Abstract
The control of messenger RNA (mRNA) function by micro RNAs (miRNAs) in animal cells requires the GW182 protein. GW182 is recruited to the miRNA repression complex via interaction with Argonaute protein, and functions downstream to repress protein synthesis. Interaction with Argonaute is mediated by GW/WG repeats, which are conserved in many Argonaute-binding proteins involved in RNA interference and miRNA silencing, from fission yeast to mammals. GW182 contains at least three effector domains that function to repress target mRNA. Here, we analyze the functions of the N-terminal GW182 domain in repression and Argonaute1 binding, using tethering and immunoprecipitation assays in Drosophila cultured cells. We demonstrate that its function in repression requires intact GW/WG repeats, but does not involve interaction with the Argonaute1 protein, and is independent of the mRNA polyadenylation status. These results demonstrate a novel role for the GW/WG repeats as effector motifs in miRNA-mediated repression.
INTRODUCTION
A key aspect of post-transcriptional regulation in eukaryotic cells is micro RNAs (miRNAs), 21–23 nt non-coding RNAs that target more than a half of all genes (1). In animals, miRNAs pair to partially complementary sites in their target messenger RNAs (mRNAs) and cause translational repression, as well as mRNA deadenylation and degradation (2–4). An unresolved issue is the mechanism by which miRNAs repress translation. Many experiments have pointed to initiation of translation as a target of repression, but there is also evidence that miRNA inhibition occurs at post-initiation steps [reviewed in (2–7)], see also (8). It is important to find out whether these disparities are artifacts of different experimental approaches, or whether miRNAs are indeed able to repress protein synthesis by different mechanisms.
miRNAs function in the form of ribonucleoprotein complexes (miRNPs), with Argonaute (AGO) proteins being the core components of miRNPs. GW182 proteins are recruited to miRNPs via interaction with AGOs, and represent another group of proteins crucial for miRNA-induced repression (9–15). Direct tethering of GW182 to an mRNA in Drosophila cells leads to translational repression and mRNA degradation, even in the absence of AGO1, arguing that GW182 functions in miRNA repression downstream of AGO proteins (14,16,17). Given this, a key issue in determining the mechanism of miRNA-mediated repression is understanding the function of GW182 proteins.
Proteins of the GW182 family are characterized by the presence of glycine-tryptophan (GW) repeats, glutamine-rich (Q-rich) regions, C-terminal DUF domains and RNA recognition motifs (RRMs), the latter two present in mammalian and Drosophila GW182 family members, but not those of Caenorhabditis elegans (18,19). The N-terminal GW repeats have been shown to interact with AGO proteins (10,14,15,20), and disruption of GW182-AGO interaction with point mutations or a peptide competing with GW182 for AGO binding also abrogated miRNA-mediated repression (13,15). RNAi depletion and in vitro experiments have demonstrated that GW182 promotes mRNA deadenylation and degradation by recruiting the CAF1:CCR4:NOT1 deadenylase complex to the target mRNA; the deadenylation is then followed by mRNA decapping by the DCP1:DCP2 decapping complex and exonucleolytic degradation by the 5′ to 3′ exonuclease Xrn1 (14,21–23).
Deletion analyses of GW182 family members in Drosophila and mammals have indicated that at least three separate domains can function in mRNA repression. Specifically, for the Drosophila family member, dGW182, tethering of the N terminal domain, the QN-rich domain and a C terminal domain including the RRM can repress expression from a reporter mRNA (17). For the mammalian GW182 family member, TNRC6C, tethering of the similar regions can repress reporter mRNA, with the major contribution of the C-terminal domain (24–26). The existence of multiple repressor domains in dGW182 could result in multiple repression mechanisms and, thus, could reconcile the variability of the current data. Recent studies (23,27,28) have demonstrated that the C-terminal domains of both mammalian and Drosophila GW182 homologs bind PABP protein, interfering with the eIF4G-PABP interaction and promoting target mRNA deadenylation. The authors hypothesize that interfering with the eIF4G-PABP interaction, and thus disrupting mRNA circularization, could also explain how the C-terminal domain inhibits translation. This model, however, cannot fully explain the repression mechanism, as mRNAs without poly(A) tails, i.e. independent of PABP, are also regulated by miRNAs and GW182 (13,17,22,29–31). In addition, it remains unknown how the N-terminal and the QN-rich domains of GW182 proteins function to repress translation.
Here, we further characterize the function of the dGW182 N-terminal effector domain, which binds AGO1 and can also repress protein synthesis (14,17). Using an mRNA–protein tethering system in Drosophila S2 cells, we mapped the N-terminal dGW182 region more precisely and identified the minimal repressor region, consisting of around 300 amino acids. Most importantly, this analysis shows that the two functions of the N-terminal region, binding to AGO1 and repression of target mRNA, reside in different domains and can be separated from each other by deletion analysis. Surprisingly, we discovered that the multiple GW/WG repeats present in the minimal N-terminal repressor domain are required for target mRNA repression. GW/WG repeats were previously thought to function in AGO binding but not repression (15,20,32). These results demonstrate a novel role for the GW/WG repeats as effector motifs in miRNA-mediated repression.
MATERIALS AND METHODS
Cell culture, transfections and RNA interference
Drosophila S2 cells were transfected in 12-well or 96-well plates with Cellfectin II and PLUS Reagents (Invitrogen), according to the manufacturer’s instructions. In tethering experiments, we transfected 40 ng FLuc-boxB plasmid, 100 ng RLuc as transfection control and 400 ng plasmid encoding NHA-fusion protein per well of a 12-well plate. For the 96-well format, the amount of plasmids was adjusted proportionally. Cells were lysed on day 3 post-transfection. In rescue experiments, transfection mixtures contained 5 ng FLuc-nerfin reporter plasmid, 25 ng RLuc as transfection control and 15 ng of either an empty vector or a plasmid encoding miRNA-9b per well of 96-well plate; plasmids encoding dGW182 fragments and lacZ were added in increasing amounts from 1 to 150 ng. RNAi experiments were performed according to ref. 33. dsRNAs were 250–500 nt long and corresponded to the 3′UTR when targeting the dGW182 gene, and to the coding sequence for control GFP dsRNA. Treatment with dsRNA was done on day 0 and repeated on day 4. Cells were transfected on day 6 and lysed on day 9. Firefly and Renilla luciferase activities were measured with the Dual-Luciferase Reporter Assay System (Promega).
DNA constructs
FLuc-boxB, FLuc-boxB-HSL, RLuc, NHA, NHA-lacZ, NHA-dGW182, NHA-1-605, NHA-490-1384, FLuc-nerfin, miR-9b and NHA-TNRC6C constructs have been described previously (17,34). Plasmids encoding deletion mutants of dGW182 and human GW182 homologs were generated by polymerase chain reaction (PCR)-amplification of the corresponding fragments of the GW182 coding region, and cloning into pAC5.1A vector containing λN and HA-tag. Point mutations in the NHA-205-490 construct were introduced by site-directed mutagenesis according to ref. 35.
Northern blotting, western blotting and immunoprecipitations
To estimate FLuc-boxB and RLuc mRNA levels, the total RNA from S2 cells was isolated with Trizol LS Reagent (Invitrogen), and reporter mRNA expression was estimated by northern blot. In brief, 15 μg total RNA was separated on a formaldehyde agarose gel, transferred to a Hybond-N+ membrane (GE Healthcare Life Sciences), and hybridized to 32P-labeled DNA probes (High Prime DNA labeling kit, Roche) in UltraHyb buffer (Ambion) according to the manufacturer's instructions, and analyzed by phosphorimaging. Quantification of FLuc-boxB and RLuc mRNA levels was done with ImageQuant software (Molecular Dynamics). To estimate the expression levels of HA-fusion proteins, cell lysates were separated on a 4–12% PAAG (Invitrogen), and western-blot probed using anti-HA antibodies (Roche 3F10). The amount of loaded lysates was normalized according to Renilla luciferase expression (transfection control). The efficiency of dGW182 knockdown was estimated by western blotting with anti-dGW182 antibodies (kindly provided by E. Izaurralde, MPI, Tuebingen). For a loading control, membranes were probed with anti-tubulin antibodies (Sigma T5168). For immunopreciptations, S2 lysates were incubated with the anti-AGO1 antibody (Abcam ab5070) bound to the protein G agarose (Invitrogen) in the binding buffer [20 mM Tris–HCl pH 7.5, 150 mM KCl, 0.5 mM DTT, 0.5% TritonX-100, protease inhibitors ‘Complete’ (Roche)] overnight at 4°C. The beads were then washed three times with the binding buffer, and proteins bound to beads were eluted by boiling in the Laemmli sample buffer and analyzed by polyacrylamide gel electrophoresis (PAGE) and western blotting with anti-HA (Roche 3F10) and anti-AGO1 antibodies (Abcam ab5070).
RESULTS
Mapping of the minimal N-terminal repressor domain of the dGW182 protein
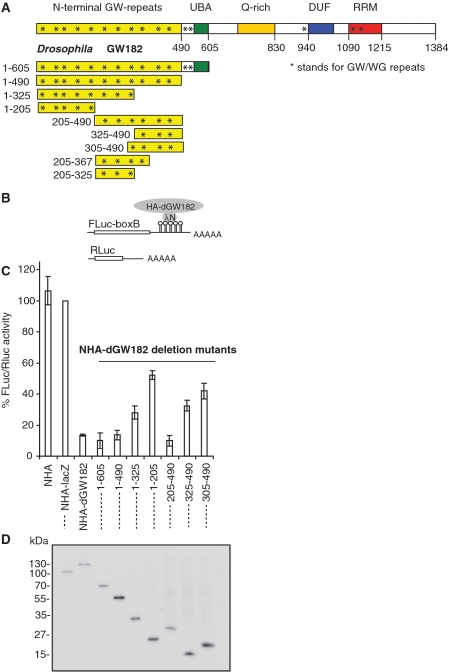
In previous work (17) we identified three regions of the Drosophila GW182 protein (dGW182) which are able to repress target mRNAs: the N-terminal part of the protein comprising amino acids 1–605, the middle Q-rich region (amino acids 605–830) and the C-terminal domain containing DUF and RRM (amino acids 940–1215). The N-terminal domain (1–605) was the most efficient in repression, and was also able to exert its repressive effect in mammalian cells (24). This domain is required for recruitment of the GW182 protein to the miRNA repression complex through an interaction with the AGO1 protein (14). We decided to perform a more detailed N-terminal domain mapping to see if we could (i) minimize the repressive domain, and (ii) separate the functions of the N-terminus (1–605) in AGO1 binding and mRNA repression.
For these experiments, we generated a series of dGW182 deletion mutants within the N-terminal domain (Figure 1A), and analyzed their effects on translation in transfected S2 cells using an RNA-protein tethering assay. Tethering was achieved by co-expressing firefly luciferase mRNA containing five boxB sites in its 3′-UTR (designated as FLuc-boxB, Figure 1B), and dGW182 deletion mutants fused with HA-tag and λN peptide, specifically recognizing the boxB hairpins (17,34,36,37). A plasmid encoding for Renilla luciferase without boxB sites was co-expressed as a transfection control. As negative controls that were not expected to repress FLuc-boxB mRNA, we used λN peptide fused to an HA-tag, either alone (NHA) or with β-galactosidase coding sequence (NHA-lacZ). As expected, we observed that NHA-dGW182 led to effective repression (∼10-fold), while none of the negative controls were able to repress FLuc-boxB; the long N-terminal fragment (1–605) had an effect similar to the full-length protein (Figure 1C). We then analyzed the effects of shorter dGW182 N-terminal fragments on expression of the tethered firefly luciferase mRNA. Of all analyzed fragments, only the ones including amino acids 205–490 were able to effectively repress tethered mRNA: the effect of the 205–490 fragment was close to that of the full-length protein. Further dissection of this region (305–490 and 325–490 fragments) decreased the repression effect. Western blot analysis of the HA-fusions showed that the differences in their repressive properties could not be explained by their expression levels (Figure 1D). We also attempted to analyze the 205–325 and 205–367 fragments, but they were not expressed, or were unstable according to the western blot analysis (data not shown). We concluded that the 205–490 region is the minimal N-terminal repressive domain and termed it NED, for the N-terminal effector domain.
Figure 1.
205–490 region of dGW182 is the minimal N-terminal effector domain (NED). (A) Schematic representation of Drosophila GW182 protein and its deletion mutants. The numbers correspond to the amino acid positions. (B) Schematic representation of reporter constructs: FLuc-boxB contains firefly luciferase (FLuc) coding sequence and 3′UTR with five boxB sites specifically binding to λN peptide; RLuc contains Renilla luciferase (RLuc) coding sequence and no boxB sites (34). (C) Repression of FLuc-boxB mRNA by NHA-dGW182 and its deletion mutants. Drosophila S2 cells were co-transfected with plasmids encoding FLuc-boxB, RLuc, and full-length NHA-dGW182 or indicated NHA-dGW182 deletion mutants. As negative controls, plasmids encoding either NHA alone or NHA fused to β-galactosidase (NHA-lacZ) were used. Expression of FLuc was normalized to that of RLuc. Values are presented as a percentage of FLuc produced in the presence of NHA-lacZ. Values represent means ±SD from at least four experiments. (D) Expression of different HA-fusion proteins was estimated by western blotting with antibodies directed against HA-peptide.
The function of the N-terminal dGW182 region (1–605) in mRNA repression can be uncoupled from its function in AGO1 binding
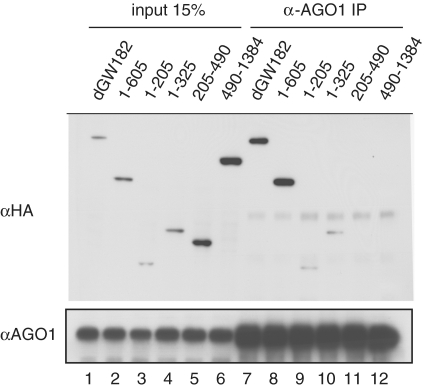
As dGW182 N-terminal GW/WG repeats are involved in the interaction with AGO1 (14,15,32), and NED contains six GW/WG repeats, we wondered if the NED is able to interact with AGO1. To test which fragments of dGW182 are sufficient for AGO1 binding, we expressed NHA-tagged dGW182 and its deletion mutants in S2 cells, immunoprecipitated AGO1 protein from the S2 extracts, and analyzed the precipitated fraction for HA-fusions (Figure 2). As expected, both full-length dGW182 and the long N-terminal fragment (1–605) efficiently interact with AGO1 (Figure 2, lanes 7 and 8, compare with the input lanes 1 and 2). The short N-terminal fragments, comprising the first 205 (1–205) or 325 amino acids (1–325), also bind AGO1, though less efficiently than the full-length GW182 and 1–605 fragment (lanes 9 and 10, compare with the input lanes 3 and 4). However, the NED and the 490-1384 GW182 fragment, which do not contain the first 205 amino acids, were not able to immunoprecipitate with AGO1, even though these fragments were intentionally expressed at higher levels than the full-length dGW182 and the 1-605 fragment (lanes 11 and 12, compare with the input lanes 5 and 6). These data show that the sequences within the first 205 amino acids of GW182 are absolutely necessary for GW182-AGO1 interaction. Thus, this experiment shows that the two functions, binding to AGO1 and repression of the target mRNA, reside in different parts of the N-terminal region and can be separated from each other by deletion analysis. These data are consistent with our previous study, which showed that repression by the tethered 1–605 dGW182 fragment does not require the AGO1 protein, i.e. it is retained in cells depleted of endogenous AGO1 (17). It should also be noted that the mapping of the AGO1 interaction to the extreme N terminus of dGW182 is in line with the work of Izaurralde and colleagues (20), showing that mutation of a single GWG repeat at the dGW182 N-terminus almost fully abrogates interaction with AGO1, and the remaining 11 GW/WG repeats in the N-terminal region only provide residual AGO1 binding. The latter observation also explains why, in our experiments, the 1–205 GW182 fragment does not bind AGO1 as efficiently as the longer 1–605 fragment: it is lacking secondary weak binding sites which are present in the longer fragment.
Figure 2.
The N-terminal effector domain (NED) does not interact with AGO1. Drosophila S2 cells were transfected with plasmids encoding the full-length NHA-dGW182 or its deletion mutants. Cell lysates were used in immunoprecipitations with anti-AGO1 antibody. Inputs and immunoprecipitates were analyzed by western blotting using anti-HA or anti-AGO1 antibodies, as indicated on the left.
Conserved residues and the GW/WG repeats are important for the function of the NED in mRNA repression
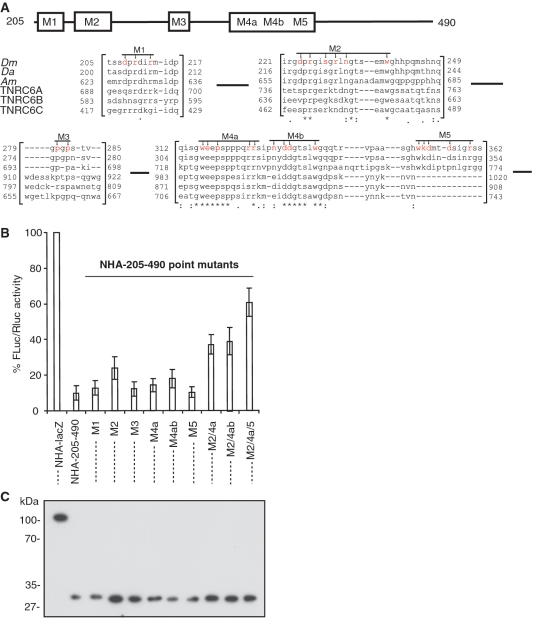
To identify the critical features of the NED domain that allow mRNA repression, we performed extensive mutagenesis of this domain, and analyzed the effects of the mutations in RNA–protein tethering assays in S2 cells (Figures 3 and 4). Comparative analysis of the NED region showed several sequence stretches conserved across insect species, and between insects and mammals (Figure 3A; see also Figure S1 for the complete alignment of the NED). We generated multiple point mutations, changing specific amino acids to alanines in the conserved regions: the mutations are depicted in Figure 3A and designated as M1, M2, M3, M4a, M4b and M5. Each of the mutants had no or only a mild effect on repression in the mRNA–protein tethering assay by the NED (Figure 3B). We then combined some of these mutations: the resulting M2/M4a/M5 mutant showed the maximum alleviation of repression (∼60% expression in the presence of M2/M4a/M5 mutant compared with ∼10% expression in the presence of the wild-type NED). Western blot analysis showed that the mutant domain is expressed as well as the wild-type, i.e. alleviated repression by the mutant is not due to its lower expression (Figure 3C). Thus, these results (i) identify conserved amino acids in the NED domain required for efficient repression, and (ii) demonstrate that no single region within this domain is responsible for mediating the repression, but rather that the repression requires multiple features of the region (see ‘Discussion’ section). We cannot exclude the possibility that multiple mutations introduced to the NED could interfere with protein folding.
Figure 3.
Mutation of the conserved amino acid residues abolish repression by NED. (A) Schematic representation of the NED region and its alignment across the species (Dm: Drosophila melanogaster, Da: Drosophila ananassae; Ap: Apis mellifera; TNRC6A, B, and C are human GW182 homologs). Alignment was performed with a T-Coffee tool [(41) see Supplementary Figure S1 for full alignment]. Mutated residues (always to alanine) are shown in red. The numbers correspond to amino acid position. Asterisks mark residues identical in all sequences, colons mark conservative substitutions, dots mark semi-conservative substitutions. (B) Repression of FLuc-boxB mRNA by the NED and its point mutants. Drosophila S2 cells were co-transfected with plasmids encoding FLuc-boxB, RLuc, and either NHA-205-490 (NED) or one of its point mutants depicted in (A). For other details, see legend to Figure 1. (C) Expression levels of HA-fusion proteins as estimated by western blotting with the anti-HA antibody.
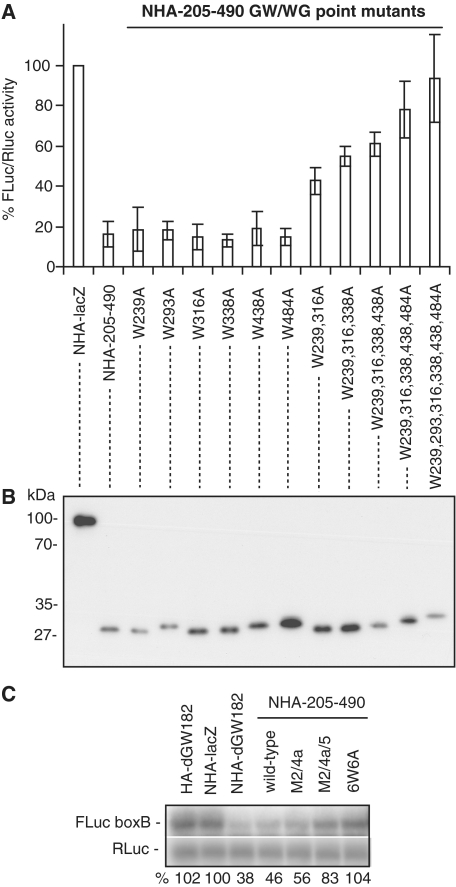
Figure 4.
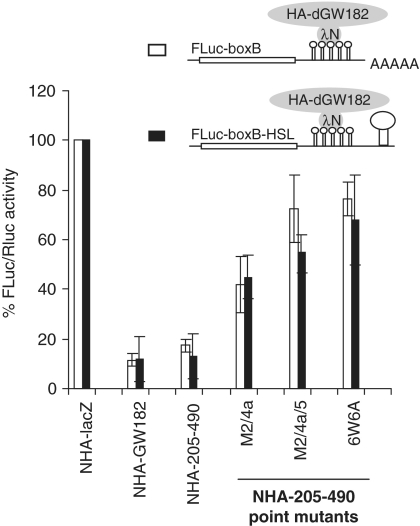
Repression by the NED requires GW/WG repeats and involves mRNA destabilization. (A) NHA-205–490, either wild-type or mutants in which one or several GW/WG repeats are mutated (always W to A), were co-transfected with FLuc-boxB and RLuc. For other details, see Figure 1. (B) The levels of HA-fusion proteins as analyzed by western blotting with the anti-HA antibody. (C) The FLuc-boxB and RLuc mRNA levels from experiments depicted in Figures 4A and 5A were analyzed by northern blot. mRNA levels were quantified by phosphoimaging; amounts of FLuc-boxB were normalized according to Renilla control and expressed as a percentage of FLuc-boxB level in the presence of NHA-lacZ protein (numbers below the figure). Lanes 1–7 show the FLuc-boxB and RLuc RNA levels in the presence of non-tethered control HA-dGW182, tethered NHA-lacZ, NHA-dGW182, NHA-NED and its point mutants: M2/4a, M2/4a/5, and 6W6A, accordingly.
The NED contains six GW/WG repeats (Figure S1). Therefore, we examined whether these GW/WG repeats have any effect on the ability of the NED to repress mRNA. We mutated each of the tryptophan residues in the GW/WG repeats to alanine, and analyzed the effects of these mutations on expression of the tethered mRNA in S2 cells (Figure 4A). Single W to A mutations had no or only a mild effect on repression, but their combination had an additive effect. Most strikingly, when all six tryptophans were mutated (6W6A), repression by the NED was almost fully alleviated. Western blot analysis of the HA-fusions showed that the differences in their repressive properties were not due to their expression levels (Figure 4B). This analysis revealed a novel role for the GW/WG repeats in GW182 function: the NED GW/WG repeats are not able to bind AGO1 (Figure 2), but are crucial to the NED function in target mRNA repression (Figure 4A).
As miRNAs and GW182 are known to repress mRNA through both increasing mRNA degradation and repressing translation (2–4), we tested whether the NED affects tethered mRNA levels (Figure 4C). By performing a northern blot analysis, we observed that the full-length dGW182 protein and the NED reduced reporter mRNA levels 2- to 3-fold (Figure 4C, lanes 3 and 4). This reduction in mRNA levels was due to tethering, as HA-dGW182 bearing no N peptide did not affect the FLuc-boxB level (Figure 4C, lane 1). In addition, tethering the NED mutants defective in repression caused no or less pronounced mRNA degradation (Figure 4C, lanes 5–7). These results are consistent with our previous findings, which showed that tethering the 1–605 dGW182 fragment leads to increased mRNA degradation (17). These results indicate that the NED not only is able to promote mRNA degradation, but also represses translation, as its overall effect on protein production (Figure 1B) is stronger than its effect on mRNA stability (Figure 4C).
The NED can function independently of the poly(A) tail
Using a reporter mRNA which lacks a poly(A) tail but has a histone H1 stem–loop structure in its 3′UTR (FLuc-boxB-HSL), we have previously shown that the extended version of the N-terminal effector domain (1–605) is able to repress tethered mRNA independently of poly(A) tails (17). As expected, a reporter having neither a poly(A) tail nor a histone stem–loop was not efficient in protein production; however, addition of the histone stem–loop structure stimulated the reporter translation, so that activity of FLuc-boxB-HSL mRNA was comparable to that of polyadenylated FLuc-boxB. We now examined whether the ability to repress protein synthesis independent of the poly(A) tail is also a feature of the isolated NED domain. In order to do this, we tethered the minimal NED domain or its mutant versions to the FLuc-boxB-HSL reporter. We observed that the NED was able to repress tethered FLuc-boxB-HSL mRNA almost as efficiently as the polyadenylated FLuc-boxB (Figure 5). This result was in line with the data produced in other laboratories, demonstrating that non-polyadenylated mRNAs bearing a 3′ histone stem–loop are repressed by miRNAs and/or tethered dGW182 similarly as their polyadenylated counterparts (13,22,30). We conclude that the NED can repress mRNAs independently of their polyadenylation status.
Figure 5.
Repression mediated by the NED is independent of the poly(A) tail. Drosophila S2 cells were transfected with the boxB-containing FLuc reporters, either FLuc-boxB (open bars) or FLuc-boxB-HSL (black bars). Unlike FLuc-boxB, FLuc-boxB-HSL lacks a polyadenylation signal and bears instead a histone H1 stem-loop (HSL) structure in its 3′UTR. FLuc reporters were co-transfected with RLuc as a normalization control, and plasmids encoding indicated NHA-fusion proteins.
NED is able to rescue knockdown of endogenous dGW182
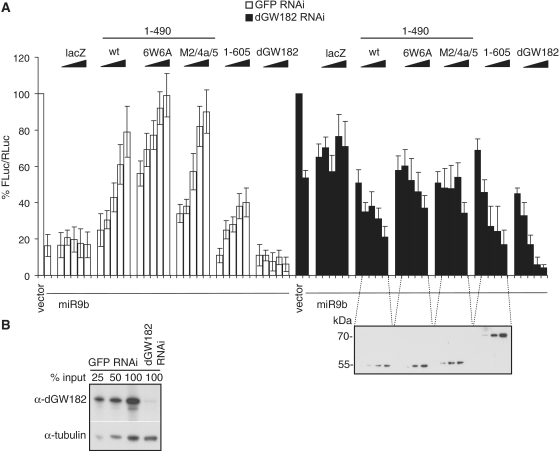
Results of the deletion analysis that mapped the AGO-binding domain to the N-terminal 205 amino acids (Figure 2) and the NED to the 205–490 region of dGW182 (Figure 1) led us to investigate whether the 1–490 dGW182 fragment would be sufficient to function in bona fide miRNA-mediated silencing and would complement the deficiency of the endogenous dGW182 protein. To test this, we depleted endogenous dGW182 by treating S2 cells with dsRNA specific for dGW182, and, as a negative control, with the GFP-specific dsRNA. As shown by western blotting, the dGW182 depletion was efficient (Figure 6B). To assess miRNA-directed silencing, we transfected cells with the FLuc-nerfin reporter construct, containing target sites for miRNA-9b. Co-expression of miRNA-9b efficiently repressed FLuc-nerfin mRNA in control GFP-treated cells (Figure 6A, first two open bars, compare empty vector and miRNA-9b), and depletion of endogenous dGW182 partially alleviated the miRNA-9b-mediated repression (Figure 6A, black bars, compare empty vector and miRNA-9b). We then analyzed whether overexpression of dGW182 and its fragments, comprising the AGO-binding domain and the NED (1–490), are able to rescue the knockdown of endogenous dGW182 (Figure 6A, black bars). As negative controls, we transfected increasing amounts of lacZ or mutant NEDs defective in repression (i.e. M2/4a/5 or 6W6A) fused to the AGO1-binding domain. We observed that transfection of a wild-type 1–490 fragment did indeed restore the miRNA-9b-directed repression in dGW182-depleted cells as efficiently as transfection of the 1–605 dGW182 fragment. Negative controls (lacZ and mutant 1–490) had no major effect on repression. It should be noted that N-terminal fragments (1–490 and 1–605) were not as efficient in the rescue of endogenous dGW182 knockdown as the full-length dGW182. This observation can be explained by the fact that the full-length dGW182 contains at least three domains contributing to mRNA repression (17), while the N-terminal fragments analyzed contained only one out of three effector domains. These results are consistent with the tethering data and our previous finding that the 1-605 N-terminal dGW182 fragment is able to rescue knockdown of the endogenous dGW182 (17). Taken together, these data indicate that the NED functions as a repressor not only in tethering experiments but also in bona fide miRNA-mediated silencing.
Figure 6.
Overexpression of the NED fused to the AGO1-binding motif rescues RNAi knockdown of endogenous dGW182. (A) Endogenous dGW182 was depleted in Drosophila S2 cells with dsRNA (black bars); a batch of cells was treated with GFP-specific dsRNA as a negative control (open bars). Cells were transfected with firefly luciferase reporter containing miRNA-9b target sites (FLuc-nerfin) and with either the miRNA-9b-encoding plasmid or the empty vector. As in all transfection experiments, RLuc was co-expressed as a transfection control; expression levels of firefly luciferase were normalized to Renilla luciferase activity and expressed as a percentage of the firefly luciferase activity in the presence of the empty vector. To rescue the knockdown of endogenous dGW182, increasing amounts of plasmids encoding NHA-dGW182 or its N-terminal fragments were co-transfected: the NED fused to the AGO1-binding motif (1–490), either wild-type or non-functional point mutants (M2/4a/5 and 6W6A), and the 1–605 construct. Expression of NHA-dGW182 fragments was estimated by western blotting with anti-HA antibody. Increasing amounts of NHA-lacZ were co-transfected for a negative control. (B) Efficiency of the endogenous dGW182 knockdown was estimated by western blot analysis. S2 cells were treated with dsRNAs, indicated above the lanes (dGW182 or GFP as a negative control), and analyzed by western blotting with the antibodies shown on the left. Expression of tubulin was estimated as a loading control.
Consistent with the previous reports (13,17), we observed that overexpression of the dGW182 N-terminal fragments in the presence of endogenous dGW182 alleviated miRNA-9b-induced repression (Figure 6A, open bars, GFP dsRNA-treated cells). One possible explanation of this result is that increasing amounts of the N-terminal dGW182 fragments compete with the endogenous dGW182 for AGO1 binding, and thereby prevent association of the endogenous dGW182 with the mRNA. The endogenous full-length dGW182, comprising all three effector domains, may be more efficient in recruiting downstream factors which mediate the repression than the 1–490 and 1–605 fragments, which contain only one effector domain. Consequently, the displaced dGW182 could prevent the assembly of the functional repression complex on the target mRNA by titrating its components.
N-terminal domains of the human GW182 paralogs, TNRC6A, B and C, also have the potential to repress mRNA
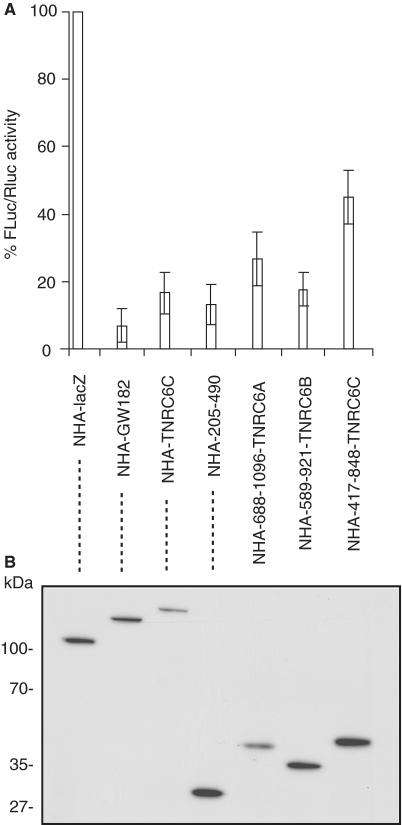
Our analysis has identified the NED as a silencing domain of the Drosophila GW182 protein. Several observations have indicated that NED-like domains are also present in GW182 family members in other organisms, and that they may function as silencing domains. First, previous results have shown that the N-terminal regions of mammalian TNRC6B and TNRC6C are able to repress protein synthesis when tethered to a reporter mRNA in S2 cells or mammalian cells (17,24,26). Second, a multiple sequence alignment shows that the most conserved motif of the Drosophila NED that we here identified as important to repression (Figure 3, M4a region) is also present in the human GW182 homologs, TNRC6A, B and C. Finally, the TNRC6A, B and C regions aligning with the Drosophila NED also contain multiple GW/WG repeats (Figure S1), consistent with the possibility that they function similarly in repression as in the Drosophila NED. Given that, we examined whether the NED-like regions within the human homologs, TNRC6A, B and C, are able to repress tethered mRNA in S2 cells (Figure 7). We observed that the N-terminal fragments of all three homologs were indeed able to downregulate FLuc-boxB reporter expression 2- to 5-fold. Hence, we suggest that the potential to repress protein synthesis is a common feature of the N-terminal domain of all GW182 family members.
Figure 7.
N-terminal domains of human GW182 homologs, TNRC6A, B and C, are able to repress tethered mRNA in Drosophila S2 cells. The assay was performed as in Figure 1. (A) The N-terminal fragments of human TNRC6A (Q8NDV7; amino acids 688–1096), TNRC6B (Q9UPQ9, amino acids 589–921), and TNRC6C (Q9HCJ0, amino acids 417–848) aligning with the Drosophila NED and enriched in GW-rich repeats (see Figure 3 and Supplementary Figure S1) were tethered to the FLuc-boxB reporter. dGW182 was tethered as a positive control, NHA-lacZ as the negative control. (B) Expression levels of NHA-fusion proteins were estimated by western blotting with anti-HA antibody.
DISCUSSION
It has been well established that proteins of the GW182 protein family are crucial to miRNA-mediated repression [reviewed in (18,19)]. However, the mechanism of GW182 function in miRNA-directed repression is not well understood. Recent results in both mammalian and Drosophila cells have shown that several different domains of GW182 family members can function in translational repression (17,24). Specifically, as shown in our previous work (17), the N-terminal domain, the central QN-rich domain, and the C-terminal region containing DUF and RRM, can all function independently to repress protein synthesis 5- to 6-fold when tethered to a reporter mRNA in Drosophila cells. The role of the three repressor domains in bona fide miRNA silencing was validated by performing complementation assays. These assays showed that in Drosophila cells in which endogenous dGW182 was knocked-down, the repression could be rescued by expressing increasing amounts of the dGW182 N-terminal fragments containing only one or two effector domains. Using the dGW182 rescue assay, Eulalio et al. (38) have also demonstrated the roles of the RRM and C-terminal regions located upstream and downstream of the RRM (20) in miRNA silencing, but failed to observe the role of the N-terminal region. This could be explained by the fact that the same amounts of the plasmids encoding the full-length dGW182 and its fragments were transfected (20). We found that the dGW182 fragments containing only one or two effector domains were less efficient in repression than the full-length protein, and needed to be expressed at higher levels to achieve notable rescue (17). Deletion analysis of human GW182 proteins not only identified their most conserved part, the C-terminal region encompassing the DUF and RRM, as the major inhibitory domain (5- to 20-fold repression), but also identified the N-terminal region as the domain able to induce moderate repression (1.5- to 2-fold; 24–26).
In this work, we provide new insight into the function of the dGW182 N-terminal domain in repression. We demonstrate that the ability of this region to silence mRNAs is independent of its ability to bind the AGO1 protein. The first 205 dGW182 residues are necessary and sufficient to bind AGO1, as based on co-immunoprecipitation assays, while the region responsible for the repression, named the NED, maps to residues 205–490, which do not significantly bind to AGO1. These results document that within the N-terminal domain of dGW182 there are separable domains, one involved in AGO1 binding and another mediating repression.
We identified sequences in the NED region that contribute to its ability to silence reporter mRNAs. First, we identified several moderately conserved regions, a combined mutation of which reduces the ability of the NED domain to silence mRNAs (Figure 3). Second, we found that mutations in the six GW/WG motifs within the NED abolish the ability of this domain to silence mRNA reporters (Figure 4). This establishes a role of these GW/WG repeats as effector motifs in mRNA silencing. Moreover, we show that N-terminal fragments of three human GW182 homologs also function in repression, suggesting that the potential to repress mRNA activity is a general characteristic of the N-terminal region of the GW182 family proteins.
There is much evidence that GW/WG motifs play a role in interactions with AGO proteins. For example, the GW182–AGO interaction occurs via GW/WG repeats present in the N-terminal region of the GW182 proteins, and mutation of those repeats leads to abrogation of AGO binding and miRNA-mediated repression (15,20). Moreover, in fission yeast, transcription silencing in centromeric regions is mediated by the RNA-induced transcriptional silencing (RITS) complex, composed of AGO1, Tas3,and Chp1 proteins (39). Tas3 binds the AGO PIWI domain via a so-called ‘AGO hook’, a motif containing GW/WG repeats (15). Mutating tryptophan residues in this motif to either alanine or phenylalanine abrogated interaction with AGO. In Arabidopsis, RNA-directed DNA methylation (RdDM) requires an interaction between AGO4 and RNA polymerase IVb (PolIVb), which also involves reiterated GW/WG repeats (32). Domain swapping experiments showed that the GW/WG-containing region of the human GW182 protein can substitute for the homologous region of PolIVb, demonstrating that GW/WG repeats are evolutionally conserved in AGO-interacting proteins.
Based on the data presented in this work, we propose that GW/WG motifs function not only as the sites of interaction with Argonaute proteins, but also play a role as effector motifs in miRNA-mediated repression. Data produced in other laboratories also support this conclusion. Extensive mutagenesis of the dGW182 GW/WG repeats showed that while mutating the very N-terminal GWG motif abrogated most of the AGO1 binding, mutating other GW/WG repeats in the N-terminal region had no effect on the interaction with AGO1 (20), suggesting that these repeats may have a different function. Similar results were reported for the human GW182 homolog, TNGW1, by Lian et al. (40). Mutating all five GW/WG repeats in the TNGW1 fragment covering amino acids 254–503 did not affect its ability to interact with AGO2, indicating a potential alternative function for these repeats. Our observations that the N-terminal fragments of all three human GW182 homologs inhibit protein synthesis upon tethering to mRNA in Drosophila cells are consistent with a repressive function of the N-terminal GW/WG-repeats in many GW182-family proteins. Indeed, by performing mutagenesis studies, Chan and colleagues have found that GW/WG motifs in a specific region of TNGW1 are important for the tethering-induced repression of protein synthesis in mammalian cells (B. Yao and E.K.L. Chan, personal communication).
An important task for the future is to understand how the NED and GW/WG repeats function in miRNA-mediated repression. It is possible that GW/WG repeats bind some protein factors mediating mRNA repression, such as translation factors or translational repressors. A more intriguing possibility is that GW/WG repeats may create an RNA binding surface, possibly with rRNA, tRNA, or mRNA 5′UTR, with the tryptophan residues engaging in stacking interactions with RNA bases. This possibility is supported by the fact that multiple GW/WG repeats had an additive effect on repression, i.e. degree of repression was proportional to the number of intact GW/WG repeats (Figure 4).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The EC FP6 ‘Sirocco’ Program; the Novartis Research Foundation; and the Human Frontier Science Programme (LT00352/2007 to M.C.). Funding for open access charges: EC FP6 ‘Sirocco’ Program; the Novartis Research Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank E. Izaurralde (MPI, Tuebingen, Germany) and A.J. Simmonds (University of Alberta, Canada) for their kind gifts of anti-dGW182 antibodies. We are grateful to Inga Loedige and Julien Bethune for fruitful discussions.
REFERENCES
- 1.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 3.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 4.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr. Opin. Cell. Biol. 2009;21:452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Nilsen TW. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007;23:243–249. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 6.Wu L, Belasco JG. Let me count the ways: mechanisms of gene regulation by miRNAs and siRNAs. Mol. Cell. 2008;29:1–7. doi: 10.1016/j.molcel.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 7.Jackson RJ, Standart N. How do microRNAs regulate gene expression? Sci. STKE. 2007;2007:re1. doi: 10.1126/stke.3672007re1. [DOI] [PubMed] [Google Scholar]
- 8.Zdanowicz A, Thermann R, Kowalska J, Jemielity J, Duncan K, Preiss T, Darzynkiewicz E, Hentze MW. Drosophila miR2 primarily targets the m7GpppN cap structure for translational repression. Mol. Cell. 2009;35:881–888. doi: 10.1016/j.molcel.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 9.Ding L, Spencer A, Morita K, Han M. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol. Cell. 2005;19:437–447. doi: 10.1016/j.molcel.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 10.Jakymiw A, Lian S, Eystathioy T, Li S, Satoh M, Hamel JC, Fritzler MJ, Chan EK. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell. Biol. 2005;7:1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- 11.Liu J, Rivas FV, Wohlschlegel J, Yates JR, III, Parker R, Hannon GJ. A role for the P-body component GW182 in microRNA function. Nat. Cell. Biol. 2005;7:1161–1166. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Luhrmann R, Tuschl T. Identification of novel argonaute-associated proteins. Curr. Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 13.Eulalio A, Huntzinger E, Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat. Struct. Mol. Biol. 2008;15:346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- 14.Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Till S, Lejeune E, Thermann R, Bortfeld M, Hothorn M, Enderle D, Heinrich C, Hentze MW, Ladurner AG. A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat. Struct. Mol. Biol. 2007;14:897–903. doi: 10.1038/nsmb1302. [DOI] [PubMed] [Google Scholar]
- 16.Li S, Lian SL, Moser JJ, Fritzler ML, Fritzler MJ, Satoh M, Chan EK. Identification of GW182 and its novel isoform TNGW1 as translational repressors in Ago2-mediated silencing. J. Cell. Sci. 2008;121:4134–4144. doi: 10.1242/jcs.036905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chekulaeva M, Filipowicz W, Parker R. Multiple independent domains of dGW182 function in miRNA-mediated repression in Drosophila. RNA. 2009;15:794–803. doi: 10.1261/rna.1364909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding L, Han M. GW182 family proteins are crucial for microRNA-mediated gene silencing. Trends Cell. Biol. 2007;17:411–416. doi: 10.1016/j.tcb.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 19.Eulalio A, Tritschler F, Izaurralde E. The GW182 protein family in animal cells: new insights into domains required for miRNA-mediated gene silencing. RNA. 2009;15:1433–1442. doi: 10.1261/rna.1703809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eulalio A, Helms S, Fritzsch C, Fauser M, Izaurralde E. A C-terminal silencing domain in GW182 is essential for miRNA function. RNA. 2009;15:1067–1077. doi: 10.1261/rna.1605509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eulalio A, Rehwinkel J, Stricker M, Huntzinger E, Yang SF, Doerks T, Dorner S, Bork P, Boutros M, Izaurralde E. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev. 2007;21:2558–2570. doi: 10.1101/gad.443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009;15:21–32. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol. Cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zipprich JT, Bhattacharyya S, Mathys H, Filipowicz W. Importance of the C-terminal domain of the human GW182 protein TNRC6C for translational repression. RNA. 2009;15:781–793. doi: 10.1261/rna.1448009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lazzaretti D, Tournier I, Izaurralde E. The C-terminal domains of human TNRC6A, TNRC6B, and TNRC6C silence bound transcripts independently of Argonaute proteins. RNA. 2009;15:1059–1066. doi: 10.1261/rna.1606309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baillat D, Shiekhattar R. Functional dissection of the human TNRC6 (GW182-related) family of proteins. Mol. Cell. Biol. 2009;29:4144–4155. doi: 10.1128/MCB.00380-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zekri L, Huntzinger E, Heimstadt S, Izaurralde E. The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of miRNA targets and is required for target release. Mol. Cell. Biol. 2009:01081–01009. doi: 10.1128/MCB.01081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jinek M, Fabian MR, Coyle SM, Sonenberg N, Doudna JA. Structural insights into the human GW182-PABC interaction in microRNA-mediated deadenylation. Nat. Struct. Mol. Biol. 2010;17:238–240. doi: 10.1038/nsmb.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by let-7 microRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 30.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl Acad. Sci. USA. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beilharz TH, Humphreys DT, Clancy JL, Thermann R, Martin DI, Hentze MW, Preiss T. microRNA-mediated messenger RNA deadenylation contributes to translational repression in mammalian cells. PLoS ONE. 2009;4:e6783. doi: 10.1371/journal.pone.0006783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El-Shami M, Pontier D, Lahmy S, Braun L, Picart C, Vega D, Hakimi MA, Jacobsen SE, Cooke R, Lagrange T. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 2007;21:2539–2544. doi: 10.1101/gad.451207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Worby CA, Simonson-Leff N, Dixon JE. RNA interference of gene expression (RNAi) in cultured Drosophila cells. Sci. STKE. 2001;2001:PL1. doi: 10.1126/stke.2001.95.pl1. [DOI] [PubMed] [Google Scholar]
- 34.Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1530–1544. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng L, Baumann U, Reymond JL. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gehring NH, Neu-Yilik G, Schell T, Hentze MW, Kulozik AE. Y14 and hUpf3b form an NMD-activating complex. Mol. Cell. 2003;11:939–949. doi: 10.1016/s1097-2765(03)00142-4. [DOI] [PubMed] [Google Scholar]
- 37.Pillai RS, Artus CG, Filipowicz W. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA. 2004;10:1518–1525. doi: 10.1261/rna.7131604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eulalio A, Tritschler F, Buttner R, Weichenrieder O, Izaurralde E, Truffault V. The RRM domain in GW182 proteins contributes to miRNA-mediated gene silencing. Nucleic Acids Res. 2009;37:2974–2983. doi: 10.1093/nar/gkp173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buhler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 40.Lian SL, Li S, Abadal GX, Pauley BA, Fritzler MJ, Chan EK. The C-terminal half of human Ago2 binds to multiple GW-rich regions of GW182 and requires GW182 to mediate silencing. RNA. 2009;15:804–813. doi: 10.1261/rna.1229409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poirot O, O'Toole E, Notredame C. Tcoffee@igs: a web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Res. 2003;31:3503–3506. doi: 10.1093/nar/gkg522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.