Abstract
Bacteria harbouring circular chromosomes have a Xer site-specific recombination system that resolves chromosome dimers at division. In Escherichia coli, the activity of the XerCD/dif system is controlled and coupled with cell division by the FtsK DNA translocase. Most Xer systems, as XerCD/dif, include two different recombinases. However, some, as the Lactococcus lactis XerS/difSL system, include only one recombinase. We investigated the functional effects of this difference by studying the XerS/difSL system. XerS bound and recombined difSL sites in vitro, both activities displaying asymmetric characteristics. Resolution of chromosome dimers by XerS/difSL required translocation by division septum-borne FtsK. The translocase domain of L. lactis FtsK supported recombination by XerCD/dif, just as E. coli FtsK supports recombination by XerS/difSL. Thus, the FtsK-dependent coupling of chromosome segregation with cell division extends to non-rod-shaped bacteria and outside the phylum Proteobacteria. Both the XerCD/dif and XerS/difSL recombination systems require the control activities of the FtsKγ subdomain. However, FtsKγ activates recombination through different mechanisms in these two Xer systems. We show that FtsKγ alone activates XerCD/dif recombination. In contrast, both FtsKγ and the translocation motor are required to activate XerS/difSL recombination. These findings have implications for the mechanisms by which FtsK activates recombination.
INTRODUCTION
Most known bacteria have circular chromosomes. This circular form facilitates the termination of replication, but renders recombination-based repair hazardous, because sister chromatid exchanges between circles may result in dimer formation [reviewed in (1,2)]. Chromosome dimers cannot be segregated to daughter cells and must therefore be resolved, to generate monomers, before cell division. Xer site-specific recombination systems are responsible for this dimer resolution.
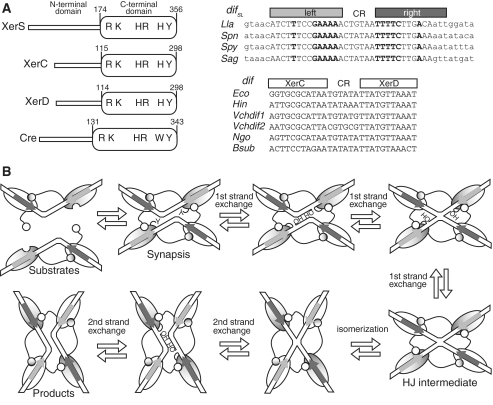
The resolution of chromosome dimers has been studied mostly in Escherichia coli [reviewed in (1,3,4)]. It involves two site-specific recombinases of the Y-recombinase family, XerC and XerD, acting at a recombination site, dif, located in the termination of replication region (Figure 1A). In contrast to the XerCD/dif system, most systems driven by Y recombinases, for example the Cre/loxP model system from plasmid P1, use a single recombinase (5). Recombination occurs in a complex containing four monomers of recombinase bound to two recombination sites synapsed in an antiparallel configuration (Figure 1B). A first pair of recombinases cuts and transfers a first pair of strands, producing a Holliday-junction (HJ)-containing intermediate. Subsequent catalysis by the second pair of recombinases resolves this intermediate to generate recombinant molecules. In the XerCD/dif system, the synaptic complex consists of two XerC and two XerD monomers bound to two dif sites. The dif site contains binding sites for XerC and XerD separated by a 6-bp central region (CR), at the edges of which, strand exchanges are catalysed (Figure 1A). XerC or XerD may catalyse the exchange of the first pair of strands, depending on external controls imposed on the synaptic complex (6,7). In vivo and in vitro studies have shown that the XerCD/dif complex naturally tends to initiate recombination by XerC-mediated strand exchange (7). This bias is thought to be due to both the asymmetry of the dif sequence and the use of two different recombinases. This “XerC-first” pattern applies to activities of XerCD other than the resolution of chromosome dimers: during the resolution of plasmid multimers at the psi or cer sites (8) and during the integration of bacteriophage genomes into their host dif sites (9). In these cases, HJ resolution by XerD catalysis is either not required (cer site and phage integration) or is induced by isomerization of the HJ intermediate (psi site, Figure 1B). In chromosomal XerCD/dif complexes, recombination is controlled by FtsK, a DNA translocase associated with the cell division septum (3). In the absence of FtsK, XerD has no catalytic activity and HJ-intermediates may be generated by XerC catalysis. However, these intermediates are not fully resolved to generate products, but are instead reconverted into substrates, by further XerC catalysis (7,10). Chromosome dimer resolution requires FtsK, which induces recombination by forcing XerD to catalyse the exchange of the first pair of strands.
Figure 1.
The FtsK-XerCD/dif and FtsK-XerS/difSL systems. (A) Left: diagram of the XerC, XerD, XerS and Cre proteins, with their length and domain organization (in amino acid). The conserved residues involved in catalysis are indicated. Right: difSL and dif sites of representative bacteria with the recombinase binding sites, separated by the central region (CR) indicated. In difSL sites, upper case bases are part of the previously defined minimal site (28) and bases shown in bold typeface are inverted repeats within this minimal site. The left-half site is indicated by the pale grey bar and the right-half site by the dark grey bar. This convention is used throughout the paper. In dif sites, the two half sites recognized by XerC and XerD, respectively, are indicated. Lla, L. lactis; Spn, Streptococcus pneumoniae; Spy, S. pyogenes; Sag, S. agalactiae; Eco, E. coli; Hin, H. influenzae; Vch, V. cholerae; Ngo, Neisseria gonorrhoeae; Bsub, B. subtilis. (B) A diagram of the mechanism of recombination by Y-recombinase based on the data obtained for the Cre/loxP system. Y indicates the catalytic tyrosine residue. OH is the 5′ hydroxyl group created by DNA cleavages.
FtsK is required for both cell division and faithful chromosome segregation [reviewed in (3)]. Its N-terminal domain is essential for growth and contains transmembrane helices that target FtsK to the division septum. Its C-terminal domain (FtsKC), which is dispensable for viability, encodes the translocation motor (7). The α and β subdomains of FtsKC carry Walker-type ATPase motifs and form a hexameric motor (11). The extreme C-terminal subdomain, FtsKγ, controls FtsK translocation and Xer recombination. FtsKγ contains a DNA-binding motif that binds to specific DNA motifs, the KOPS (5′-GGGNAGGG). KOPSs are preferentially oriented towards the dif site, thereby orienting translocation towards dif (12–17). Translocation stops when the XerCD/dif complex is reached, and this termination of translocation is FtsKγ-independent (18). Recombination is then induced by a mechanism thought to involve a particular interaction of FtsKC with the DNA in the immediate vicinity of dif (19) and a direct interaction between FtsKγ and XerD (20). This second interaction induces XerD-mediated strand exchange in vitro, suggesting it induces a recombination pathway in which XerD catalyses the exchange of the first pair of strands, leading to productive recombination and dimer resolution (20).
Homologs of FtsK are found in most bacteria, in which they are involved in DNA transfer during conjugation, sporulation and chromosome segregation (21–25). Y-recombinases are also widespread and may play a role in various DNA transactions (5,26). Homologs of XerC and XerD, together with a potential dif site opposite the origin of replication, are found in most proteobacteria and firmicutes (27–29). Other families of bacteria use alternative systems to resolve chromosome dimers (28,29). In streptococci/lactococci, a family of firmicutes, the dimer resolution system consists of a single Y-recombinase, XerS, which is responsible for recombination at a specific recombination site, difSL (Figure 1A) (28). The XerS/difSL system is the only single-recombinase Xer system functionally studied to date. However, it is certainly not the only such system. It has recently been reported that a subgroup of ε-proteobacteria harbour only one conserved Y-recombinase, XerH, together with a potential dif site resembling difSL and located within the putative terminus regions of these bacteria (29). In evolutionary terms, XerH is only distantly related to XerCD and XerS, consistent with the acquisition of Xer systems on several occasions during evolution (29).
The Lactococcus lactis XerS/difSL system resolves chromosome dimers in an FtsKC-dependent manner if it is used to replace the XerCD/dif system in E. coli (28). This observation seems to go against the highly accurate control of chromosome dimer resolution in E. coli, which involves specific interactions between partners, and raises questions about the possible control of dimer resolution by a single recombinase. In this study, we investigated the molecular mechanism of XerS/difSL recombination and its control by FtsK. We show that XerS binds and recombines difSL sites in vitro. Both binding and strand exchange were asymmetric. We showed, using in vivo assays, that the FtsK-dependent coupling of chromosome segregation with cell division is conserved in the FtsK/Xer system of L. lactis. However, the mechanism by which recombination was activated differed from that in the E. coli system. FtsKγ can activate XerCD/dif recombination on its own, whereas the translocation motor is also required to activate XerS/difSL recombination. These findings have implications for the control of recombination in the two Xer systems.
EXPERIMENTAL PROCEDURES
Strains and plasmids
Strains used were derived from E. coli K12 strain LN2666 [W1485 F- leu thyA thi deoB or C supE rpsL (StR)], referred as the wt strain (30). Unless indicated, recombinant DNA was constructed in transgenesis vectors of the pFC13 series by standard cloning techniques, and inserted into the chromosome as previously described (30,31). The dif-lacI-dif and difSL-lacI-difSL cassettes were inserted in place of the Δ(dif)58 deletion (30). Three 5′-GGGCAGGG KOPS motifs separated by 6-bp random sequences (5′-TTCATT and 5′-TAGCAT) were eventually inserted in a non-permissive orientation, 10 bp from the XerD binding site of the dif site on one side of the dif-lacI-dif cassette or at the equivalent position in the difSL-lacI-difSL cassette. The Δ(lacI) deletion removes bp −37 to 1075 of the lacI gene. In the Δ(ftsKC)::Tc allele, the residues from position 814 to the end of FtsK are deleted. The Δ(ftsKγ)-Cm, ftsKKOPSblind-Cm and Δ(xerC)::Gm alleles have been described elsewhere (32). Resistance gene-tagged alleles were transferred by P1 transduction. For protein production and purification, xerS and the E. coli and L. lactis ftsKγ subdomains (corresponding to the sequence from residue 1253 of E. coli FtsK and 682 of L. lactis FtsK to the ends of these proteins) were inserted into pFSKB3X (GTP Technology, France) to obtain his-flag-gene constructs. We then mutated xerS, using mutagenic oligonucleotides, to obtain the xerSY341F allele. For in vivo expression of the recombinases, genes were inserted into a pGB2 derivative carrying the araC-ARAp expression cassette, yielding plasmids pFC241 (pXerC), pCP56 (pXerS) and pFC250 (pXerC-XerD). For FtsKγ and FtsKC production, the pBAD18 derivatives, pFX150 (pFtsKC) (7), pCL379 (p-γE.coli) and pCL378 (p-γL.lactis) were used. For subsequent in vitro analysis, a synthetic minimal difSL site, 5′ATCTTTCCGAAAAACTGTAATTTTCTTGACA and its variants lacking the right-half site, 5′ATCTTTCCGAAAAACTGTAAGGCTACGTCAT, or the left-half site, 5′CTGAGCTAGTCACACTGTAATTTTCTTGACA, were inserted into pGhost9 (33).
Purification of XerS
For XerS and XerSY341F purification, pCL255 (XerS) and pCL360 (XerSY341F) were transferred to E. coli strain BL21 (DE3; Novagen). The resulting strains were grown in L broth at 42°C, to an OD600 of 0.6. We then added IPTG (0.1 mM) to the medium and incubated the culture at 25°C for 3 h. Cells were recovered by centrifugation, resuspended in buffer [50 mM phosphate buffer pH 8, 500 mM NaCl, 10 mM imidazole, 10% glycerol, 1 mg/ml lysosyme, 230 µg/ml RNaseA and EDTA-free protease inhibitor cocktail (Roche)], sonicated and the lysate was cleared by centrifugation. His-FLAG-tagged XerS and XerSY341F were purified on nickel resin (1 ml His-trap HP, GE Healthcare) followed by heparin (1 ml High-Trap HP, GE Healthcare) and gel filtration (HiLoad 16/60 Superdex 200, GE Healthcare) columns (Supplementary Figure S1). Purified recombinases were concentrated with an ICON concentrator (Thermoscientific) and stored at −80°C in buffer containing 40 mM Hepes (pH 7.7), 400 mM KCl, 1 mM DTT, 0.5 mM EDTA and 10% glycerol.
In vitro experiments
For electrophoretic mobility shift assays (EMSAs), 142-bp fragments carrying the site of recombination at their centre were amplified by PCR and purified by gel electrophoresis. These substrates were 5′ end-labelled with [γ-32P] ATP and T4 DNA polynucleotide kinase. Binding reactions were carried out in a buffer containing 25 mM Hepes (pH 7.7), 50 mM KCl, 0.25 mM EDTA, 0.5 mM DTT, 10 µg/ml BSA, 10 mM MgCl2 and 10% glycerol, in the presence of 10 000 c.p.m. of labelled DNA (<1 nM), 1 µg of poly(dI-dC) and the indicated protein concentrations. The reactions were incubated at 30°C for 30 min and analysed by electrophoresis in 5% polyacrylamide native gels run in TGE. Gels were dried and analysed with a Fuji PhosphorImager.
For DNAse1 footprinting analysis, various amounts of XerS were mixed with 226-bp difSL-carrying DNA fragments labelled with 32P at the 5′ end of either the top or the bottom strand (BS). Reactions were performed as for EMSA. After 30 min of incubation at 30°C, we added 7 mM MgCl2 and 3 mM CaCl2, and partial digestion of the DNA was initiated by adding 1 µl of an empirically determined dilution (typically 10−3) of a DNase I stock solution. The mixture was incubated for 2 min at 30°C and the reaction was stopped by adding 1 µl EDTA (0.5 M). The DNA was then precipitated in ethanol, re-suspended in 6 µl of loading buffer and separated by electrophoresis in an 8% polyacrylamide denaturing sequencing gel run in TBE. A sequencing reaction was carried out with the Thermo Sequenase Cycle Sequencing Kit (USB) and was run in parallel, as a size marker. Gels were dried and analysed with a PhosphorImager.
For recombination experiments, the HJ substrate (Figure 3) was constructed by annealing four oligonucleotides:
(I) 5′-CTGCCGTGATCACGCTGAACGCGTTTTAGCATCTTTCCGAAAAACTGTAATTTTCTTGACAATTGGAGGCCTAACGCCTAAAGCGGCCGCCTAGTCC;
(II) 5′-GGACTAGGCGGCCGCTTTAGGCGTTAGGCCTCCAATTGTCAAGAAAATTACAGTTTTTCGGAAAGATCGACCGTTGCCGGATCCGCTGC;
(III) 5′-GGATTCGAGATCTCAGGATGTCTCCAATTGTCAAGAAAATTACAGTTTTTCGGAAAGATGCTAAAACGCGTTCAGCGTGATCACGGCAG;
(IV) 5′-GCAGCGGATCCGGCAACGGTCGATCTTTCCGAAAAACTGTAATTTTCTTGACAATTGGAGACATCCTGAGATCTCGAATCC.
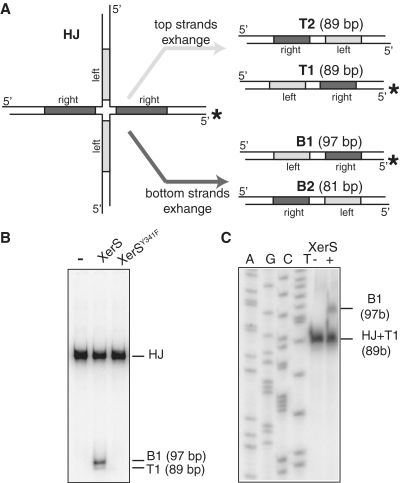
Figure 3.
Strand exchange by XerS. (A) Left: diagram of the HJ substrate with the labelled strand indicated by asterisk. The left and right difSL half sites are indicated by pale grey and dark grey, respectively. Right: resolution products (T1, T2 and B1, B2; with their respective sizes indicated) obtained by exchange of either the top or bottom pairs of strands (defined in Figure 2). Labelled strands are indicated by asterisk. (B) PAGE analysis of the recombination products with positions of the substrate (HJ), B1 and T1 products indicated. (−) lane, no XerS; XerS and XerSY341F lanes, reactions contained 200 mM of the protein indicated. (C) Analysis of the individual DNA strands by denaturing PAGE. Positions of the labelled strands are indicated. (AGCT) lanes, size ladder (‘Experimental procedures’ section). (−) lane, no XerS; (+) lane: reaction contained 200 nM XerS.
The indicated oligonucleotide (20 pmol) was 5′ end-labelled with [γ-32P] ATP and T4 DNA polynucleotide kinase and was mixed with the other, unlabelled oligonucleotides (30 pmol each). The oligonucleotides were annealed by heating to 100°C and slow cooling. The HJ-containing molecules were purified by PAGE in a 5% polyacrylamide gel, recovered from excised gel slices by incubation in elution buffer [10 mM Tris–HCl (pH 8), 1 mM EDTA, 0.2% SDS, 0.3 M NaCl, 1 mg/ml glycogen], precipitated in ethanol and re-suspended in deionized water. Reactions were performed as described earlier for EMSA (except that the reaction buffer contained 5 mM spermidine) and were stopped by adding 1% SDS and 1 mM EDTA. The samples were then treated with proteinase K (1 h at 37°C) and the DNA was analysed by PAGE in 5% polyacylamide native gels in (Tris/glycin/EDTA) TGE or in 8% polyacrylamide denaturing gels (containing 7 M urea) in 8% TBE.
Co-culture experiments
Co-culture experiments were conducted as previously described (34). Briefly, the strains to be analysed were mixed with the parental strains [either wt or Δ(dif)::difSL pXerS strains]. In the case of strains containing the pFtsKC plasmid, 0.025% arabinose was added during the whole course of the experiment. The mixture of strains was grown in serial cultures, subcultured by dilution every 10 generations and plated every 20 generations on appropriate medium to determine the relative frequencies of the two strains. Frequencies of unresolved dimers were calculated from the slope of strain ratio plotted against generation (34). The means of at least three independent measurements, with SDs, are presented.
XerCD/dif and XerS/difSL in vivo recombination assay
Strains carrying the Δ(lacI) and xerC::Gm mutations, either the dif-lacI-dif or the difSL-lacI-difSL cassette in place of the dif site and the indicated ftsK alleles and plasmids were grown in L broth, rendered competent and transformed with pXerC or pXerS. Transformants were plated on L agar containing 20 µg/ml spectinomycin and grown overnight at 37°C. Five independent transformants were re-suspended in L broth plus spectinomycin and, in some cases, arabinose (0.025% for FtsKC production and 0.1% for FtsKγ production), grown 5 h, diluted and plated on L broth plus X-gal (40 µg/ml). This entire procedure corresponds to ∼20 generations before plating on L agar plus Xgal. The ratio of dark blue to total colonies was used to calculate the frequency of lacI loss per cell, per generation. The mean and SD of five independent measurements are shown in the figures.
Western-blot analysis
Strains were grown in the assay conditions, harvested at an OD600 of 0.1 and resuspended in sample buffer (1% SDS, 10% glycerol, 0.1 M DTT, 0.2% bromophenol blue, 50 mM Tris–HCl, pH 6.8). Samples were boiled for 5 min. We used ∼0.04 OD units for each sample. Proteins were separated by SDS–PAGE and transferred to nitrocellulose membranes (Hybond-C Extra, GE Healthcare). Membranes were then incubated with mouse anti-FLAG primary antibodies (Sigma), followed by goat anti-mouse IgG peroxidase-linked secondary antibodies (Sigma), as indicated by manufacturer, and antibody binding was detected with an ECL kit (Thermo Scientific) and a BioImager (LAS-4000 FUJIFILM).
RESULTS
Asymmetric binding of XerS to difSL
The potential difSL site has been defined as a minimal 31-bp site, on the basis of comparative genomics studies of streptococci and in vivo functional assays of recombination activity (Figure 1A) (28). This site contains two imperfect inversely repeated sequences of different sizes that are potential binding sites for XerS (the right and left-half sites in Figure 1), separated by a CR. For studies of XerS binding to difSL, we constructed a his-flag-xerS allele. We produced the protein encoded by this allele (herein referred to as XerS), purified it (Experimental procedures section, Supplementary Figure S1) and used it in EMSA experiments with various difSL-containing DNA fragments. We found that the previously defined minimal difSL site bound XerS poorly, with efficient binding requiring additional nucleotides flanking the right arm (data not shown). Nucleotides other than those naturally flanking difSL could give efficient binding, demonstrating that these nucleotides are not specific, accounting for their lack of conservation among difSL sites from different streptococci [Figure 1A; (28)].
At low-XerS concentration, a first XerS/difSL complex (C1) is detected, which is transformed into a second lower mobility complex (C2) with increasing XerS concentration (Figure 2A). Considered together with data from other Y recombinase systems, these data suggest that the C1 complex consists of a single XerS monomer bound to one half site of difSL, with the binding of a second monomer to the second half site giving rise to the C2 complex. We then studied the location of XerS binding in DNase1 footprinting experiments (Figure 2B and Supplementary Figure S2). In these experiments, XerS protected both inverted repeats, together with the CR and few base pairs adjacent to the right arm. Thus, the non-specific base pairs next to the right-half site required for efficient binding are also protected from DNase1 cleavage. XerS binding also increased sensitivity to DNAse1 cleavage on either side of difSL, suggesting that the DNA is distorted at these positions, which thus probably correspond to the first unbound bases on either side of the XerS/difSL complex (Figure 2B and Supplementary Figure S2). Thus, the difSL site is longer than previously thought and the left and right XerS binding sites are of similar lengths.
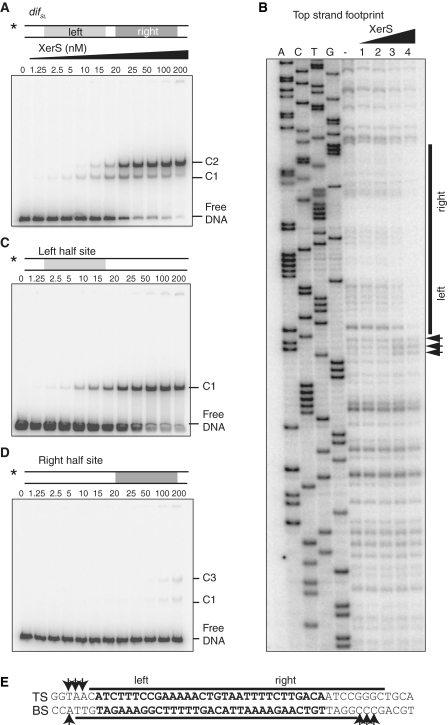
Figure 2.
Asymmetric binding of XerS to difSL. (A) Titration of a 142-bp DNA fragment containing difSL by increasing concentrations of XerS in an EMSA experiment. The DNA substrate is shown at the top. XerS concentrations are given in nano molar and the positions of the free DNA probes and the C1, and C2 XerS/DNA complexes are indicated. The asterisk indicates the 5′-labelled strand. (B) DNaseI footprinting of the top strand (TS) of difSL. Experiments were carried out with increasing XerS concentrations, as described in the ‘Experimental procedures’ section. A, C, T and G, ladder sequence of the difSL-containing substrate; (−) lane, no XerS; Lanes 1, 2, 3 and 4: reactions contained 12.5, 25, 50 and 100 nM XerS, respectively. The region protected from cleavage is indicated by the black bars and positions of increased cleavage by arrows. (C and D) Same experiments as in (A), with substrates containing mutated right- and left-half sites, respectively. (E) Summary of the data for the difSL sequence. The footprint of the BS is shown in Supplementary Figure S2. Black bars, protected regions; Arrows, positions hypersensitive to cleavage.
The left and right-half sites of difSL are not perfect inverted repeats (Figure 1A). We therefore studied XerS binding to the two half sites separately. We did this by using difSL variants in which one of the half sites had been mutated. Both half sites formed a complex equivalent to the C1 complex (Figure 2C and D) in these experiments, consistent with our hypothesis that this complex consists of a single XerS monomer bound to one half site. XerS bound the left arm with a much higher affinity than the right arm (compare Figure 2C and D). This suggests that XerS binds preferentially to the left arm and co-operatively to the right arm of difSL. At high-XerS concentration, a second complex is detected with DNA carrying the low affinity right-half site (C3 in Figure 2D). XerS binding to the right-half site may thus promote the binding of a second XerS monomer to non-specific adjacent sequences. However, this complex appeared to be different from the C2 complex, indicating that the left arm DNA is required for the formation of a C2 complex in the correct conformation.
Strand preference in XerS/difSL recombination
As XerS belongs to the Y-recombinase family, the recombination it mediates should involve a HJ-containing intermediate, produced by the exchange of a first pair of strands by a pair of recombinases and with product resolution subsequently achieved by the exchange of the second pair of strands by the other pair of recombinases [reviewed in (5); Figure 1B]. The catalytic state of pairs of recombinases thus switches from active to inactive and vice versa, during the HJ step of the reaction, making this intermediate a target of choice for control of the reaction. This led us to assay XerS-mediated cleavage and strand exchange with HJ-containing substrates. We constructed a HJ-containing substrate that mimicked classical intermediates of Y recombinase-driven recombination (35–37) (Figure 3A). The incubation of this HJ substrate with XerS yielded linear products of the expected size following cleavage by both pairs of recombinases (B1 and T1 in Figure 3B). Thus, XerS is able to cleave both the difSL strands. Covalent complexes between XerS and the DNA were detected in the absence of proteinase K treatment (Supplementary Figure S3). These complexes are intermediate between the cleavage and the ligation steps during strand exchange (Figure 1B). The small quantity of these covalent complexes suggested that religation occurs efficiently to give rise to exchanged strands. We assessed strand transfer directly, by analysing HJ substrates by electrophoresis in denaturing gels after incubation with XerS (Figure 3C and Supplementry Figure S3). The predicted recombinant strands were readily detected in these experiments, showing that XerS does indeed transfer and religate DNA strands. The two pairs of strands are not exchanged equally efficiently, with the BSs clearly being exchanged preferentially (Figure 3B). The two pairs of recombinases therefore do not have equivalent activities at this step in the recombination reaction, demonstrating a marked asymmetry of cleavage-competent XerS/difSL complexes.
Alignment of the amino acid sequence of XerS with those of other known Y recombinases led to the identification of conserved residues involved in catalysis, including Y341, which was identified as the residue likely to be responsible for DNA cleavage (Supplementary Figure S4A). We used mutagenesis to replace this residue by a phenylalanine residue (‘Experimental procedures’ section). The resulting protein was purified and its difSL binding and cleavage were assessed. XerSY341F bound difSL, albeit slightly less efficiently than wild type (wt) XerS (Supplementary Figure S4B). As predicted, XerSY341F was unable to cleave the HJ-containing substrate (Figure 3B), demonstrating the involvement of Y341 in catalysis.
Taken together, these results show that XerS binds difSL and exchanges its DNA strands in vitro. Both activities were found to be highly asymmetric: binding to the left-half site was more efficient than binding to the right-half site and a preferential exchange of the BSs was observed. As other known bacterial Y recombinases cleave DNA in cis (Figure 1B) (5), we would expect the BSs of difSL to be cleaved by recombinases bound to its right-half site, which interacts poorly with XerS.
Translocation by septum-borne FtsK controls dimer resolution by XerS/difSL
We studied the control of chromosome dimer resolution by XerS/difSL, using E. coli strains harbouring difSL in place of the dif site and measuring the capacity of these strains to resolve dimers, by co-culture with a parental reference strain [‘Experimental procedures’ section; (34)]. The Δ(dif)::difSL strain producing XerS from a plasmid (pXerS) displayed a clear growth advantage over the dif+ Δ(xerC) strain, which is unable to resolve dimers (Figure 4B and C). On the other hand, this strain displayed no viability difference with the wt strain, which resolves dimers by XerCD/dif recombination (Figure 4B and C). Thus XerS/difSL is as efficient as XerCD/dif for the resolution of chromosome dimers in E. coli. Mutant alleles of ftsK (Figure 4A) were then transferred to the Δ(dif)::difSL pXerS strain. The deletion of FtsKC or the mutation of its ATP hydrolysis motif both inactivate dimer resoltion in E. coli (38,39). These mutations also inactivated dimer resolution by the XerS/difSL system, demonstrating a dependence of the dimer resolution activity of XerS/difSL on FtsKC and its translocation activity. We then produced the C-terminal domain of FtsK from a plasmid in Δ(ftsKC) strains, in expression conditions that support XerCD/dif recombination (see below and ‘Experimental procedures’ section). This form of FtsK that lacks the N-terminal domain and is thus not bound to the division septum does not support dimers resolution by the XerCD/dif system (10,40). Although coculture experiments with strains containing pFtsKC showed important variations and high standard deviations (SD), it was clear that FtsKC did not support significant rates of dimer resolution (Figure 4B). Thus, dimer resolution by the XerS/difSL systems requires a septum-borne FtsK.
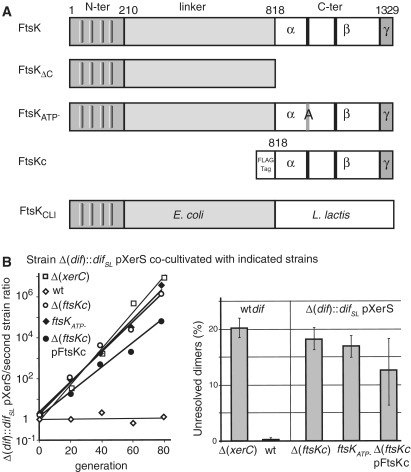
Figure 4.
Resolution of chromosome dimers by XerS/difSL. (A) Diagram of the FtsK mutants used. Top line, wt E. coli FtsK with the three domains and the C-terminal subdomains indicated (α,β,γ). Black bars represent the A and B Walker-type motifs. Co-ordinates at the top are in amino acid. The grey bar labelled (A) indicates the mutation of the FtsKATP- mutant. The FtsKC domain was produced from a plasmid (pFtsKC; ‘Experimental procedures’ section) under the control of the pBAD promoter and was used in Δ(ftsKC) strains in the presence of 0.025% arabinose. (B) Left: typical co-culture experiments. A Δ(dif)::difSL strain carrying the pXerS plasmid was co-cultured with strains carrying either the dif site [Δ(xerC) and wt strains] or the difSL site in place of dif, the pXerS plasmid and the indicated ftsK allele [Δ(ftsKC), ftsKATP- and Δ(ftsKC) pFtsKC strains]. Right: frequencies of unresolved dimers in the indicated strains, calculated from experiments in B (34). The mean results of at least three independent experiments with standard deviations are shown.
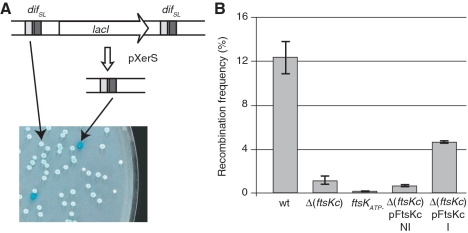
We assayed recombination directly, by constructing strains carrying two directly repeated difSL sites flanking the lacI gene (Figure 5A; ‘Experimental procedures’ section). This difSL-lacI-difSL cassette was inserted in place of the dif site, in a strain carrying a deletion of lacI. This made possible the accurate assessment of recombination events, evaluated by scoring the number of blue colonies after transformation with pXerS. As previously reported, XerS/difSL recombined at high frequency (∼12% per generation) when inserted at the dif position, and recombination was found to be dependent on FtsKC (Figure 5B) (28). As previously shown for XerCD/dif (10), inactivation of the ATPase activity of FtsK inactivated XerS/difSL recombination, showing that FtsK translocation is a prerequisite for the activation of XerS/difSL recombination. The production of FtsKC in the Δ(ftsKC) strains significantly increased the frequency of XerS/difSL recombination (Figure 5B). Thus, septum-independent FtsKC is sufficient to induce XerS/difSL recombination, but only septum-tethered FtsKC can induce chromosome dimer resolution.
Figure 5.
XerS/difSL recombination depends on the translocation activity of FtsKC. (A) The recombination reporter cassettes used consist of two directly repeated difSL, separated by the lacI gene, inserted at the natural position of dif. Recombination deletes lacI, giving rise to blue colonies on indicator medium. An example of assessments made by plating is shown. (B) Recombination frequencies in strains carrying the difSL-lacI-difSL cassette and the indicated ftsK alleles (‘Experimental procedures’ section). Proteins encoded by the Δ(ftsKC) and ftsKATP− mutations and the pFtsKC plasmid are shown in Figure 4A. Strains containing pFtsKC were grown in absence of inducer (NI) or in presence (I) of 0.025% arabinose.
FtsKγ activates XerS/difSL recombination via a local effect
The data reported above suggest that recombination is controlled in similar ways in the L. lactis and E. coli Xer systems: both the XerS/difSL and XerCD/dif systems resolve chromosome dimers in an FtsKC-dependent manner and, in both cases, recombination can be induced by septum-independent FtsKC, but dimer resolution can only occur if FtsKC is tethered to the septum [Figures 4 and 5; (10,38,40,41)]. This suggests that the L. lactis FtsK homologue, FtsKLl, exerts the same regulatory activity as the E. coli FtsK. We therefore replaced the C-terminal domain of E. coli FtsK by its homologue in FtsKLl (‘Experimental Procedures’ section) and measured its capacity to induce XerS/difSL and XerCD/dif recombination. We used strains carrying equivalent difSL-lacI-difSL or dif-lacI-dif cassettes and producing the cognate recombinase from plasmids (‘Experimental procedures’ section). The resulting ftsKCLl chimera supported high frequencies of recombination by both the XerS/difSL and XerCD/dif systems (Figure 6B). Thus, either the activities of FtsKC are conserved between these two bacteria or some of these activities are dispensable for normal levels of recombination.
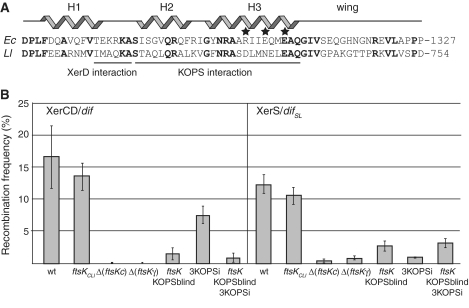
Figure 6.
FtsK must reach difSL to induce recombination. (A) Alignment of the FtsKγ subdomains of E. coli (Ec) and L. lactis (Ll). Structural features of the Ec FtsKγ are indicated: the H1-3 helices and the wing of the w-helix DNA-binding domain. Identical residues are shown in bold and residues for which mutation leads to the KOPS-blind phenotype are indicated by stars. The regions thought to interact with XerD and the KOPS motif are indicated. The co-ordinate of the last residue aligned here is given to the right. (B) Recombination frequencies in strain carrying either a dif-lacI-dif (left, XerCD/dif) or a difSL-lacI-difSL (right, XerS/difSL) recombination cassette, the pXerC or pXerS plasmids, respectively, and the indicated ftsK alleles and/or insertion of non-permissive KOPS motifs close to one of the recombination sites (3KOPSi; ‘Experimental procedures’ section). The Δ(ftsKC)::ftsKCLl allele carries the whole C-terminal domain of L. lactis FtsK in place of the E. coli domain (‘Experimental procedures’ section; Figure 4A). The ftsKKOPSblind allele carries mutations of the three residues indicated by stars in (A).
In E. coli, the FtsKγ subdomain is responsible for the recognition of both KOPS and XerD. The level of identity of the E. coli and L. lactis FtsKγ subdomains (41% over 61 residues; Figure 6A) suggests that these activities may be conserved in FtsKLl. However, the presence of several changes in important residues is not consistent with this hypothesis. The TEKRKA motif of FtsK, which appears to interact with XerD (20,39,42), is only weakly conserved, and changes in the H2 and H3 helices of the W-helix motif may have made it possible for FtsKLl to recognize DNA motifs other than the E. coli KOPS motif (Figure 6A). We compared the roles of FtsKγ activities in XerS/difSL and XerCD/dif recombination, by first using a Δ(ftsKγ) allele. As previously reported for the XerCD/dif system (32), the deletion of FtsKγ had the same effect on XerS/difSL recombination as deletion of the entire FtsKC (Figure 6B). Thus, the translocation activity of FtsK is not sufficient to induce XerS/difSL recombination, with at least one of the FtsKγ activities being required. We then inactivated the KOPS recognition activity of FtsK, using the ftsKKOPSblind allele (Figure 6A) (16,32). This almost completely inactivated XerS/difSL recombination, demonstrating a requirement for FtsK to translocate to difSL to activate recombination. It follows that, as the FtsKCLl chimera supports recombination, FtsKLl either recognizes the E. coli KOPSs or a motif with an orientation sufficiently biased in the ter region of the E. coli chromosome to direct translocation towards the dif site. We investigated whether FtsK needed to reach the XerS/difSL complex to induce recombination, by inserting three KOPS motifs (5′-GGGCAGGG motifs separated by 6-bp random sequences; ‘Experimental procedures’ section), 10 bp from the recombination cassettes in a non-permissive orientation, thereby preventing FtsK from reaching one side of the cassette (12). This significantly reduced XerS/difSL recombination in ftsKwt conditions but had no effect in ftsKKOPSblind conditions (Figure 6B). The same effects were observed in equivalent strains carrying the dif-lacI-dif cassette (Figure 6B). We conclude that FtsK must reach the XerS/difSL complex for the induction of XerS/difSL recombination.
A differential role for FtsKγ
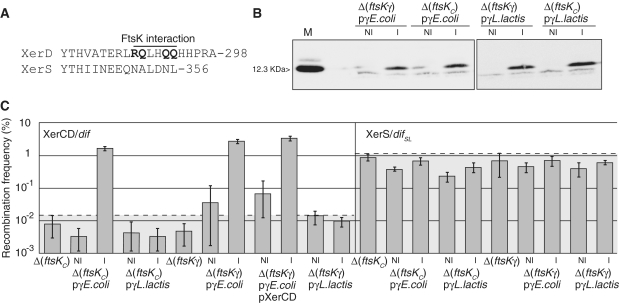
The data presented above and previous reports suggest that FtsK must deliver FtsKγ to the recombination complex to induce recombination through a direct interaction with the recombinases. However, for XerS/difSL, this appears to be inconsistent with the poor conservation of the TEKRKA motif in FtsKLl (Figure 6A). Furthermore, the XerD residues identified as potentially interacting with FtsKγ are not conserved in XerS (Figure 7A). To investigate the mechanisms underlying the activation of recombination, we first attempted to set up in vitro recombination using purified FtsK50C (7) and XerS proteins. We repeatedly failed to observe XerS/difSL recombination in conditions supporting XerCD/dif recombination (data not shown), which further suggested a difference in the mechanism of XerCD/dif and XerS/difSL recombination induction. We then assayed the FtsKγ-mediated activation of recombination in vivo. To this aim, we assessed the effect of ftsKγ expression in strains with deletions of either ftsKC or ftsKγ carrying the recombination cassettes. Control experiments in these mutant strains showed that XerS supported higher levels of FtsKC- and FtsKγ-independent recombination than XerCD (1 and 0.01%, respectively; Figure 7B). The E. coli and L. lactis FtsKγ genes were cloned and fused to a FLAG tag for subsequent western analysis and plasmids producing one or other of these FtsKγ proteins were introduced into the Δ(ftsKγ) and Δ(ftsKC) strains. Western analysis showed that E. coli FtsKγ and L. lactis FtsKγ were produced to similar levels in the assay conditions used (Figure 7B; ‘Experimental procedures’ section).
Figure 7.
FtsKγ induces XerCD/dif but not XerS/difSL recombination. (A) Alignment of the FtsK interaction region of XerD with its corresponding region in XerS. Residues inferred to interact with FtsK are indicated and shown in bold typeface [R1300A, E1303A, E1306A; (21)]. The co-ordinate of the last residue is given to the right. (B) Western-blot analysis of FtsKγ production. Cells were collected from the indicated strains during the course of the experiments shown in (C). His-Flag-tagged E. coli and L. lactis FtsKγ were detected with an anti-FLAG antibody (‘Experimental procedures’ section). M, Purified E. coli His-FLAG-FtsKγ, with its size indicated. The faint bands that migrate faster than the FtsKγ peptides are background detection in the migration front of the gel. (C) Recombination frequencies in strains carrying either a dif-lacI-dif (left, XerCD/dif) or a difSL-lacI-difSL (right, XerS/difSL) recombination cassette and the indicated ftsK alleles and FtsKγ-coding plasmid. Recombination was induced by transformation with the pXerC plasmid (all strains in the XerCD/dif panel except strains labelled pXerCD for which the pXerC-XerD plasmid was used; ‘Experimental procedures’ section), or the pXerS plasmids (XerS/difSL panel). NI, absence of inducer; I, FtsKγ production was induced by adding 0.1% arabinose. The dotted lines and grey zones indicate the FtsKC-independent recombination background for the two systems. In both cases, the recombination background in the absence of the cognate recombinase is below 10−3 (data not shown).
Escherichea coli FtsKγ supported recombination by the E. coli XerCD/dif system in both Δ(ftsKC) and Δ(ftsKγ) strains (Figure 7C). Thus, the translocation activity of FtsK is not required for the induction of recombination and it is not essential for FtsKγ to be tethered to FtsKC for the induction of recombination. In contrast, E. coli FtsKγ did not support recombination by the XerS/difSL system (Figure 7C). The L. lactis FtsKγ did not support recombination by either of the Xer systems (Figure 7C). In these experiments, XerS is produced from a plasmid, while XerCD/dif recombination is analysed in strains containing normal levels of XerD, making it possible that the difference observed between the two Xer systems is due to a titration of FtsKγ by excess XerS (‘Experimental procedures’ section). We ruled out this possibility by co-producing XerD and XerC from the same plasmid and in the same conditions as XerC or XerS alone (‘Experimental procedures’ section). This yielded equivalent levels of FtsKγ-induced XerCD/dif recombination as production of XerC alone (Figure 7B), showing that FtsKγ titration by excess recombinase does not explains the observed effects. Taken together, these data strongly suggest that there is no FtsKγ-XerS interaction functionally equivalent to the FtsKγ-XerD interaction in the L. lactis FtsK-XerS system.
DISCUSSION
We have shown in this study that the control of chromosome dimer resolution and its coupling with cell division are mostly conserved between evolutionary remote FtsK-Xer systems involving either one or two recombinases. Indeed, despite this important difference, the E. coli XerCD/dif and the L. lactis XerS/difSL systems have several important characteristics in common, in terms of their recombination mechanism and its control by the FtsK translocase. The mode of binding of XerS to difSL is similar to the mode of binding of XerCD to dif. Both recombinase/DNA complexes are highly asymmetric, with one of the two half sites bound with higher affinity [Figure 2; (43)]. Similarly, cleavage and strand exchange are also asymmetric in these two systems. XerS preferentially exchanged the BSs of difSL on HJ-substrates, which, assuming XerS cleaves DNA in cis, would be cleaved by the monomer bound with weaker affinity to the right-half site (Figure 3). Similarly, XerC displays less efficient binding but more efficient strand exchange than XerD, with dif-containing HJ substrates (35,44). The control of recombination by FtsK is also similar in the two systems. Both Xer systems require FtsKC translocation activity for efficient recombination, and a physical tethering of FtsKC to the division septum for the resolution of chromosome dimers [Figures 4 and 5; (10,38,41)]. The XerCD/dif system also appears to be faithfully controlled by the L. lactis FtsKC and the XerS/difSL system is controlled by E. coli FtsK in assays in vivo in E. coli [Figure 6; (28)]. These results unambiguously show that the mode of chromosome dimer resolution and its coupling with cell division by FtsK described in E. coli can operate in Xer systems including only one recombinase. We therefore cannot account for the selection of two-recombinase systems in most bacteria due to a need for two enzymes for the control of recombination. Importantly, these controls also apply to non rod-shaped bacteria from phyla other than Proteobacteria. These findings conflict with those obtained for Bacillus subtilis, which, like L. lactis, belongs to the Firmicute phylum. Bacillus subtilis encodes a ‘classical’ XerCD/dif chromosome resolution system, the activity of which has been reported to be independent of both the FtsK homologues present in this bacterium, SpoIIIE and SftA (formerly YtpT) (45). However, it has recently been reported that the inactivation of SftA results in a high frequency of septum closure over the nucleoids and that this effect depends on RecA (24). This suggests that SftA may control chromosome dimer resolution in B. subtilis, although it may not directly control XerCD/dif recombination.
A key conserved feature of the XerCD/dif and XerS/difSL systems is that efficient recombination, in addition to efficient dimer resolution, requires an activation step dependent on FtsKC. Efficient XerCD/dif recombination has been shown to occur at a time corresponding to septation—long after the replication of dif—and to depend on the activities of FtsZ and FtsI (46,47). Thus, the observed dependence on FtsKC certainly ensures that recombination occurs only at the very end of the segregation process, when FtsK reaches the zone of converging KOPS containing the dif site, thereby preventing undesirable recombination events creating dimers from monomers or producing other harmful rearrangements. This notion is supported by the data obtained with strains in which chromosome dimers are resolved by the Cre/loxP system (48). In such strains, recombination between loxP sites inserted in place of dif does not depend on FtsK, but the resolution of dimers requires FtsKC activity (38). The inactivation of FtsKC leads to a viability defect significantly greater than that observed in strains carrying the XerCD/dif or XerS/difSL system [(32), compare with Figure 4B]. This strongly suggests that Cre/loxP recombination readily creates dimers in the absence of FtsKC activity. We thus assume that dependence on FtsKC prevents harmful recombination events, including dimer formation.
The requirement for an activation step in XerCD/dif and XerS/difSL recombination indicates that the recombination synaptic complexes do not assemble efficiently in the absence of FtsKC or that they do not give rise to productive recombination. A feature clearly linked to this control is the asymmetry of the recombination complexes. In Y-recombinase systems, asymmetry is required for the correct assembly of the synaptic complexes [reviewed in (5)]. The use of two different recombinases in the XerCD/dif system clearly adds an additional level of asymmetry. This certainly contributes to the weak activity of the XerCD/dif complex in the absence of FtsKC (Figure 7). In this view, the conformation that activates XerD catalysis is highly improbable at both the synapsis and HJ intermediate steps, so productive recombination occurs only when imposed by FtsK. A similar control mechanism seems to operate in the XerS/difSL system, although may be less stringent, hence the higher recombination activity in the absence of FtsKC of the XerS/difSL compared to the XerCD/dif system. Notably, this difference is not due to different levels of recombinase production (Figure 7C). Thus, the use of two recombinases, incorporating an additional level of asymmetry, may allow tighter control of recombination than is possible with a single recombinase. The control exerted over recombination events may also involve a differencial control of individual strand exchanges. In the XerCD/dif system, the exchange of a first pair of strands can occur in the absence of FtsKC, producing the HJ intermediate in vitro and in vivo (10). It may be essential to keep XerCD/dif complexes active for XerC-mediated strand exchange, for the integration of foreign DNA by XerCD recombination. This is the case during integration of the Vibrio cholerae CTX prophage, in which XerC catalyses the formation of an integration intermediate that is not further processed by XerD-mediated catalysis (9). This mechanism may be general to proteobacteria, in which an increasing number of mobile elements have been shown to integrate into the host dif site (49–51). Tight control of the exchange of the two pairs of strands may not be possible in single-recombinase Xer systems. Two recombinase systems may therefore have been selected due to their role in horizontal gene transfer. Consistent with this idea, no mobile genetic element integrated into the streptococcal or lactococcal difSL sites has been reported to date.
The mechanism by which FtsK activates recombination remains unclear. Our data show that FtsKγ is sufficient to induce XerCD/dif recombination independently of FtsKC (Figure 7). In these conditions, FtsKγ interacts with XerD and induces the XerD-mediated exchange of either the first or the second pair of strands. Whatever the mechanism involved in these recombination events, it contrasts with the lack of recombination observed with the ftsKATP- allele (39), which encodes an intact FtsKγ subdomain and may thus induce recombination in the absence of translocation. In addition, an FtsKCATP- mutant protein produced in conditions equivalent to those used here for FtsKγ production did not induce XerCD/dif recombination (10). These data suggest that, when tethered to FtsKαβ, FtsKγ cannot induce recombination in the absence of translocation. FtsKγ may thus be inactivated by an interaction with FtsKαβ, which is released during translocation, or when FtsK reaches the XerCD/dif complex. Lactococcus lactis FtsKγ cannot induce recombination by itself, even through the XerS/difSL system. There is therefore no interaction equivalent to the E. coli FtsKγ-XerD interaction in the L. lactis system (Figure 7). Nevertheless, the FtsKCLl chimera induces high levels of recombination by both systems, strongly suggesting that the specific FtsKγ-XerD interaction is also dispensable for the induction of XerCD/dif recombination (Figure 6). This finding contrasts with a previous report that an FtsK chimera carrying the C-terminal domain of Haemophilus influenzae FtsK is deficient in XerD activation, resulting in a large deficit in XerCD/dif recombination in E. coli (39). However, further analysis of this FtsK allele suggests that this chimeric protein is also deficient for the recognition of KOPS motifs, which certainly accounts for the recombination deficiency observed in strains carrying this chimera (C. Pages and F. Cornet, unpublished data).
So, what is the likely mechanism for recombination induction common to E. coli and L. lactis? The observation that the insertion of non-permissive KOPS motifs next to a difSL site strongly inhibits XerS/difSL recombination strongly suggests that FtsK must come into close contact with the XerS/difSL complex to activate recombination (Figure 6). We thus hypothesize that recombination induction involves a special activity of the FstKαβ translocation motor on reaching the recombination complex (18). FtsK stops translocating at this point. This cessation of translocation is not dependent on FtsKγ and is not highly sequence-specific, since translocation also stops with Cre/loxP and XerCD/dif complexes (18). It is accompanied by a particular interaction with bases immediately flanking the dif site and, thus, also those flanking the difSL site (19). This suggests a special action of FtsK on the DNA at this step, which may be the major factor indicating an active conformation to the XerCD/dif or XerS/difSL complexes. The specific interaction of FtsKγ with XerD would then be an additional but dispensable control, potentially helping to stablize the active conformation and/or inducing XerD catalysis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Cnetre National de la Recherche Scientifique; University Paul Sabatier; Fondation pour la Recherche Medicale (FRM, to S.N.); Agence Nationale de la Recherche (ANR; contract BLAN06-2 134012). Funding for open access charge: ANR (contract BLAN06-2 134012).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jean-Yves Bouet for technical assistance and critical reading of the article and the members of the Cornet and Ritzenthaler groups for assistance and helpful discussions.
REFERENCES
- 1.Lesterlin C, Barre F, Cornet F. Genetic recombination and the cell cycle: what we have learned from chromosome dimers. Mol. Microbiol. 2004;54:1151–1160. doi: 10.1111/j.1365-2958.2004.04356.x. [DOI] [PubMed] [Google Scholar]
- 2.Sherratt D. Bacterial chromosome dynamics. Science. 2003;301:780–785. doi: 10.1126/science.1084780. [DOI] [PubMed] [Google Scholar]
- 3.Bigot S, Sivanathan V, Possoz C, Barre F, Cornet F. FtsK, a literate chromosome segregation machine. Mol. Microbiol. 2007;64:1434–1441. doi: 10.1111/j.1365-2958.2007.05755.x. [DOI] [PubMed] [Google Scholar]
- 4.Sherratt D, Søballe B, Barre F, Filipe S, Lau I, Massey T, Yates J. Recombination and chromosome segregation. Philos. Trans Roy. Soc. Lond. B Biol. Sci. 2004;359:61–69. doi: 10.1098/rstb.2003.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grindley N, Whiteson K, Rice P. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 6.Bregu M, Sherratt D, Colloms S. Accessory factors determine the order of strand exchange in Xer recombination at psi. EMBO J. 2002;21:3888–3897. doi: 10.1093/emboj/cdf379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aussel L, Barre F, Aroyo M, Stasiak A, Stasiak A, Sherratt D. FtsK Is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell. 2002;108:195–205. doi: 10.1016/s0092-8674(02)00624-4. [DOI] [PubMed] [Google Scholar]
- 8.Colloms S, McCulloch R, Grant K, Neilson L, Sherratt D. Xer-mediated site-specific recombination in vitro. EMBO J. 1996;15:1172–1181. [PMC free article] [PubMed] [Google Scholar]
- 9.Val M, Bouvier M, Campos J, Sherratt D, Cornet F, Mazel D, Barre F. The single-stranded genome of phage CTX is the form used for integration into the genome of Vibrio cholerae. Mol. Cell. 2005;19:559–566. doi: 10.1016/j.molcel.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Barre F, Aroyo M, Colloms S, Helfrich A, Cornet F, Sherratt D. FtsK functions in the processing of a Holliday junction intermediate during bacterial chromosome segregation. Genes Dev. 2000;14:2976–2988. doi: 10.1101/gad.188700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Massey T, Mercogliano C, Yates J, Sherratt D, Löwe J. Double-stranded DNA translocation: structure and mechanism of hexameric FtsK. Mol. Cell. 2006;23:457–469. doi: 10.1016/j.molcel.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 12.Bigot S, Saleh O, Lesterlin C, Pages C, El Karoui M, Dennis C, Grigoriev M, Allemand J, Barre F, Cornet F. KOPS: DNA motifs that control E. coli chromosome segregation by orienting the FtsK translocase. EMBO J. 2005;24:3770–3780. doi: 10.1038/sj.emboj.7600835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levy O, Ptacin J, Pease P, Gore J, Eisen M, Bustamante C, Cozzarelli N. Identification of oligonucleotide sequences that direct the movement of the Escherichia coli FtsK translocase. Proc. Natl Acad. Sci. USA. 2005;102:17618–17623. doi: 10.1073/pnas.0508932102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bigot S, Saleh O, Cornet F, Allemand J, Barre F. Oriented loading of FtsK on KOPS. Nat. Struct. Mol. Biol. 2006;13:1026–1028. doi: 10.1038/nsmb1159. [DOI] [PubMed] [Google Scholar]
- 15.Ptacin J, Nöllmann M, Bustamante C, Cozzarelli N. Identification of the FtsK sequence-recognition domain. Nat. Struct. Mol. Biol. 2006;13:1023–1025. doi: 10.1038/nsmb1157. [DOI] [PubMed] [Google Scholar]
- 16.Sivanathan V, Allen M, de Bekker C, Baker R, Arciszewska L, Freund S, Bycroft M, Löwe J, Sherratt D. The FtsK gamma domain directs oriented DNA translocation by interacting with KOPS. Nat. Struct. Mol. Biol. 2006;13:965–972. doi: 10.1038/nsmb1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Löwe J, Ellonen A, Allen M, Atkinson C, Sherratt D, Grainge I. Molecular mechanism of sequence-directed DNA loading and translocation by FtsK. Mol. Cell. 2008;31:498–509. doi: 10.1016/j.molcel.2008.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graham J, Sivanathan V, Sherratt D, Arciszewska L. FtsK translocation on DNA stops at XerCD-dif. Nucleic Acids Res. 2010;38:72–81. doi: 10.1093/nar/gkp843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonné L, Bigot S, Chevalier F, Allemand J, Barre F. Asymmetric DNA requirements in Xer recombination activation by FtsK. Nucleic Acids Res. 2009;37:2371–2380. doi: 10.1093/nar/gkp104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yates J, Zhekov I, Baker R, Eklund B, Sherratt D, Arciszewska L. Dissection of a functional interaction between the DNA translocase, FtsK, and the XerD recombinase. Mol. Microbiol. 2006;59:1754–1766. doi: 10.1111/j.1365-2958.2005.05033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Possoz C, Ribard C, Gagnat J, Pernodet J, Guérineau M. The integrative element pSAM2 from Streptomyces: kinetics and mode of conjugal transfer. Mol. Microbiol. 2001;42:159–166. doi: 10.1046/j.1365-2958.2001.02618.x. [DOI] [PubMed] [Google Scholar]
- 22.Wu L, Errington J. Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science. 1994;264:572–575. doi: 10.1126/science.8160014. [DOI] [PubMed] [Google Scholar]
- 23.Val M, Kennedy S, El Karoui M, Bonné L, Chevalier F, Barre F. FtsK-dependent dimer resolution on multiple chromosomes in the pathogen Vibrio cholerae. PLoS Genet. 2008;4:e1000201. doi: 10.1371/journal.pgen.1000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biller S, Burkholder W. The Bacillus subtilis SftA (YtpS) and SpoIIIE DNA translocases play distinct roles in growing cells to ensure faithful chromosome partitioning. Mol. Microbiol. 2009;74:790–809. doi: 10.1111/j.1365-2958.2009.06893.x. [DOI] [PubMed] [Google Scholar]
- 25.Kaimer C, González-Pastor J, Graumann P. SpoIIIE and a novel type of DNA translocase, SftA, couple chromosome segregation with cell division in Bacillus subtilis. Mol. Microbiol. 2009;74:810–825. doi: 10.1111/j.1365-2958.2009.06894.x. [DOI] [PubMed] [Google Scholar]
- 26.Hallet B, Sherratt D. Transposition and site-specific recombination: adapting DNA cut-and-paste mechanisms to a variety of genetic rearrangements. FEMS Microbiol. Rev. 1997;21:157–178. doi: 10.1111/j.1574-6976.1997.tb00349.x. [DOI] [PubMed] [Google Scholar]
- 27.Recchia G, Sherratt D. Conservation of xer site-specific recombination genes in bacteria. Mol. Microbiol. 1999;34:1146–1148. doi: 10.1046/j.1365-2958.1999.01668.x. [DOI] [PubMed] [Google Scholar]
- 28.Le Bourgeois P, Bugarel M, Campo N, Daveran-Mingot M, Labonté J, Lanfranchi D, Lautier T, Pagès C, Ritzenthaler P. The unconventional Xer recombination machinery of Streptococci/Lactococci. PLoS Genet. 2007;3:e117. doi: 10.1371/journal.pgen.0030117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carnoy C, Roten C. The dif/Xer recombination systems in proteobacteria. PLoS ONE. 2009;4:e6531. doi: 10.1371/journal.pone.0006531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cornet F, Mortier I, Patte J, Louarn J. Plasmid pSC101 harbors a recombination site, psi, which is able to resolve plasmid multimers and to substitute for the analogous chromosomal Escherichia coli site dif. J. Bacteriol. 1994;176:3188–3195. doi: 10.1128/jb.176.11.3188-3195.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cornet F, Louarn J, Patte J, Louarn J. Restriction of the activity of the recombination site dif to a small zone of the Escherichia coli chromosome. Genes Dev. 1996;10:1152–1161. doi: 10.1101/gad.10.9.1152. [DOI] [PubMed] [Google Scholar]
- 32.Sivanathan V, Emerson J, Pages C, Cornet F, Sherratt D, Arciszewska L. KOPS-guided DNA translocation by FtsK safeguards Escherichia coli chromosome segregation. Mol. Microbiol. 2009;71:1031–1042. doi: 10.1111/j.1365-2958.2008.06586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maguin E, Prévost H, Ehrlich S, Gruss A. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 1996;178:931–935. doi: 10.1128/jb.178.3.931-935.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pérals K, Cornet F, Merlet Y, Delon I, Louarn J. Functional polarization of the Escherichia coli chromosome terminus: the dif site acts in chromosome dimer resolution only when located between long stretches of opposite polarity. Mol. Microbiol. 2000;36:33–43. doi: 10.1046/j.1365-2958.2000.01847.x. [DOI] [PubMed] [Google Scholar]
- 35.Arciszewska L, Sherratt D. Xer site-specific recombination in vitro. EMBO J. 1995;14:2112–2120. doi: 10.1002/j.1460-2075.1995.tb07203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Massy B, Dorgai L, Weisberg R. Mutations of the phage lambda attachment site alter the directionality of resolution of Holliday structures. EMBO J. 1989;8:1591–1599. doi: 10.1002/j.1460-2075.1989.tb03543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee J, Jayaram M. Junction mobility and resolution of Holliday structures by Flp site-specific recombinase. Testing partner compatibility during recombination. J. Biol. Chem. 1995;270:19086–19092. doi: 10.1074/jbc.270.32.19086. [DOI] [PubMed] [Google Scholar]
- 38.Capiaux H, Lesterlin C, Pérals K, Louarn J, Cornet F. A dual role for the FtsK protein in Escherichia coli chromosome segregation. EMBO Rep. 2002;3:532–536. doi: 10.1093/embo-reports/kvf116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bigot S, Corre J, Louarn J, Cornet F, Barre F. FtsK activities in Xer recombination, DNA mobilization and cell division involve overlapping and separate domains of the protein. Mol. Microbiol. 2004;54:876–886. doi: 10.1111/j.1365-2958.2004.04335.x. [DOI] [PubMed] [Google Scholar]
- 40.Dubarry N, Barre F. Fully efficient chromosome dimer resolution in Escherichia coli cells lacking the integral membrane domain of FtsK. EMBO J. 2010;29:597–605. doi: 10.1038/emboj.2009.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pérals K, Capiaux H, Vincourt J, Louarn J, Sherratt D, Cornet F. Interplay between recombination, cell division and chromosome structure during chromosome dimer resolution in Escherichia coli. Mol. Microbiol. 2001;39:904–913. doi: 10.1046/j.1365-2958.2001.02277.x. [DOI] [PubMed] [Google Scholar]
- 42.Yates J, Aroyo M, Sherratt D, Barre F. Species specificity in the activation of Xer recombination at dif by FtsK. Mol. Microbiol. 2003;49:241–249. doi: 10.1046/j.1365-2958.2003.03574.x. [DOI] [PubMed] [Google Scholar]
- 43.Blakely G, May G, McCulloch R, Arciszewska L, Burke M, Lovett S, Sherratt D. Two related recombinases are required for site-specific recombination at dif and cer in E. coli K12. Cell. 1993;75:351–361. doi: 10.1016/0092-8674(93)80076-q. [DOI] [PubMed] [Google Scholar]
- 44.Arciszewska L, Grainge I, Sherratt D. Effects of Holliday junction position on Xer-mediated recombination in vitro. EMBO J. 1995;14:2651–2660. doi: 10.1002/j.1460-2075.1995.tb07263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sciochetti S, Piggot P, Blakely G. Identification and characterization of the dif Site from Bacillus subtilis. J. Bacteriol. 2001;183:1058–1068. doi: 10.1128/JB.183.3.1058-1068.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steiner W, Kuempel P. Cell division is required for resolution of dimer chromosomes at the dif locus of Escherichia coli. Mol. Microbiol. 1998;27:257–268. doi: 10.1046/j.1365-2958.1998.00651.x. [DOI] [PubMed] [Google Scholar]
- 47.Kennedy S, Chevalier F, Barre F. Delayed activation of Xer recombination at dif by FtsK during septum assembly in Escherichia coli. Mol. Microbiol. 2008;68:1018–1028. doi: 10.1111/j.1365-2958.2008.06212.x. [DOI] [PubMed] [Google Scholar]
- 48.Leslie N, Sherratt D. Site-specific recombination in the replication terminus region of Escherichia coli: functional replacement of dif. EMBO J. 1995;14:1561–1570. doi: 10.1002/j.1460-2075.1995.tb07142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin N, Chang R, Lee S, Tseng Y. Plasmids carrying cloned fragments of RF DNA from the filamentous phage (phi)Lf can be integrated into the host chromosome via site-specific integration and homologous recombination. Mol. Genet. Genomics. 2001;266:425–435. doi: 10.1007/s004380100532. [DOI] [PubMed] [Google Scholar]
- 50.Gonzalez M, Lichtensteiger C, Caughlan R, Vimr E. Conserved filamentous prophage in Escherichia coli O18:K1:H7 and Yersinia pestis biovar orientalis. J. Bacteriol. 2002;184:6050–6055. doi: 10.1128/JB.184.21.6050-6055.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hamilton H, Domínguez N, Schwartz K, Hackett K, Dillard J. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol. Microbiol. 2005;55:1704–1721. doi: 10.1111/j.1365-2958.2005.04521.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.