Abstract
The DNA-binding protein RdgC has been identified as an inhibitor of RecA-mediated homologous recombination in Escherichia coli. In Neisseria species, RdgC also has a role in virulence-associated antigenic variation. We have previously solved the crystal structure of the E. coli RdgC protein and shown it to form a toroidal dimer. In this study, we have conducted a mutational analysis of residues proposed to mediate interactions at the dimer interfaces. We demonstrate that destabilizing either interface has a serious effect on in vivo function, even though a stable complex with circular DNA was still observed. We conclude that tight binding is required for inhibition of RecA activity. We also investigated the role of the RdgC finger domain, and demonstrate that it plays a crucial role in the binding of circular DNA. Together, these data allow us to propose a model for how RdgC loads onto DNA. We discuss how RdgC might inhibit RecA-mediated strand exchange, and how RdgC might be displaced by other DNA metabolism enzymes such as polymerases and helicases.
INTRODUCTION
The rdgC gene encodes a 34-kDa nucleoid-associated protein (1), which was originally identified as a factor essential for growth in recombination-deficient mutants of Escherichia coli (2). More recent studies show it to be required for viability in strains lacking the primosome assembly protein, PriA, and to counter RecFOR-mediated loading of RecA (3). Biochemical studies revealed that RdgC binds non-specifically to linear and circular DNA (3,4), and inhibits RecA-mediated strand exchange reactions in vitro (5). Taken together, these data are consistent with the proposed cellular function of RdgC as a modulator of RecA activity.
RecA is essential for homologous recombination in prokaryotes, which provides a generally error-free pathway for the repair of DNA double-strand breaks and for the rescue of damaged replication forks. RecA, and the archaeal and eukaryotic homologues RadA and Rad51 respectively, catalyse the key process of homologous DNA strand exchange (6,7). RecA assembles on ssDNA to form a helical filament which repeatedly binds non-specifically to dsDNA, searching for a region of homology. Once a homologous region is found, RecA promotes switching of base pairs between the unwound duplex and the RecA-coated ssDNA. The resulting heteroduplex intermediates can then be processed by a variety of pathways resulting in a repaired break or a rescued replication fork.
Homologous recombination also promotes the biological fitness of a species by increasing population genetic variation (8). For example, Neisseria gonorrhoeae (9) and Neisseria meningitidis (10) use a RecA-mediated recombination process to alter the sequence of the main structural component of the type IV pili. It is thought that the high rate of pilin antigenic variation contributes to evasion of the host immune response, explaining the failure of pilus-based vaccine trials (11). Interestingly, RdgC has been shown to promote recombination-dependent antigenic variation in Neisseria sp. (12). RecA-mediated homologous recombination is also thought to be involved in antigenic variation and virulence in other prokaryotic pathogens, such as Helicobacter pylori (13), Campylobacter fetus (14), Pseudomonas tolaasii (15) and Haemophilus influenzae (16), while Rad51-mediated homologous recombination is thought to play an equivalent role in the eukaryotic parasite, Trypanosoma brucei (17).
Since recombination plays such an important part in DNA repair and genome maintenance, it is essential to control where, when and how recombination occurs. When unregulated, recombination can lead to genome instability and carcinogenesis. The RecA family of recombinases provide an obvious target for regulation (18,19). In E. coli, RecA function is regulated at many levels (18). Expression from the recA gene is controlled within the SOS response, which provides the first level of regulation. RecA itself provides another level of control, as the C-terminal region has been shown to have an autoregulatory function. Additionally, many other proteins have been shown to modulate RecA activity. The SSB protein competes with RecA for ssDNA, and can block RecA filament formation if it binds to ssDNA before the addition of RecA. However, SSB can also remove secondary structure from ssDNA, so promoting the formation of the RecA filament. The RecF, RecO and RecR proteins have been shown to provoke both the assembly and disassembly of RecA filaments, and to be necessary for the loading of RecA onto SSB-coated DNA. RecBCD actively loads RecA onto the regions of ssDNA it creates as it resects a DNA end. The DinI and RecX proteins are antagonists, acting to either stabilize or destabilize RecA filaments, respectively. The PsiB protein inhibits nucleofilament assembly by binding free RecA, blocking the site necessary for SSB displacement as well as reducing DNA-binding affinity (20). RdgC is another protein that modulates homologous recombination, acting as a negative regulator of RecA activity.
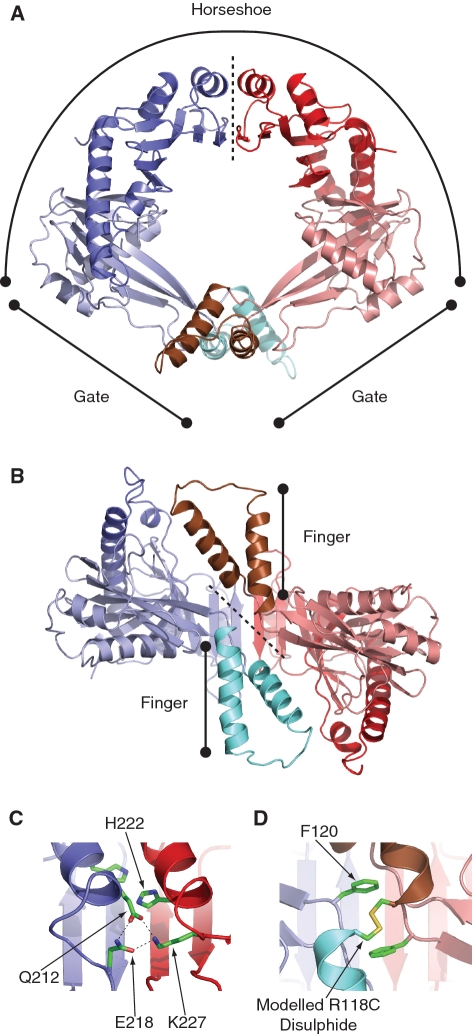
In order to better understand how RdgC could inhibit RecA activity, we determined the crystal structure of RdgC from E. coli (21). This structure shows RdgC to form a dimeric ring or toroid, with a head-to-head, tail-to-tail organization (Figure 1A). Electropositive ‘finger’ domains project on either side of the ring face (Figure 1B). The 30 Å diameter hole at the centre of the ring is lined with positively charged residues and provides the likely site for DNA binding. A similar structure was seen for RdgC from Pseudomonas aeruginosa (22). Although advantageous for a protein that binds to and/or moves along DNA (23), a toroidal shape poses a problem when binding circular DNA, as this requires the ring to open.
Figure 1.
Structure of RdgC. (PDB co-ordinates 2OWL). (A) A ribbon diagram showing the ring structure of RdgC and identifying the horseshoe and gate regions. The dotted line indicates the horseshoe domain interface. (B) A rotated ribbon diagram illustrating projection of the finger domains. The dotted line indicates the gate domain interface. (C) Detail of one of the hydrogen bond networks at the horseshoe interface between the Q212 and E218 residues on one dimer and K227 on the other. The conserved H222 residues are also shown. (D) Detail of the gate interface interactions, showing the conserved F120 residues and the modelled disulphide formed in the R118C derivative.
Other toroidal proteins, e.g. the β-clamp (24) and PCNA (25) sliding clamp processivity factors and the DnaB (26) and BLM (27) hexameric helicases, achieve ring-opening with the assistance of accessory proteins. The sliding clamps are assembled on DNA by clamp loader proteins in an ATP-dependent reaction (28), while some hexameric helicases have accessory proteins necessary for efficient DNA binding. In E. coli, the DnaC protein facilitates assembly of the replicative helicase, DnaB, on DNA (29). Likewise, the Cdc6 protein is implicated in the loading of the eukaryotic replicative MCM helicase complex (30). However, accessory proteins are not required in all cases; the T7 gp4A’ replicative helicase has an additional N-terminal primase domain which provides the DNA loading activity (31). Our studies, reported here, confirm that RdgC can also load onto circular DNA without the requirement for accessory proteins, and we provide evidence that the ‘finger’ domains provide the required DNA-loading activity. We propose a model explaining how the finger domain opens the RdgC ring, allowing it to load onto circular DNA. We also identify residues at the domain interfaces that are necessary for forming a stable RdgC-DNA complex in vitro, and demonstrate that the in vivo function of RdgC is dependent on the ability to form a stable tight-binding protein–DNA complex. This provides an insight into the mechanism of how RdgC could modulate RecA-mediated recombination.
MATERIALS AND METHODS
Bacterial strains and plasmids
Mutant rdgC genes encoding RdgC with specific substitutions and deletions were made by site-directed mutagenesis using either the Quikchange (Stratagene) or the Phusion (Finnzymes) mutagenesis procedures. All plasmid constructs (Table 1) were based on the pT7-7 derivative containing the rdgC gene, pGS583 (3) or the pET22b derivative, pYJ001, containing the NdeI–HindIII rdgC gene fragment from pGS583. All mutations were confirmed by DNA sequencing.
Table 1.
Plasmid constructs used
| Amino acid Substitution | pT7-7 derivative | pET22b derivative | pET22b-cat derivative |
|---|---|---|---|
| Wild-type | pGS583 | pYJ001 | pAM447 |
| R118A | pGB049 | pGB050 | pAM450 |
| R118C | pGB045 | pGB047 | pAM441 |
| F120S | pGB051 | pGB052 | pAM465 |
| F120T | pGB053 | pGB054 | pAM466 |
| R118C, F120T | pGB065 | pYJ019 | |
| Q212A | pYJ017 | pYJ020 | |
| E218R | pYJ013 | pAM448 | |
| H222A | pYJ003 | pYJ006 | pAM443 |
| K227A | pYJ007 | pAM444 | |
| F120T, K227A | pGB084 | ||
| Δfinger (P76G, Δ77–115, L116T) | pGB043 | pAM434 | pAM437 |
| Δfinger, R118C | pGB046 | pGB048 | pAM442 |
| Fingertip (R97S, K98Q, K100Q, K101E) | pGB055 | pGB056 | pAM468 |
All strains used for the synthetic lethality assay (Table 2) are derivatives of E. coli MG1655 ΔlacIZYA, TB28 (32) carrying a priA+ derivative of pRC7, pAM374 (33). A cat expression cassette was placed immediately downstream of either native or mutant rdgC in the pET22b-derivative expression plasmid. The rdgC-cat cassette was used to replace the native rdgC from the start to the stop codon in TB28 by chromosome engineering (34) and confirmed by sequencing. These mutations were combined with either ΔpriA or ΔpriA dnaC810 by P1 transduction to the appropriate recipient (35). Synthetic lethality assays were performed as described previously (33), using either LB media (LB) or 56/2 minimal media (MA) (35). The MG1655 ΔrecA strain, N7358, was constructed by replacing the recA gene (deleting residues G16 to S330) with a spectinomycin resistance cassette by chromosome engineering.
Table 2.
Escherichia coli strains used
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| N3072 | W3110 recA269::Tn10 | (2) |
| DIM167 | DM4000 priA2::kan dnaC810 zji-202::Tn10 | (3) |
| YJ014 | BL21 (DE3) plysS ΔrdgC::dhfr | This work |
| MG1655 derivatives | ||
| MG1655 | (44) | |
| PN105 | priA2::kan sulA | (45) |
| N5521 | priA300 dnaC810 zji-202::Tn10 | P1·DIM167 × N5500 to Tcr |
| N5539 | priA2::kan dnaC810 zji-202::Tn10 | P1·PN105 × N5521 to Kmr |
| N4586 | ΔrdgC::dhfr | (3) |
| N7358 | ΔrecA::spc | This work |
| TB28 | ΔlacIZYA | (32) |
| TB28 derivatives | ||
| AM1833 | rdgCR118C-cat | This work |
| AM1834 | rdgCP76G,Δ(77-115),L116T-cat | This work |
| AM1835 | rdgCP76G,Δ(77-115),L116T,R118C-cat | This work |
| AM1836 | rdgCH222A-cat | This work |
| AM1837 | rdgCK227A-cat | This work |
| AM1838 | rdgCR118A-cat | This work |
| AM1840 | rdgCE218R-cat | This work |
| AM1887 | rdgCwt-cat | This work |
| AM1905 | rdgCF120T-cat | This work |
| AM1917 | rdgCF120S-cat | This work |
| AM1929 | ΔrdgC2::dhfr | This work |
| AM1950 | rdgCR97S,K98Q,K100Q,K101E-cat | This work |
| AM2029 | rdgCQ212A-cat | This work |
| YJ025 | rdgCR118C,F120T-cat | This work |
| TB28 derivatives containing pAM374 (pRC7-priA+) | ||
| N5936 | (33) | |
| N5972 | ΔpriA::apra | (33) |
| N6021 | ΔpriA::apra dnaC810 zji-202::Tn10 | P1·N5539 × N5972 to Tcr |
| N6038 | ΔrdgC::dhfr | P1·N4586 × N5936 to Tmr |
| N6040 | ΔpriA::apra ΔrdgC::dhfr | P1·N4586 × N5972 to Tmr |
| N6111 | ΔpriA::apra dnaC810 zji-202::Tn10 ΔrdgC::dhfr | P1·N4586 × N6021 to Tmr |
| N7701 | ΔpriA::apra dnaC810 zji-202::Tn10 ΔrdgC::dhfr ΔrecA::spc | P1·N7358 × N6111 to Spr |
| N7709 | ΔpriA::apra ΔrdgC::dhfr recA269::Tn10 | P1·N3072 × N6040 to Tcr |
| AM1853 | ΔpriA::apra dnaC810 zji-202::Tn10 rdgCR118C-cat | P1·AM1833 × N6021 to Cmr |
| AM1854 | ΔpriA::apra dnaC810 zji-202::Tn10 | P1·AM1834 × N6021 to Cmr |
| rdgCP76G,Δ(77-115),L116T-cat | ||
| AM1855 | ΔpriA::apra dnaC810 zji-202::Tn10 | P1·AM1835 × N6021 to Cmr |
| rdgCP76G,Δ(77-115),L116T,R118C-cat | ||
| AM1856 | ΔpriA::apra dnaC810 zji-202::Tn10 rdgCH222A-cat | P1·AM1836 × N6021 to Cmr |
| AM1857 | ΔpriA::apra dnaC810 zji-202::Tn10 rdgCK227A-cat | P1·AM1837 × N6021 to Cmr |
| AM1858 | ΔpriA::apra dnaC810 zji-202::Tn10 rdgCR118A-cat | P1·AM1838 × N6021 to Cmr |
| AM1860 | ΔpriA::apra dnaC810 zji-202::Tn10 rdgCE218R-cat | P1·AM1840 × N6021 to Cmr |
| AM1911 | ΔpriA::apra rdgCF120T-cat | P1·AM1905 × N5972 to Cmr |
| AM1912 | ΔpriA::apra dnaC810 zji-202::Tn10 rdgCF120T-cat | P1·AM1905 × N6021 to Cmr |
| AM1918 | ΔpriA::apra ΔrdgC2::dhfr | P1·AM1929 × N5972 to Tmr |
| AM1919 | ΔpriA::apra dnaC810 zji-202::Tn10 ΔrdgC2::dhfr | P1·AM1929 × N6021 to Tmr |
| AM1948 | ΔpriA::apra rdgCR97S,K98Q,K100Q,K101E-cat | P1·AM1950 × AM1918 to Cmr |
| AM1949 | ΔpriA::apra dnaC810 zji-202::Tn10 rdgCR97S,K98Q,K100Q,K101E-cat | P1·AM1950 × AM1919 to Cmr |
| AM1963 | ΔpriA::apra rdgCF120S-cat | P1·AM1917 × AM1918 to Cmr |
| AM1964 | ΔpriA::apra dnaC810 zji-202::Tn10 rdgCF120S-cat | P1·AM1917 × AM1919 to Cmr |
| YJ041 | ΔpriA::apra rdgCQ212A-cat | P1·AM2029 × AM1918 to Cmr |
| YJ043 | ΔpriA::apra rdgCR118C,F120T-cat | P1·YJ025 × AM1918 to Cmr |
| YJ044 | ΔpriA::apra dnaC810 zji-202::Tn10 rdgCQ212A-cat | P1·AM2029 × AM1919 to Cmr |
| YJ046 | ΔpriA::apra dnaC810 zji-202::Tn10 rdgCR118C,F120T-cat | P1·YJ025 × AM1919 to Cmr |
| YJ047 | ΔpriA::apra rdgCwt-cat | P1·AM1887 × AM1918 to Cmr |
| YJ048 | ΔpriA::apra dnaC810 zji-202::Tn10 rdgCwt-cat | P1·AM1887 × AM1919 to Cmr |
Expression and purification of RdgC
Native RdgC was expressed from E. coli BL21(DE3) pLysS containing pGS583 as described previously (3). Mutant RdgC proteins were expressed in YJ014, a ΔrdgC::dhfr version of BL21(DE3) pLysS, containing the relevant pT7-7 or pET22b derivative of rdgC, as described for native RdgC. Native and mutant RdgC proteins were purified using an identical protocol as described previously (21). Induced cells from 1 l of culture were resuspended in buffer A (50 mM Tris–HCl, pH 7.5, 1 mM EDTA, 1 mM DTT), lysed by sonication on ice, and cell debris was removed by centrifugation. RdgC was recovered from the supernatant by 40–65% ammonium sulphate precipitation and the pellet was resuspended in buffer A containing 0.2 M NaCl. This was purified by passage through a 5 ml Hitrap Heparin HP column (GE Healthcare), a 5 ml Hitrap Q-sepharose HP column (GE Healthcare) and a HiPrep 16/60 Sephacryl S200 HR gel filtration column (GE Healthcare). Fractions containing pure RdgC were dialysed against buffer A containing 150 mM NaCl and 50% glycerol before storage at −80°C. A Superdex 75 10/300 GL column (GE Healthcare) was used for analytical gel filtration, where a 100 μl sample of 10 μM RdgC (dimer concentration) was eluted in gel filtration buffer (50 mM Tris, pH 7.5, 100 mM NaCl) at 0.5 ml/min. All chromatography steps where performed at 4°C.
Oligonucleotide and plasmid binding assays
The ssDNA substrate was made by labelling the oligonucleotide RGL13 (5′-GAC GCT GCC GAA TTC TGG CTT GCT AGG ACA TCT TTG CCC ACG TTG ACC C-3′) at the 5′-end with [α32P] ATP using T4 polynucleotide kinase (New England Biolabs). Unincorporated ATP was removed using a Biospin P6 spin column (Biorad).The 49-bp duplex substrate, RGL13/17, was made by annealing labelled RGL13 to RGL17 (5′-GGG TCA ACG TGG GCA AAG ATG TCC TAG CAA GCC AGA ATT CGG CAG CGT C-3′). The duplex was purified by gel electrophoresis and eluted into 10 mM Tris–HCl, pH 8.5.
Binding of RdgC to either RGL13 or RGL13/17 was measured using a band-shift assay, as described previously (36). Briefly, RdgC and 32P-labelled substrate DNA were mixed in binding buffer [50 mM Tris–HCl, pH 8.0, 5 mM EDTA, 1 mM DTT, 100 μg/ml BSA, 6% (v/v) glycerol] and incubated on ice for 15 min before electrophoresis at 160 V on a prechilled 4% native polyacrylamide gel in low ionic strength buffer (6.7 mM Tris–HCl, pH 8.0, 3.3 mM sodium acetate, 2 mM EDTA). Binding was quantified by calculating the amount of free (unbound) oligonucleotide using the software package ImageJ (37). Apparent dissociation constants were calculated as described previously (38) using the curve-fitting module in the software package Prism (GraphPad Software). Band-shift assays with plasmid DNA were performed using circular pGEM-7 Zf(+) plasmid prepared using a Qiaprep spin column (Qiagen). Plasmid and protein were mixed and incubated using the same conditions as for oligonucleotide band-shift assays before loading onto a 1% agarose gel. After electrophoresis at 80 V for 90 min, gels were stained with SYBR Green (Molecular Probes) and visualized under UV light.
RESULTS
ΔpriA strains can be used to measure rdgC function
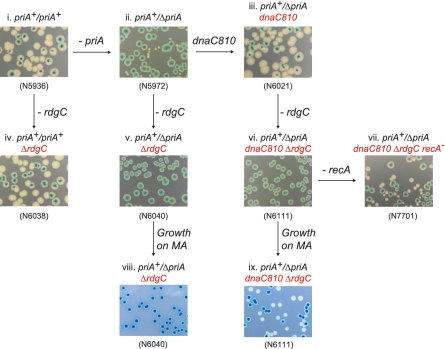
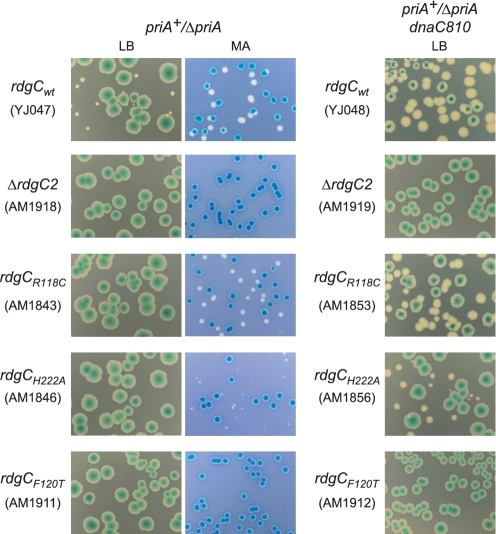
To characterize the effects of mutations on the RdgC protein, we needed an assay for in vivo function. Previous studies have revealed that a ΔpriA ΔrdgC double mutant is inviable under normal growth conditions (3). The poor viability of ΔpriA mutants, and how this can be suppressed by mutations in dnaC such as dnaC810, dnaC212 and dnaC809,820 is well documented (40–42), and the introduction of dnaC212 to a ΔpriA ΔrdgC strain partly recovers viability, resulting in a slow-growth phenotype (3). We established a synthetic lethality assay (32) to study the effect of rdgC mutations in ΔpriA and ΔpriA dnaC810 backgrounds. The assay is based on a pRC7, a lac+ mini-F derivative that is rapidly lost. The loss of the plasmid is revealed in Δlac strains by the appearance of white or sectored colonies on plates containing the β-galactosidase indicator, X-gal. By using a derivative of the plasmid that contains the priA+ gene to cover a chromosomal deletion of priA (priA+/ΔpriA), the viability of cells containing ΔpriA in combination with mutations in dnaC and rdgC can be determined, and so provide a measure of in vivo RdgC activity.
When a ΔpriA (rdgC+) strain of E. coli containing the priA+ derivative of pRC7 is grown on LB agar plates containing X-gal and IPTG, two colony types can be observed: large blue colonies formed by cells retaining the priA+ plasmid and small white colonies formed by cells that have lost the plasmid and therefore are priA− (Figure 2ii). Certain mutations in dnaC suppress this poor-growth phenotype, and a priA+/ΔpriA dnaC810 strain gives large white colonies and sectored blue colonies, demonstrating the suppression of ΔpriA by dnaC810 (Figure 2iii). When ΔrdgC is introduced into a priA+/ΔpriA strain, only blue colonies are seen when grown on LB agar (Figure 2v). Slow growth conditions, such as growth on minimal media, is known to promote viability of ΔpriA mutants (43), but growth of priA+/ΔpriA ΔrdgC on 56/2 minimal agar (MA) yielded only blue colonies (Figure 2viii), demonstrating that priA is essential for viability in a ΔrdgC background. A priA+/ΔpriA dnaC810 ΔrdgC strain also yielded blue colonies on LB agar, with no white colonies (Figure 2vi). However, it segregates white colonies on MA (Figure 2ix), indicating that RdgC is dispensable under these conditions. Viability could also be recovered in a ΔpriA dnaC810 ΔrdgC strain by eliminating recA (Figure 2vii), enabling robust growth on LB agar and providing strong evidence that the in vivo function of RdgC is to modulate RecA activity. The use of the synthetic lethality assay, as described here, provides us with a method for measuring in vivo RdgC functionality.
Figure 2.
The effect of ΔrdgC on the viability of ΔpriA and ΔpriA dnaC810 strains. Photographs of plates illustrating the results of synthetic lethality assays. All constructs contained a priA+ derivative of the pRC7 plasmid, and white colonies indicate the segregation of plasmid-free cells. Cells were plated on either LB agar (rich medium) or MA (56/2 minimal agar medium) as indicated.
The design of mutations to destabilize dimer interfaces
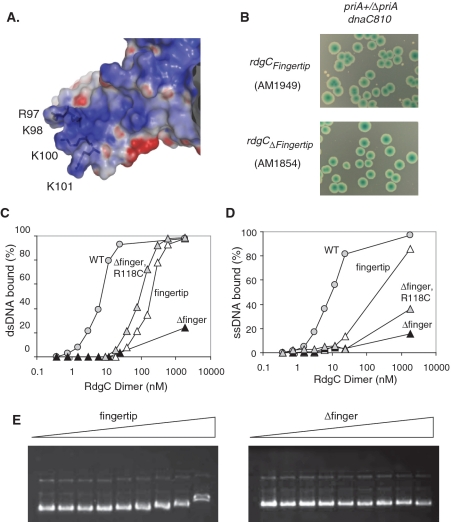
Previous analysis of the RdgC crystal structure identified six residues as targets for mutation studies (21). Conserved residues Q212 and E218 are positioned at one end of the α6-helix, with a conserved K227 residue at the other. When opposing α6-helices come together at the ‘horseshoe’ domain boundary, these residues form two sets of hydrogen bond networks across the horseshoe dimer interface (Figure 1A and C). We attempted to disrupt this network with Q212A, E218R and K227A substitutions. A conserved residue H222 is positioned in the middle of these α6-helices, and may also be important for the stability of the interface. This was investigated by a H222A substitution. The ‘gate’ interface (Figure 1B) appears to be stabilized by hydrogen bonding between opposing R118 residues, and by the interactions between F120 and a hydrophobic pocket on the opposing monomers. We examined the importance of these with R118A, F120S and F120T substitutions. We also looked at the effect of increasing the stability of the gate interface by an R118C substitution, both in isolation and in combination with an F120T mutation, as modelling studies using DSDBASE (39) had suggested that opposing R118C residues could form a disulphide bond across the dimer interface (Figure 1D).
Mutant proteins purify identically to the wild-type protein
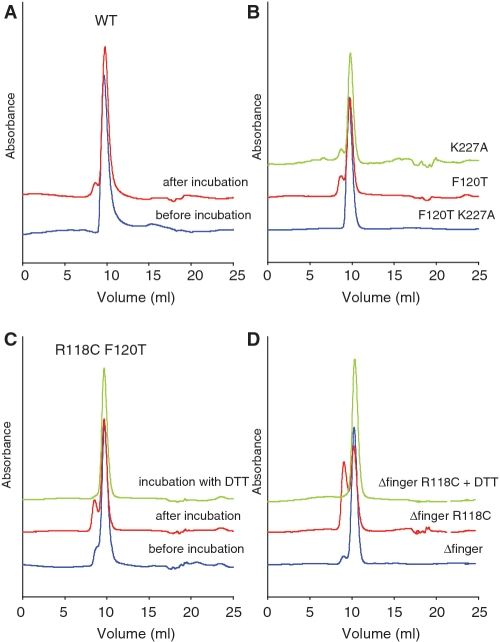
In addition to looking at the in vivo effects of mutations at the dimer interfaces, we also wanted to investigate the in vitro activities of the purified proteins, to compare and correlate with the synthetic lethality data. RdgC binds tightly to small oligonucleotides, with a slight preference for dsDNA over ssDNA (3). We purified each of our mutant RdgC proteins so that we could look at the DNA binding activity. All behaved identically to the wild-type protein throughout the purification procedure, and both the wild-type protein and the mutants eluting as a single peak during gel filtration with an estimated size of ∼60 kDa. This agrees with previous work reporting that RdgC elutes as a dimer by gel filtration (3,5), as the calculated molecular weight of the RdgC monomer is 34 kDa. However, more detailed analysis showed the emergence of a secondary peak or shoulder, corresponding to a larger protein species with an estimated size of ∼120 kDa, after incubation of RdgC samples in gel filtration buffer (50 mM Tris, pH 7.5, 100 mM NaCl) at both room temperature and at 4°C (Figure 3A and B and data not shown). This secondary peak was particularly pronounced in mutants containing the R118C mutation, indicating disulphide-bound RdgC oligomers (Figure 3C). The addition of 5 mM DTT to this disulphide-bound species resulted in a loss of the secondary peak and an equivalent increase in the main peak. These results indicate that, due to its non-compact structure, monomeric RdgC elutes at the position of a 60 kDa protein, whereas the dimer elutes as a 120 kDa protein. These results also suggest that RdgC was monomeric when purified, and the secondary peak is evidence of the protein dimerizing after purification. The purification procedure contains several steps that involve exposure to highly ionic conditions (ammonium sulphate precipitation, elution from heparin and Q-sepharose columns at high NaCl concentration) which could affect the interactions at the dimer interfaces. Gel filtration analysis of pre-incubated RdgC in either 1 M NaCl or after ammonium sulphate precipitation confirmed the loss of the secondary peak consistent with the dimeric protein becoming monomeric (data not shown); suggesting the monomeric state of purified RdgC may be an artefact of the purification procedure. CD analysis demonstrated that the mutant proteins contained similar proportions of secondary structure as the native protein (data not shown). From this, we concluded that all of our mutants had folded correctly and had broadly equivalent structures and characteristics to that of the native protein. Consequently, we could investigate the effect of substitutions at the dimer interfaces using in vitro DNA binding studies.
Figure 3.
Analytical gel filtration analysis of RdgC proteins. (A) Gel filtration of purified WT RdgC protein. Protein was diluted to a final concentration of 10 μM (dimer) in gel filtration buffer and either analysed immediately or incubated overnight at 4°C before analysis on a Superdex 75 10/300 column (GE Healthcare). (B) Gel filtration of purified RdgC proteins. Protein was diluted to a final concentration of 10 μM (dimer) in gel filtration buffer and incubated overnight at 4°C before analysis on a Superdex 75 10/300 column (GE Healthcare). (C) Gel filtration of purified RdgC R118C F120T protein. Protein was diluted to a final concentration of 10 μM (dimer) in gel filtration buffer and either analysed immediately or incubated overnight at 4°C before analysis on a Superdex 75 10/300 column (GE Healthcare). An additional sample was incubated and analysed in gel filtration buffer containing 5 mM DTT. (D) Gel filtration of purified RdgC Δfinger proteins. Proteins were diluted to a final concentration of 10 μM (dimer) in gel filtration buffer and incubated overnight at 4°C before analysis on a Superdex 75 10/300 column (GE Healthcare). An additional sample of Δfinger R118C was incubated and analysed in gel filtration buffer containing 5 mM DTT.
A stable dimer interface is essential for in vivo and in vitro RdgC activity
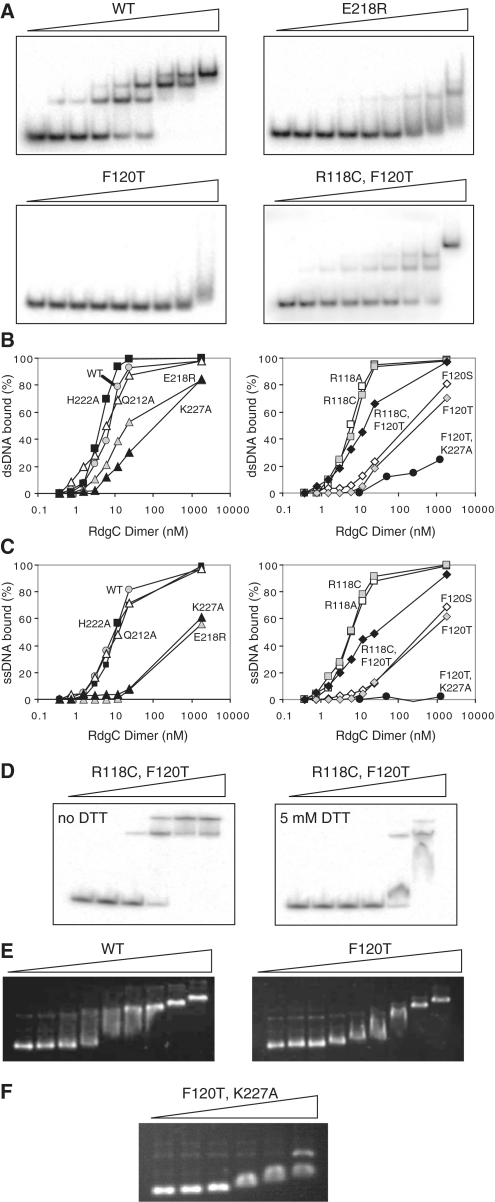
We performed band-shift assays with RdgC and either 49-bp dsDNA oligonucleotide, RGL13/17, (Figure 4A and B) or a 49-nt ssDNA oligonucleotide, RGL13, (Figure 4C). Native RdgC gave multiple bands with the 49-bp dsDNA oligonucleotide at high protein concentrations, suggesting two or more molecules of the RdgC dimer were able to bind to each oligonucleotide. Previous studies have also described the formation of higher-order complexes (3–5), and analysis of the crystal structure suggests that a single dimer of RdgC could interact with ∼15–20 bp (21), so the higher order complexes would be consistent with a 49-bp oligonucleotide containing 2–3 binding sites. To account for the presence of multiple bands, we quantified the reduction in intensity of the band correlating to the unbound oligonucleotide rather than those associated with the formation of protein-oligonucleotide complexes. Analysis of the resulting binding curves, assuming that the concentration of oligonucleotide (0.2 nM) is sufficiently lower than the dissociation constant (Kd), can provide a measure of binding affinity by the calculation of approximate Kd values (38). These were estimated as 7 nM for the RdgC dimer when bound to dsDNA and 9 nM when bound to ssDNA. Pre-incubation of RdgC, to change the proportions of primary and secondary forms as observed by gel filtration, had no effect on the binding properties of RdgC (data not shown). Band-shift assays using our mutant proteins showed that the DNA binding of proteins with either a R118A or R118C substitution at the gate interface was largely indistinguishable to that of the native protein (Figure 4B and C), and analysis of binding curves gave calculated Kd values of 6 nM for both proteins for both ssDNA and dsDNA. Surprisingly, synthetic lethality assays, where the chromosomal copy of rdgC is replaced with mutated copies, revealed that the R118A and R118C mutations (Figure 5 and data not shown) did have a weak effect on functionality. Only blue colonies appear when combined with priA+/ΔpriA on LB agar, although we see large white and sectored colonies in a priA+/ΔpriA dnaC810 background, or when cells are plated on MA. However, as this effect was relatively mild, and in conjunction with the DNA binding data, this implies the hydrogen-bond network between opposing R118 residues at the gate interface is not important for dimer stability. Additionally, the R118C disulphide bond, if formed in vivo, does not seriously inhibit or promote function.
Figure 4.
Oligonucleotide binding by RdgC proteins. (A) Representative band-shift assays with dsDNA. Reactions used the RdgC proteins indicated at 0, 0.375, 0.75, 1.5, 3, 6, 12, 24 and 1850 nM (of dimer) and 32P-labelled RGL13/17 DNA at 0.2 nM. Assays with R118A, R118C, Q212A and H222A gave the same band pattern as the WT protein. Assays with F120S and K227A gave the same band pattern as F120T. (B) Quantification of dsDNA binding. RGL13/17 DNA was used at 0.2 nM. Data are means of at least two band-shift experiments. (C) Quantification of ssDNA binding. 32P-labelled RGL13 DNA was used at 0.2 nM. Data are means of at least two band-shift experiments. (D) Effect of DTT on duplex binding by RdgC R118C F120T. Reactions used mutant RdgC protein at 0, 1, 5, 25, 125 and 625 nM (of dimer) and 32P-labelled RGL13/17 DNA at 0.2 nM. (E) Plasmid binding by RdgC proteins. Reactions used the RdgC proteins indicated at 0, 34, 68, 136, 272, 544, 1088, 2176 and 23 400 nM (of dimer) and pGEM-7 Zf(+) circular dsDNA at 2.2 nM plasmid (6480 nM nucleotide pairs). (F) Plasmid binding by the F120T K227A double mutant. Reactions used mutant RdgC at 0, 1000, 2000, 5000, 10 000 and 20 000 nM (of dimer) and pGEM-7 Zf(+) circular dsDNA at 2.2 nM plasmid (6480 nM nucleotide pairs).
Figure 5.
The effect of rdgC mutations on the viability of ΔpriA and ΔpriA dnaC810 strains. Photographs of plates illustrating the results of synthetic lethality assays. All constructs contained a priA+ derivative of the pRC7 plasmid, and white colonies indicate the segregation of plasmid-free cells. Colonies were grown on either LB agar or MA as indicated.
Similar results were seen with the H222A substitution at the horseshoe interface. Binding to both ssDNA and dsDNA was largely indistinguishable to that of the wild-type protein (Figure 4B and C), with estimated Kd values of 5 nM for ssDNA and 12 nM for dsDNA. Synthetic lethality assays showed this mutation was slightly more deleterious than the R118 mutations (Figure 5), as the white colonies observed in a priA+/ΔpriA dnaC810 background, or when plated on MA in a priA+/ΔpriA background, were significantly smaller than those observed for R118A or R118C. However, the cells were viable after the loss of the priA+ plasmid, suggesting H222 is not a major factor in stabilizing the horseshoe dimer interface.
In contrast, strains containing derivatives of RdgC with Q212A, E218R or K227A mutations at the horseshoe interface, or with F120S or F120T mutations at the gate interface all gave only blue colonies in both the priA+/ΔpriA and priA+/ΔpriA dnaC810 backgrounds when grown on LB agar (Figure 5 and data not shown). In addition, growth on MA could not recover viability in the priA+/ΔpriA background. This suggested that each of these residues was essential for functionality, and formed stabilizing interactions across the dimer interfaces. However, interpretation of the in vitro binding data shows that the situation is more complex. DNA binding by the K227A, F120T and F120S mutants is particularly poor (Figure 4A–C), which agrees well with the results of the synthetic lethality assays. All have estimated Kd values of between 100 and 500 nM with dsDNA and 500–1500 nM with ssDNA. In these cases, it is impossible to get an accurate Kd value as the complex is evidently falling apart as it undergoes electrophoresis, as indicated by the broadening and smearing of bands observed on the gels (Figure 4A). Even at high concentrations of protein (>1 μM), we did not see 100% binding. These data suggest that disruption of the domain interfaces has a severe effect on the ability of RdgC to stably bind DNA, and that binding of ssDNA is affected more than the binding of dsDNA.
The juxtaposition of binding dsDNA with binding ssDNA is clearly shown with the E218R mutant. Binding to ssDNA is extremely weak (Figure 4C), of the same order as that of K227A, F120S and F120T. However, it appears to bind dsDNA reasonably well (with an estimated Kd of 22 nM), although this is still weaker than the native protein, and the smearing observed on the gel indicates the complex is dissociating during electrophoresis (Figure 4A and B). The Q212A mutant is even more surprising, as both ssDNA and dsDNA binding appeared to be equivalent to the native protein, with estimated Kds of between 5 and 15 nM. This is at odds with the in vivo data, and implies that RdgC functionality reflects more than just the ability to bind short oligonucleotide DNA. The differences between DNA binding by the E218R and Q212A proteins may also suggest that the cross-domain interaction between K227 and E218 at the horseshoe interface is more important to domain stability than that between K227 and Q212.
Overall, these data may help to explain the differences in binding of dsDNA and ssDNA observed for many of the mutants and for the native protein. Binding dsDNA in the central pore allows interactions between the protein and the DNA across the whole surface of the pore, which may assist in pulling and holding the domain together, meaning the cross-interface interactions becomes less important. Because ssDNA is unlikely to interact with all the residues lining the central pore, it is less likely to pull the domains together and so the stability of the dimer becomes more dependent on the interactions at the domain interfaces.
To check whether a R118C disulphide could substitute for the F120 hydrophobic interaction at the gate dimer interface, we combined these two mutations to form an R118C F120T double mutant. Synthetic lethality assays indicated that the viability of this double mutant was identical to that of the F120T single mutant in both the priA+/ΔpriA and priA+/ΔpriA dnaC810 backgrounds, suggesting either that the disulphide bond does not form in vivo, or that it cannot compensate for the F120 hydrophobic interaction. However, the analytical gel filtration data suggests that the R118C disulphide bond is present in vitro and the DNA binding data suggests it can partially overcome the loss of the F120-mediated interaction. Improved binding of both dsDNA and ssDNA is observed when comparing the double mutant with the F120T single mutant (Figure 4A–C), with the estimated Kd values changing from >500 nM to <20 nM. Additionally, no band smearing or broadening (attributable to complex dissociation) was observed, and the formation of multiple bands associated with higher order complex formation was comparable to that for the native protein. To confirm whether the increased stability was due to the formation of a R118C-mediated disulphide bond, we repeated the binding experiments in binding buffer containing either no DTT, or 5 mM DTT (Figure 4D). This clearly demonstrated that the increased stability of the R118C F120T double mutant relative to the F120T single mutant is sensitive to DTT and so is due to the formation of a disulphide bond. This suggests that either the recovery of in vitro DNA binding activity is not enough to recover in vivo viability or the disulphide bond does not form in vivo.
As F120 appeared to be the key residue at the gate interface and K227 appeared to be the key residue at the horseshoe interface, we constructed an F120T K227A double mutant to determine the effect of weakening both interfaces simultaneously with the expectation that this protein would not be able to dimerize. As with the single mutant derivatives, the double mutant purified using the same methodology as the wild-type RdgC, although it eluted from the Heparin-HP column at significantly lower NaCl concentrations than wild-type RdgC or any of the other mutants. Analytical gel filtration demonstrated that it eluted in the same volume as the wild-type protein, with an estimated molecular weight of 60 kDa. However, the secondary peak, with estimated MW of 120 kDa, was not seen after incubation at room temperature or at 4°C. This would suggest that combining the F120T and the K227A mutations was successful in limiting the ability of RdgC to dimerize.
As expected, dsDNA binding by the F120T K227A double mutant was very poor, and far weaker than that observed for either the F120T or the K227A single mutant (Figure 4B). Significant smearing was observed on the band-shift gels, indicative of complex dissociation, and the very weak binding meant it was impossible to calculate a Kd value. More surprising was the complete lack of binding seen with ssDNA (Figure 4C), with no complex formation observed even at a 5000-fold excess of protein over oligonucleotide. These results are consistent with the double mutant being a monomer in solution, and the weak binding observed with dsDNA is likely to be due to the opportune transient binding of two monomers in close proximity on the DNA helix, allowing residual cross-domain interactions to form and hold the protein in a dimeric conformation. Evidently, ssDNA cannot promote dimerization in this manner. This highlights the role that interactions between the DNA duplex and the residues lining the inside surface of the ring can have on dimer stability. These results also demonstrate that the monomeric state of RdgC cannot stably bind to small oligonucleotides, particularly with ssDNA. If the complexes observed with wild-type RdgC at low protein concentration were with a monomeric species, then we would expect equivalent complexes with the F120T K227A mutant. From this, we conclude that the primary complexes observed must be between the dimeric protein and the oligonucleotide, and the higher order complexes seen at higher protein concentrations are likely to be due to the subsequent addition of RdgC dimers.
RdgC modifications appear to have no effect on plasmid binding
The in vitro oligonucleotide binding studies are broadly in agreement with the in vivo synthetic lethality studies, but they do not tell the whole story. All mutations had a negative effect on viability, even when little or no effect on oligonucleotide binding was seen. Does this reflect the use of small oligonucleotides rather than a large circular DNA molecule corresponding to a chromosome? It is possible that an RdgC dimer does not have to open and close its ring structure to bind to an oligonucleotide; rather it can simply slide on from the end. In order to determine the effects of domain boundary mutations on the binding of molecules when the opening and closing is essential, we looked at the binding of RdgC to a circular DNA molecule.
Band-shift studies with native RdgC and the circular plasmid, pGEM-7 Zf(+), confirmed that RdgC can load onto and bind circular DNA (Figure 4E). However, the results with the single mutation proteins, and the R118C F120T double mutant, were largely indistinguishable from those with native protein and all appeared to be able to load onto and bind circular DNA with the same affinity as the native protein. In all cases, binding could be observed with 100–200 nM RdgC (dimer) and 2.2 nM plasmid DNA (equivalent to ∼300 nM RdgC binding sites). This was completely unexpected and is in variance with both the in vivo viability data and the oligonucleotide binding data. In contrast, plasmid binding by the F120T K227A double mutant was at least an order of magnitude weaker (Figure 4F). Importantly, this implies there is a specific requirement for RdgC to be in a dimeric form before it can efficiently bind circular DNA. An explanation for the difference between the plasmid and oligonucleotide binding observed is that the mutant proteins can load onto DNA, but do not bind tightly once they are loaded. If the DNA is in the form of a short oligonucleotide, then the protein simply slides along the DNA and falls off the end. This would also explain the smearing seen in the band-shift assays (Figure 4A), which is indicative of complex dissociation. When the DNA is circular, then the loaded mutant RdgC can slide along the DNA molecule without falling off and so the complex, as observed by the band-shift assay, remains intact. However, this complex is not functional, as demonstrated by our synthetic lethality assays. For RdgC to function, it is not enough to be simply associated with the DNA: there is a specific requirement for a tight-binding complex. This has implications for its mode of activity, and provides insight into how RdgC modulates RecA-mediated homologous recombination.
The role of the finger domain
We have previously speculated on how RdgC may load onto circular DNA, and proposed that this was a property of the electropositive finger domain (21). We investigated the role of the finger domain by constructing two mutant proteins. The first construct, named the fingertip mutation, had all four highly conserved basic residues located at the end of the finger (Figure 6A) mutated to R97S, K98Q, K100Q and K101E. The second construct had a deletion of residues 77–115, and substitutions of P76G and L116T to allow the formation of a type II’ β-hairpin turn. This Δfinger construct removes the whole of the finger domain, while allowing the rest of the protein to fold correctly. In addition, this construct retains the R118 and F120 residues at the gate interface, although it may disrupt some of the hydrophobic binding pocket. To overcome this, we also made a Δfinger construct which incorporated the R118C mutation, as we postulated that any loss of dimer interface stability caused by disruption of the hydrophobic pocket may be overcome by the addition of a disulphide bond.
Figure 6.
Properties of the finger domain. (A) Electrostatic surface representation of a finger domain, illustrating its electropositive nature and the position of the four conserved basic residues modified in the RdgCfingertip derivative. (B) Synthetic lethality assays showing the effect of finger domain mutations on ΔpriA dnaC810 strain viability. Colonies were grown on LB agar. (C) Quantification of dsDNA binding. Reactions used the RdgC proteins indicated and 32P-labelled RGL13/17 DNA at 0.2 nM. Data are means of at least two band-shift assays. (D) Quantification of ssDNA binding. Reactions used the RdgC proteins indicated and 32P-labelled RGL13 DNA at 0.2 nM. Data are means of at least two band-shift assays. (E) Plasmid binding. Reactions used the RdgC proteins indicated at 0, 34, 68, 136, 272, 544, 1088, 2176 and 23 400 nM (of dimer) and pGEM-7 Zf(+) DNA at 2.2 nM plasmid.
When the chromosomal copy of rdgC was replaced with our finger-mutation versions of rdgC in the synthetic lethality assay, we saw that the Δfinger derivative of RdgC gave only blue colonies in the priA+/ΔpriA background on LB agar and MA and in the priA+/ΔpriA dnaC810 background on LB agar (Figure 6B and data not shown). This indicates that the Δfinger protein is functionally inactive. The addition of the R118C mutation appeared to have no effect, as this also gave only blue colonies. The fingertip derivative also gave only large blue colonies in a priA+/ΔpriA background on both LB agar and MA, but a few small white colonies were observed in a priA+/ΔpriA dnaC810 background on LB agar (Figure 6B), suggesting some functionality had been retained. Taken together, these data suggest that the finger domain is critical to the in vivo role of RdgC.
We purified the three proteins, and found that they behaved identically to the native protein throughout the purification, although the Δfinger constructs eluted slightly later from the gel filtration column with an estimated molecular weight of 45 kDa. As this construct contained a deletion of 39 residues and had a calculated molecular weight of 28.8 kDa, this was consistent with the aberrant elution profile of the full-length native protein. After incubation of the purified proteins in gel filtration buffer (50 mM Tris, pH 7.5, 100 mM NaCl), analytical gel filtration of the Δfinger protein showed the emergence of a secondary peak with an estimated MW of 90 kDa, while the Δfinger R118C construct displayed a DTT-sensitive peak of the same size, indicative of disulphide-bound oligomers (Figure 3D). CD analysis suggested the mutant proteins contained similar proportions of secondary structure to the native protein (data not shown), suggesting that they had folded correctly.
Band-shift assays using the small double-stranded and single-stranded oligonucleotides (Figure 6C and D) indicated that the Δfinger construct was completely unable to stably bind to either oligonucleotide, and the data were such that it was impossible to calculate a value for the Kd. Some formation of complex was observed with a 5000-fold excess of protein, but the smearing seen is indicative of this complex falling apart during electrophoresis. In contrast, both the Δfinger R118C protein and the fingertip protein showed some binding to both ssDNA and dsDNA, with the formation of a stable complex with dsDNA at very high concentrations of protein. This was different to the dsDNA binding seen with several of the dimer interface mutations where, even though the complex could form, it was less stable and appeared to dissociate readily. Further analysis of dsDNA binding by the Δfinger R118C and the fingertip proteins, using an extended protein concentration, gave a sigmoid binding curve reminiscent of that seen with the native protein, though with a much reduced affinity. The calculated Kd values for the fingertip protein with dsDNA and ssDNA were 170 nM and 240 nM, respectively, whereas those for Δfinger R118C were 80 nM with dsDNA and ∼6000 nM with ssDNA. The large difference between the activities of the Δfinger and the Δfinger R118C proteins would suggest that the engineered R118C disulphide observed by analytical gel filtration stabilizes the dimer interface. It is possible that the formation of the disulphide is enhanced by dsDNA binding, where the interactions around the RdgC central pore pulls the domains together into a conformation conducive to disulphide bond formation. This may explain the large difference in affinities of the Δfinger R118C protein for dsDNA over ssDNA. This would also suggest that the gate interface is compromised by the deletion of the finger region, such that the F120-mediated interaction is destabilized, which is not surprising as some of the residues forming the hydrophobic binding pocket are at the base of the finger domain.
These data provide insight into the role of the finger domain. It appears to be involved in the initial DNA binding, but to be less important in the stability of the complex once it is formed. This agrees with our hypothesis that the main function of the finger domain is the loading of RdgC onto the DNA. If this is true, we would expect to see a marked difference in the ability of the finger mutants to bind to circular DNA molecules, where loading is essential. Band-shift assays with the finger mutants and pGEM-7 Zf(+) confirmed that this was the case (Figure 6E). In stark contrast to the domain boundary mutants, which were indistinguishable from native RdgC, each of our finger mutants were severely compromised for plasmid binding. We saw no complex formation with the Δfinger protein, even when the protein concentration was increased 100-fold over that required to show a complete shift. For native RdgC, some binding was observed with the other two mutants at these very high concentrations, but the degree of band-shift when compared to the results from the native protein was indicative that very few RdgC molecules had actually bound. This confirms that the finger domain, and specifically the basic nature of the finger, is essential for the efficient loading of RdgC onto circular DNA and by extrapolation, the binding of the bacterial chromosome.
DISCUSSION
The crystal structures of RdgC from E. coli (21) and P. aeruginosa (22) showed it to form a ring that was proposed to encircle the DNA, so stabilizing the DNA to unwinding. However, this left unanswered the question of how this ring could be loaded onto a circular DNA molecule. Does RdgC assemble on the DNA, with sequential binding by monomers, or does the ring open at one or both of the domain interfaces, allowing it to slip over the DNA duplex? To investigate this, we studied the effect of destabilizing the domain boundaries using site-directed mutagenesis and a combination of genetic and biochemical assays.
Our initial discovery that native RdgC purified as a monomer, rather than a dimer as reported previously (3,4), was surprising. However, the previous reports are from before the structure was known, and the non-globular nature of RdgC would account for the anomalous migration on gel filtration which had been used to determine the oligomeric state. This does not inform us about the in vivo state of E. coli RdgC, as we have shown that incubation in a high ionic strength buffer causes the protein to monomerize, and such a step is reported in all previous purification protocols. Ha et al. (22) report that RdgC from P. aeruginosa purifies as a dimer, although data to demonstrate this is not shown, and their purification procedure also contains highly ionic step. However, it is worth noting that RdgC from both E. coli and P. aeruginosa crystallized as a dimer in the absence of an oligonucleotide. Indeed, Drees et al. (5) suggest that RdgC is in equilibrium between monomer–dimer–trimer states in solution, although they also suggest a monomer–dimer–tetramer model would fit their data. Subsequent knowledge of the crystal structure suggests the second model is more likely.
Our experiments demonstrate that weakening either the gate or the horseshoe interface by removing potential residue-mediated interactions has a severe effect on the binding of small DNA oligonucleotides. This adds weight to our assumption that the DNA must go through the central pore, as a weakened dimer interface would only effect binding if the DNA was encircled. Interestingly, these modifications appeared to have no effect on the ability of RdgC to bind to circular plasmid DNA. We propose that a tight-binding complex does not form, and the modified RdgC is sliding along the DNA without dissociating. This is consistent with gel filtration data showing that the mutant proteins can still form stable dimers in solution. Presumably, even with weakened domain interfaces, RdgC can adopt an open or closed conformation, both of which would stay associated with a circular molecule. However, the loss of in vivo activity observed in our synthetic lethality assays suggests that remaining associated with DNA is not sufficient for function: RdgC is only functional when it is bound tightly to the DNA.
We have also shown that the loading of RdgC onto circular DNA can be attributed to the finger domain, as modification or deletion of this domain eliminated plasmid binding. It is possible that the deletion of the entire finger domain (Δ76–115) also disrupts the hydrophobic F120-binding pocket, but this would not be enough to explain the complete lack of plasmid binding observed, as the F120S and F120T proteins were seen to load normally. That the ‘loading’ activity is largely due to the electropositive nature of the finger domain was confirmed by the observed effect of substituting the basic residues at the fingertip. Additionally, the electropositive nature of the finger domain seems to be involved in the initial recognition and binding of the DNA molecule by RdgC, as mutations appear to affect the ability to form the protein-DNA complex, but contribute little to the stability of this complex once it has formed. Analysis of the F120T K227A double mutant, which retains the native electropositive finger, supports this, as it is incredibly poor at binding any of the three DNA substrates assayed. If the finger contributed significantly to the stability of the RdgC–DNA complex, then the F120T K227A monomer would be expected to bind to DNA. As it is, we see no evidence of the monomer forming a stable complex with the DNA.
These results, coupled with the crystal structures, provide compelling evidence that RdgC is bound to DNA as a dimer, but doesn’t address the mechanism of how RdgC binds to DNA. It is possible that the RdgC monomers bind sequentially, with the dimer assembling on the DNA. However, if this was the case, then we would expect to see some binding by the RdgC monomer. In addition, it would be difficult to explain why mutating the fingertip residues has such a dramatic effect on DNA binding activity, as the electropositive residues on the inside of the ring would be accessible to the DNA and the residues required for dimerization are intact. In contrast, if RdgC is a dimer in solution, such that the ring is already closed, then the electropositive residues inside the ring are not accessible for DNA binding, and an additional solvent-accessible structure is necessary for the initial location and interaction with the DNA. We suggest the finger domain forms this solvent-accessible structure and we propose a model for how this could facilitate DNA binding by the RdgC dimer in vivo.
The first step involves RdgC locating the DNA by means of the electropositive finger domain, which is solvent exposed and accessible. The DNA then binds across the surface of the finger domain and across the face of the RdgC ring (Figure 7A). This causes a conformational rotation in the finger domain, which in turn disrupts the F120-mediated interaction at the gate interface (Figure 7B). Disruption of the interface allows the gate to open, flexing at the linker region between the gate and horseshoe domains (residues S160 to N170), and the RdgC slides over the DNA (Figure 7C). Once the DNA is properly located in the central binding channel, the electropositive residues lining the channel pull the dimer together. The finger domain returns to its original conformation and lies parallel to the bound DNA duplex, allowing the gate to close; such that the ring structure is reformed (Figure 7D). This leaves RdgC clamped tightly around the DNA, stabilizing the encircled duplex to unwinding.
Figure 7.
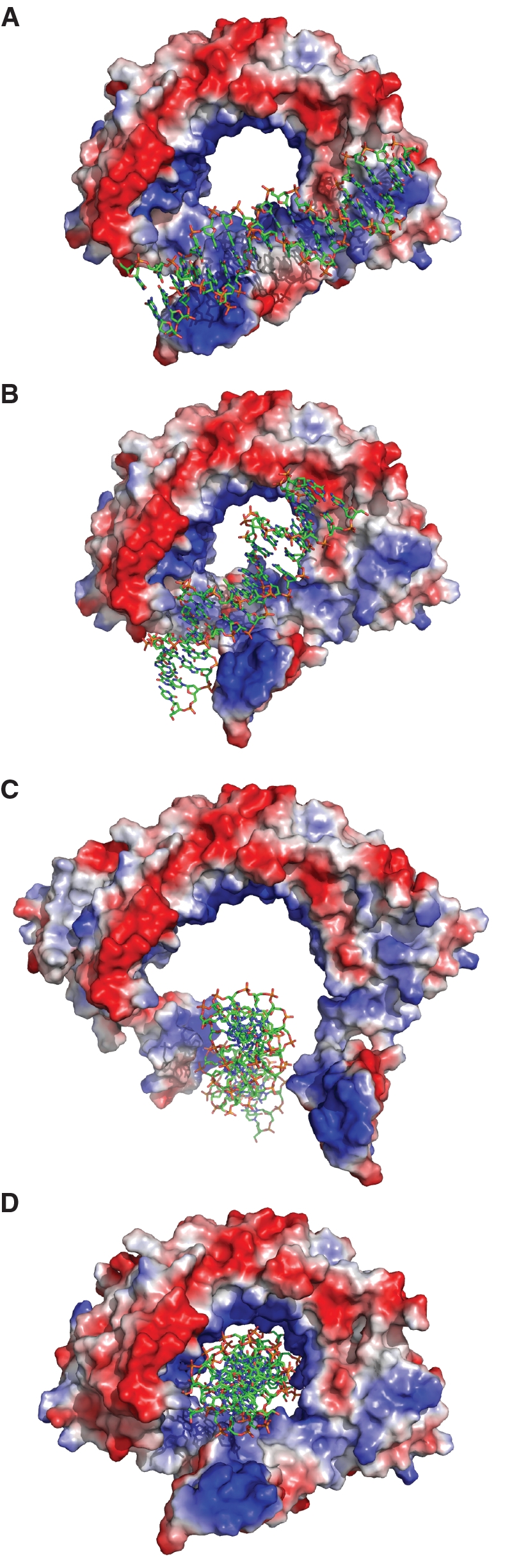
Model illustrating how RdgC may load onto DNA. (A) DNA binds across an exposed positive face, interacting with the finger domain. (B) The finger domain rotates, disrupting the gate dimer interface. (C) The gate opens, and RdgC slides over the DNA. (D) The gate closes, encircling the DNA within the ring.
Such a tight-binding complex could readily block RecA mediated strand exchange. If RdgC is bound near the site of a double-strand break, it would inhibit RecA filament assembly by preventing extension of the filament from the ssDNA region into the duplex. Alternatively, if bound near a potential site of recombination, RdgC would either restrict the unwinding of the duplex required for strand invasion, or would limit branch migration and so the completion of strand exchange.
This model does highlight a potential problem: if RdgC can bind so tightly to DNA that it can efficiently block RecA activity, then why doesn’t it affect other processes that require DNA strand separation like replication and transcription? It is possible that this is another important role for the finger domain. As the finger domains project perpendicular to the ring, they will lie along the length of, and parallel to, the encircled DNA strand. This would mean that any protein translocating along the DNA, e.g. a helicase or polymerase, would make an initial protein–protein interaction with the finger domain. This could then induce a conformational shift in the finger domain similar to that proposed on DNA loading, causing the gate to open, and allowing the translocating protein to efficiently displace RdgC from the DNA duplex.
FUNDING
Medical Research Council UK (G9806167, G0800970). Funding for open access charge: Medical Research Council UK.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Laura Tye and Mark Searle for the CD analysis, Ian Kerr for advice on determining dissociation constants, and Karen Bunting for useful discussions. We are grateful to Carol Buckman and Lynda Harris for providing outstanding technical assistance.
REFERENCES
- 1.Murphy LD, Rosner JL, Zimmerman SB, Esposito D. Identification of two new proteins in spermidine nucleoids isolated from Escherichia coli. J. Bacteriol. 1999;181:3842–3844. doi: 10.1128/jb.181.12.3842-3844.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ryder L, Sharples GJ, Lloyd RG. Recombination-dependent growth in exonuclease-depleted recBC sbcBC strains of Escherichia coli K-12. Genetics. 1996;143:1101–1114. doi: 10.1093/genetics/143.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore T, McGlynn P, Ngo HP, Sharples GJ, Lloyd RG. The RdgC protein of Escherichia coli binds DNA and counters a toxic effect of RecFOR in strains lacking the replication restart protein PriA. EMBO J. 2003;22:735–745. doi: 10.1093/emboj/cdg048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moore T, Sharples GJ, Lloyd RG. DNA binding by the meningococcal RdgC protein, associated with pilin antigenic variation. J. Bacteriol. 2004;186:870–874. doi: 10.1128/JB.186.3.870-874.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drees JC, Chitteni-Pattu S, McCaslin DR, Inman RB, Cox MM. Inhibition of RecA protein function by the RdgC protein from Escherichia coli. J. Biol. Chem. 2006;281:4708–4717. doi: 10.1074/jbc.M513592200. [DOI] [PubMed] [Google Scholar]
- 6.Cox MM. Motoring along with the bacterial RecA protein. Nat. Rev. Mol. Cell Biol. 2007;8:127–138. doi: 10.1038/nrm2099. [DOI] [PubMed] [Google Scholar]
- 7.Galletto R, Kowalczykowski SC. RecA. Curr. Biol. 2007;17:R395–R397. doi: 10.1016/j.cub.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 8.Vos M. Why do bacteria engage in homologous recombination? Trends Microbiol. 2009;17:226–232. doi: 10.1016/j.tim.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 9.Hill SA, Davies JK. Pilin gene variation in Neisseria gonorrhoeae: reassessing the old paradigms. FEMS Microbiol. Rev. 2009;33:521–530. doi: 10.1111/j.1574-6976.2009.00171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andrews TD, Gojobori T. Strong positive selection and recombination drive the antigenic variation of the PilE protein of the human pathogen Neisseria meningitidis. Genetics. 2004;166:25–32. doi: 10.1534/genetics.166.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boslego JW, Tramont EC, Chung RC, McChesney DG, Ciak J, Sadoff JC, Piziak MV, Brown JD, Brinton CC, Jr, Wood SW, et al. Efficacy trial of a parenteral gonococcal pilus vaccine in men. Vaccine. 1991;9:154–162. doi: 10.1016/0264-410x(91)90147-x. [DOI] [PubMed] [Google Scholar]
- 12.Mehr IJ, Long CD, Serkin CD, Seifert HS. A homologue of the recombination-dependent growth gene, rdgC, is involved in gonococcal pilin antigenic variation. Genetics. 2000;154:523–532. doi: 10.1093/genetics/154.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pride DT, Blaser MJ. Concerted evolution between duplicated genetic elements in Helicobacter pylori. J. Mol. Biol. 2002;316:629–642. doi: 10.1006/jmbi.2001.5311. [DOI] [PubMed] [Google Scholar]
- 14.Dworkin J, Blaser MJ. Molecular mechanisms of Campylobacter fetus surface layer protein expression. Mol. Microbiol. 1997;26:433–440. doi: 10.1046/j.1365-2958.1997.6151958.x. [DOI] [PubMed] [Google Scholar]
- 15.Sinha H, Pain A, Johnstone K. Analysis of the role of recA in phenotypic switching of Pseudomonas tolaasii. J. Bacteriol. 2000;182:6532–6535. doi: 10.1128/jb.182.22.6532-6535.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoiseth SK, Moxon ER, Silver RP. Genes involved in Haemophilus influenzae type b capsule expression are part of an 18-kilobase tandem duplication. Proc. Natl Acad. Sci. USA. 1986;83:1106–1110. doi: 10.1073/pnas.83.4.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCulloch R, Barry JD. A role for RAD51 and homologous recombination in Trypanosoma brucei antigenic variation. Genes Dev. 1999;13:2875–2888. doi: 10.1101/gad.13.21.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cox MM. Regulation of bacterial RecA protein function. Crit. Rev. Biochem. Mol. Biol. 2007;42:41–63. doi: 10.1080/10409230701260258. [DOI] [PubMed] [Google Scholar]
- 19.West SC. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 20.Petrova V, Chitteni-Pattu S, Drees JC, Inman RB, Cox MM. An SOS inhibitor that binds to free RecA protein: the PsiB protein. Mol. Cell. 2009;36:121–130. doi: 10.1016/j.molcel.2009.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Briggs GS, McEwan PA, Yu J, Moore T, Emsley J, Lloyd RG. Ring structure of the Escherichia coli DNA-binding protein RdgC associated with recombination and replication fork repair. J. Biol. Chem. 2007;282:12353–12357. doi: 10.1074/jbc.C700023200. [DOI] [PubMed] [Google Scholar]
- 22.Ha JY, Kim HK, Kim do J, Kim KH, Oh SJ, Lee HH, Yoon HJ, Song HK, Suh SW. The recombination-associated protein RdgC adopts a novel toroidal architecture for DNA binding. Nucleic Acids Res. 2007;35:2671–2681. doi: 10.1093/nar/gkm144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hingorani MM, O'Donnell M. A tale of toroids in DNA metabolism. Nat. Rev. Mol. Cell Biol. 2000;1:22–30. doi: 10.1038/35036044. [DOI] [PubMed] [Google Scholar]
- 24.Kong XP, Onrust R, O'Donnell M, Kuriyan J. Three-dimensional structure of the beta subunit of E.coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 25.Krishna TS, Kong XP, Gary S, Burgers PM, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 26.Bailey S, Eliason WK, Steitz TA. Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase. Science. 2007;318:459–463. doi: 10.1126/science.1147353. [DOI] [PubMed] [Google Scholar]
- 27.Karow JK, Newman RH, Freemont PS, Hickson ID. Oligomeric ring structure of the Bloom's syndrome helicase. Curr. Biol. 1999;9:597–600. doi: 10.1016/s0960-9822(99)80264-4. [DOI] [PubMed] [Google Scholar]
- 28.O'Donnell M, Kuriyan J. Clamp loaders and replication initiation. Curr. Opin. Struct. Biol. 2006;16:35–41. doi: 10.1016/j.sbi.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 29.Davey MJ, Fang L, McInerney P, Georgescu RE, O'Donnell M. The DnaC helicase loader is a dual ATP/ADP switch protein. EMBO J. 2002;21:3148–3159. doi: 10.1093/emboj/cdf308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 31.Ahnert P, Picha KM, Patel SS. A ring-opening mechanism for DNA binding in the central channel of the T7 helicase-primase protein. EMBO J. 2000;19:3418–3427. doi: 10.1093/emboj/19.13.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernhardt TG, de Boer PA. Screening for synthetic lethal mutants in Escherichia coli and identification of EnvC (YibP) as a periplasmic septal ring factor with murein hydrolase activity. Mol. Microbiol. 2004;52:1255–1269. doi: 10.1111/j.1365-2958.2004.04063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahdi AA, Buckman C, Harris L, Lloyd RG. Rep and PriA helicase activities prevent RecA from provoking unnecessary recombination during replication fork repair. Genes Dev. 2006;20:2135–2147. doi: 10.1101/gad.382306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Al-Deib AA, Mahdi AA, Lloyd RG. Modulation of recombination and DNA repair by the RecG and PriA helicases of Escherichia coli K-12. J. Bacteriol. 1996;178:6782–6789. doi: 10.1128/jb.178.23.6782-6789.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Briggs GS, Mahdi AA, Wen Q, Lloyd RG. DNA binding by the substrate specificity (wedge) domain of RecG helicase suggests a role in processivity. J. Biol. Chem. 2005;280:13921–13927. doi: 10.1074/jbc.M412054200. [DOI] [PubMed] [Google Scholar]
- 37.Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophot. Int. 2004;11:36–42. [Google Scholar]
- 38.Szwajkajzer D, Dai L, Fukayama JW, Abramczyk B, Fairman R, Carey J. Quantitative analysis of DNA binding by the Escherichia coli arginine repressor. J. Mol. Biol. 2001;312:949–962. doi: 10.1006/jmbi.2001.4941. [DOI] [PubMed] [Google Scholar]
- 39.Vinayagam A, Pugalenthi G, Rajesh R, Sowdhamini R. DSDBASE: a consortium of native and modelled disulphide bonds in proteins. Nucleic Acids Res. 2004;32:D200–D202. doi: 10.1093/nar/gkh026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sandler SJ, Samra HS, Clark AJ. Differential suppression of priA2::kan phenotypes in Escherichia coli K-12 by mutations in priA, lexA, and dnaC. Genetics. 1996;143:5–13. doi: 10.1093/genetics/143.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sandler SJ, Marians KJ, Zavitz KH, Coutu J, Parent MA, Clark AJ. dnaC mutations suppress defects in DNA replication- and recombination-associated functions in priB and priC double mutants in Escherichia coli K-12. Mol. Microbiol. 1999;34:91–101. doi: 10.1046/j.1365-2958.1999.01576.x. [DOI] [PubMed] [Google Scholar]
- 42.Gregg AV, McGlynn P, Jaktaji RP, Lloyd RG. Direct rescue of stalled DNA replication forks via the combined action of PriA and RecG helicase activities. Mol. Cell. 2002;9:241–251. doi: 10.1016/s1097-2765(02)00455-0. [DOI] [PubMed] [Google Scholar]
- 43.Masai H, Asai T, Kubota Y, Arai K, Kogoma T. Escherichia coli PriA protein is essential for inducible and constitutive stable DNA replication. EMBO J. 1994;13:5338–5345. doi: 10.1002/j.1460-2075.1994.tb06868.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bachmann BJ. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12. In: Neidardt FCEA, editor. Escherichia coli and Salmonella Cellular and Molecular Biology. 2nd edn. Vol. 2. Washington DC: ASM Press; 1996. pp. 2460–2488. [Google Scholar]
- 45.Nurse P, Zavitz KH, Marians KJ. Inactivation of the Escherichia coli PriA DNA replication protein induces the SOS response. J. Bacteriol. 1991;173:6686–6693. doi: 10.1128/jb.173.21.6686-6693.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]