Abstract
Mammalian mitochondria contain full-length genome and a single-stranded 7S DNA. Although the copy number of mitochondrial DNA (mtDNA) varies depending on the cell type and also in response to diverse environmental stresses, our understanding of how mtDNA and 7S DNA are maintained and regulated is limited, partly due to lack of reliable in vitro assay systems that reflect the in vivo functionality of mitochondria. Here we report an in vitro assay system to measure synthesis of both mtDNA and 7S DNA under a controllable in vitro condition. With this assay system, we demonstrate that the replication capacity of mitochondria correlates with endogenous copy numbers of mtDNA and 7S DNA. Our study also shows that higher nucleotide concentrations increasingly promote 7S DNA synthesis but not mtDNA synthesis. Consistently, the mitochondrial capacity to synthesize 7S DNA but not mtDNA noticeably varied along the cell cycle, reaching its highest level in S phase. These findings suggest that syntheses of mtDNA and 7S DNA proceed independently and that the mitochondrial capacity to synthesize 7S DNA dynamically changes not only with cell-cycle progression but also in response to varying nucleotide concentrations.
INTRODUCTION
Most animal cells contain two sets of genetic information, one in the nucleus and the other in mitochondria. Although human mitochondria harbor a relatively small genome of only 16 569 bp (1), a high incidence of diverse metabolic diseases and various cancers are associated with alterations in the mitochondrial genome (2–4), clearly indicating the significance of its stable maintenance. Mammals typically contain ∼103–104 copies of mitochondrial DNA (mtDNA) per cell (5–7). Early studies, using electron microscopy, indicated that mammalian mtDNA contains an unwound or displaced single-stranded region, called the D-loop (8,9). The 5′-end of the D-loop region coincides with the replication origin of the H-strand (OH), while the 3′-end contains the termination-associated sequence (TAS) (10). Replication of mammalian mtDNA initiates at the OH and frequently terminates at the TAS site, generating a short single-stranded DNA (ssDNA) fragment, called 7S DNA (9,10).
Several studies have reported in vitro synthesis of mtDNA and 7S DNA in isolated mitochondria, through which a rapid turnover of 7S DNA as well as direct uptake of dNTP by mitochondria and subsequent utilization for mtDNA synthesis have been demonstrated (11–15). Also, Mitra and Bernstein (14) proposed for the first time the conversion of thymidine to TTP within mitochondria (i.e. the presence of the mitochondrial nucleotide salvage pathway). These previous studies testify to the potential of an in vitro assay system to gain insights into the mechanism of mtDNA replication. However, whether mtDNA synthesis measured in vitro is catalyzed by mitochondrial DNA polymerase γ, reflecting the in vivo replication capacity of mitochondria, and whether in vitro assay systems are suitable to study the regulation of mtDNA copy number remain unexplored.
Recently, while establishing an in vitro assay system for human DNA replication, we observed de novo DNA synthesis in reactions that were not provided with exogenous DNA template. Subsequent analysis revealed that the de novo synthesized DNA is the human full-length mitochondrial genome. In this report, we describe that mtDNA synthesis measured in vitro is resistant to aphidicolin but sensitive to dideoxynucleotide, indicating that mitochondria-specific DNA polymerase γ is responsible for the observed mtDNA synthesis. Using this assay system, we observed that the capacity of mitochondria to synthesize 7S DNA but not mtDNA changes with cell-cycle progression and with varying nucleotide concentrations. Thus we conclude that syntheses of 7S DNA synthesis and mtDNA occur independently of each other.
MATERIALS AND METHODS
Preparation of replication–competent cytoplasmic and nuclear extracts
HeLa cells were grown in 5–20, 150 mm culture plates to ∼80% confluence. On the day of harvesting, cells were replenished with fresh media and ∼6 h later were collected in conical tubes. After washing with ice-cold PBS, cells were treated with a hypotonic buffer [10 mM HEPES–HCl, pH 7.4, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol (DTT) and 5 mM β-glycerophosphate] on ice for 15 min and homogenized with 10 stokes using a type B pestle (Kontes). Homogenates were cleared by centrifugation for 15 min at 2000g, and the nuclear pellet was salt-extracted as described previously (16). The resultant nuclear extracts (NEs) were dialyzed against buffer G, consisting of 20 mM Hepes–HCl, pH 7.4, 0.1 M NaCl, 0.5 mM DTT, 0.2 mM ethylenediamine tetra-acetate, 5 mM β-glycerophosphate and 20% glycerol, and cleared by centrifugation at 25 000g for 30 min (N1) or 100 000g for 1 h (N2). On the other hand, the homogenate supernatants were dialyzed for 4 h against buffer G lacking β-glycerophosphate (buffer A) and the dialysates (C4) were stored at −70°C in aliquots. The dialysates were further subjected to centrifugation at 100 000g for 1 h (C1), 25 000g (C2) or 10 000g for 15 min (C3) and the clear supernantants were stored at −70°C in aliquots. Throughout the presented study for mtDNA replication and 7S DNA synthesis in vitro, replication–competent cytoplasmic extracts (CEs) were prepared the same as C4.
Cell synchronization
HeLa cells were seeded to 10% confluence in three groups (A–C) on 100-mm culture plates for determination of endogenous levels of mtDNA and 7S DNA or in three groups of ten 150-mm culture plates for the preparation of replication competent CE. On the following day, cells were replenished with fresh DME medium containing 2 mM thymidine and 7% fetal bovine serum (FBS). After 16 h-incubation, culture plates were washed twice with serum-free DME, then supplemented with 24 µM deoxycytidine and 7% FBS. After incubation for 9 h at 37°C, cells were washed twice, provided with DME containing 2 mM thymidine and 7% FBS, and then continuously incubated for 20 h. Cells in group A were harvested immediately but cells in group B were incubated for an additional 7 h in DME containing 7% FBS and those in group C were grown for 18 h in DME containing 7% FBS and 0.2 mM nocodazole (Sigma). Cells were collected using policeman and washed twice with ice-cold 1× PBS. Cells obtained from 100 mm culture plates were split into two 15 ml conical tubes. Cells in one tube were re-suspended in buffer G (0.5 ml) and stored at −70°C in aliquots. Cells in another tube were stained with propidium iodide (Invitrogen) and subjected to standard fluorescence activated cell sorter (FACS) analysis (BD Biosciences FACSCaliburs) to determine the extent of their synchrony at a specific stage of the cell cycle. On the other hand, cells harvested from 150 mm plates were processed to prepare the replication–competent CE, as described earlier.
Western blot analysis
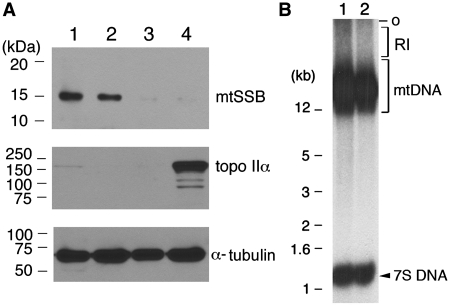
Aliquots of cytoplasmic fractions enriched in mitochondria and other subcellular fractions, indicated in Figure 3, were mixed with one-fifth volume of 5× sodium dodecylsulfate (SDS) sample buffer and heated at 95°C for 5 min. Subsequently, they were separated by 10% SDS–PAGE and proteins were transferred to PVDF filter (Schleicher and Schuell) using the SemiDry system (Bio-Rad). Western blot analysis was performed using an ECL reagent (Amersham Biosciences) and indicated antibodies (Ab) including, mouse anti-topoisomerase IIα Ab (1 : 1000 dilution) (BD Biosciences), mouse anti-α-tubulin Ab (1 : 2000 dilution) (Santa Cruz) and rabbit anti-mtSSB Ab (1 : 300 dilution) (a kind gift of Dr M. Zeviani).
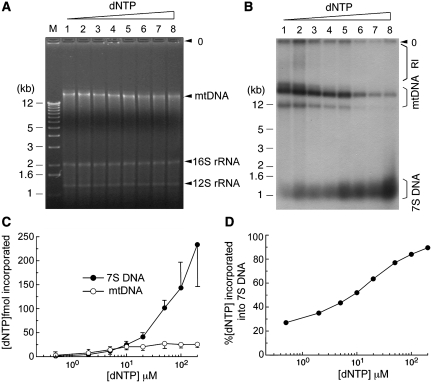
Figure 3.
Influence of nucleotide concentration on in vitro synthesis of 7S DNA and mtDNA. Replication reactions were performed with HeLa CE (∼1.5 × 105 cell equivalent) in the presence of variable equimolar dNTP levels: lane 1, 0.5 µM dNTP/5 µCi α-32P-dATP; lanes 2–4, 2, 5 or 10 µM dNTP/10 µCi α-32P-dATP; lanes 5–7, 20, 50 or 100 µM dNTP/20 µCi α-32P-dATP; lane 8, 200 µM dNTP/40 µCi α-32P-dATP. For each reaction, the specific radioactivity per µM dNTP is estimated as follows: lane 1, 2.2 × 107 cpm; lane 2, 1.1 × 107 c.p.m.; lane 3, 4.4 × 106 c.p.m.; lanes 4–5, 2.2 × 106 c.p.m.; lane 6, 8.8 × 105 c.p.m.; lanes 7–8, 4.4 × 105 c.p.m. After 2 h incubation, reaction mixtures were resolved on a 1% agarose gel, as described in ‘Materials and Methods' section. Position of mtDNA and two mitochondrial ribosomal DNAs, 16S and 12S rRNA, are indicated in (A). Immediately after photography, the gel was dried and then subjected to autoradiography to detect newly synthesized 7S DNA, mtDNA and replication intermediates (RI), as shown in (B). Nucleotide incorporation into mtDNA and 7S DNA was quantified as described in 'Materials and methods' section and average values obtained from four independent experiments are presented in (C). (D) Nucleotide incorporation into 7S DNA is presented relative to total dNTP incorporation (i.e. 7S DNA + full-length mtDNA).
In vitro mtDNA synthesis
Unless otherwise indicated, the reaction mixture (20 µl) contained 30 mM Tris–HCl, pH 8.5, 0.1 M potassium acetate, 7 mM MgCl2, 0.5 mM dithiothreitol (DTT), 4 mM ATP, 0.2 mM CTP/UTP/GTP, 0.1 mM dCTP/dTTP/dGTP, 20 µM dATP/2 µCi α-32P-dATP (3000 Ci/mmol), 6 mM creatine phosphate (CP), 1.25 µg of creatine phosphokinase (CPK), 0.1 mg/ml bovine serum albumin and ∼50 µg of CE. In Figure 1, however, the reaction mixture was constituted with 100 µg of CE in a final volume of 50 µl. After incubation at 37°C for 2 h, the reaction mixture was centrifuged at 10 000g for 1 min four times, while washing with buffer G (∼2 ml). The final pellet, enriched in mitochondria, was re-suspended in a mixture (12 µl) of SDS (0.5%) and proteinase K (0.25 mg/ml). Subsequent to incubation at 37°C for an additional 1 h, the lysate was mixed with 3 µl of 5× gel loading buffer (50% glycerol, 0.1% xylene cyanol and 0.1% bromophenol blue) and resolved on a 1% agarose gel in 1× TAE buffer using a 1-kb ladder (Invitrogen) as a molecular standard. The gel was electrophoresed at constant 80 V for 2.5 h, washed with deionized water, and dried under vacuum on Whatman DE-81 paper. De novo synthesized DNA was visualized by autoradiography and quantified using a PhosphorImager (Molecular Dynamics).
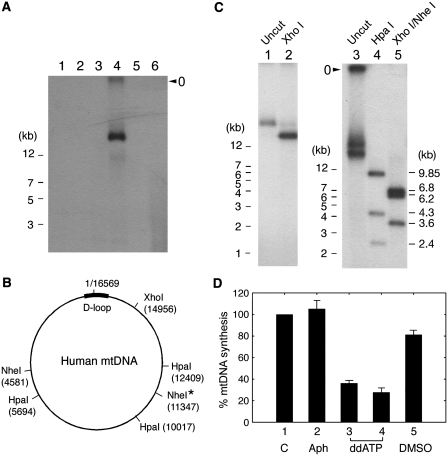
Figure 1.
Analysis of de novo synthesized DNA. (A) Control replication reactions were carried out in the standard reaction mixture (50 µl), as described in ‘Materials and Methods' section, with either 100 µg of cytoplasmic extract (C1–C4) (lanes 1–4, respectively) or 50 µg of nuclear extract (N1 & N2) (lanes 5 and 6), but omitting the template DNA, pSLVD (Supplementary Figure S1). Procedures employed for the preparation of C1-4 and N1-2 are detailed in ‘Material and Methods’ section. DNA, isolated by conventional phenol/chloroform extraction and ethanol precipitation, was resolved on a 1% agarose gel. Subsequently, de novo synthesized DNA was visualized by autoradiography. (B) Predicted cleavage sites for the indicated restriction enzymes in human mitochondrial DNA (GenBank AC000021). Asterisk indicates a single nucleotide polymorphism at position 11 347, which is found in 3 out of 2704 mtDNA sequences (http://www.genpat.uu.se/mtDB/). (C) DNA was digested for 1 h with Xho I (lane 2), Hpa I (lane 4) or Xho I and Nhe I (lane 5). In (A) and (C), the gel well is indicated as ‘o’. (D) Reactions (in triplicate) for mtDNA synthesis were carried out using the standard reaction mixture provided with 100 µg of HeLa CE in the presence of aphidicolin (100 µg/ml, lane 2), ddATP (20 or 100 µM, lanes 3–4) or DMSO (4%, lane 5). De novo synthesized DNA was quantified using a PhosphorImager. In the control reaction in (lane 1), 100% indicates the incorporation of 34 fmol α-32P-dATP into full-length mtDNA.
Real-time quantitative PCR
For experiments in Figure 4, the LightCycler (Roche) was used to measure copy number of full-length mtDNA, 7S DNA and β-actin DNA. Forward primer a, 5′-gtggctttggagttgcagtt-3′, is derived from nucleotide position 16 240–16 259 within the D-loop of mtDNA sequence. Sequences of reverse primers b1 and b2 are 5′-cagccaccatgaatattgtac-3′ and 5′-gaagcagatttgggtaccac-3′ that map to positions 16 108–16 128 and 16 036–16 055, respectively. Forward and reverse primers for β-actin DNA are derived from nucleotide positions 1315–1337 (5′-agaaaaatctggcaccacaccttc-3′) and 1547–1570 (5′-gcagagttccaaaggagactcagg-3′) (Genbank CCDS 5341.1). Each reaction was provided with 1× LightCycler® FastStart DNA Master HybProbe, 300 nM each of the forward and reverse primer, 200 nM probe and 0.1 unit heat-labile uracil-DNA glycosylase (Roche) in a total volume of 10 µl. In each run, pKW125 (17), a plasmid DNA containing amplicon representing full-length mtDNA, was run as an external control. The probe for the ensuing amplicon is located at nucleotide position 13 461–13 480 and is tagged with the reporter FAM (6-carboxy fluorescein, λmax 518 nm) at the 5′ end and the quencher BHQ1 at the 3′ end. The copy numbers of mtDNA and 7S DNA per cell was determined from the plasmid control copy number and its threshold cycle following the procedure previously described (17). For experiments in Figure 5, RT–qPCR was carried out using Applied Biosystem 7500 system in a mixture (20 µl) containing ∼500–1000 (reaction for mtDNA or 7S DNA) or ∼10 000 HeLa cells (reaction for β-actin DNA), 1× SYBR® Green Master Mix and each of the forward and reverse primers (200 nM).
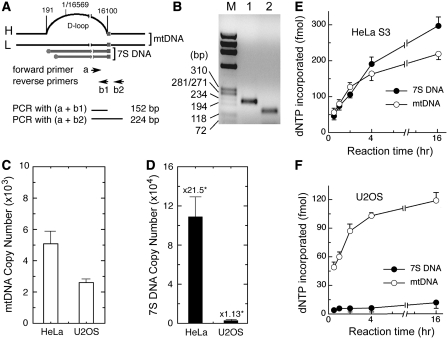
Figure 4.
Comparative analysis of endogenous levels of mtDNA/7S DNA and the mitochondrial capacity to synthesize mtDNA/7S DNA measured in vitro. (A) RT–qPCR was performed to measure endogenous levels of mtDNA and 7S DNA using the primer pairs (a+b1) and (a+b2), respectively. Schematic diagram indicates relative positions of PCR primers and expected PCR products. Numbers on top of mtDNA show nucleotide positions. (B) An aliquot (5 µl) of a representative PCR reaction provided with the primer pair (a+b2) (lane 1) or (a+b1) (lane 2) was analyzed on a 2% agarose gel and DNA was visualized by EtBr staining. ϕX174 DNA-Hae III digests, used as a molecular standard, are indicated (M). (C and D) RT–qPCR was performed with aliquots of diluted CE, equivalent to ∼300 HeLa cells or ∼59 U2OS cells. Average copy numbers of mtDNA (C) and 7S DNA (D) were obtained from two independent duplicate experiments. In (D), numbers on top of each bar indicate fold-abundance of 7S DNA relative to mtDNA. (E–F) Replication reactions were carried out with aliquots of HeLa CE (∼1.5 × 105 cell equivalents) or U2OS CE (∼5 × 105 cell equivalents). The extent of dNTP incorporation into mtDNA and 7S DNA, obtained from two independent duplicate experiments, is presented.
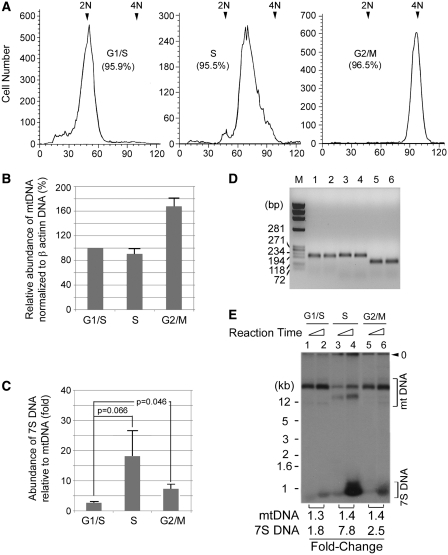
Figure 5.
Comparative analysis of mtDNA replication and 7S DNA synthesis in vivo. (A) Aliquots of HeLa cells (∼1 × 104 cells) synchronized at the indicated stage of the cell cycle were subjected to FACS analysis. The extent of cell synchrony is presented as percent in parenthesis. (B–D) The copy number of β-actin gene was determined by RT–qPCR, to which mtDNA copy number was normalized (B). Fold ratio of 7S DNA to mtDNA at different stages of the cell cycle, as determined by qPCR, is presented in (C) where SD and the two-tailed Student’s t-test probabilities of three independent experiments are shown. (D) Aliquots (5 µl) of two representative RT–qPCR reactions provided with the primer pair for mtDNA (lanes 1–2), β-actin gene (lanes 3–4), or 7S DNA (lanes 5–6), were analyzed on a 2% agarose gel. (E) The mitochondrial capacity to synthesize mtDNA and 7S DNA was determined with aliquots (5 µl) of replication competent CE prepared from HeLa cells synchronized at the indicated stage of the cell cycle. After 2 h or 14 h incubation, reaction mixtures were resolved on a 1% agarose gel. Positions of mtDNA and 7S DNA are indicated.
RESULTS
De novo synthesis of mtDNA in vitro catalyzed by DNA polymerase γ
During efforts to develop an in vitro assay system that supports replication of human DNA, we found that a CE fraction supported DNA synthesis in the absence of an added template (Figure 1A, lane 4). Since DNA was synthesized by CE without any added template, we suspected that it might be de novo synthesized mtDNA in vitro. To test this possibility, de novo synthesized DNA was subjected to restriction enzyme analysis using Xho I, Hpa I and Nhe I. Digestion of mtDNA with Xho I should yield a full-length linear fragment of ∼16.5 kb and Hpa I should generate three DNA fragments, 9.85, 4.3 and 2.4 kb in length, as depicted in Figure 1B. Indeed, Xho I digestion produced a single DNA fragment (Figure 1C, lane 2) and Hpa I digestion generated three fragments of the expected size (Figure 1C, lane 4). In the case of the Xho I/Nhe I double digestion, we initially predicted the production of two DNA fragments (∼14.2 and 2.4 kb) since most human mtDNAs contain a single cleavage site for each enzyme (Figure 1B). Unexpectedly, three DNA fragments (∼6.8, 6.2 and 3.6 kb) were detected (Figure 1C, lane 5), suggesting the presence of an additional cleavage site. In essence, these results support that ∼16 kb DNA synthesized in vitro is mtDNA.
Of a total of 14 human DNA polymerases identified to date, three enzymes (pol α, pol δ and pol ε) take part in genomic DNA replication and one polymerase (pol γ) is involved in mtDNA replication (18–21). Pol α, pol δ and pol ε are all sensitive to aphidicolin but relatively resistant to dideoxynucleotides (ddNTPs) such as ddATP (22). Conversely, pol γ is highly sensitive to ddNTPs but resistant to aphidicolin (23). Thus, by determining the sensitivity of this reaction to aphidicolin and ddNTPs, we explored whether nuclear or mitochondrial DNA polymerases catalyzed the de novo synthesis of the 32P-labeled mtDNA. Simian virus 40 (SV40) DNA replication was chosen as a control reaction since it requires at least two replicative DNA polymerases (pol α and pol δ) (24,25). In contrast to SV40 DNA replication, as summarized in Supplementary Data, mtDNA synthesis was severely impaired by relatively low levels of ddATP (20–100 µM) (Figure 1D, lanes 3–4) but not by aphidicolin (100 µg/ml) (Figure 1D, lane 2). Taken together, these results demonstrate that de novo DNA synthesis observed with CE in the absence of an added DNA template represents mtDNA synthesis catalyzed by DNA pol γ.
Stable association of de novo synthesized 7S DNA and mtDNA with mitochondrial factors
As shown in Figure 1A, only full-length mtDNA synthesis was observed but not 7S DNA synthesis. In those experiments, de novo synthesized DNA was isolated from reaction mixtures through phenol/chloroform extraction following treatment with SDS and proteinase K. Since association of certain mitochondrial factors with mtDNA is sustained even after SDS and proteinase K treatment (26–28), we decided to omit the phenol/chloroform extraction step in analyzing de novo synthesized DNA. Instead, reactions were subjected to repeated centrifugations at 10 000g. Under this condition, only intact mitochondria but neither cytoplasmic RNA nor free DNA could be recovered. After the final wash, pelleted sample as well as supernatant, and total cytoplasmic extract were analyzed by western blotting. Results show that mitochondrial single-stranded DNA-binding protein (mtSSB) was detected in the total cytoplasmic extract and pellet (Figure 2A, lanes 1–2) but not in the supernatant (Figure 2A, lane 3) or nuclear extract (Figure 2A, lane 4). By contrast, the nuclear protein topoisomerase IIα was present only in the nuclear fraction (Figure 2A, lane 4). Throughout the fractionation steps, membrane detergents were not used, which might explain the presence of α-tubulin in all fractions (Figure 2A, lanes 1–4). α-Tubulin is an abundant protein that interacts with organelle membranes through hydrophobic interactions (29). Detection of mtSSB in the pellet fraction, but not in the clear cytoplasmic supernatant or NE, demonstrates efficient recovery of mitochondria after repeated centrifugations.
Figure 2.
An optimal reaction condition for 7S DNA synthesis in vitro. (A) Replication–competent CE (lane 1) was subjected to four consecutive centrifugations at 10 000g for 1 min. The clear supernatant (lane 3) obtained after the first centrifugation was stored, while the pellet (lane 2), enriched in mitochondria, was re-suspended in buffer G after four rounds of centrifugation. Subcellular fractions, including NE (lane 4), were analyzed by western blotting for the indicated protein with the following aliquots: lane 1, 15 µg of CE; lane 2, 4.2 µg of mitochondria fraction; lane 3, 15 µg of clear CE; lane 4, 10.8 µg of NE. (B) A duplicate set of replication reactions was performed in the standard mixture provided with replication–competent HeLa CE. After 2 h incubation, reaction mixtures were subjected to repeated centrifugations, as described in (A). After the final centrifugation, the pellets were treated with SDS (0.5%) and proteinase K (0.25 mg/ml) in a final volume of 12 µl. Following 30 min incubation at 37°C, the lysates were directly loaded on an agarose gel, and de novo synthesized DNAs were resolved by electrophoresis and visualized by autoradiography. O, the gel well. RI, replication intermediates.
Next, the pellets enriched in mitochondria were treated with SDS (0.25%) and proteinase K (0.25 mg/ml) in a final volume of 12 µl. After incubation for 30 min at 37°C, the lysates were mixed with 3 µl of 5× gel loading buffer and directly analyzed on an agarose gel (Figure 2B). Indeed, omission of phenol/chloroform extraction enabled us to detect not only de novo synthesized mtDNA but also 7S DNA (Figure 2B). This result is consistent with reports that mtDNA and 7S DNA stably associate with mitochondrial factors such as mtSSB (27) and ATAd3p (26). It should be noted that although the quantification of in vitro DNA synthesis was presented only for the full-length mtDNA and 7S DNA, replication intermediates, indicated as RI, were estimated to be as much as 25% of the total dNTP incorporation.
Influence of nucleotide concentrations on in vitro synthesis of mtDNA and 7S DNA
With the exception of ATP and GTP, all other nucleotides required for mtDNA synthesis must be imported from the cytoplasm or generated through the nucleotide salvage pathway within mitochondria (30–35). Although nucleotide depletion predictably leads to a decrease in the mtDNA copy number (32–34,36,37), the impact of the cytoplasmic nucleotide pool on mtDNA replication has not been directly investigated. We explored this issue by measuring synthesis of both mtDNA and 7S DNA using α-32P-dATP as tracer in the presence of increasing concentrations (0.5–200 µM) of dNTP. It should be noted that though partially adjusted, the specific radioactivity of dATP in each reaction generally decreased with higher dNTP concentrations, as described in the legend of Figure 3.
One representative set of reactions resolved on an agarose gel is shown in Figure 3A. Staining with ethidium bromide (EtBr) revealed no significant change in overall levels of mtDNA and mitochondrial rRNAs with varying dNTP concentrations. Autoradiography, however, unveiled differential changes in mtDNA replication and 7S DNA synthesis with increasing dNTP concentration (Figure 3B–D). In brief, synthesis of mtDNA marginally increased, reaching a plateau at 10 µM of dNTPs (Figure 3C), while 7S DNA synthesis continued to increase at higher levels of dNTPs (Figure 3C), accounting for as much as 90% of the total observed DNA synthesis (Figure 3D). This result suggests that 7S DNA synthesis and mtDNA synthesis may not be coupled but instead proceed independently in response to varying nucleotide concentrations. Consistent with this possibility, our result has also shown that the in vivo level of 7S DNA relative to mtDNA fluctuates with progression of the cell cycle (see below).
Synthesis of mtDNA and 7S DNA in vitro reflects the in vivo functionality of mitochondria
Next, we asked whether synthesis of mtDNA and 7S DNA measured in vitro reflects the in vivo functionality of mitochondria. In this study, we utilized HeLa (a cervical carcinoma line) and U2OS (an osteosarcoma line) cells since they exhibited marked differences in their endogenous levels of 7S DNA among all cells tested. First, we determined the copy number of endogenous mtDNA and 7S DNA employing real-time quantitative PCR (RT–qPCR) using a forward primer (primer a) and two reverse primers (primers b1 and b2) (Figure 4A), as described in ‘Materials and Methods' section. The 152 bp PCR product was derived from both mtDNA and 7S DNA, while the 224 bp PCR product was produced only from mtDNA (Figure 4B). Copy numbers of mtDNA and 7S DNA were determined by comparing the abundance of PCR product obtained with mitochondria relative to that with pKW125, a control plasmid containing the entire mtDNA, as described previously (17). It was estimated that HeLa and U2OS cells contained ∼5100 and 2300 copies of mtDNA per cell, respectively (Figure 4C). The ratios of 7S DNA to mtDNA in HeLa and U2OS cells were ∼21.5 and 1.13, respectively (Figure 4D).
We also tested whether the replication capacity of mitochondria, measured in vitro, reflects endogenous levels of mtDNA and 7S DNA in HeLa and U2OS cells. When analyzed between 0.5 and 16 h of in vitro reaction, mtDNA synthesis in both HeLa and U2OS mitochondria increased linearly up to 2 h and then slowly plateaued (Figure 4E–F, open circles). During the first 2 h of reaction, the extent of mtDNA synthesis measured with HeLa CE (Figure 4E, open circles) was significantly higher than that with U2OS CE (Figure 4F, open circles), consistent with their relative levels of endogenous mtDNA (Figure 4C). A similar pattern was observed for the 7S DNA synthesis (Figure 4E–F, filled circles).
Differential synthesis of mtDNA and 7S DNA with cell-cycle progression
It is generally believed that mtDNA replication occurs throughout the cell cycle (38,39). Accordingly, it can be predicted that the replication capacity of mitochondria may not significantly vary with cell-cycle progression. In the above, we provided evidence that mtDNA synthesis measured in vitro using the newly established assay system reflect the in vivo functionality of mitochondria. Therefore, using the newly established assay system, we tested how the replication capacity of mitochondria changes with cell-cycle progression. For this purpose, we synchronized HeLa cells at different stages of the cell cycle and comparatively determined the replication capacity of mitochondria and endogenous levels of mtDNA and 7S DNA. The extent of cell synchronization, measured by FACS analysis, was estimated to be ∼95%, as shown in Figure 5A.
RT–qPCR analysis was performed with aliquots of mitochondria obtained from synchronous HeLa cells, which revealed that the ratio of mtDNA to β-actin genomic DNA remains unchanged during a 4-h transition from G1/S to S. Considering that β-actin genomic DNA replicates only in S phase, this result implies that mtDNA copy number increases in proportion with genomic DNA content. When cells were released from the G1/S phase and allowed to progress through S phase and to reach the next cell-cycle checkpoint, G2/M, by incubating for an additional 14 h in the presence of nocodazole, mtDNA content increased up to ∼160% relative to β-actin (Figure 5B). This result suggests that in contrast to genomic DNA replicating only in S phase, mtDNA continuously increased during the 14 h transition from G1/S to G2/M. By simply subtracting the extent of increase during the first 4 h in S phase, the mtDNA content is estimated to increase by additional 60% during prolonged incubation beyond the first 4 h of S phase up until G2/M phase. Overall, this result is consistent with an earlier observation reporting the continuous mode of mtDNA replication in S phase through G2/M phase of the cell cycle (38). In contrast to mtDNA, however, endogenous levels of 7S DNA noticeably varied with cell-cycle progression. The ratio of 7S DNA to mtDNA increased almost 7-fold when cells entered into S phase from the G1/S boundary (Figure 5C). Aliquots (5 µl) of the two representative reactions were subjected to agarose gel electrophoresis, which confirmed that expectedly, RT–qPCR reactions yielded amplicons of 224 bp from mtDNA (Figure 5D, lanes 1–2), 256 bp from β-actin gene (Figure 5D, lanes 3–4), and 152 bp from both 7S DNA and mtDNA (Figure 5D, lanes 5–6).
We asked whether the changes in endogenous levels of mtDNA and 7S DNA reflect the varying functionality of mitochondria along the cell cycle. Our study, performed with heterogeneous HeLa CE prepared from an exponentially growing population, revealed that during the transition from 2 to 14 h incubation newly synthesized mtDNA and 7S DNA increased about 1.4- and 2.6-fold, respectively (Figure 4E). Taking this observation into consideration, we compared the replication capacity of mitochondria in HeLa cells synchronized at different stages of the cell cycle by determining the fold-change in mtDNA and 7S DNA newly synthesized in vitro during the transition from 2 to 14 h incubation. Regardless of the cell cycle, newly synthesized mtDNA increased about 1.3- to 1.4-fold (Figure 5E), reminiscent of the result shown in Figure 4E. This result suggests that the mitochondrial capacity to synthesize mtDNA remains relatively unchanged throughout the cell cycle. However, it should be noted that a significant fraction of newly synthesized mtDNA in S phase mitochondria exhibits a different migration pattern (lanes 3–4). Therefore, unlike the capacity to synthesize mtDNA, mitochondria might undergo as yet unknown functional changes in S phase to catalyze a faster introduction of negative superhelicity into newly synthesized mtDNA. Changes in mitochondrial functionality along the cell cycle can be clearly seen from its capacity to synthesize 7S DNA that significantly fluctuates with cell-cycle progression: newly synthesized 7S DNA increased about 1.8-, 7.8- and 2.5-fold in mitochondria at G1/S, S and G2/M phase, respectively (Figure 5E). This result well reflects endogenous levels of mtDNA and 7S DNA measured by RT–qPCR (Figure 5C). Thus we conclude that mitochondrial functionality to synthesize 7S DNA but not mtDNA changes along the cell cycle.
DISCUSSION
In this report, we describe an in vitro assay system with which both mtDNA and 7S DNA are synthesized in vitro in such a way to reflect the in vivo replication capacity of mitochondria. Also, we provide experimental evidence for the first time that the replication capacity of mitochondria correlates with the endogenous copy number of both mtDNA and 7S DNA and that 7S DNA synthesis highly fluctuates with cell-cycle progression, whereas mtDNA synthesis occurs throughout the cell cycle. Consistently, our study illustrates that 7S DNA synthesis is highly influenced by varying nucleotide concentrations.
Prior to our observation, there have been several reports on in vitro synthesis of mtDNA (11–15,40). In 1991, Jui and Wong (13) reported for the first time in vitro synthesis of mtDNA using HeLa mitochondria permeabilized with Triton X-100. In their study, the use of the TCA precipitation method led them to score ∼4–5 pmol of TTP incorporation into acid-insoluble DNA polymer for 1 h in a reaction (100 µl) provided with permeabilized mitochondria (300 µg). However, when DNA was isolated from the reaction mixture by phenol/chloroform extraction followed by ethanol precipitation and subsequently resolved on an agarose gel, only heterogeneous DNAs were detected (13). In 1994, Enriquez et al. (11) reported that an in vitro assay system was established for highly efficient mtDNA synthesis using mitochondria isolated from rat liver. However, from a reaction (0.5 ml) provided with relatively high levels of mitochondria (1 mg), only <2 fmol dNTP was incorporated into full-length mtDNA for 2 h under the optimal condition (11). More recently, using essentially the same assay system, Wiesner and colleagues reported that 7S synthesis was enhanced when exogenous TFAM was imported into rat mitochondria permeabilized with Triton X-100 (1%) (12). However, de novo synthesized 7S DNA was not quantified and full-length mtDNA replication was not detected. These somewhat variable observations on mtDNA replication and 7S DNA synthesis may be due to a poor synthesis of both mtDNA and 7S DNA in the presence of Triton X-100 (data not shown) and also a poor recovery of both mtDNA and 7S DNA after phenol/chloroform extraction. By contrast, the in vitro assay system described in this study allowed us to detect both full-length mtDNA and 7S DNA with relatively higher efficiency: ∼34 fmol dATP was incorporated into full-length mtDNA during 2 h incubation in a reaction (20 µl) provided with ∼100 µg HeLa CE (Figure 1D, lane 1).
Reactions for SV40 DNA replication (Supplementary Data) was provided with 0.5 µg of the template pSVLD (10 kb), which is equivalent to ∼2.3 × 1011 molecules of the template DNA. Incorporation of 143 fmol dATP into SV40 DNA equals production of ∼1.7 × 107 daughter molecules; one copy per every 13 530 template DNA. In case of mtDNA replication (Supplementary Figure S1D, lane 1), considering the size (16 569 bp) of mtDNA and its endogenous copy number (5100/cell) (Figure 4C), incorporation of 34 fmol dATP corresponds to production of ∼2.5 × 106 molecules of full-length mtDNA on ∼7.7 × 108 molecules of endogenous template. Although the mtDNA synthesis observed in vitro was only ∼0.3% of the input template (one copy per every 308 molecules of template DNA), mtDNA synthesis in vitro can be considered fairly efficient as compared to in vitro SV40 DNA replication.
Our results showing that endogenous levels of mtDNA and 7S DNA do not correlate to each other, nor the extent of their synthesis, appears to be at odds when considering the following observations that implicate 7S DNA in mtDNA replication. (i) The 7S DNA is present in the D-loop region where de novo H-strand synthesis initiates (41–43). (ii) The 5′-end of 7S DNA coincides with the OH (10). (iii) The 7S DNA possesses a 3′-OH end that can be extended by DNA polymerase β in vitro (40), suggesting that 7S DNA has a potential to serve as a primer for the synthesis of full-length mitochondrial DNA. (iv) In addition, it is possible that through the formation or maintenance of the D-loop, the 7S DNA may facilitate the replication complex to assemble on the OH.
Although the presence of 7S DNA in the D-loop region can be a hallmark of replicating mitochondrial DNA, all of the above observations, implicating 7S DNA in mtDNA replication, have yet to be experimentally verified. In addition, although the proposed functions for 7S DNA presuppose its physical association with mtDNA how and when 7S DNA associates with mtDNA remains poorly understood. An earlier study by Clayton and colleagues made an intriguing observation that a large fraction of newly synthesized 7S DNA is associated with non-replicating mitochondrial DNA (44). In a subsequent study, they found that ∼20% of mitochondrial nucleotides are utilized for the synthesis of 7S DNA but over 95% of newly synthesized 7S DNA are rapidly lost with an estimated half-life of 70 min (45). Based on the high turnover rate of 7S DNA in isolated rat mitochondria, Gensler and colleagues argued that 7S DNA is not an intermediate of leading strand synthesis during replication (12). In support of this view, Attardi and coworkers reported that different replication origins are utilized for the synthesis of 7S DNA and the H-strand of full-length mtDNA (46). They proposed that mitochondria harbor two types of mtDNA replications, one for mtDNA maintenance under steady-state conditions and another for accelerating mtDNA replication in response to certain physiological demands such as recovery after mtDNA depletion. In the proposed model, they reasoned the involvement of 7S DNA in the latter (46). Although this is an attractive possibility, the function of 7S DNA under normal physiological conditions remains largely unknown.
While nuclear DNA replication takes place only in S phase of the cell cycle, mtDNA replication is believed to occur throughout the cell cycle (38,39,47,48). Unlike the nucleus where nucleotides are rapidly depleted upon completion of DNA replication (39), mitochondria may have evolved to maintain their own nucleotide pool to support mtDNA replication throughout the cell cycle. In support of this possibility, Bogenhagen and Clayton (30) reported that treatment of mouse LMTK− cells with methotrexate, an inhibitor of de novo nucleotide synthesis, inhibited over 96% of nuclear DNA synthesis but under the same condition, mtDNA synthesis continued at 50–60% of the control level for at least 10 h. In this regard, the present study may suggest a novel regulatory pathway for mtDNA synthesis, which consists of 7S DNA synthesis and its degradation. In brief, we show that 7S DNA synthesis is highly stimulated in S phase when nucleotide de novo synthesis actively takes place. Considering the relatively short half-life of 7S DNA (<70 min) (12,44,45), it is plausible that the mitochondrial nucleotide pool can be significantly influenced by degradation of 7S DNA relative to its synthesis, particularly outside of S phase when the dNTP supply is limited. Since mitochondria are well equipped with salvage enzymes such as TK2 and dGK (34,49), free dNMP, generated through 7S DNA degradation, could be converted to dNTP, and subsequently utilized for mtDNA replication.
In non-dividing cells that lack de novo synthesis of nucleotides, the salvage enzymes are expected to play key roles in the maintenance of both the nuclear genome and mtDNA, by providing necessary nucleotides under dynamically changing nutrient conditions. It would be very informative to determine how endogenous levels of 7S DNA and the extent of 7S DNA synthesis relative to mtDNA replication fluctuate under diverse growth and differentiation conditions. This would help establish whether 7S DNA synthesis and degradation affect mtDNA replication and the maintenance of the mitochondrial genome. Such studies will be greatly facilitated if cellular factors are identified that influence 7S DNA synthesis/degradation. Those factors could function in conjunction with replication factors and/or modulate the degradation of 7S DNA. Existence of such factors has been reported in a recent study, led by I. Holt (50), which describes that POLGβ, the accessory subunit of pol γ, positively regulates the 7S DNA level relative to mtDNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors thank Drs D. Clayton, J. Hurwitz, L. Kaguni and D. Bohenhagen, for their helpful comments on the manuscript, and Ms D. Stein and Mr S. Ragavendra for their technical support in using a FACSCaliburs and fluorescence microscope, respectively. The authors also thank Dr C. Reichman for editing the manuscript, and Drs T. Valeria and M. Zevani for providing rabbit polyclonal anti-mtSSB Ab.
Conflict of interest statement. None declared.
REFERENCES
- 1.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 2.Copeland WC. Inherited mitochondrial diseases of DNA replication. Annu. Rev. Med. 2008;59:131–146. doi: 10.1146/annurev.med.59.053006.104646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schapira AH. Mitochondrial disease. Lancet. 2006;368:70–82. doi: 10.1016/S0140-6736(06)68970-8. [DOI] [PubMed] [Google Scholar]
- 4.Schon EA. Mitochondrial genetics and disease. Trends Biochem. Sci. 2000;25:555–560. doi: 10.1016/s0968-0004(00)01688-1. [DOI] [PubMed] [Google Scholar]
- 5.Takamatsu C, Umeda S, Ohsato T, Ohno T, Abe Y, Fukuoh A, Shinagawa H, Hamasaki N, Kang D. Regulation of mitochondrial D-loops by transcription factor A and single-stranded DNA-binding protein. EMBO Rep. 2002;3:451–456. doi: 10.1093/embo-reports/kvf099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shmookler Reis RJ, Goldstein S. Mitochondrial DNA in mortal and immortal human cells. Genome number, integrity, and methylation. J. Biol. Chem. 1983;258:9078–9085. [PubMed] [Google Scholar]
- 7.Bogenhagen D, Clayton DA. The number of mitochondrial deoxyribonucleic acid genomes in mouse L and human HeLa cells. Quantitative isolation of mitochondrial deoxyribonucleic acid. J. Biol. Chem. 1974;249:7991–7995. [PubMed] [Google Scholar]
- 8.Kasamatsu H, Robberson DL, Vinograd J. A novel closed-circular mitochondrial DNA with properties of a replicating intermediate. Proc. Natl Acad. Sci. USA. 1971;68:2252–2257. doi: 10.1073/pnas.68.9.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robberson DL, Kasamatsu H, Vinograd J. Replication of mitochondrial DNA. Circular replicative intermediates in mouse L cells. Proc. Natl Acad. Sci. USA. 1972;69:737–741. doi: 10.1073/pnas.69.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doda JN, Wright CT, Clayton DA. Elongation of displacement-loop strands in human and mouse mitochondrial DNA is arrested near specific template sequences. Proc. Natl Acad. Sci. USA. 1981;78:6116–6120. doi: 10.1073/pnas.78.10.6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enriquez JA, Ramos J, Perez-Martos A, Lopez-Perez MJ, Montoya J. Highly efficient DNA synthesis in isolated mitochondria from rat liver. Nucleic Acids Res. 1994;22:1861–1865. doi: 10.1093/nar/22.10.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gensler S, Weber K, Schmitt WE, Perez-Martos A, Enriquez JA, Montoya J, Wiesner RJ. Mechanism of mammalian mitochondrial DNA replication: import of mitochondrial transcription factor A into isolated mitochondria stimulates 7S DNA synthesis. Nucleic Acids Res. 2001;29:3657–3663. doi: 10.1093/nar/29.17.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jui HY, Wong TW. In vitro replication of heavy strand DNA in permeabilized human mitochondria. Nucleic Acids Res. 1991;19:905–911. doi: 10.1093/nar/19.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitra RS, Bernstein IA. Thymidine incorporation into deoxyribonucleic acid by isolated rat liver mitochondria. J. Biol. Chem. 1970;245:1255–1260. [PubMed] [Google Scholar]
- 15.Parsons P, Simpson MV. Deoxyribonucleic acid biosynthesis in mitochondria. Studies on the incorporation of labeled precursors into mitochondrial deoxyribonucleic acid. J. Biol. Chem. 1973;248:1912–1919. [PubMed] [Google Scholar]
- 16.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pogozelski WK, Hamel CJ, Woeller CF, Jackson WE, Zullo SJ, Fischel-Ghodsian N, Blakely WF. Quantification of total mitochondrial DNA and the 4977-bp common deletion in Pearson's syndrome lymphoblasts using a fluorogenic 5′-nuclease (TaqMan) real-time polymerase chain reaction assay and plasmid external calibration standards. Mitochondrion. 2003;2:415–427. doi: 10.1016/S1567-7249(03)00033-3. [DOI] [PubMed] [Google Scholar]
- 18.Graziewicz MA, Longley MJ, Copeland WC. DNA polymerase gamma in mitochondrial DNA replication and repair. Chem Rev. 2006;106:383–405. doi: 10.1021/cr040463d. [DOI] [PubMed] [Google Scholar]
- 19.Hubscher U, Maga G, Spadari S. Eukaryotic DNA polymerases. Annu. Rev. Biochem. 2002;71:133–163. doi: 10.1146/annurev.biochem.71.090501.150041. [DOI] [PubMed] [Google Scholar]
- 20.Weill JC, Reynaud CA. DNA polymerases in adaptive immunity. Nat. Rev. Immunol. 2008;8:302–312. doi: 10.1038/nri2281. [DOI] [PubMed] [Google Scholar]
- 21.Kaguni LS. DNA polymerase gamma, the mitochondrial replicase. Annu. Rev. Biochem. 2004;73:293–320. doi: 10.1146/annurev.biochem.72.121801.161455. [DOI] [PubMed] [Google Scholar]
- 22.Weiser T, Gassmann M, Thommes P, Ferrari E, Hafkemeyer P, Hubscher U. Biochemical and functional comparison of DNA polymerases alpha, delta, and epsilon from calf thymus. J. Biol. Chem. 1991;266:10420–10428. [PubMed] [Google Scholar]
- 23.Naviaux RK, Markusic D, Barshop BA, Nyhan WL, Haas RH. Sensitive assay for mitochondrial DNA polymerase gamma. Clin. Chem. 1999;45:1725–1733. [PubMed] [Google Scholar]
- 24.Waga S, Bauer G, Stillman B. Reconstitution of complete SV40 DNA replication with purified replication factors. J. Biol. Chem. 1994;269:10923–10934. [PubMed] [Google Scholar]
- 25.Lee SH, Eki T, Hurwitz J. Synthesis of DNA containing the simian virus 40 origin of replication by the combined action of DNA polymerases alpha and delta. Proc. Natl Acad. Sci. USA. 1989;86:7361–7365. doi: 10.1073/pnas.86.19.7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holt IJ, He J, Mao CC, Boyd-Kirkup JD, Martinsson P, Sembongi H, Reyes A, Spelbrink JN. Mammalian mitochondrial nucleoids: organizing an independently minded genome. Mitochondrion. 2007;7:311–321. doi: 10.1016/j.mito.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 27.Van Tuyle GC, Pavco PA. Characterization of a rat liver mitochondrial DNA-protein complex. Replicative intermediates are protected against branch migrational loss. J. Biol. Chem. 1981;256:12772–12779. [PubMed] [Google Scholar]
- 28.Van Tuyle GC, Pavco PA. The rat liver mitochondrial DNA-protein complex: displaced single strands of replicative intermediates are protein coated. J. Cell Biol. 1985;100:251–257. doi: 10.1083/jcb.100.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sonesson A, Berglund M, Staxen I, Widell S. The characterization of plasma membrane-bound tubulin of cauliflower using Triton X-114 fractionation. Plant Physiol. 1997;115:1001–1007. doi: 10.1104/pp.115.3.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bogenhagen D, Clayton DA. Thymidylate nucleotide supply for mitochondrial DNA synthesis in mouse L-cells. Effect of 5-fluorodeoxyuridine and methotrexate in thymidine kinase plus and thymidine kinase minus cells. J. Biol. Chem. 1976;251:2938–2944. [PubMed] [Google Scholar]
- 31.Dolce V, Fiermonte G, Runswick MJ, Palmieri F, Walker JE. The human mitochondrial deoxynucleotide carrier and its role in the toxicity of nucleoside antivirals. Proc. Natl Acad. Sci. USA. 2001;98:2284–2288. doi: 10.1073/pnas.031430998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franco M, Johansson M, Karlsson A. Depletion of mitochondrial DNA by down-regulation of deoxyguanosine kinase expression in non-proliferating HeLa cells. Exp. Cell Res. 2007;313:2687–2694. doi: 10.1016/j.yexcr.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 33.Kaukonen J, Juselius JK, Tiranti V, Kyttala A, Zeviani M, Comi GP, Keranen S, Peltonen L, Suomalainen A. Role of adenine nucleotide translocator 1 in mtDNA maintenance. Science. 2000;289:782–785. doi: 10.1126/science.289.5480.782. [DOI] [PubMed] [Google Scholar]
- 34.Saada A, Shaag A, Mandel H, Nevo Y, Eriksson S, Elpeleg O. Mutant mitochondrial thymidine kinase in mitochondrial DNA depletion myopathy. Nat. Genet. 2001;29:342–344. doi: 10.1038/ng751. [DOI] [PubMed] [Google Scholar]
- 35.Kibbey RG, Pongratz RL, Romanelli AJ, Wollheim CB, Cline GW, Shulman GI. Mitochondrial GTP regulates glucose-stimulated insulin secretion. Cell Metab. 2007;5:253–264. doi: 10.1016/j.cmet.2007.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bourdon A, Minai L, Serre V, Jais JP, Sarzi E, Aubert S, Chretien D, de Lonlay P, Paquis-Flucklinger V, Arakawa H, et al. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat. Genet. 2007;39:776–780. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- 37.Ashley N, Adams S, Slama A, Zeviani M, Suomalainen A, Andreu AL, Naviaux RK, Poulton J. Defects in maintenance of mitochondrial DNA are associated with intramitochondrial nucleotide imbalances. Hum. Mol. Genet. 2007;16:1400–1411. doi: 10.1093/hmg/ddm090. [DOI] [PubMed] [Google Scholar]
- 38.Bogenhagen D, Clayton DA. Mouse L cell mitochondrial DNA molecules are selected randomly for replication throughout the cell cycle. Cell. 1977;11:719–727. doi: 10.1016/0092-8674(77)90286-0. [DOI] [PubMed] [Google Scholar]
- 39.Newlon CS, Fangman WL. Mitochondrial DNA synthesis in cell cycle mutants of Saccharomyces cerevisiae. Cell. 1975;5:423–428. doi: 10.1016/0092-8674(75)90061-6. [DOI] [PubMed] [Google Scholar]
- 40.Eichler DC, Wang TS, Clayton DA, Korn D. In vitro replication of mitochondrial DNA. Elongation of the endogenous primer sequence in D loop mitochondrial DNA by human DNA polymerase beta. J. Biol. Chem. 1977;252:7888–7893. [PubMed] [Google Scholar]
- 41.Brown WM, Shine J, Goodman HM. Human mitochondrial DNA: analysis of 7S DNA from the origin of replication. Proc. Natl Acad. Sci. USA. 1978;75:735–739. doi: 10.1073/pnas.75.2.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang DD, Clayton DA. Priming of human mitochondrial DNA replication occurs at the light-strand promoter. Proc. Natl Acad. Sci. USA. 1985;82:351–355. doi: 10.1073/pnas.82.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang DD, Hauswirth WW, Clayton DA. Replication priming and transcription initiate from precisely the same site in mouse mitochondrial DNA. EMBO J. 1985;4:1559–1567. doi: 10.1002/j.1460-2075.1985.tb03817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robberson DL, Clayton DA. Pulse-labeled components in the replication of mitochondrial deoxyribonucleic acid. J. Biol. Chem. 1973;248:4512–4514. [PubMed] [Google Scholar]
- 45.Bogenhagen D, Clayton DA. Mechanism of mitochondrial DNA replication in mouse L-cells: kinetics of synthesis and turnover of the initiation sequence. J. Mol. Biol. 1978;119:49–68. doi: 10.1016/0022-2836(78)90269-3. [DOI] [PubMed] [Google Scholar]
- 46.Fish J, Raule N, Attardi G. Discovery of a major D-loop replication origin reveals two modes of human mtDNA synthesis. Science. 2004;306:2098–2101. doi: 10.1126/science.1102077. [DOI] [PubMed] [Google Scholar]
- 47.Posakony JW, England JM, Attardi G. Mitochondrial growth and division during the cell cycle in HeLa cells. J. Cell. Biol. 1977;74:468–491. doi: 10.1083/jcb.74.2.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Volpe P, Eremenko T. Nuclear and cytoplasmic DNA synthesis during the mitotic cycle of HeLa cells. Eur. J. Biochem. 1973;32:227–232. doi: 10.1111/j.1432-1033.1973.tb02600.x. [DOI] [PubMed] [Google Scholar]
- 49.Wang L, Eriksson S. Mitochondrial deoxyguanosine kinase mutations and mitochondrial DNA depletion syndrome. FEBS Lett. 2003;554:319–322. doi: 10.1016/s0014-5793(03)01181-5. [DOI] [PubMed] [Google Scholar]
- 50.Di Re M, Sembongi H, He J, Reyes A, Yasukawa T, Martinsson P, Bailey LJ, Goffart S, Boyd-Kirkup JD, Wong TS, et al. The accessory subunit of mitochondrial DNA polymerase gamma determines the DNA content of mitochondrial nucleoids in human cultured cells. Nucleic Acids Res. 2009;37:5701–5713. doi: 10.1093/nar/gkp614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.