Abstract
While a number of aminoacyl tRNA synthetase (aaRS):tRNA pairs have been engineered to alter or expand the genetic code, only the Methanococcus jannaschii tyrosyl tRNA synthetase and tRNA have been used extensively in bacteria, limiting the types and numbers of unnatural amino acids that can be utilized at any one time to expand the genetic code. In order to expand the number and type of aaRS/tRNA pairs available for engineering bacterial genetic codes, we have developed an orthogonal tryptophanyl tRNA synthetase and tRNA pair, derived from Saccharomyces cerevisiae. In the process of developing an amber suppressor tRNA, we discovered that the Escherichia coli lysyl tRNA synthetase was responsible for misacylating the initial amber suppressor version of the yeast tryptophanyl tRNA. It was discovered that modification of the G:C content of the anticodon stem and therefore reducing the structural flexibility of this stem eliminated misacylation by the E. coli lysyl tRNA synthetase, and led to the development of a functional, orthogonal suppressor pair that should prove useful for the incorporation of bulky, unnatural amino acids into the genetic code. Our results provide insight into the role of tRNA flexibility in molecular recognition and the engineering and evolution of tRNA specificity.
INTRODUCTION
Engineering the genetic code has recently emerged as a method to potentially create novel ‘allo-proteins’ which could have a myriad of applications in the pharmaceutical and biotechnology industries as well as serve as novel components for synthetic biology. To facilitate the site-specific incorporation of novel amino acids into proteins, additional engineered orthogonal aminoacyl tRNA synthetase (aaRS):tRNA pairs can be expressed in cells [reviewed in (1,2)]. Orthogonal aaRS–tRNA pairs frequently take advantage of the cross-species differences in the recognition elements of aaRSs and their cognate tRNAs (3), and have utilized nonsense codons (3) or frameshift codons (4) to expand the canonical genetic code.
To date, there have been only eight completely unique orthogonal pairs reported for use in the prokaryotic (Escherichia coli) translational system. These orthogonal pairs include the aspartic acid (5), glutamine (6), and phenylalanine (7) aaRS/tRNA pairs from Saccharomyces cerevisiae; the glutamic acid pair from Methanosarcina mazei (8); the leucine pair from Methanobacterium thermoautotrophicum (9); the lysine pair from Pyrococcus horikoshii (4); the pyrrolysine pair from Methanosarcinia barkeri (10); and the tyrosyl pair from Methanococcus jannaschii (3). However, only the TyrRS-tRNATyr pair from M. jannaschii has been used extensively to expand the genetic code of bacteria (2).
The tryptophanyl tRNA synthetase (WRS) is part of the class Ic subclass of aaRSs that also includes the evolutionarily related tyrosyl tRNA synthetase (11). Based on the structural and phylogenetic studies of the tryptophanyl and the tyrosyl tRNA synthetases, it is generally believed that the WRS was the last synthetase to evolve (12–14). Both the tyrosyl tRNA and WRSs lack any sort of an editing domain. Instead, they rely on the interaction of conserved active site residues to specifically recognize and position their substrates into their binding pockets (15).
The lack of an editing domain in WRS and its capacious binding pocket for the largest natural amino acid have led to its use for the incorporation of amino acid analogs into proteins (16,17). The expanded substrate flexibility of WRS has even supported adaptive evolution of whole bacteria and bacteria phage proteomes to tryptophan analogs (18–20). However, the engineering of the WRS for the site-specific insertion of large, unnatural amino acids has largely been ignored, especially when compared with tyrosyl tRNA synthetase (2). Zhang et al. (21) have used the Bacillus subtilis WRS and an opal suppressor (anticodon UCA) variant of the B. subtilis tRNATrp for the site-specific incorporation of 5-hydroxytryptophan into proteins in mammalian cells, but this orthogonal pair cannot be used in E. coli since they have overlapping recognition elements.
Therefore, to further explore the types of unnatural amino acids that can be site-specifically encoded in bacteria and to expand the potential utility of synthetic orthogonal pairs both in vivo and in vitro, we have sought to develop a new tryptophanyl orthogonal pair. The E. coli tRNATrp has a major identity element at the G73 discriminator base and the anticodon CCA sequence, and additional weak identity elements in the first 3 bp of the acceptor stem (22). In contrast, yeast tRNATrp has an adenosine residue at position 73, and the identity elements in the first 3 bp of the acceptor stem also differ from those found in the E. coli tRNATrp [(23); Figure 1A]. Himeno et al. have shown that mutations at A73 and the G1-C72 base pair of the E. coli tRNATrp that make it more similar to the yeast tRNATrp also lead to inactivity with the E. coli WRS. Moreover, amber suppressor versions of E. coli tRNATrp (anticodon CUA) lose their ability to be recognized by the E. coli TrpRS and are instead aminoacylated by the E. coli GlnRS (24). However, this is not the case for the yeast tRNATrp, which not only maintains its tryptophan identity in yeast cells but also is an efficient amber suppressor (25,26). Taken together, these results suggest that it should be possible to develop a new orthogonal pair for use in E. coli based on the WRS and the amber suppressor variant of its cognate tRNA from S. cerevisiae.
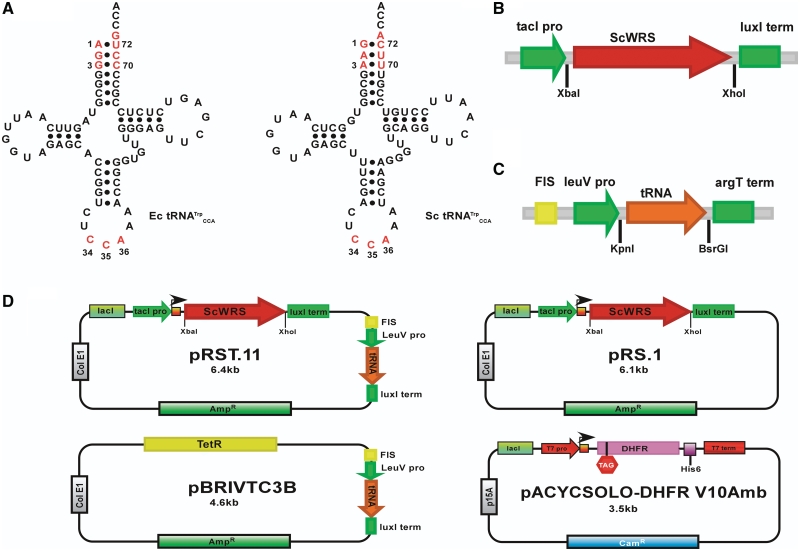
Figure 1.
Tryptophan tRNAs and DNA expression constructs. (A) Cloverleaf representations of E. coli tRNATrp and S. cerevisiae tRNATrp. The known identity elements for each tRNA are shown in red. (B) Expression construct for ScWRS in plasmids pRS.1 and pRST.11B. (C) Expression construct for the Sc-tRNATrpAmb and its variants in plasmids pBRIVTC3B and pRST.11B. (D) Plasmid constructs. pRST.11B is the dual tRNA/aaRS expression vector containing both of the expression constructs shown in (B and C). Plasmid pRS.1 expresses only the ScWRS and is similar to pRST.11B except that it lacks the tRNA expression construct shown in (B). Plasmid pBRIVTC3B expresses only the tRNA. Plasmid pACYCSOLO-DHFR V10Amb is the target protein (DHFR) expression vector. The encoded DHFR gene contains an in-frame amber (TAG) mutation at the 10th amino acid position and a C-terminal HisTag.
MATERIALS AND METHODS
Materials
E. coli strain, CSH108 (Genotype: F′ [F128: LacZ8(Am), LacI373] Δ(gpt-lac)5, λ-,ara(FG),gyrA-(NAlR),argE-(Am),rpoB-(rifR), thi-1) was obtained from the E. coli Genetic Stock Center (Strain # 8081) http://cgsc.biology.yale.edu/. Top10 (for routine cloning) and BL21(DE3)Star chemically competent cells were from Invitrogen (Carlsbad, CA, USA). Terrific Broth Powder was from VWR Scientific (West Chester, PA, USA).
All restriction enzymes, T4 DNA ligase and vector pBR322 were from New England Biolabs (Ipswich, MA, USA). Vector pACYCDuet-1 was obtained from Novagen (San Diego, CA, USA). Vector pET28b and KOD polymerase were from Novagen (Gibbstown, NJ, USA). Wizard SV Gel and PCR cleanup kit were from Promega (Madison, WI, USA). Oligonucleotides were obtained from Integrated DNA Technologies (IDT; Coralville, IA, USA). The Klenow fragment polymerase, SimplyBlue Safestain and NuPAGE 4–12% Bis–Tris precast gels were from Invitrogen (Carlsbad, CA, USA). Quickchange site-directed mutagensis kits and Pfu polymerase were from Stratagene (La Jolla, CA, USA).
Protease inhibitor cocktail was from Roche (Indianapolis, IN, USA). Ni-NTA resin was obtained from Qiagen (Germantown, MD, USA). The anti-polyhistidine alkaline phosphatase primary conjugate antibody was from Sigma-Aldrich (St. Louis, MO, USA). All buffers and chemicals were obtained from Sigma-Aldrich (St. Louis, MO, USA) or Fisher Scientific (Waltham, MA, USA).
Cloning of the S. cerevisiae WRS gene
The coding sequence for the WRS from S. cerevisiae (ScWRS) (27) was amplified using the PCR and the primers ScWRS.f-3 5′-TCGAAAAGCTTCCATGAGCAACGACGAAACTGTAGAG-3′ (HindIII), ScWRS.r-3 5′-GCAGCCTCGAGTTACTTCTTTTCTTGCTTAGTTTTTGGC-3′ (XhoI) from a glass-bead lysed yeast cell extract. The restriction sites introduced are underlined and specified in parentheses. The resulting approximately 1.3 kb PCR product was digested with XhoI and HindIII and ligated into a similarly digested pET-28b vector using T4 DNA ligase following the conditions recommended by the enzyme supplier. The ligation mixture was transformed into Top10 cells and the resulting kanamycin resistant-colonies were screened via colony PCR for the ScWRS insert. Positive clones were verified by DNA sequencing from purified clonal plasmids using the primers T7 Terminator Primer 5′-GCTAGTTATTGCTCAGCGG-3′ and T7 Promoter Primer 5′-TAATACGACTCACTATAGGG-3′. The resulting sequence-confirmed plasmid was termed pET28b-ScWRS. This expression vector expresses ScWRS from the T7 promoter with an N-terminal His-tag.
Design and construction of the pRS.1 tRNA synthetase expression vector
In order to express the ScWRS gene in E. coli, the expression construct (module) shown in Figure 1B was constructed. This expression construct was designed to be modular in that different components of the construct could be replaced using unique restriction sites flanking each of the three portions of the construct (i.e. promoter-RBS, aaRS, transcription terminator). For example, the gene for the ScWRS shown in green in Figure1B could be replaced by restriction digestion with XbaI and XhoI. Since successful nonsense suppression is the result of an acylated suppressor tRNA successfully competing with endogenous release factors for termination codons, maintenance of high levels of acylated suppressor tRNAs within the cell is important, therefore we expressed ScWRS from the strong, synthetic tacI promoter (28) and the strong Shine Dalgarno sequence (AAGGAG) from the expression vector pET14b. Transcription termination was mediated by the luxICDABEG terminator sequence (Biobrick part: BBa_B0021) obtained from the Biobrick parts registry at MIT (http://partsregistry.org/Main_Page). The overall construct was assembled by overlap extension PCR.
The plasmid construct for expressing the ScWRS is shown in Figure 1D. The aforementioned synthetase expression construct was cloned into the HindIII and EagI sites of pBR322 to yield the vector pRS-ScWRS. This cloning step removes the majority of the tetracycline resistance element found in pBR322, but leaves the ampicillin resistance element. This vector contains the ColE1 origin of replication which maintains the plasmid at 15–30 copies per cell (29). To help regulate the expression of the ScWRS from the strong tacI promoter, the expression sequence for LacIq was cloned into the ZraI and HindIII sites of this vector to yield vector pRS.1. The IPTG-induced expression of the ScWRS gene from this plasmid was verified by a western blot assay by probing for the C-terminal His-Tag in a His tagged version of ScWRS (in vector pRS.1ht) using an alkaline phosphatase conjugated anti-polyhistidine antibody (Sigma-Aldrich; St. Louis, MO, USA). As can be seen from Supplementary Figure S1, the ScWRS is efficiently expressed from this construct, though some aggregation in the pellet occurs due to overexpression. A more detailed description of the construction of pRS.1 and related vectors can be found in the Supplementary Data.
Design and construction of the IVTC3 tRNA expression plasmids
To express the yeast tRNATrp amber suppressor, the synthetic construct shown in Figure 1C was designed and built from overlapping oligonucleotides. This synthetic construct expresses the tRNA under the control of the tRNA leuV 5′-UTR (region from −112 to +33), which contains the stable RNA promoter −35 and −10 regions and an upstream FIS element (30,31). The FIS element was included in the 5′-UTR as it is known to enhance transcription initiation from this promoter (32). In addition, the +1 to +33 sequences from the leuV 5′-UTR was maintained as it contains the natural tRNA processing sequence. The tRNA sequence is flanked by unique KpnI and BsrGI sites to facilitate cloning of different tRNA sequences into the expression construct. Transcription termination is carried out by the rho independent terminator contained in the argT 3′-UTR (33). This construct was cloned into vector pBR322 or vector pRS.1 to yield the tRNA expression vectors pBRIVTC3 and pRST.11, respectively (Figure 1D). The expression and maturation of the yeast tRNATrp amber suppressor from these plasmid constructs in vivo was verified via northern blotting with a Sc-tRNATrp-specific DNA oligonucleotide probe (Supplementary Figure S2). While the recombinant Sc-tRNATrpAmb was cleaved from the expressed pre-tRNA transcript at sufficient levels (∼20-30%) to observe nonsense suppression in vivo, future improvements to the processing efficiency of tRNAs expressed from this construct could lead to increased nonsense suppression efficiency in vivo. A thorough description of the design, construction and assay of these constructs and vectors can be found in the Supplementary Data.
Construction of vector pACYCSolo
Vector pACYCDuet-1 was digested with EcoNI and BsrGI to remove the MCS1 and redundant T7promoter. The double-digested plasmid band was gel-purified from a low-melt agarose gel using the Promega (Madison, WI, USA) PCR and gel purification kit. The linearized vector was ‘blunted’ by reaction with the Klenow fragment polymerase using the following conditions: linearized plasmid, 0.5 mM each dNTP, 1× React2 Buffer (Klenow reaction buffer, supplied with enzyme) and 1.5 U Klenow fragment. The elongation reaction was carried out at room temperature for 30 min, followed by agarose gel purification. The blunted vector was closed with T4 DNA ligase under the following conditions: linearized vector, 1× T4 DNA ligase buffer (50 mM Tris pH 7.5, 10 mM MgCl2, 10 mM DTT, 1 mM ATP) and 20 U T4 DNA ligase. The ligation mixture was transformed into Top10 cells and colonies were screened via colony PCR using the primers pACYCDuet-f and pACYCDuet-r. Plasmids that appeared to contain the correct insert were submitted for sequencing at the ICMB DNA Core Facility. The sequence verified vector was named pACYCSolo. This vector contained a multiple cloning site sequence flanked by the T7lac promoter and a transcription terminator. It also contained the coding sequences for the Lac repressor and chloramphenicol acetyl transferase. This vector is a pACYC184 derivative and contains the P15A origin which is compatible with other vectors containing ColE1 origins.
Construction of vectors pACYCSolo-DHFR and pACYCSolo-DHFR_V10Amb
The coding sequence for E. coli dihydrofolate reductase (DHFR) and DHFR_V10Amb was amplified from the plasmids pIVEX1.4WG-DHFR or pIVEX1.4WG-DHFR_V10Amb (R. A. Hughes and A. D. Ellington, unpublished results), respectively, with primers DHFR-pivex1.4-f-1 5′-GTTACTTTCACATATGATCAGTCTGATTGCGGCGTTAGC (NdeI), DHFR-HIS6-r 5′-GCTGCTCGAGTTAATGATGATGATGATGATGGCTGCCCCGCCGCTCCAGAATCTC-3′(XhoI). The restriction sites used for cloning are underlined. The flanking primer, DHFR-His6-r, encoded a C-terminal His-tag (amino acid sequence GSGHHHHHH). The amplified genes were digested with NdeI and XhoI and cloned into similarly digested pACYCSolo. Cloned sequences were screened by colony PCR for insertion of the gene sequence and verified by sequencing with primers pACYCDuet-f 5′-TTGCGCCATTCGATGGTGTC and pACYCDuet-r 5′-AAAACCCCTCAAGACCCGTT. Sequence verified clones were designated pACYCSolo-DHFR and pACYCSolo-DHFR_V10 (Figure 1D) for the expression constructs containing C-terminal His-tagged DHFR and DHFR_V10Amb, respectively.
DHFR nonsense suppression assay
Plasmids pBRIVTC3B and pRST.11B containing Sc-tRNATrpAmb or both ScWRS and Sc-tRNATrpAmb, respectively, were co-expressed with pACYCSolo-DHFR_V10Amb in BL21(DE3) or BL21(DE3)Star cells. A control strain containing pACYCSolo-DHFR_V10Amb and pBR322, which lacks both ScWRS and Sc-tRNATrpAmb, was used to determine background suppression rates. The strains containing these plasmids were grown in Terrific Broth at 37°C to OD600 ∼0.7–0.8 at which point the expression of DHFR and ScWRS was induced by the addition of IPTG to 1 mM. Cells were grown overnight at 37 or 30°C. Following induction and co-expression, the cells were collected by centrifugation and resuspended in binding buffer (50 mM Tris, pH 8.0, 0.5 M NaCl, 5 mM imidazole) containing 1 × protease inhibitor cocktail and 1 mg/ml lysozyme. The resuspended cells were lysed by sonication on ice using 30% probe amplitude and five pulse cycles (30 s ON, 15 s OFF). Following sonication, the cell debris was pelleted via centrifugation at 10 000g for 20 min. The cleared lysate was transferred to a new centrifuge tube and centrifuged again at 11 000g for 15 min to remove any remaining insoluble material.
The His-tagged DHFR was purified via immobilized metal affinity chromatography (IMAC) (34). The cleared lysate was applied to a 1 ml (bead volume) Ni-NTA gravity column pre-incubated with 4× column volumes of binding buffer and allowed to enter the column by gravity flow. The column was washed once with 10× column volumes of Binding Buffer, once with 3× column volumes of Wash Buffer 1 (50 mM Tris, pH 8.0, 0.5 M NaCl, 20 mM imidazole) and once with 3× column volumes of Wash Buffer 2 (50 mM Tris, pH 8.0, 0.5 M NaCl, 30 mM imidazole). DHFR-His6 was eluted from the Ni-NTA column by the addition of 4× column volumes of Elution Buffer (50 mM Tris, pH 8.0, 0.5 M NaCl, 250 mM imidazole). Protein fractions were analyzed by SDS–PAGE on a 4–12% Bis–Tris NuPAGE developed in MES SDS buffer (50 mM MES pH 7.3, 50 mM Tris, 3.5 mM SDS, 1 mM EDTA) under reduced, denaturing conditions and stained using SimplyBlue Safestain. Each fraction was normalized to the wet cell pellet weight prior to loading on the gel. Purified proteins were quantitated using a modified Bradford Assay (Bio-Rad; Hercules, CA, USA).
Edman N-terminal protein sequencing
Proteins (usually ∼300 pmol) to be sequenced were separated on a 4–12% NuPAGE Bis–Tris gradient gels in MES SDS buffer and transferred to a Polyvinylidene fluoride (PVDF) membrane using the Bio-Rad semidry transfer apparatus (Biorad; Hercules, CA, USA) in NuPAGE transfer buffer (25 mM Bis–Tris, pH7.2, 25 mM Bicine, 1 mM EDTA, 2 mM DTT, 20% methanol). The transfer was performed at 25 V for 1 h. Following transfer, the membrane was washed with HPLC grade water containing 2.5 mM DTT for 2 min. The membrane was then stained in 0.1% Coomassie Blue + 2mM DTT until the protein bands were visible. The membrane was destained via multiple washes in 50% methanol, 10% acetic acid, and 1 mM DTT. The destained membrane was washed in HPLC grade water containing 1 mM DTT for 5–10 min. Destained blots were submitted to the Protein Core Facility (Institute for Cellular and Molecular Biology, The University of Texas at Austin) where the first 15–20 amino acids of the membrane-bound 18 kDa DHFR band were excised and sequenced according to Edman degradation protocols on an Applied Biosystems model 477 protein sequencer (ABI; Carlsbad, CA, USA). Sequenced amino acids were identified and quantitated using reference standards. Identification of the amino acid substituted at amber codon 10 of the modified DHFR protein was done by subtractive comparison of the chromatographs produced for position 9 from those produced at position 10. The identity of the amino acid incorporated at position 10 within DHFR was defined as the most abundant (in pmol) amino acid in the subtracted spectrum.
Construction and cloning of Sc-tRNATrp variants
All of the tRNA gene sequences were designed to be cloned between the KpnI and BsrGI sites of the tRNA expression cassette in vectors pBRIVTC3B and pRST.11B. The constructs contained tRNA processing sequences (following the KpnI site and preceding the BsrGI site).
The construct sequences were assembled from four overlapping oligonucleotides according to the gene assembly PCR procedure developed by Stemmer (35). The component ∼60 nt oligonucleotides were designed using DNAWorks software (available at: http://helixweb.nih.gov/dnaworks/) (36). The oligonucleotides necessary to assemble the tRNA sequences are shown in Table 1. Each tRNA variant was assembled from four overlapping oligonucleotides (numbered XXX.1–XXX.4). The oligonucleotides were resuspended in sterilized water to a final concentration of 250 µM, and were mixed together to give a 2.5 µM oligonucleotide mixture of each oligonucleotide. An initial assembly PCR was carried out under the following conditions: 1 µl 2.5 µM oligo mix, 5 µl 10× PfuTurbo buffer, 2.5 µl 4 mM dNTP mix, 1 µl Pfu polymerase (added after an initial denaturation step) and water to 50 µl total volume. The assembly reactions were thermally cycled under the following regime: (1) 95°C-5min, (2) 95°C-30sec, (3) 50°C-30sec, (4) 72°C-60sec, (5) Go to step (2) 25 times and (6) 72°C-10min. Full-length genes were subsequently amplified by PCR: 1 µM assembly PCR, 5 µl 10× PfuTurbo buffer, 2.5 µl 4 mM dNTP mix, 1 µl of each flanking primer (20 µM; Table 1), 1 µl Pfu polymerase (added after an initial denaturation step) and water to 50 µl total volume. The amplification reactions were thermally cycled: (1) 95°C-5min, (2) 95°C-30sec, (3) 62°C-30sec, (4) 72°C-60sec, (5) Go to step (2) 25 times and (6) 72°C-10min. The final PCR was separated on agarose gels to verify the presence of full-length product.
Table 1.
Oligonucleotides used to construct suppressor tRNAs
| Oligonucleotide name | Sequence (5′–3′) |
|---|---|
| RH-ScWA_U30G40.1 | GCGCCTCCGTGGTACCAATTCTTTTTAAGAATTGATGGTATGAAGCGGTGGC |
| RH-ScWA_U30G40.2 | CCTGCAACCCTTCCATTTAGAGTAGAAAGCTCTACCATTGAGCCACCGCTTCATACCATC |
| RH-ScWA_U30G40.3 | TCTAAATGGAAGGGTTGCAGGTTCAATTCCTGTCCGTTTCACCAATTTTGAACCCCGCTT |
| RH-ScWA_U30G40.4 | GTGACGAAATGTACAGAAAACAAAAAACCCCGCCGAAGCGGGGTTCAAAATTGGT |
| RH-ScWA_C70.1 | GCGCCTCCGTGGTACCAATTCTTTTTAAGAATTGATGGTATGAAGCGGTGG |
| RH-ScWA_C70.2 | GCAACCCTTCGATTTAGAGTCGAAAGCTCTACCATTGAGCCACCGCTTCATACCATCAAT |
| RH-ScWA_C70.3 | GACTCTAAATCGAAGGGTTGCAGGTTCAATTCCTGTCCGCTTCACCAATTTTGAACCCCG |
| RH-ScWA_C70.4 | GTGACGAAATGTACAGAAAACAAAAAACCCCGCCGAAGCGGGGTTCAAAATTGGTGAAG |
| RH-ScWA_C70U30G40.1 | GCGCCTCCGTGGTACCAATTCTTTTTAAGAATTGATGGTATGAAGCGGTGGC |
| RH-ScWA_C70U30G40.2 | CCTGCAACCCTTCCATTTAGAGTAGAAAGCTCTACCATTGAGCCACCGCTTCATACCATC |
| RH-ScWA_C70U30G40.3 | TCTAAATGGAAGGGTTGCAGGTTCAATTCCTGTCCGCTTCACCAATTTTGAACCCCGCTT |
| RH-ScWA_C70U30G40.4 | GTGACGAAATGTACAGAAAACAAAAAACCCCGCCGAAGCGGGGTTCAAAATTGGT |
| RH-ScWA_AS1.1 | GCGCCTCCGTGGTACCAATTCTTTTTAAGAATTGATGGTATGAAGCGGTGGC |
| RH-ScWA_AS1.2 | ACCTGCAACCCGCCGGTTTAGAGCCGGCAGCTCTACCATTGAGCCACCGCTTCATACCAT |
| RH-ScWA_AS1.3 | CGGCGGGTTGCAGGTTCAATTCCTGTCCGTTTCACCAATTTTGAACCCCGCTTCGGCGGG |
| RH-ScWA_AS1.4 | GTGACGAAATGTACAGAAAACAAAAAACCCCGCCGAAGCGGG |
| RH-ScWA_AS2.1 | GCGCCTCCGTGGTACCAATTCTTTTTAAGAATTGATGGTATGAAGCGGTG |
| RH-ScWA_AS2.2 | CAACCCCGCGCTTTAGAGGCGCGAGCTCTACCATTGAGCCACCGCTTCATACCATCAATT |
| RH-ScWA_AS2.3 | CTCTAAAGCGCGGGGTTGCAGGTTCAATTCCTGTCCGTTTCACCAATTTTGAACCCCGCT |
| RH-ScWA_AS2.4 | GTGACGAAATGTACAGAAAACAAAAAACCCCGCCGAAGCGGGGTTCAAAATTGGT |
| RH-ScWA_C70AS1.1 | GCGCCTCCGTGGTACCAATTCTTTTTAAGAATTGATGGTATGAAGCGGTGGC |
| RH-ScWA_C70AS1.2 | ACCTGCAACCCGCCGGTTTAGAGCCGGCAGCTCTACCATTGAGCCACCGCTTCATACCAT |
| RH-ScWA_C70AS1.3 | CGGCGGGTTGCAGGTTCAATTCCTGTCCGCTTCACCAATTTTGAACCCCGCTTCGGCGGG |
| RH-ScWA_C70AS1.4 | GTGACGAAATGTACAGAAAACAAAAAACCCCGCCGAAGCGGG |
| RH-ScWA_C70AS2.1 | GCGCCTCCGTGGTACCAATTCTTTTTAAGAATTGATGGTATGAAGCGG |
| RH-ScWA_C70AS2.2 | CCGCGCTTTAGAGGCGCGAGCTCTACCATTGAGCCACCGCTTCATACCATCAATTCTTAA |
| RH-ScWA_C70AS2.3 | CGCCTCTAAAGCGCGGGGTTGCAGGTTCAATTCCTGTCCGCTTCACCAATTTTGAACCCC |
| RH-ScWA_C70AS2.4 | GTGACGAAATGTACAGAAAACAAAAAACCCCGCCGAAGCGGGGTTCAAAATTGGTGAAGC |
| RH-ScWA_AS3.1 | TAGAGCTGCCGCCTCTAAAGCGGCGGGTTGC |
| RH-ScWA_AS3.2 | GCAACCCGCCGCTTTAGAGGCGGCAGCTCTA |
| RH-ScWA_AS3.2.1 | GCGCCTCCGTGGTACCAATTCTTTTTAAGAATTGATGGTATGAAGCGGTGG |
| RH-ScWA_AS3.2.2 | TGCAACCCGGCGGTTTAGAGCCGCCAGCTCTACCATTGAGCCACCGCTTCATACCATCAA |
| RH-ScWA_AS3.2.3 | AACCGCCGGGTTGCAGGTTCAATTCCTGTCCGTTTCACCAATTTTGAACCCCGCTTCGGC |
| RH-ScWA_AS3.2.4 | GTGACGAAATGTACAGAAAACAAAAAACCCCGCCGAAGCGGGGTTCA |
| RH-ScWA_AS3.3.1 | GCGCCTCCGTGGTACCAATTCTTTTTAAGAATTGATGGTATGAAGCGGTGG |
| RH-ScWA_AS3.3.2 | TGCAACCCCCCGGTTTAGAGCCGGGAGCTCTACCATTGAGCCACCGCTTCATACCATCAA |
| RH-ScWA_AS3.3.3 | TAAACCGGGGGGTTGCAGGTTCAATTCCTGTCCGTTTCACCAATTTTGAACCCCGCTTCG |
| RH-ScWA_AS3.3.4 | GTGACGAAATGTACAGAAAACAAAAAACCCCGCCGAAGCGGGGTTCAAAATTG |
| RH-ScWA_AS3.4.1 | GCGCCTCCGTGGTACCAATTCTTTTTAAGAATTGATGGTATGAAGCGGTGG |
| RH-ScWA_AS3.4.2 | TGCAACCCGGCGCTTTAGAGGCGCCAGCTCTACCATTGAGCCACCGCTTCATACCATCAA |
| RH-ScWA_AS3.4.3 | AAGCGCCGGGTTGCAGGTTCAATTCCTGTCCGTTTCACCAATTTTGAACCCCGCTTCGGC |
| RH-ScWA_AS3.4.4 | GTGACGAAATGTACAGAAAACAAAAAACCCCGCCGAAGCGGGGTTCA |
| RH-ScWA_AS3.5.1 | GCGCCTCCGTGGTACCAATTCTTTTTAAGAATTGATGGTATGAAGCGGTG |
| RH-ScWA_AS3.5.2 | CAACCCCCCGCTTTAGAGGCGGGAGCTCTACCATTGAGCCACCGCTTCATACCATCAATT |
| RH-ScWA_AS3.5.3 | CTCTAAAGCGGGGGGTTGCAGGTTCAATTCCTGTCCGTTTCACCAATTTTGAACCCCGCT |
| RH-ScWA_AS3.5.4 | GTGACGAAATGTACAGAAAACAAAAAACCCCGCCGAAGCGGGGTTCAAAATTGGT |
| RH-ScWA_AS3.6.1 | GCGCCTCCGTGGTACCAATTCTTTTTAAGAATTGATGGTATGAAGCGGTG |
| RH-ScWA_AS3.6.2 | CAACCCCGCGGTTTAGAGCCGCGAGCTCTACCATTGAGCCACCGCTTCATACCATCAATT |
| RH-ScWA_AS3.6.3 | CTCTAAACCGCGGGGTTGCAGGTTCAATTCCTGTCCGTTTCACCAATTTTGAACCCCGCT |
| RH-ScWA_AS3.6.4 | GTGACGAAATGTACAGAAAACAAAAAACCCCGCCGAAGCGGGGTTCAAAATTGGT |
The assembled tRNA constructs were digested with KpnI and BsrGI and ligated into similarly digested pBRIVTC3B and pRST.11B with T4 DNA ligase. Clones were screened via colony PCR and sequences were confirmed by sequencing with primers pRS1871-94 5′-AACCCTTGGCAGAACATATCCATC-3′ or pRS2022-47r 5′-CTCGCGTATCGGTGATTCATTCTGCT-3′. Plasmids containing the tRNA variants are designated by the following nomenclature: plasmid name-tRNA variant (i.e. pBRIVTC3B-AS3.4).
β-galactosidase colony spot assay
Plasmids pBRIVTC3B (tRNA only) and pRST.11B (ScWRS and tRNA) containing Sc-tRNATrpAmb variants were transformed into strain CSH108 via electroporation. Transformed colonies were picked and suspended in 10 µl of LB broth. Two microliter aliquots of this mixture were spotted in triplicate onto LB agar plates containing 50 µg/mL ampicillin, 1 mM IPTG and 0.2 mg/mL X-gal. The spotted plates were incubated at 37°C overnight.
LB broth cultures (3 ml) containing 50 µg/ml ampicillin and 1 mM IPTG were also inoculated (10 µl) and grown overnight at 37°C. Some 2 ml of each of the cultures was spun down and the cells lysed in 300 µl of B-PER reagent containing 1 × protease inhibitor and 200 µg/ml lysozyme. The cell debris was removed by centrifugation at 15 000g for 15 min. The protein concentration in the lysate was determined using a modified Bradford Assay. The β-galactosidase activity in the lysate samples was assayed using the high sensitivity β-galactosidase assay kit from Stratagene (Cedar Creek, TX, USA). Triplicate samples from each culture were measured by optical absorbance (at 570 nm) of the chromogenic product, chlorophenol red.
RESULTS
Assaying the functionality of the ScWRS/Sc-tRNATrpAmb pair
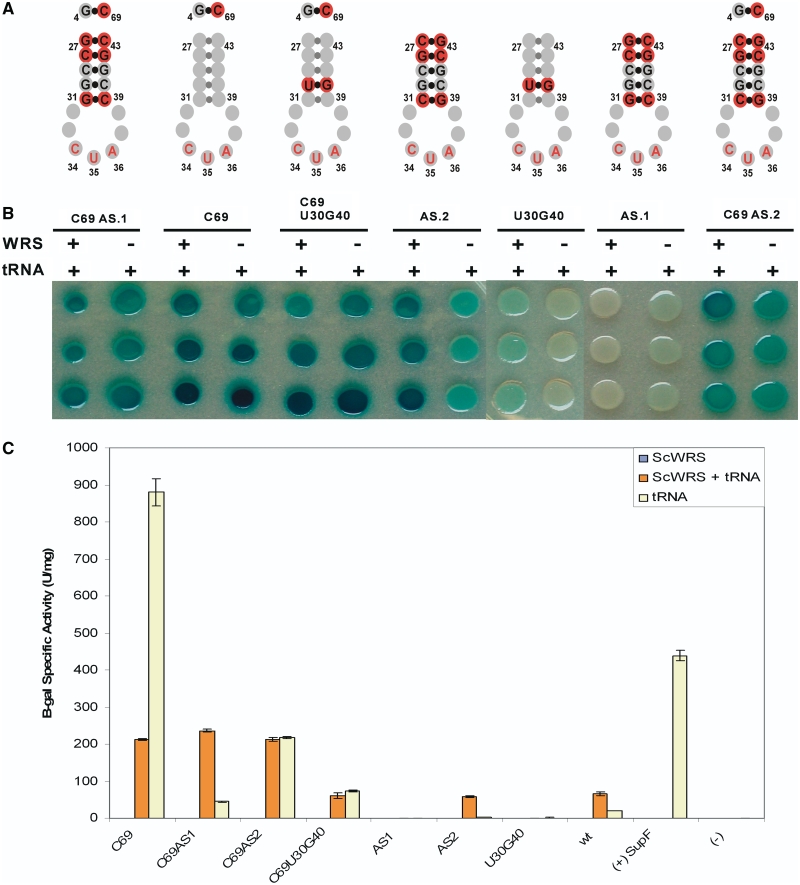
In order to test the functionality of the ScWRS/Sc-tRNATrpAmb pair, the plasmids pBRIVTC3B, pRS.1 and pRST.11B which express only the Sc-tRNATrpAmb, only the ScWRS or both the ScWRS and Sc-tRNATrpAmb together, respectively, were transformed into E. coli strain CSH108. This strain contains an episomal lacZ gene with an amber nonsense codon (TAG) in a position that has previously been shown to be tolerant of a variety of amino acid substitutions. If a suppressor tRNA is active in vivo, then full-length β-galactosidase should be produced resulting in a blue-colored colony upon cleavage of the galactoside analog, 5-bromo-4-chloro-3-indoyl-β-D-galactopyranoside (X-gal). A translationally inactive suppressor or an orthogonal suppressor in the absence of its cognate aaRS will yield in a white colored colony due to in-frame translation termination.
As can be seen in Figure 2, when the ScWRS/tRNATrpAmb pair was expressed in CSH108, a blue colony was formed, indicating that the heterologous yeast suppressor tRNA was active in the bacterial translational system. However, a blue colony was also obtained when the Sc-tRNATrpAmb was expressed by itself in CSH108, indicating that this tRNA was not completely orthogonal to the set of E. coli tRNA synthetases. That said, when these strains were grown in solution, the tryptophanyl pair produced 66 Miller units of β-galactosidase per milligram of total protein (U/mg), whereas the strain carrying only Sc-tRNATrpAmb suppressor produced 21 U/mg of enzyme. While it was likely that the ScWRS was being expressed in an active form and was able to aminoacylate its cognate suppressor tRNA in E. coli, the background aminoacylation activity of the Sc-tRNATrpAmb required additional modification in order to create an orthogonal suppressor.
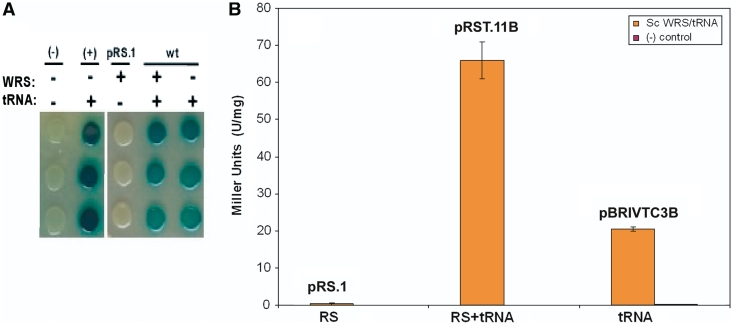
Figure 2.
LacZ suppression assay with Sc-tRNATrpamb. (A) β-galactosidase suppression assay for ScWRS/tRNATrpAmb pair activity. Triplicate spots from the same initial culture are shown in column orientation. Cultures transformed with (+) or without (−) the Sc-tRNATrpAmb or ScWRS are shown in the right panel. The (−) negative control culture represents the background level of β-galactosidase activity in the absence of a suppressor tRNA. The (+) control represents β-galactosidase activity in the presence of a known amber suppressor tRNA [SupF, (Ec-tRNATyrAmb)] which is expressed from the pBRIVTC3B plasmid similar to the experimental suppressor tRNAs. (B) Quantitation of β-galactosidase activity produced by the suppression assay shown in A. pRS.1 expresses just the ScWRS, pRST.11B expresses the ScWRS/tRNATrpAmb pair and pBRIVTC3B expresses just the Sc-tRNATrpAmb suppressor. Error bars represent 1 SD (n = 3).
Identifying background mischarging
To determine which aaRS was responsible for misacylating the yeast tryptophanyl suppressor tRNA (Sc-tRNATrpAmb), the plasmid pBRIVTC3B that expresses the Sc-tRNATrpAmb was co-transformed with pACYCSolo-DHFR_V10Amb, a plasmid that expresses the protein DHFR. The DHFR protein sequence contains an amber nonsense codon (TAG) at amino acid position 10 (V10Amber). Therefore, only in the presence of an amber suppressor tRNA is full-length DHFR translated (Figure 3A). Expression of the pACYCSolo-DHFR_V10Amb plasmid in the absence of the Sc-tRNATrpAmb suppressor does not yield any DHFR as determined by a Commassie stained SDS–PAGE gel (control (−) bands, Figure 3B). The DHFR expressed in the presence of the suppressor was purified via its C-terminal HisTag and submitted for N-terminal Edman peptide sequencing (37). As can be seen in Figure 3C, the Sc-tRNATrpAmb suppressor mediated the incorporation of lysine (K) at position 10 of DHFR. Indicating that the Sc-tRNATrpAmb was being misacylated by the E. coli lysine tRNA synthetase (EcKRS) when expressed alone in E. coli. In contrast, when pACYCSolo-DHFR_V10Amb was co-transformed with plasmid, pRST.11B, which expresses both the ScWRS and the Sc-tRNATrpAmb suppressor DHFR contained primarily tryptophan at position 10 of DHFR (Figure 3D), again confirming the activity of ScWRS in E. coli.
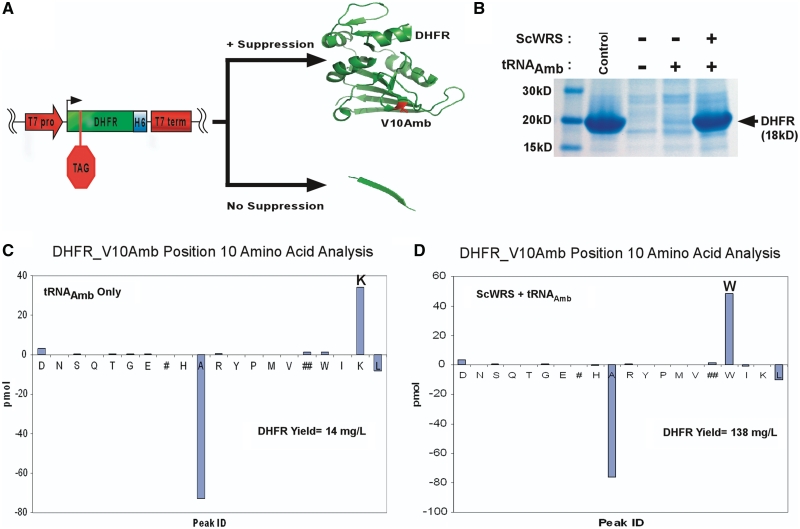
Figure 3.
DHFR suppression assay. (A) The gene for DHFR containing a V10Amber mutation on plasmid pACYCSolo-DHFR_V10Amb produces full-length protein when a functional nonsense suppressor is co-expressed with it. (B) Coomassie stained SDS–PAGE gel showing purification fractions from an immobilized nickel chromatography column for DHFR_V10Amb expressed without Sc-tRNATrpAmb or ScWRS (−) with Sc-tRNATrpAmb only (tRNAamb) or with both ScWRS and Sc-tRNATrpAmb (RS +tRNAamb). M = marker, W = wash fractions and E = elution fractions. Full length DHFR appears as an 18 kDa protein band. (C) N-terminal sequencing results for DHFR position 10 from a culture containing pBRIVTC3B (Sc-tRNATrpAmb only). The bar graph represents the detected chromatographic amino acid abundances for position 10 of DHFR minus those from position 9. Positive abundance represents occupation at position 10 of the DHFR sample, whereas the negatively abundant peaks are carried over from the previous round of sequencing (position 9). (D) N-terminal sequencing results for DHFR position 10 from a culture containing pRST.11B (ScWRS and Sc-tRNATrpAmb). The yields of purified DHFR produced via suppression of the V10Amber codon are given for each sample.
As was the case with β-galactosidase assay, when the strains were grown in solution the suppressor tRNA led to tryptophan being incorporated preferentially to lysine (actually exclusively, within limits of detection for the assay). The ScWRS/Sc-tRNATrpAmb pair produced 138 milligrams of DHFR per liter of bacterial culture (mg/l), versus 14 mg/l for the Sc-tRNATrpAmb suppressor alone and 418 mg/l in the absence of the stop codon. Thus, the suppression efficiency of the ScWRS/Sc-tRNATrpAmb pair can be estimated to be about 33% versus 3% for the Sc-tRNATrpAmb alone. The greater discrimination against lysine in the DHFR expression assay relative to the β-galactosidase expression assay may be due to the fact that protein expression alone was monitored, as opposed to enzymatic activity.
Rational design of an orthogonal tRNATrp amber suppressor
Since the EcKRS was determined to be responsible for causing the background aminoacylation of the Sc-tRNATrpAmb, we compared the Sc-tRNATrpAmb and the E. coli tRNALys (Ec-tRNALys) sequences to determine what features these tRNAs might have in common. Figure 4 shows the cloverleaf secondary structure of both tRNAs with conserved residues colored in green (identical between Ec-tRNALys and Sc-tRNATrpAmb) or blue (universally conserved amongst tRNAs). Surprisingly, the yeast tRNATrpAmb suppressor shares 73% sequence identity with the Ec-tRNALys. More importantly, Sc-tRNATrpAmb shares several key lysyl identity determinants with the Ec- tRNALys. In particular, the A73 discriminator base is a major identity element for both the EcKRS (38,39) and the S. cerevisiae WRS (40). In addition to this discriminator base, the anticodon sequence UUU for Ec-tRNALys (38,39) and CCA for Sc-tRNATrp (23) are important identity elements for recognition by their cognate aaRSs. In the case of the yeast amber suppressor Sc-tRNATrpAmb, since the CCA anticodon sequence has been changed to CUA, the critical lysine identity element U35 (39) is inadvertently added to the anticodon sequence of the suppressor tRNA. While the U34 anticodon base in Ec-tRNALys is normally modified to 5-[(methylamino)-methyl]-2-thiouridine (mnm5s2U) to help read the rare AAG lysine codon (since E. coli lacks an isoacceptor with a CUU anticodon), the C34 in the amber suppressor is accommodated equally well (41).
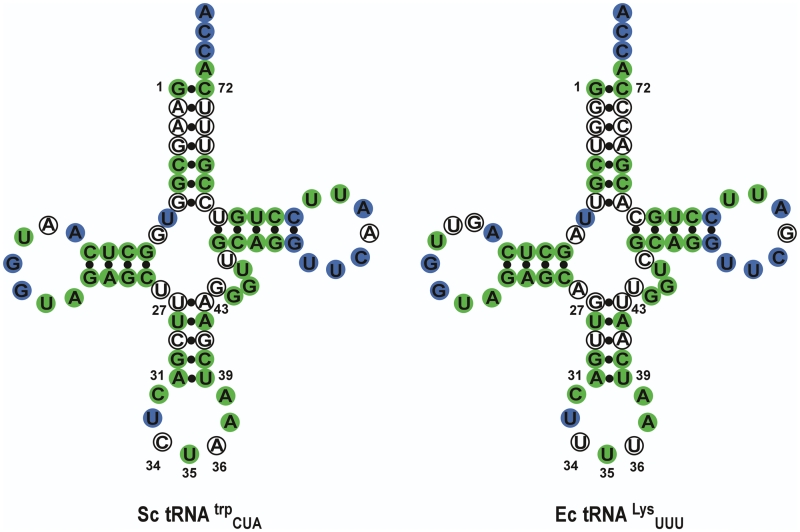
Figure 4.
Comparing Sc-tRNATrpAmb and Ec-tRNALysUUU. Cloverleaf structures of both tRNAs showing the universally conserved residues in blue and residues conserved between the two tRNAs in green, for a total of 73% sequence identity overall.
The fact that the CUA amber suppressor anticodon in Sc-tRNATrpAmb is cross-reactive is not unique. In fact, the importance of the A73 discriminator and U35 anticodon bases for lysine recognition leads amber suppressor tRNAs derived from yeast tRNAs for the other aromatic amino acids (Phe, Tyr) to be cross-reactive to the EcKRS in vivo (42,43). These tRNAs also all share a common G-C base pair between positions 1 and 72 in the acceptor stem, and an anticodon loop sequence that is similar to that of Ec-tRNALys.
Since the anticodon loop and the 1–72 base pair sequences are required for translation activity in most tRNAs, it is unlikely that they can be altered in order to enhance discrimination and orthogonality between the Sc-tRNATrpAmb and Ec-tRNALysUUU. Instead, it has previously been hypothesized that cross recognition may also be due to the structural plasticity of amber suppressor tRNAs to adapt to the KRS binding site (44). A number of facts support this hypothesis. While the crystal structure of the KRS–tRNALys complex has not been determined in its entirety, the anticodon binding domain has been studied in some detail (41,45–47). These structural and biochemical analyses of KRS reveals that it undergoes a dramatic change in conformation upon binding lysine as well as upon the formation of the Lys-AMP adenylate (41,48). Recently, it has been shown that the anticodon binding domain of KRS can enhance the binding efficiency of the lysyl-adenylate in the active site (49).
The hypothesis that the amber suppressor may be flexible enough to be charged by lysyl tRNA synthetase in turn suggests how the suppressor might be rationally engineered to avoid cross-reaction. The anticodon stem of both the E. coli lysine tRNA and the yeast suppressor tRNA share a relatively A-U rich (3/5 bp) stem sequence. Indeed, the Ec-tRNALys has only one G-C pair (G30-C40) in the anticodon stem, while most E. coli tRNA sequences contain three or more G-C pairs in this stem. We therefore hypothesized that ‘stiffening’ the anticodon stem of the suppressor by making it more G-C rich should lead to a reduction in background aminoacylation by EcKRS. This hypothesis was further supported by the fact that most of the known orthogonal suppressors in E. coli have relatively G-C rich anticodon stems (Supplementary Figure S3) and that Fukunaga et al. (43) have reported that the misacylation of the Sc-tRNATyr amber suppressor by the EcKRS can also be reduced by G-C enrichment of the anticodon stem.
Other substitutions have also been shown to reduce background charging by EcKRS. A U30-G40 wobble pair has previously been shown to eliminate the cross reactivity of the EcKRS with the yeast tRNAIle amber suppressor (50). This same negative discriminator was also inserted into the anticodon stem of the Sc-tRNAPhe amber suppressor which led to a reduction in the background misacylation caused by the EcKRS (7).
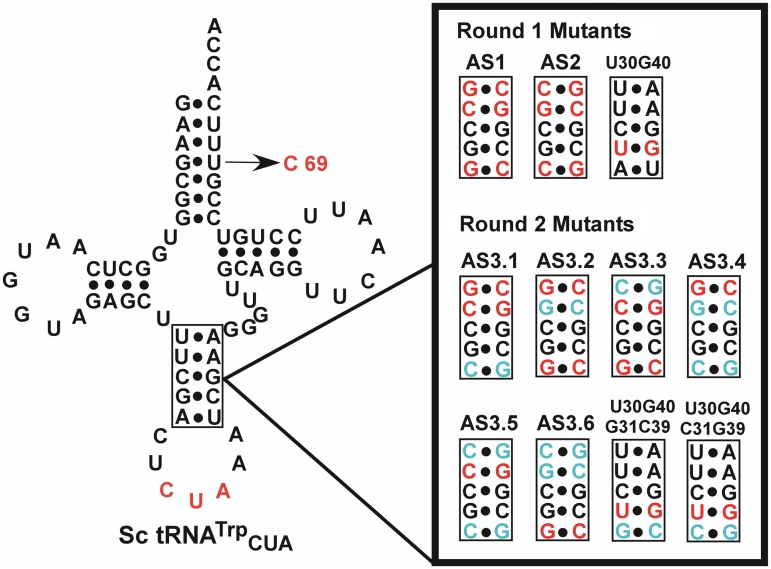
Based on this analysis, parallel paths were devised to eliminate the EcKRS-catalyzed background misacylation of Sc-tRNATrp. First, we constructed G-C rich anticodon stem variants of Sc-tRNATrp (Figure 5, Round 1 Mutants). Secondly, we introduced the negative identity determinant U30-G40 wobble pair into the anticodon stem of Sc-tRNATrp. Finally, a U69C substitution was introduced into the acceptor stem, since replacement of a similar wobble pair (U4-G69) in yeast tRNATyr had previously been shown to increase the suppression activity of this tRNA. Combinations of these changes were also generated.
Figure 5.
Rational Sc-tRNATrpCUA mutants. Designed anticodon stem sequences are shown. The first round of mutations from the original suppressor tRNA are shown in red. The anticodon stem mutants were also made in combination with the U69C mutation. The second round anticodon stem mutations that differ from the first set of suppressor designs are shown in cyan. Mutations preserved from the first round are shown in red. Unchanged nucleotides from the original suppressor tRNA are in black.
These suppressor variants were assayed either alone or in combination with the U69C substitution in the acceptor stem (Figure 6). The suppressor tRNA variants were cloned into the pBRIVTC3B or pRST.11B expression vectors and transformed into E. coli strain CSH108 with or without the cognate ScWRS. Specific suppression of the amber nonsense mutation in the β-galactosidase gene by strains containing the ScWRS/Sc-tRNATrpAmb pair was compared with background suppression from the strains only containing the suppressor tRNA variants (Figure 6). The AS.1 variant shows no suppression activity with or without the ScWRS. The U30G40 substitution not only had greatly reduced activity with ScWRS but also showed no background suppression activity. The AS.2 variant shows roughly equivalent suppression activity to its parent, but had 13-fold reduced background suppression activity (Figure 6).
Figure 6.
Suppression activity of rational suppressor tRNA mutants. (A) Structures of tRNA mutants anticodon stems. Sequence changes from Sc-tRNATrpAmb are denoted by red circles. (B) β-galactosidase colony spot activity assay. Each mutant tRNA was expressed with (+) or without (−) the ScWRS to test for orthogonality. Each sample was taken from the same culture and spotted in triplicate in column format. (C) Quantitation of suppression activity with β-galactosidase. Error bars represent 1 SD (n = 3).
Interestingly, all of the variants that contained the U69C substitution in the acceptor stem showed increased background suppression relative to the original Sc-tRNATrpAmb suppressor. N-terminal sequencing of the DHFR protein produced from the DHFR_V10amb construct co-expressed with this suppressor revealed that the U69C substitution changes the identity of the Sc-tRNATrp suppressor from tryptophan to histidine (data not shown). Therefore, the G4-C69 base pair introduced by this substitution appears to be a heretofore unknown identity element for the E. coli histidine tRNA synthetase (EcHRS) and when combined with the known G1-C72 and A73 identity elements that are also fortuitously in Sc-tRNATrp leads to efficient aminoacylation by EcHRS.
Optimization of an orthogonal tRNATrp amber suppressor
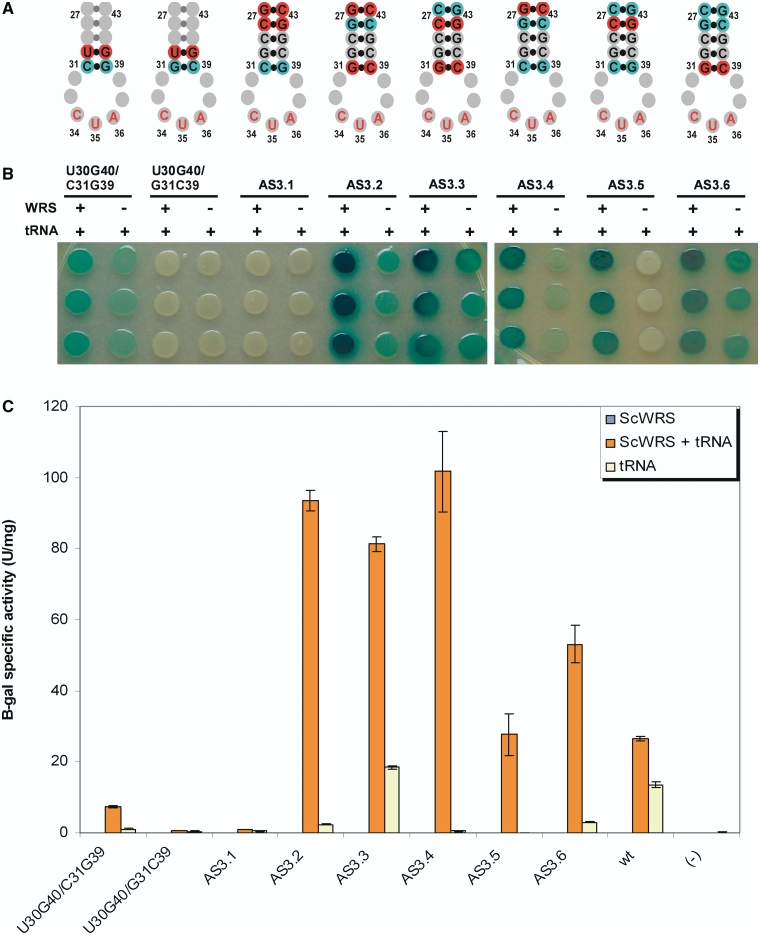
Since initial results indicated that ‘stiffening’ the anticodon stem yielded improved suppressor function, each of the A-U base pairs in the original tRNA anticodon stem was systematically replaced with G-C base pairs (Figure 5, Round 2 Mutants). In addition, since the U30G40 variant eliminated background activity this substitution was also combined with G-C base pairs (Figure 5, Round 2 Mutants). These variants were again screened in the presence and absence of ScWRS (Figure 7).
Figure 7.
Suppression activity of optimized, rational suppressor tRNA mutants. (A) Mutant tRNA anticodon stem structures with mutations shown in red or cyan. (B) β-galactosidase colony spot activity assay. Each mutant tRNA was expressed with (+) or without (−) the ScWRS to test for orthogonality. Each sample was taken from the same culture and spotted in triplicate in column format. (C) Quantitation of suppression activity with β-galactosidase. Error bars represent 1 SD (n = 3).
The G-C replacements showed a range of activities with and without the ScWRS. Variant AS3.1 was translationally inactive, whereas variants AS3.2, AS3.3 and AS3.6 were active yet still showed significant background suppression activity. However, variants AS3.4 and AS3.5 showed a marked reduction in background suppression activity while maintaining activity in the presence of ScWRS. AS3.4 reduced background activity by 28-fold over the original Sc-tRNATrp suppressor, while AS3.5 showed a reduction of nearly 50-fold over the Sc-tRNATrp suppressor. All of the variants containing U30G40 were either inactive or showed residual background activity.
Verifying the orthogonality of suppressors AS3.4 and AS3.5
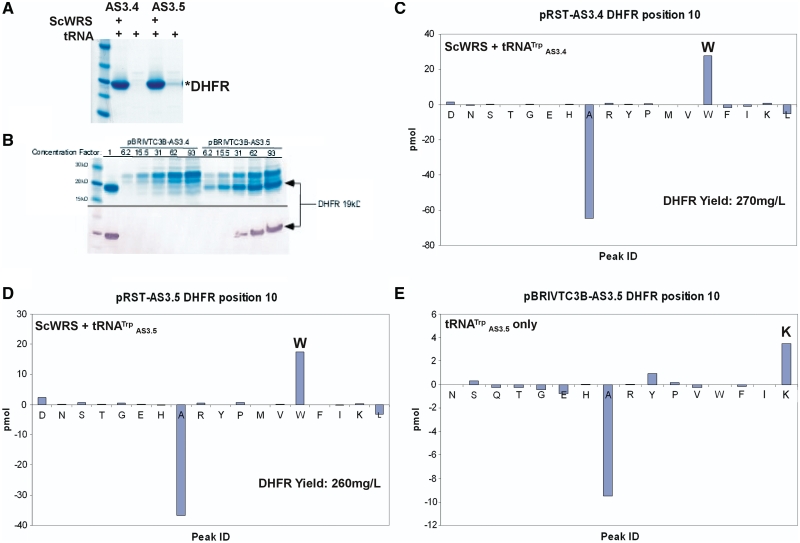
To verify the orthogonality of the two promising suppressor candidates (AS3.4, AS3.5) from the LacZ screen, these suppressors were co-expressed with and without the ScWRS in the presence of the DHFR_V10Amber plasmid. Full-length DHFR with a C-terminal histidine tag was isolated by IMAC, yielding 270 mgs/l DHFR for AS3.4 or 260 mgs/l DHFR for AS3.5 [roughly two-thirds the yield from the construct lacking the amber (TAG) stop codon (pACYCSolo-DHFR-wt)].
However, as can be seen in Figure 8A, the AS3.5 suppressor still produces a faint DHFR band on a Coomassie-stained gel. Concentration of these protein samples cells that only contained the suppressor tRNA clearly reveals that DHFR is still being expressed in the presence of the AS3.5 suppressor but not the AS3.4 suppressor (93-fold concentration; Figure 8B). N-terminal peptide sequencing of the DHFR samples isolated from the strains expressing the AS3.4 and the AS3.5 suppressors with the ScWRS revealed that both incorporate tryptophan in DHFR in response to the amber nonsense codon at position 10 (Figure 8C and D). However, N-terminal sequencing of the DHFR band from the AS3.5 suppressor-only sample revealed that lysine was still being misincorporated at a low level by the Sc-tRNATrp-AS3.5 suppressor (Figure 8E).
Figure 8.
Verifying the orthogonality of AS3.4 and AS3.5. (A) Coomassie stained SDS–PAGE gel of IMAC purified DHFR expressed with and without ScWRS and with the AS3.4 or the AS3.5 suppressor reveals that low level background suppression (AS3.5 suppressor tRNA without ScWRS) still occurs with AS3.5. (B) Concentration of the suppressor tRNA-only (expressed from plasmid pBRIVTC3B) samples confirms the presence of the 18.7 kDa DHFR band in the AS3.5 fraction, but not the AS3.4 fraction. The lane next to the marker is wild-type DHFR run with no additional concentration. The elution fractions from IMAC columns for AS3.4 and AS3.5 expressed without ScWRS but with plasmid pACYCSOLO-DHFR_V10Amb were concentrated and run on the gel at fractions representing concentration factors of 6.2-, 15.5-, 31-, 62- and 93-fold over the initially isolated protein concentration (Top panel). Western blot probing of the C-terminal HisTag on DHFR by a primary AP-conjugated anti-polyhistidine antibody confirms that DHFR is expressed at low levels in the AS3.5 sample but not in the AS3.4 sample (Bottom panel). N-terminal sequencing of the purified DHFR samples reveals that both incorporate tryptophan when the ScWRS is co-expressed with the suppressor tRNA (C and D). However, the AS3.5 suppressor is also misacylated by the E. coli KRS when expressed by itself (E).
Pathways for the acquisition of suppressor activity
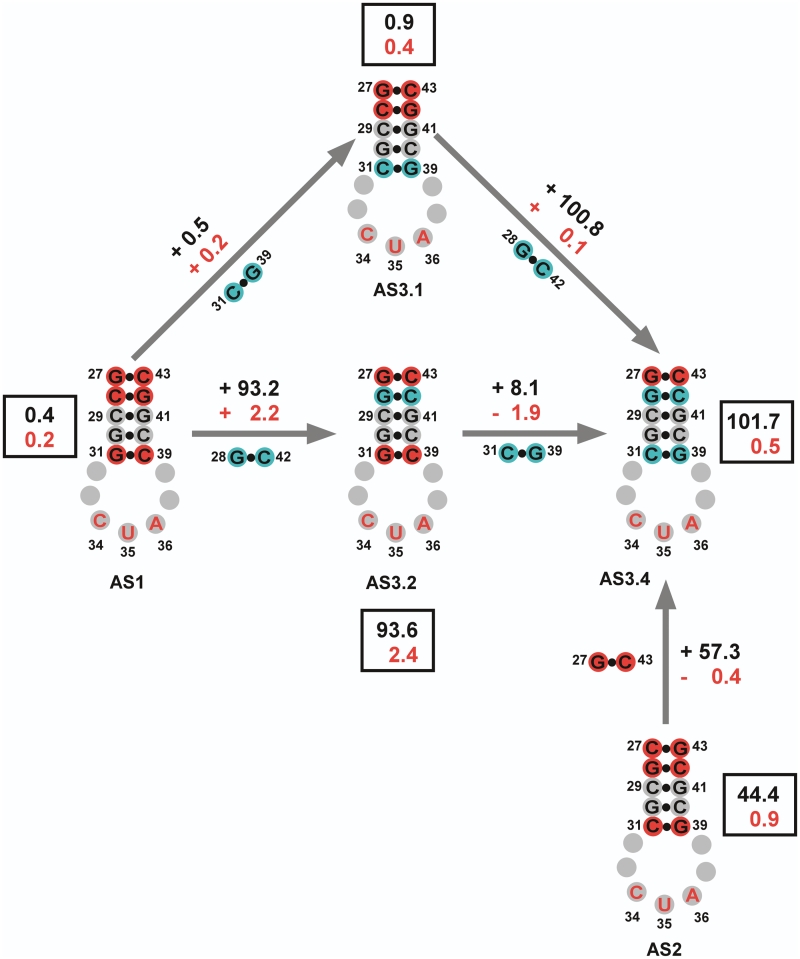
Several functional patterns emerge from these results. If the anticodon stem contains the G27-C43/C28-G42 pairs, the suppressors are non-functional (see variants AS.1 and AS3.1). If the anticodon stem contains the G31-C39 base pair, these tRNAs are functional but show varying levels of background suppression (see variants AS3.2, AS3.3 and AS3.6). If the base of the anticodon stem contains the C31-G39 base pair, the tRNA demonstrates reduced background suppression levels (see variants AS2, AS3.4 and AS3.5).
The contributions of individual base pairs towards charging and orthogonality can be better determined by comparing individual variants (Figure 9). Starting with the inactive AS1 variant, if the C28-G42 base pair is changed to G28-C42 base pair found in variant AS3.2, a significant restoration of suppression activity is restored in the presence of ScWRS (Δ 93.2 U/mg of β-galactosidase activity) but an increase in the background from AS1 is also seen (Δ 2.2 U/mg).
Figure 9.
Dissecting the contribution of mutations to suppressor activity. Anticodon representations of the designed suppressors are shown with the mutations made in the first and second round of designs shown in red and cyan, respectively. Grey spheres represent positions unchanged from the original Sc-tRNATrpAmb suppressor. The suppressor activities were calculated from the average values obtained from the β-galactosidase assay. The top numbers (black) represent the change in activity between any two variants in the presence of the ScWRS. The bottom number (red) represents the change in the background suppressor activity (tRNA only) between any two variants. The boxed values are the raw averaged suppressor activities obtained for each variant, with the top number representing the activity with the ScWRS and the bottom number the activity without the ScWRS.
Continuing to compare substitutions, if the G31-C39 base pair at the base of the stem in variant AS3.2 is then changed to C31-G39 to yield variant AS3.4, we lose most of the background activity seen in AS3.2 (Δ −1.9 U/mg) but gain a small amount of activity (Δ 8.1 U/mg) in the presence of ScWRS. A similar effect is seen if we go from variant AS1 to AS3.1 and then to AS3.4. If the G31-C39 base pair in AS1 is first changed to the C31-G39 base pair to yield variant AS3.1, only a modest gain in activity is seen (Δ 0.5 U/mg) but the suppressor remains largely inactive and produces a white colony in the spotting assay with or without the ScWRS. When the C28-G42 base pair in AS3.1 is finally changed to G28-C42 to yield variant AS3.4, there is a quite significant gain in suppression activity in the presence of the ScWRS (Δ 100.8 U/mg) but little in the way of background activity (Δ 0.1 U/mg). In other words, both mutational routes from AS1 to AS3.4 yield sequence intermediates whose activities can be readily rationalized.
We can also calculate the contribution of the G27-C43 base pair to suppression activity by comparing the activity of variant AS2 with variant AS3.4. In this case, the C27-G43 base pair at the top of the acceptor stem in AS2 is changed to a G27-C43 base pair found in AS3.4, and this more than doubles the activity of the suppressor with ScWRS (Δ 57.3 U/mg), while reducing the background at the same time (Δ −0.4 U/mg).
Overall from these results, it is apparent that the G28-C42 base pair is responsible for increasing the overall activity of the suppressor, but especially the activity associated with the ScWRS. The G27-C43 base pair not only contributes to the overall activity of the suppressor in the presence of the ScWRS, but also contributes in part to reducing the background activity associated with the E. coli KRS. The C31-G39 base pair does not contribute greatly to the overall activity of the suppressor with the ScWRS, but is the main contributor to the reduction of background activity with the E. coli KRS. It is remarkable that these contributions are apparently modular, and not highly context-dependent.
DISCUSSION
One of the requirements for developing an orthogonal aaRS -tRNA pair is that the tRNA works independently of the host aaRSs and tRNAs. This means that the tRNA must not interact with the endogenous aaRSs because this could result in the insertion of more than one amino acid at a given codon. The aaRS -tRNA recognition is governed by interactions between a given aaRS and identity elements on its cognate, the so-called ‘second genetic code’. These interactions are not only unique to each pair, but also typically exhibit species-specific differences, as well, which is one reason that heterologous pairs can often be engineered to function as orthogonal pairs (40). In the case of tryptophanyl tRNAs, the relevant identity elements are thought to be discriminator bases at position 73, either A73 in bacteria or G73 in eukaryotes (40,51).
In order to introduce multiple unnatural amino acids into proteins and whole organisms, it will be useful to have multiple orthogonal tRNA synthetase:tRNA pairs. In particular, we have previously attempted to evolve E. coli to completely utilize the unnatural amino acid 4-fluorotryptophan (20). Insights from these experiments revealed that ‘top-down’ evolution of an organism’s genetic code is not a practical approach to augment the encoded amino acid content of proteins in a living cell (52). These efforts were only partially successful, owing in part to the fact that we did not attempt to pre-engineer the aaRS -tRNA constituents of the cell. Over the last decade, several groups have demonstrated the utility of using mutated or evolved aaRSs and their cognate tRNAs to expand the genetic code and add additional amino acids into proteins (2,53). These ‘bottom-up’ approaches to evolving the genetic code when combined with the cellular adaptation protocols used in the ‘top-down’ approaches may some day lead to artificial cells that have refracted genetic codes which simultaneously encode for multiple unnatural amino acids (52). In order to expand the amino acid diversity of a cell, multiple distinct aaRS–tRNA orthogonal pairs will be required to accommodate the structural diversity multiple unnatural amino acids while maintaining the selectivity of the translation process. To date, there are relatively few orthogonal tRNA synthetase:tRNA pairs available for use in E. coli, therefore in order to generate an ‘unColi’ with an alternative genetic code and a proteome that is completely augmented with an unnatural amino acid, we have developed a new orthogonal tRNA synthetase:tRNA pair based on the S. cerevisiae WRS and tRNA.
We chose the WRS and its cognate tRNA as a starting point for constructing an orthogonal pair due to a number of features which make it particularly attractive for potentially adding unnatural amino acids into proteins. These features include the lack of an editing domain in the WRS and its capacious binding pocket for the largest natural amino acid which has previously led to its use for incorporating tryptophan analogs into proteins (16,17). The expanded substrate flexibility of WRS has even supported adaptive evolution of whole bacteria and bacteria phage proteomes to tryptophan analogs (18–20). However, the engineering of the WRS (and its cognate tRNA) for the site-specific insertion of large, heterocyclic unnatural amino acids has not been sufficiently explored, especially when compared with commonly used tyrosyl tRNA synthetase (54). Zhang et al. (21) have used the WRS from E. coli and an opal (TGA) nonsense suppressor version of its cognate tRNA for the site-specific incorporation of 5-hydroxytryptophan into proteins in mammalian cells, but this orthogonal pair cannot be used in E. coli since it is of bacterial origin and has overlapping recognition features which eliminates its orthogonality. Additionally, screening of synthetic suppressor tRNAs from E. coli has revealed that the tRNATrp is the most efficient tRNA for incorporating large unnatural amino acids such as fluorophores into proteins in E. coli in vitro (55,56). Therefore, we sought to develop an orthogonal tryptophanyl pair for use in the E. coli genetic background.
We anticipated that the yeast tryptophanyl tRNA would be orthogonal to E. coli WRS based upon known differences relative to the E. coli tRNATrp (22,23). The conversion of the CCA anticodon sequence to CUA to make an amber nonsense suppressor was also expected to further enhance the anti-discrimination of the E. coli WRS for the yeast tRNATrpAmb, as this mutation in the E. coli tRNATrp changes its identity from tryptophan to glutamine (24). However, when the yeast tRNATrpAmb was initially expressed in E. coli, it was shown to be functional but not orthogonal to the E. coli translational system (Figure 2). Further analysis revealed that EcKRS was responsible for the misacylation of the original Sc-tRNATrpAmb suppressor in the absence of the ScWRS (Figure 3C). The misacylation of amber suppressor tRNAs by the EcKRS is a widely reported phenomenon, so much so that nearly a third of the amber suppressors derived from different E. coli tRNAs (Ile, Arg, Met (elongation), Asp, Val) all encode lysine in vivo (57). Other amber suppressors including those derived from the yeast tRNAs for phenylalanine (7), isoleucine (50) and tyrosine (43) have also been shown to be misacylated by the EcKRS in vivo.
The major identity elements of the Ec-tRNALys have been previously determined to be the anticodon nucleotides mnm5s2U34, U35, U36 and the discriminator nucleotide A73 (38,39). Since EcKRS can bind the mnm5s2U34 modified nucleotide or cytosine equally well this means that three of the four major identity elements are shared between the yeast tRNATrpamb suppressor and Ec-tRNALys (Figure 4). Making mutations in the anticodon stems of amber suppressor tRNAs derived from the yeast phenylalanine, tyrosine and isoleucine, tRNAs has been previously reported to reduce the mischarging of the suppressor tRNAs by the EcKRS (7,43,50). Fukunaga et al. (43) reported that G-C enrichment of the anticodon stem of Sc-tRNATyrAmb led to significant reduction in misacylation by EcKRS. Based on these findings, these authors hypothesized that the interaction between EcKRS and Ec-tRNALys requires a structural element in the anticodon stem which provides flexibility during the aaRS/tRNA recognition process (44). G-C enrichment of the anticodon stem presumably reduces its flexibility and thereby reduces its interaction with EcKRS. This hypothesis is anecdotally supported by comparison of the anti-codon stems of all of the known E. coli orthogonal tRNAs (Supplementary Figure S3), all of which have relatively G-C rich anticodon stems. In comparison, the tRNATrpAmb contains an A-U rich anticodon stem, much like the Ec-tRNALys.
Building on these findings, we made a series of anticodon stem mutations in Sc-tRNATyrAmb aimed at disrupting the structural plasticity of this stem and thus the aberrant interaction with EcKRS. In order to better parse out the individual effects of G-C substitutions, 16 different Sc-tRNATyrAmb anticodon stem mutants were made and screened via a β-galactosidase suppression assay. Of these 16 mutants, one variant, AS3.4, demonstrated a completely orthogonal phenotype. This variant contained a highly G-C enriched anticodon stem (G28-C42, C31-G39 and G27-C43). The G28-C42 and G27-C43 base pairs at the top of the anticodon stem were found to enhance suppression activity in the presence of the ScWRS, while the C31-G39 base pair at the bottom of the anticodon stem led to the greatest reduction in background suppression by EcKRS (Figure 9). In agreement with these results, this latter mutation has also been shown to reduce the mischarging of the yeast tRNATyr by EcKRS (44).
Interestingly, the C31-G39 base pair in yeast tRNATyrAmb also led to an increase in misacylation by E. coli glutamine tRNA synthetase. This result was similar to the results reported by Normanly et al. (57) two decades ago for amber suppressors derived from two isoacceptors for E. coli tRNAIle. In this case, when tRNAIle1 was made into an amber suppressor it was aminoacylated with glutamine, whereas the tRNAIle2 amber suppressor was charged with lysine. One of the sequence differences between these two tRNAIle isoacceptors was the 31–39 base pair which was C31-G39 in tRNAIle1 and A31-Ψ39 for tRNAIle2, confirming the importance of the C31-G39 base pair for anti-discrimination by the EcKRS. These results suggest that the EcKRS could utilize a tRNA recognition and catalysis mechanism similar to the arginyl-tRNA synthetase which requires some flexibility in its cognate tRNA to undergo its induced-fit mode of catalysis (58). The reduction in the flexibility of the anticodon stem in mutant AS3.4 may have abolished recognition by the EcKRS.
Unfortunately, the previous attempts to rationally design an orthogonal suppressor tRNA based on the yeast tRNATyr were ultimately unsuccessful, even though the methodology was similar to that reported herein. This could be due to subtle structural differences between the engineered yeast tRNATyr anticodon stem relative to the engineered yeast tRNATrp anticodon stem, as none of the mutants assayed were identical. That said, the most orthogonal tRNATyr suppressor that was reported had an anticodon stem sequence that was very similar to our AS3.5 tRNATrp variant, differing at only a single base pair (G29-C41 for the tRNATyr variant versus C29-G41 for AS3.5).
It is especially interesting that the impact of individual G-C substitutions on both charging by the ScWRS and mischarging by the Ec-LysRS appeared to be modular and additive (Figure 9). This was not necessarily to have been expected, since many protein:RNA interactions require precise conformational fits in which any perturbation will significantly decrease affinity and activity. The apparent additivity of the interactions implies that it may be possible in the future to rationally engineer the flexibility of tRNAs to achieve novel specificities. More intriguingly, these results suggest how changes in the genetic code may have evolved. Changes in tRNA anticodons are thought to yield quantized changes in the genetic code, with new amino acids being lost or acquired at particular codons. However, additional mutations in tRNAs that lead to ambiguous charging specificities could modulate the adoption of a new code, moving through an intermediate state in which more than one amino acid was proportionately introduced across from a given codon (59–61). The fact that changes in stem flexibility can gradually and additively lead to alterations in this ambiguity (in ‘mischarging’) implies that evolutionary routes to new codes could lead gradually through ambiguously recoded proteomes.
Previously reported refinements of orthogonal suppressor tRNAs were largely done by directed evolution from a library of mutations that tended to focus any mutations made from a proposed suppressor in the acceptor stem (4), or the loop regions of the tRNA sequence (62) to screen for mutations that enhanced the orthogonal character of a mutant tRNA. These regions of the tRNA are logical starting points for incorporating additional discrimination elements into tRNAs as these regions are commonly accessed by aaRSs. However, in light of the results presented herein, modification of the structural features of tRNAs should also be considered when designing/evolving aaRS interactions. Due to tRNA’s central role in the translation system, tRNA molecules have to interact with multiple proteins (aaRSs, EFTu) and the ribosome itself. The evolved flexibility of the tRNA molecule enables it to interact with multiple binding partners making this feature of tRNAs one of the most essential for translation (63). While interactions between tRNA identity elements and aaRSs (64), and between the tRNA acceptor stem and EFTu (65) have been extensively explored, the role of structural flexibility in fine-tuning these interactions is still largely unknown. Recent and future molecular dynamic simulations of tRNAs during translation will hopefully help to illuminate this largely overlooked property of tRNAs (63). In this regard, mutant tRNAs such as those reported herein could be useful for studying the structural dynamics of tRNA interactions between cognate and non-cognate aaRSs, much as the Hirsh suppressor tRNATrp (66) was useful for decoding the dynamics of codon–anticodon interactions on the ribosome (67).
The Sc-tRNATrp-AS3.4 and ScWRS orthogonal pair reported herein is to our knowledge the only reported tryptophanyl-based orthogonal pair available for use in prokaryotes and should be of great use in expanding efforts to evolve proteins that contain unnatural amino acids. In addition to potentially being used for changing the genetic code of a cell, this orthogonal suppressor system could also be used for recoding in vitro. For example, recombinant translation systems [similar to the PURE system (68)] have been used by Forster and colleagues (69,70) to introduce chemically acylated tRNAs containing unnatural amino acids into peptides. Similarly, Szostak and colleagues (71,72) have made peptides containing multiple unnatural amino acid analogs by taking advantage of the natural substrate flexibility of aaRSs. In either instance, the new orthogonal pair could be introduced in place of tRNA synthetase and its cognate tRNA, or in addition to the standard complement of synthetases and tRNAs in order to more efficiently generate proteins with altered compositions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: National Science Foundation (grant number MCB-0943383); National Institutes of Health/National Institute of General Medical Sciences (grant number R01GM084703); The Welch Foundation (grant number F-1654).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Kiga D, Sakamoto K, Kodama K, Kigawa T, Matsuda T, Yabuki T, Shirouzu M, Harada Y, Nakayama H, Takio K, et al. An engineered Escherichia coli tyrosyl-tRNA synthetase for site-specific incorporation of an unnatural amino acid into proteins in eukaryotic translation and its application in a wheat germ cell-free system. Proc. Natl Acad. Sci. USA. 2002;99:9715–9720. doi: 10.1073/pnas.142220099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xie J, Schultz PG. A chemical toolkit for proteins–an expanded genetic code. Nat. Rev. Mol. Cell Biol. 2006;7:775–782. doi: 10.1038/nrm2005. [DOI] [PubMed] [Google Scholar]
- 3.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 4.Anderson JC, Wu N, Santoro SW, Lakshman V, King DS, Schultz PG. An expanded genetic code with a functional quadruplet. Proc. Natl Acad. Sci. USA. 2004;101:7566–7571. doi: 10.1073/pnas.0401517101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pastrnak M, Mageliery TM, Schultz PG. A new orthogonal suppressor tRNA/Aminoacyl-tRNA synthetase pair for evolving an organism with an expanded genetic code. Helv. Chim. Acta. 2000;83:2277–2286. [Google Scholar]
- 6.Liu DR, Schultz PG. Progress toward the evolution of an organism with an expanded genetic code. Proc. Natl Acad. Sci. USA. 1999;96:4780–4785. doi: 10.1073/pnas.96.9.4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwon I, Wang P, Tirrell DA. Design of a bacterial host for site-specific incorporation of p-bromophenylalanine into recombinant proteins. J. Am. Chem. Soc. 2006;128:11778–11783. doi: 10.1021/ja0626281. [DOI] [PubMed] [Google Scholar]
- 8.Santoro SW, Anderson JC, Lakshman V, Schultz PG. An archaebacteria-derived glutamyl-tRNA synthetase and tRNA pair for unnatural amino acid mutagenesis of proteins in Escherichia coli. Nucleic Acids Res. 2003;31:6700–6709. doi: 10.1093/nar/gkg903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson JC, Schultz PG. Adaptation of an orthogonal archaeal leucyl-tRNA and synthetase pair for four-base, amber, and opal suppression. Biochemistry. 2003;42:9598–9608. doi: 10.1021/bi034550w. [DOI] [PubMed] [Google Scholar]
- 10.Nguyen DP, Garcia Alai MM, Kapadnis PB, Neumann H, Chin JW. Genetically encoding N(epsilon)-methyl-L-lysine in recombinant histones. J. Am. Chem. Soc. 2009;131:14194–14195. doi: 10.1021/ja906603s. [DOI] [PubMed] [Google Scholar]
- 11.Landes C, Perona JJ, Brunie S, Rould MA, Zelwer C, Steitz TA, Risler JL. A structure-based multiple sequence alignment of all class I aminoacyl-tRNA synthetases. Biochimie. 1995;77:194–203. doi: 10.1016/0300-9084(96)88125-9. [DOI] [PubMed] [Google Scholar]
- 12.Yang XL, Otero FJ, Skene RJ, McRee DE, Schimmel P, Ribas de Pouplana L. Crystal structures that suggest late development of genetic code components for differentiating aromatic side chains. Proc. Natl Acad. Sci. USA. 2003;100:15376–15380. doi: 10.1073/pnas.2136794100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Praetorius-Ibba M, Stange-Thomann N, Kitabatake M, Ali K, Soll I, Carter CW, Jr, Ibba M, Soll D. Ancient adaptation of the active site of tryptophanyl-tRNA synthetase for tryptophan binding. Biochemistry. 2000;39:13136–13143. doi: 10.1021/bi001512t. [DOI] [PubMed] [Google Scholar]
- 14.Ribas de Pouplana L, Frugier M, Quinn CL, Schimmel P. Evidence that two present-day components needed for the genetic code appeared after nucleated cells separated from eubacteria. Proc. Natl Acad. Sci. USA. 1996;93:166–170. doi: 10.1073/pnas.93.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doublie S, Bricogne G, Gilmore C, Carter CW., Jr Tryptophanyl-tRNA synthetase crystal structure reveals an unexpected homology to tyrosyl-tRNA synthetase. Structure. 1995;3:17–31. doi: 10.1016/s0969-2126(01)00132-0. [DOI] [PubMed] [Google Scholar]
- 16.Budisa N. Engineering the Genetic Code. Weinheim: Wiley-VCH; 2006. [Google Scholar]
- 17.Budisa N, Rubini M, Bae JH, Weyher E, Wenger W, Golbik R, Huber R, Moroder L. Global replacement of tryptophan with aminotryptophans generates non-invasive protein-based optical pH sensors. Angew. Chem. Int. Ed. Engl. 2002;41:4066–4069. doi: 10.1002/1521-3773(20021104)41:21<4066::AID-ANIE4066>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 18.Wong JT. Membership mutation of the genetic code: loss of fitness by tryptophan. Proc. Natl Acad. Sci. USA. 1983;80:6303–6306. doi: 10.1073/pnas.80.20.6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bacher JM, Bull JJ, Ellington AD. Evolution of phage with chemically ambiguous proteomes. BMC Evol. Biol. 2003;3:24. doi: 10.1186/1471-2148-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bacher JM, Ellington AD. Selection and characterization of Escherichia coli variants capable of growth on an otherwise toxic tryptophan analogue. J. Bacteriol. 2001;183:5414–5425. doi: 10.1128/JB.183.18.5414-5425.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Z, Alfonta L, Tian F, Bursulaya B, Uryu S, King DS, Schultz PG. Selective incorporation of 5-hydroxytryptophan into proteins in mammalian cells. Proc. Natl Acad. Sci. USA. 2004;101:8882–8887. doi: 10.1073/pnas.0307029101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Himeno H, Hasegawa T, Asahara H, Tamura K, Shimizu M. Identity determinants of E. coli tryptophan tRNA. Nucleic Acids Res. 1991;19:6379–6382. doi: 10.1093/nar/19.23.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yesland KD, Johnson JD. Anticodon bases C34 and C35 are major, positive, identity elements in Saccharomyces cerevisiae tRNA(Trp) Nucleic Acids Res. 1993;21:5079–5084. doi: 10.1093/nar/21.22.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yaniv M, Folk WR, Berg P, Soll L. A single mutational modification of a tryptophan-specific transfer RNA permits aminoacylation by glutamine and translation of the codon UAG. J. Mol. Biol. 1974;86:245–260. doi: 10.1016/0022-2836(74)90016-3. [DOI] [PubMed] [Google Scholar]
- 25.Yesland KD, Nelson AW, Six Feathers DM, Johnson JD. Identity of Saccharomyces cerevisiae tRNA(Trp) is not changed by an anticodon mutation that creates an amber suppressor. J. Biol. Chem. 1993;268:217–220. [PubMed] [Google Scholar]
- 26.Atkin AL, Riazi MA, Greer CL, Roy KL, Bell JB. The functional analysis of nonsense suppressors derived from in vitro engineered Saccharomyces cerevisiae tRNA(Trp) genes. Gene. 1993;134:57–65. doi: 10.1016/0378-1119(93)90174-2. [DOI] [PubMed] [Google Scholar]
- 27.John TR, Ghosh M, Johnson JD. Identification and expression of the Saccharomyces cerevisiae cytoplasmic tryptophanyl-tRNA synthetase gene. Yeast. 1997;13:37–41. doi: 10.1002/(SICI)1097-0061(199701)13:1<37::AID-YEA55>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 28.de Boer HA, Comstock LJ, Vasser M. The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc. Natl Acad. Sci. USA. 1983;80:21–25. doi: 10.1073/pnas.80.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balbas P, Soberon X, Merino E, Zurita M, Lomeli H, Valle F, Flores N, Bolivar F. Plasmid vector pBR322 and its special-purpose derivatives–a review. Gene. 1986;50:3–40. doi: 10.1016/0378-1119(86)90307-0. [DOI] [PubMed] [Google Scholar]
- 30.Duester G, Campen RK, Holmes WM. Nucleotide sequence of an Escherichia coli tRNA (Leu 1) operon and identification of the transcription promoter signal. Nucleic Acids Res. 1981;9:2121–2139. doi: 10.1093/nar/9.9.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bauer BF, Kar EG, Elford RM, Holmes WM. Sequence determinants for promoter strength in the leuV operon of Escherichia coli. Gene. 1988;63:123–134. doi: 10.1016/0378-1119(88)90551-3. [DOI] [PubMed] [Google Scholar]
- 32.Ross W, Salomon J, Holmes WM, Gourse RL. Activation of Escherichia coli leuV transcription by FIS. J. Bacteriol. 1999;181:3864–3868. doi: 10.1128/jb.181.12.3864-3868.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsu LM, Klee HJ, Zagorski J, Fournier MJ. Structure of an Escherichia coli tRNA operon containing linked genes for arginine, histidine, leucine, and proline tRNAs. J. Bacteriol. 1984;158:934–942. doi: 10.1128/jb.158.3.934-942.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith MC, Furman TC, Ingolia TD, Pidgeon C. Chelating peptide-immobilized metal ion affinity chromatography. A new concept in affinity chromatography for recombinant proteins. J. Biol. Chem. 1988;263:7211–7215. [PubMed] [Google Scholar]
- 35.Stemmer WP, Crameri A, Ha KD, Brennan TM, Heyneker HL. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene. 1995;164:49–53. doi: 10.1016/0378-1119(95)00511-4. [DOI] [PubMed] [Google Scholar]
- 36.Hoover DM, Lubkowski J. DNAWorks: an automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic Acids Res. 2002;30:e43. doi: 10.1093/nar/30.10.e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reim DF, Speicher DW. N-terminal sequence analysis of proteins and peptides. Curr. Protoc. Protein Sci. 2001 doi: 10.1002/0471140864.ps1110s08. Chapter 11, Unit 11 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McClain WH, Foss K, Jenkins RA, Schneider J. Nucleotides that determine Escherichia coli tRNA(Arg) and tRNA(Lys) acceptor identities revealed by analyses of mutant opal and amber suppressor tRNAs. Proc. Natl Acad. Sci. USA. 1990;87:9260–9264. doi: 10.1073/pnas.87.23.9260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamura K, Himeno H, Asahara H, Hasegawa T, Shimizu M. In vitro study of E.coli tRNA(Arg) and tRNA(Lys) identity elements. Nucleic Acids Res. 1992;20:2335–2339. doi: 10.1093/nar/20.9.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu F, Chen X, Xin L, Chen L, Jin Y, Wang D. Species-specific differences in the operational RNA code for aminoacylation of tRNA(Trp) Nucleic Acids Res. 2001;29:4125–4133. doi: 10.1093/nar/29.20.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cusack S, Yaremchuk A, Tukalo M. The crystal structures of T. thermophilus lysyl-tRNA synthetase complexed with E. coli tRNA(Lys) and a T. thermophilus tRNA(Lys) transcript: anticodon recognition and conformational changes upon binding of a lysyl-adenylate analogue. EMBO J. 1996;15:6321–6334. [PMC free article] [PubMed] [Google Scholar]
- 42.Furter R. Expansion of the genetic code: site-directed p-fluoro-phenylalanine incorporation in Escherichia coli. Protein Sci. 1998;7:419–426. doi: 10.1002/pro.5560070223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fukunaga J, Yokogawa T, Ohno S, Nishikawa K. Misacylation of yeast amber suppressor tRNA(Tyr) by E. coli lysyl-tRNA synthetase and its effective repression by genetic engineering of the tRNA sequence. J. Biochem. 2006;139:689–696. doi: 10.1093/jb/mvj078. [DOI] [PubMed] [Google Scholar]
- 44.Fukunaga J, Ohno S, Nishikawa K, Yokogawa T. A base pair at the bottom of the anticodon stem is reciprocally preferred for discrimination of cognate tRNAs by Escherichia coli lysyl- and glutaminyl-tRNA synthetases. Nucleic Acids Res. 2006;34:3181–3188. doi: 10.1093/nar/gkl414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brevet A, Chen J, Commans S, Lazennec C, Blanquet S, Plateau P. Anticodon recognition in evolution: switching tRNA specificity of an aminoacyl-tRNA synthetase by site-directed peptide transplantation. J. Biol. Chem. 2003;278:30927–30935. doi: 10.1074/jbc.M302618200. [DOI] [PubMed] [Google Scholar]
- 46.Commans S, Lazard M, Delort F, Blanquet S, Plateau P. tRNA anticodon recognition and specification within subclass IIb aminoacyl-tRNA synthetases. J. Mol. Biol. 1998;278:801–813. doi: 10.1006/jmbi.1998.1711. [DOI] [PubMed] [Google Scholar]
- 47.Commans S, Plateau P, Blanquet S, Dardel F. Solution structure of the anticodon-binding domain of Escherichia coli lysyl-tRNA synthetase and studies of its interaction with tRNA(Lys) J. Mol. Biol. 1995;253:100–113. doi: 10.1006/jmbi.1995.0539. [DOI] [PubMed] [Google Scholar]
- 48.Onesti S, Desogus G, Brevet A, Chen J, Plateau P, Blanquet S, Brick P. Structural studies of lysyl-tRNA synthetase: conformational changes induced by substrate binding. Biochemistry. 2000;39:12853–12861. doi: 10.1021/bi001487r. [DOI] [PubMed] [Google Scholar]
- 49.Saruwatari Y, Wada T, Takita T, Inouye K. Substrate-induced conformational changes of the truncated catalytic domain of Geobacillus stearothermophilus lysyl-tRNA synthetase as examined by fluorescence. Biochim. Biophys. Acta. 2008;1784:1633–1640. doi: 10.1016/j.bbapap.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 50.Buttcher V, Senger B, Schumacher S, Reinbolt J, Fasiolo F. Modulation of the suppression efficiency and amino acid identity of an artificial yeast amber isoleucine transfer RNA in Escherichia coli by a G-U pair in the anticodon stem. Biochem. Biophys. Res. Commun. 1994;200:370–377. doi: 10.1006/bbrc.1994.1458. [DOI] [PubMed] [Google Scholar]
- 51.Guo Q, Gong Q, Tong KL, Vestergaard B, Costa A, Desgres J, Wong M, Grosjean H, Zhu G, Wong JT, et al. Recognition by tryptophanyl-tRNA synthetases of discriminator base on tRNATrp from three biological domains. J. Biol. Chem. 2002;277:14343–14349. doi: 10.1074/jbc.M111745200. [DOI] [PubMed] [Google Scholar]
- 52.Bacher JM, Hughes RA, Tze-Fei Wong J, Ellington AD. Evolving new genetic codes. Trends Ecol. Evol. 2004;19:69–75. doi: 10.1016/j.tree.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 53.Wang Q, Parrish AR, Wang L. Expanding the genetic code for biological studies. Chem. Biol. 2009;16:323–336. doi: 10.1016/j.chembiol.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xie J, Schultz PG. An expanding genetic code. Methods. 2005;36:227–238. doi: 10.1016/j.ymeth.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 55.Taira H, Matsushita Y, Kojima K, Hohsaka T. Development of amber suppressor tRNAs appropriate for incorporation of nonnatural amino acids. Nucleic Acids Symp. Ser. 2006:233–234. doi: 10.1093/nass/nrl116. [DOI] [PubMed] [Google Scholar]
- 56.Taira H, Matsushita Y, Kojima K, Shiraga K, Hohsaka T. Comprehensive screening of amber suppressor tRNAs suitable for incorporation of non-natural amino acids in a cell-free translation system. Biochem. Biophys. Res. Commun. 2008;374:304–308. doi: 10.1016/j.bbrc.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 57.Normanly J, Kleina LG, Masson JM, Abelson J, Miller JH. Construction of Escherichia coli amber suppressor tRNA genes. III. Determination of tRNA specificity. J. Mol. Biol. 1990;213:719–726. doi: 10.1016/S0022-2836(05)80258-X. [DOI] [PubMed] [Google Scholar]
- 58.Guigou L, Mirande M. Determinants in tRNA for activation of arginyl-tRNA synthetase: evidence that tRNA flexibility is required for the induced-fit mechanism. Biochemistry. 2005;44:16540–16548. doi: 10.1021/bi051575h. [DOI] [PubMed] [Google Scholar]
- 59.Schultz DW, Yarus M. Transfer RNA mutation and the malleability of the genetic code. J. Mol. Biol. 1994;235:1377–1380. doi: 10.1006/jmbi.1994.1094. [DOI] [PubMed] [Google Scholar]
- 60.Schultz DW, Yarus M. On malleability in the genetic code. J. Mol. Evol. 1996;42:597–601. doi: 10.1007/BF02352290. [DOI] [PubMed] [Google Scholar]
- 61.Yarus M, Schultz DW. Further comments on codon reassignment. Response. J. Mol. Evol. 1997;45:3–6. doi: 10.1007/pl00006171. [DOI] [PubMed] [Google Scholar]
- 62.Wang L, Schultz PG. A general approach for the generation of orthogonal tRNAs. Chem. Biol. 2001;8:883–890. doi: 10.1016/s1074-5521(01)00063-1. [DOI] [PubMed] [Google Scholar]
- 63.Alexander RW, Eargle J, Luthey-Schulten Z. Experimental and computational determination of tRNA dynamics. FEBS Lett. 2010;584:376–386. doi: 10.1016/j.febslet.2009.11.061. [DOI] [PubMed] [Google Scholar]
- 64.Giege R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sanderson LE, Uhlenbeck OC. Exploring the specificity of bacterial elongation factor Tu for different tRNAs. Biochemistry. 2007;46:6194–6200. doi: 10.1021/bi602548v. [DOI] [PubMed] [Google Scholar]
- 66.Hirsh D. Tryptophan transfer RNA as the UGA suppressor. J. Mol. Biol. 1971;58:439–458. doi: 10.1016/0022-2836(71)90362-7. [DOI] [PubMed] [Google Scholar]
- 67.Cochella L, Green R. An active role for tRNA in decoding beyond codon: anticodon pairing. Science. 2005;308:1178–1180. doi: 10.1126/science.1111408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, Ueda T. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 69.Forster AC, Tan Z, Nalam MN, Lin H, Qu H, Cornish VW, Blacklow SC. Programming peptidomimetic syntheses by translating genetic codes designed de novo. Proc. Natl Acad. Sci. USA. 2003;100:6353–6357. doi: 10.1073/pnas.1132122100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Forster AC, Cornish VW, Blacklow SC. Pure translation display. Anal Biochem. 2004;333:358–364. doi: 10.1016/j.ab.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 71.Josephson K, Hartman MC, Szostak JW. Ribosomal synthesis of unnatural peptides. J. Am. Chem. Soc. 2005;127:11727–11735. doi: 10.1021/ja0515809. [DOI] [PubMed] [Google Scholar]
- 72.Hartman MC, Josephson K, Lin CW, Szostak JW. An expanded set of amino acid analogs for the ribosomal translation of unnatural peptides. PLoS ONE. 2007;2:e972. doi: 10.1371/journal.pone.0000972. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.