISTLE, JMJD1C, shWHISTLE, si-JMJD1C or si-SF-1) evidenced their anticipated effects in TM3 cells (Figure 5F and Supplementary Figure S5). Taken together, these findings demonstrate that WHISTLE may repress the early stages of steroidogenesis by inhibiting the SF-1-mediated transactivation of target genes, and also that JMJD1C induces the transcriptional activation of SF-1 in later stages of steroidogenesis.
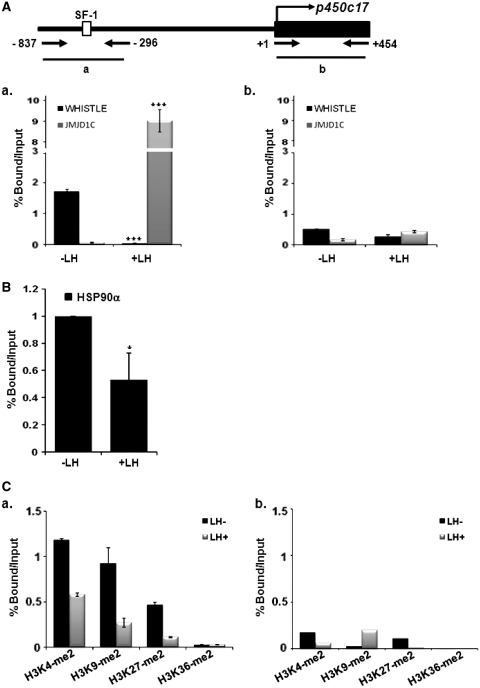
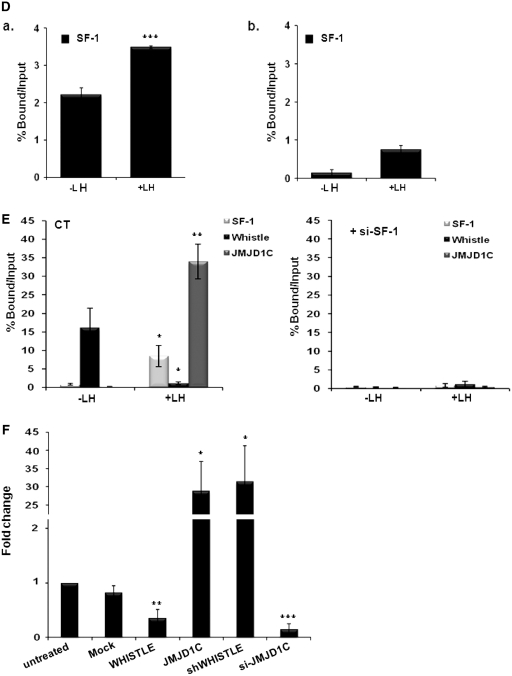
ChIP analysis with real-time PCR using testicular cells revealed that WHISTLE and JMJD1C were recruited to specific response elements of p450c17 at different times during steroidogenesis. We noted that WHISTLE is specifically recruited to the p450c17 promoter region prior to LH treatment, and that during steroidogenesis by LH treatment, the binding of WHISTLE to the promoter was abolished (Figure 6A, a). On the other hand, JMJD1C did not occupy the p450c17 promoter prior to LH treatment, and then evidenced significantly elevated recruitment afterward (Figure 6A, a). ChIP with real-time PCR analysis showed stronger recruitment of the WHISTLE cofactor, HSP90α, on the p450c17 promoter prior to LH treatment; this is consistent with the stronger recruitment of WHISTLE observed prior to LH treatment (Figure 6B). Comparable results were noted when we assessed histone methylation status on the p450c17 promoter region. Real-time PCR with the ChIP assay showed that histone H3K4-me2 and H3K27-me2 methylation levels on the p450c17 promoter region were significantly downregulated after LH treatment (Figure 6C). The methylation level of histone H3K9-me2 was also reduced, thus reflecting the increased level of JMJD1C after LH treatment (Figure 6C, a). These results are consistent with our data, which revealed strong recruitment of the H3K4-me2 and H3K27-me2-methylating HMTase WHISTLE to the p450c17 promoter before LH signaling and subsequent replacement by JMJD1C. We then confirmed that both WHISTLE and JMJD1C regulate steroidogenic markers via SF-1-mediated transcription. The results of quantitative PCR after ChIP analysis demonstrated that SF-1 is strongly recruited to the p450c17 promoter both prior to and after LH treatment, although recruitment was stronger after LH treatment. This indicates that both WHISTLE and JMJD1C require the presence of SF-1 for its role in steroidogenesis (Figure 6D, a). On the contrary, no changes were detected in WHISTLE/JMJD1C, SF-1 recruitment or histone methylation levels in the exonic region of p450c17 (Figure 6A, C and D, b). Consistent with the results obtained in the SF-1 response element reporter assay with si-SF-1, the depletion of SF-1 by si-SF-1 in TM3 cells also resulted in failure to recruit WHISTLE and JMJD1C to the p450c17 promoter prior to and after LH treatment, respectively (Figure 6E). Having established the observation that WHISTLE and JMJD1C were recruited to the p450c17 promoter and regulated p450c17 transcription during SF-1-mediated steroidogenesis, we attempted to determine whether WHISTLE and JMJD1C can regulate the expression of p450c17. As anticipated, the overexpression of WHISTLE downregulated the expression of p450c17, and JMJD1C significantly enhanced the expression of p450c17 (Figure 6F). Knockdown by siRNA clearly confirmed the endogenous role of WHISTLE and JMJD1C on p450c17 transcription (Figure 6F). Together, these results demonstrated that SF-1 performs a crucial function in the recruitment of WHISTLE and JMJD1C to the target promoter p450c17 during the development of mouse steroidogenesis.
Figure 6.
SF-1-mediated recruitments of WHISTLE and JMJD1C in p450c17 promoter. (A) Schematic diagram of primer pairs in ChIP analysis (upper panel). Arrows indicate the primers for real-time PCR amplification. ChIP analyses of the p450c17 with or without LH-treated TM3 cells were conducted using α-WHISTLE and α-JMJD1C antibodies and examined by real-time PCR (lower panel). The recruitments of WHISTLE and JMJD1C in the p450c17 promoter region (a) and p450c17 exonic region (b) were normalized by input. The results are expressed as the means ± SD; n = 3. ***P < 0.001. (B) ChIP analyses of the p450c17 with or without LH-treated TM3 cells were conducted using α-HSP90α antibodies. The immunoprecipitated DNA fragments were real-time PCR amplified using primers for the p450c17 promoter (a). The results are expressed as the means ± SD; n = 3. *P < 0.05. (C) ChIP assay and real-time PCR for the methylation status of histone H3K4-me2, K9-me2, K27-me2 and K36-me2 in the p450c17 promoter (a) and p450c17 exonic region (b) of TM3 cells treated prior to and after LH treatment. Histone methylation levels were normalized by input. The results are expressed as means ± SD; n = 3. (D) Real-time PCR analysis following ChIP assays for SF-1 recruitment in p450c17 promoter prior to and after LH treatments. SF-1 recruitment levels in the p450c17 promoter (a) and exonic region (b) were normalized by input. Results were expressed as the means ± SD; n = 3. ***P < 0.001. (E) Recruitment of WHISTLE and JMJD1C in the p450c17 promoter were analyzed via ChIP assay and real-time PCR using si-SF-1 treated TM3 cells in the absence of SF-1 prior to and after LH treatments. The results are expressed as means ± SD; n = 3. *P < 0.05 and **P < 0.01. (F) The expression levels of p450c17 in each construct-transfected cell via real-time PCR. Results are expressed as means ± SD; n = 3. *P < 0.05, **P < 0.01 and ***P < 0.001.
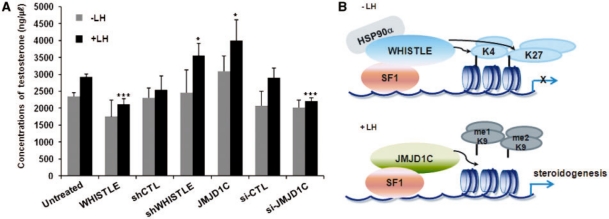
In an effort to elucidate in greater detail the regulatory roles of WHISTLE and JMJD1C in mouse testis development, TM3 cells were transiently transfected with WHISTLE or JMJD1C and subjected to LH treatment; following LH treatment, the testosterone concentrations were assayed via RIA. The treatment of TM3 cells with LH increased testosterone concentration to some degree in the control cells. When the WHISTLE-overexpressing cells were treated with LH, testosterone expression was reduced (Figure 7A). Consistent with the above results, when LH treatment was administered in conjunction with JMJD1C, steroidogenesis was induced to a significant degree in the TM3 cells (Figure 7A). The RNAi-mediated knockdowns of both WHISTLE and JMJD1C relieved the effects of the overexpressed and endogenous proteins. The effects of each transfection proved to be marginal in the absence of LH treatment. These results indicate that WHISTLE and JMJD1C contribute to the transcriptional regulation of SF-1-mediated steroidogenesis via the switching of methylation marks on the promoter of the target gene, and are involved in the development of mouse testes (Figure 7B).
Figure 7.
WHISTLE and JMJD1C perform a regulatory role in SF-1-mediated mouse steroidogenesis. (A) TM3 cells were cultured in the absence or presence of LH (200 ng/ml) and transfected with each of the indicated constructs. Cultured media were collected and the testosterone concentrations (ng/µl) were determined via RIA. Data are representative of three independent experiments. Results are expressed as means ± SD; n = 3. *P < 0.05 and ***P < 0.001. (B) Model of the regulatory role of WHISTLE and JMJD1C in mouse steroidogenesis.
DISCUSSION
Since the identification of the H3K4-specific LSD1, a growing number of histone demethylases with different specificities have been characterized. Classes of demethylases that harbor the JmjC domain have recently been the focus of active investigations. Approximately 28 human Jmj-harboring proteins that appear relevant to the dynamic regulation of histone methylation have been identified thus far (5). The aberrant regulation of histone demethylases causes a variety of human diseases, including certain cancers and developmental defects (5,24). JMJD1C was originally referred to as thyroid hormone receptor-interacting protein 8 (TRIP8) (25). The other JMJD1 family demethylases, testis-specific gene A (TSGA, JMJD1A) and JMJD1B (5qNCA), were also identified and implicated as playing a role in mouse spermatogenesis and human hematopoiesis, respectively (21,26). The JMJD1C variant s-JMJD1C was identified previously as an androgen receptor-interacting coactivator, and its observed reduced expression in human breast cancer is suggestive of its functions in tumor suppression (27).
In this study, we purified the HMTase WHISTLE interacting complex via an affinity purification TAP strategy, and isolated proteins with a variety of biological functions, including transcriptional regulation. Among them, we identified HSP90α as a co-factor for the full H3K4- and H3K27-methylating activity of WHISTLE. A recent report has demonstrated that the carcinogenesis-related HMTase, SMYD3, requires the HSP90α protein to exert its biological activities, which include histone methylation (28). Via different analysis methods, we further identified the JMJD1 family demethylase, JMJD1C, as an H3K9-me1/-me2 specific demethylase. The differential expression patterns of WHISTLE and JMJD1C during the developmental stages of the mouse testis were also identified in greater detail in this study. Consistent results were noted when we induced steroidogenesis in mouse testis TM3 cells with LH. We then demonstrated that HMTase WHISTLE and demethylase JMJD1C differentially regulated the transcription of a mouse steroidogenic target gene, p450c17, during testis development. These results strongly suggest that WHISTLE may perform a repressive regulatory role during the early stages of steroidogenesis, whereas JMJD1C exerts positive effects as steroidogenesis proceeds. Finally, we identified the SF-1-mediated steroidogenesis regulatory network via the addition and removal of specific methylation marks by HMTase WHISTLE and demethylase JMJD1C.
Based on our results, we propose that SF-1 is able to bind to H3K4, H3K27 methyltransferase WHISTLE and H3K9 demethylase JMJD1C. During the early stages of steroidogenesis, H3K4 and H3K27 methyltransferase WHISTLE binds to SF-1 and negatively regulates the transcription of the steroidogenic marker, p450c17. Upon the induction of steroidogenesis by LH, the interaction between SF-1 and WHISTLE is significantly reduced. On the other hand, the recruitment of H3K9 demethylase JMJD1C via SF-1 begins to increase with progressing development of the mouse testes (Figure 7B). The presence of SF-1 at the p450c17 promoter prior to LH treatment warrants further studies designed to identify in detail the mechanisms underlying SF-1-mediated p450c17 transcriptional regulation by LH signaling and histone modification marks. Consistent with our results, coordinate regulations of histone methylation and demethylation for the fine control of target gene transcription have been recently reported by other researchers. The differential regulation of hematopoietic cell development by histone demethylase LSD1 and methyltransferase hSET1 have both been previously described (29). H3K27 methylating Polycomb-Repressive Complex 2 (PRC2) and H3K4 demethylase Rbp2 (Jarid1a) have been proposed to perform a role in mouse ES cell differentiation (10). The importance of T-box-mediated interactions between H3K4 methyltransferase and H3K27 demethylase activities were identified via the association of human developmental defects with disruptions of the interaction (11). Interestingly, a recent finding has demonstrated that the proper balance between Set1-mediated H3K4 methylation and H3K4 demethylation by Jhd2 is regulated by another histone modification—namely, polyubiquitination mediated by the E3 ubiquitin ligase, Not4 (30). Collectively, our results provide evidence for the HMTase WHISTLE-interacting complex, including H3K9-me1/2 demethylase JMJD1C and WHISTLE cofactor HSP90α. More importantly, two distinct and antagonistic activities function as transcriptional activators or repressors of the steroidogenesis target gene, p450c17, and demonstrate their transcriptional regulatory functions in the development of mouse testes via interactions with SF-1. Further studies will be necessary to provide information regarding physiological implications of the delicate balances pertaining under various epigenetic modifications, including histone methylation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Mid-career Researcher Program, National Research Foundation of Korea grant, Ministry of Education, Science and Technology (R01-2008-000-20358-0); Korea Healthcare Technology R&D Project, Ministry for Health, Welfare & Family Affairs, Republic of Korea (A084869). Funding for open access charge: Mid-career Researcher Program, National Research Foundation of Korea grant, Ministry of Education, Science and Technology (R01-2008-000-20358-0).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Professors Kee-Sook Lee and Heung-Sik Choi of Chonnam National University for the TM3 cells and pcDNA3-SF-1 clones, respectively.
REFERENCES
- 1.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 2.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 3.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 4.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 5.Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol. Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 6.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 8.Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, Di Croce L, Shiekhattar R. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 9.Shirato H, Ogawa S, Nakajima K, Inagawa M, Kojima M, Tachibana M, Shinkai Y, Takeuchi T. A jumonji (Jarid2) protein complex represses cyclin D1 expression by methylation of histone H3-K9. J. Biol. Chem. 2009;284:733–739. doi: 10.1074/jbc.M804994200. [DOI] [PubMed] [Google Scholar]
- 10.Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, Helin K. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and polycomb-repressive complex 2. Genes Dev. 2008;22:1345–1355. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller SA, Huang AC, Miazgowicz MM, Brassil MM, Weinmann AS. Coordinated but physically separable interaction with H3K27-demethylase and H3K4-methyltransferase activities are required for T-box protein-mediated activation of developmental gene expression. Genes Dev. 2008;22:2980–2993. doi: 10.1101/gad.1689708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parker KL, Schimmer BP. Steroidogenic factor 1: a key determinant of endocrine development and function. Endocr. Rev. 1997;18:361–377. doi: 10.1210/edrv.18.3.0301. [DOI] [PubMed] [Google Scholar]
- 13.Toppari J, Kaleva M, Virtanen HE, Main KM, Skakkebaek NE. Luteinizing hormone in testicular descent. Mol. Cell Endocrinol. 2007;269:34–37. doi: 10.1016/j.mce.2006.10.021. [DOI] [PubMed] [Google Scholar]
- 14.Kim SM, Kee HJ, Eom GH, Choe NW, Kim JY, Kim YS, Kim SK, Kook H, Kook H, Seo SB. Characterization of a novel WHSC1-associated SET domain protein with H3K4 and H3K27 methyltransferase activity. Biochem. Biophys. Res. Commun. 2006;345:318–323. doi: 10.1016/j.bbrc.2006.04.095. [DOI] [PubMed] [Google Scholar]
- 15.Kim SM, Kee HJ, Choe N, Kim JY, Kook H, Kook H, Seo SB. The histone methyltransferase activity of WHISTLE is important for the induction of apoptosis and HDAC1-mediated transcriptional repression. Exp. Cell Res. 2007;313:975–983. doi: 10.1016/j.yexcr.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 16.Knuesel M, Wan Y, Xiao Z, Holinger E, Lowe N, Wang W, Liu X. Identification of novel protein-protein interactions using a versatile mammalian tandem affinity purification expression system. Mol. Cell. Proteomics. 2003;2:1225–1233. doi: 10.1074/mcp.T300007-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 18.Mendis-Handagama SM, Ariyaratne HB. Differentiation of the adult Leydig cell population in the postnatal testis. Biol. Reprod. 2001;65:660–671. doi: 10.1095/biolreprod65.3.660. [DOI] [PubMed] [Google Scholar]
- 19.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 20.Loh YH, Zhang W, Chen X, George J, Ng HH. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21:2545–2557. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knebel J, De Haro L, Janknecht R. Repression of transcription by TSGA/Jmjd1a, a novel interaction partner of the ETS protein ER71. J. Cell. Biochem. 2006;99:319–329. doi: 10.1002/jcb.20945. [DOI] [PubMed] [Google Scholar]
- 22.Okada Y, Scott G, Ray MK, Mishina Y, Zhang Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature. 2007;450:119–123. doi: 10.1038/nature06236. [DOI] [PubMed] [Google Scholar]
- 23.Sugawara T, Saito M, Fujimoto S. Sp1 and SF-1 interact and cooperate in the regulation of human steroidogenic acute regulatory protein gene expression. Endocrinology. 2000;141:2895–2903. doi: 10.1210/endo.141.8.7602. [DOI] [PubMed] [Google Scholar]
- 24.Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22:1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katoh M. Identification and characterization of TRIP8 gene in silico. Int. J. Mol. Med. 2003;12:817–821. [PubMed] [Google Scholar]
- 26.Hu Z, Gomes I, Horrigan SK, Kravarusic J, Mar B, Arbieva Z, Chyna B, Fulton N, Edassery S, Raza A, et al. A novel nuclear protein, 5qNCA (LOC51780) is a candidate for the myeloid leukemia tumor suppressor gene on chromosome 5 band q31. Oncogene. 2001;20:6946–6954. doi: 10.1038/sj.onc.1204850. [DOI] [PubMed] [Google Scholar]
- 27.Wolf SS, Patchev VK, Obendorf M. A novel variant of the putative demethylase gene, s-JMJD1C, is a coactivator of the AR. Arch. Biochem. Biophys. 2007;460:56–66. doi: 10.1016/j.abb.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 28.Hamamoto R, Furukawa Y, Morita M, Iimura Y, Silva FP, Li M, Yagyu R, Nakamura Y. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat. Cell. Biol. 2004;6:731–740. doi: 10.1038/ncb1151. [DOI] [PubMed] [Google Scholar]
- 29.Hu X, Li X, Valverde K, Fu X, Noguchi C, Qiu Y, Huang S. LSD1-mediated epigenetic modification is required for TAL1 function and hematopoiesis. Proc. Natl Acad. Sci. USA. 2009;106:10141–10146. doi: 10.1073/pnas.0900437106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mersman DP, Du HN, Fingerman IM, South PF, Briggs SD. Polyubiquitination of the demethylase Jhd2 controls histone methylation and gene expression. Genes Dev. 2009;23:951–962. doi: 10.1101/gad.1769209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.