Abstract
Many genes exist in the form of families; however, little is known about their size variation, evolution and biology. Here, we present the size variation and evolution of the nucleotide-binding site (NBS)-encoding gene family and receptor-like kinase (RLK) gene family in Oryza, Glycine and Gossypium. The sizes of both families vary by numeral fold, not only among species, surprisingly, also within a species. The size variations of the gene families are shown to correlate with each other, indicating their interactions, and driven by natural selection, artificial selection and genome size variation, but likely not by polyploidization. The numbers of genes in the families in a polyploid species are similar to those of one of its diploid donors, suggesting that polyploidization plays little roles in the expansion of the gene families and that organisms tend not to maintain their ‘surplus’ genes in the course of evolution. Furthermore, it is found that the size variations of both gene families are associated with organisms’ phylogeny, suggesting their roles in speciation and evolution. Since both selection and speciation act on organism’s morphological, physiological and biological variation, our results indicate that the variation of gene family size provides a source of genetic variation and evolution.
INTRODUCTION
A significant finding of modern genome research is that many of the genes in a genome exist in multiple copies or the form of families (1–4) that are often defined groups of homologous or paralogous genes that may have similar functions (2); however, studies in their size variation, evolution and biology are very limited (5). It has been shown that gene families exist commonly in the life kingdom including both prokaryotes (1,6–8) and eukaryotes (2–4,9,10). They, as individual genes, could be born or dead in whole and contract or expand dramatically in size during a course of organismal evolution through a process called the genomic ‘revolving door’ of gene gain (through duplication) and loss (through deletion and/or pseudogenization) (2,3,5,11,12). Consequently, the size of or the number of genes within a gene family has been found to vary significantly among different species (2,9,10,13–16). Nevertheless, it is unknown what the variation of gene family sizes implies with regard to morphology, physiology and complexity of the organisms. It has been hypothesized that the birth and death or expansion and contraction of gene families may play a significant role in the observed difference between organisms in morphology, physiology and complexity (1–3,8–10,17,18), but this hypothesis is untested. A crucial step to testing the hypothesis is to have knowledge of whether the number of genes in a gene family varies among closely related species, particularly within a species, and what factors may shape the fate of a gene family in the course of speciation and evolution. However, there is little such knowledge due to the fact that most studies in the field were restricted to diverged species (2,8–10,13,15,17,18). There have been few studies reported in the variation and evolution of gene family size in plants (15). No studies have been reported in the variation and evolution of a gene family size within a species.
In this study, we address the question of variation and evolution of gene family sizes within a species and among congeneric species in plants using one monocotyledonous genus, Oryza L., and two dicotyledonous genera, Gossypium L. and Glycine Willd., and two gene families, the nucleotide-binding site (NBS)-encoding gene family and the receptor-like kinase (RLK) gene family. Genome sequence analyses showed that the NBS family has 500–600 members in O. sativa ssp. japonica cv. Nipponbare (15,19–21), while the RLK family has 1147 members in O. sativa ssp. indica cv. 93-11 (22). The NBS family contributes ∼80% of genes conferring plant defense (23,24) and the RLK genes play important roles in plant growth, development and defense (25,26). We first scrutinized the numbers of genes in each of the families in the genomes of 187 accessions or cultivars (hereafter referred to as lines) randomly selected from 57 species of the three genera. Then, we analyzed the size variation of the gene families among lines of a species and among congeneric species, and estimated the roles of polyploidization, genome size variation, natural selection, artificial selection including domestication, breeding and cultivation, and gene family interaction in the gene family size variation and evolution. Finally, we determined the evolution of the gene family sizes in context of organismal speciation and evolution. This represents the first comprehensive study of the size variation and evolution of gene families within a species and among closely related species in plants. These findings provide novel insights into the molecular basis of genetic variation and evolution of plants in morphology, physiology and complexity.
MATERIALS AND METHODS
Detailed materials and methods, the comparative analyses of the methods that have been previously used to estimate the number of genes in a gene family and the pilot experiment results are provided in ‘Materials and Methods’ section in Supplementary Data.
Plant materials, DNA preparation and methodology of gene copy number estimation
A total of 187 lines randomly selected from 57 species of Oryza, Glycine and Gossypium were used in this study, with 1–18 lines per species (Supplementary Table S1, Materials and Methods in Supplementary Data). Nuclear DNA was isolated from 1–5 plants of each line and purified, and their concentrations were determined and verified. To assay the number of genes in the NBS and RLK families in the genome of each line, we first comparatively analyzed the methods that have been used to estimate the number of genes in a gene family in a genome (2,3,27–31). These methods included whole-genome sequence blast analysis (WSBA), membrane array (MA), microarray (M), random genomic clone sequencing (RGCS), quantitative real-time PCR (qrtPCR) and small-insert DNA library screening (SDLS) (Supplementary Table S2). Then, we conducted pilot experiments to test the three methods, MA, SDLS and WSBA, selected from the analysis for this study (Materials and Methods in Supplementary Data). As comparable results were obtained among the three methods (Supplementary Tables S3–S5), the MA method is practically the most feasible, simplest and most economical among the six methods for our research purposes (Supplementary Table S2) and well reproducible among different plants within a rice cultivar (Table 1), we chose to use the MA method to assay the number of genes in the NBS and RLK families in all 187 lines of the species selected.
Table 1.
ANOVA in log10-transformed number of genes in the NBS and RLK families within a cultivar, within a subspecies, within a species, among subspecies and among species in Oryza, Glycine and Gossipium
| Genome | No. of lines | df | NBS |
RLKa |
|||
|---|---|---|---|---|---|---|---|
| F-value | P-value | F-value | P-value | ||||
| Oryza | |||||||
| Different plants within a cultivar | |||||||
| cv. Teqing | AA | 23 | 0.715 | 0.554 | 0.587 | 0.650 | |
| cv. Nipponbare | AA | 29 | 1.085 | 0.385 | 1.297 | 0.305 | |
| Different cultivars within a subspecies | |||||||
| ssp. indica | AA | 6 | 53 | 7.872*** | 0.000 | 4.000** | 0.009 |
| ssp. japonica | AA | 6 | 59 | 10.718*** | 0.000 | 1.459 | 0.280 |
| ssp. javanica | AA | 6 | 35 | 12.366*** | 0.000 | 2.013 | 0.162 |
| Among subspecies of a species | |||||||
| Ind–jap–jav | AA | 18 | 149 | 12.840*** | 0.000 | 9.477*** | 0.000 |
| Different cultivars or accessions within a species | |||||||
| O. sativa | AA | 18 | 149 | 11.731*** | 0.000 | 3.676*** | 0.000 |
| O. rufipogon | AA | 3 | 17 | 0.193 | 0.826 | 1.053 | 0.429 |
| O. glaberrima | AA | 7 | 41 | 4.986*** | 0.001 | 0.869 | 0.549 |
| O. barthii | AA | 4 | 23 | 15.611*** | 0.000 | 8.984** | 0.003 |
| O. punctata | BB | 5 | 29 | 3.059* | 0.035 | 0.671 | 0.629 |
| O. eichingeri | CC | 5 | 29 | 1.277 | 0.306 | 1.595 | 0.257 |
| O. officinalis | CC | 4 | 23 | 4.794* | 0.011 | 14.323*** | 0.001 |
| O. rhizomatis | CC | 3 | 17 | 3.176 | 0.071 | 4.531 | 0.075 |
| O. punctata | BBCC | 5 | 29 | 5.679** | 0.002 | 6.363* | 0.017 |
| O. minuta | BBCC | 5 | 29 | 1.926 | 0.137 | 6.446** | 0.008 |
| O. alta | CCDD | 2 | 11 | 12.910** | 0.005 | 37.584*** | 0.001 |
| Among different species | |||||||
| Diploids | 49 | 305 | 97.356*** | 0.000 | 29.909*** | 0.000 | |
| Polyploids | BBCC | 10 | 59 | 5.663* | 0.021 | 5.379* | 0.029 |
| Glycine | |||||||
| Different cultivars or accessions within a species | |||||||
| G. max | GG | 11 | 87 | 25.007*** | 0.000 | 5.534*** | 0.000 |
| G. soja | GG | 9 | 71 | 20.867*** | 0.000 | 4.920*** | 0.000 |
| Among different species | |||||||
| Diploids | 10 | 223 | 18.667*** | 0.000 | 11.408*** | 0.000 | |
| Gossypium | |||||||
| Different cultivars or accessions within a species | |||||||
| G. herbaceum | A1A1 | 10 | 59 | 19.395*** | 0.000 | ||
| G. arboreum | A2A2 | 4 | 23 | 9.326*** | 0.000 | ||
| G. armourianum | D2-1D2-1 | 2 | 10 | 0.245 | 0.632 | ||
| G. klotzchianum | D3-kD3-k | 3 | 14 | 0.168 | 0.847 | ||
| G. raimondii | D5D5 | 3 | 17 | 0.302 | 0.744 | ||
| G. gossypioides | D6D6 | 2 | 11 | 0.429 | 0.527 | ||
| G. trilobum | D8D8 | 3 | 16 | 0.026 | 0.974 | ||
| G. hirsutum | (AADD)1 | 9 | 53 | 6.114*** | 0.000 | ||
| G. barbadense | (AADD)2 | 5 | 29 | 3.018* | 0.037 | ||
| G. tomentosum | (AADD)3 | 12 | 77 | 1.404 | 0.192 | ||
| G. mustelinum | (AADD)4 | 3 | 17 | 2.312 | 0.133 | ||
| G. darwinii | (AADD)5 | 2 | 17 | 2.038 | 0.173 | ||
| Among different species | |||||||
| Diploids | 65 | 372 | 4.737*** | 0.000 | |||
| Polyploids | 31 | 197 | 5.204*** | 0.000 | |||
aThe variation of the RLK family size was not studied in Gossypium.
The variation is significant in two-tailed test at *P ≤ 0.05, **P ≤ 0.01 and ***P ≤ 0.001.
Experimental design
Membrane arrays were prepared by printing 320–2000 ng of purified nuclear DNA per dot onto nylon membrane (for detail, see Materials and Methods in Supplementary Data). To remove the potential noise background and determine the numbers of NBS and RLK genes in the genomes, positive and negative controls were included in the arrays, with each positive control having three levels of copy numbers of the target genes. The three levels of positive controls were used to optimize the assay of gene copy number and the negative controls were used to minimize the potential influences of the background noises on the assay. To minimize the influence of gene member sequence homology on gene copy number assay, NBS- or RLK-specific degenerate overgos or the combined NBS gene members representing the NBS family were used as the positive controls on the arrays and the probes in the MA hybridization (see below). In such cases, each member of the gene families would have an equal ability to hybridize with the probes. So, the possibility of identifying all members of the gene families could be maximized. To estimate the potential influence of the hybridization stringencies on the assay, we tested high-, moderate- and low-hybridization stringencies. Identical or extremely similar results were obtained from the three stringencies; therefore, the moderate stringency was used in the MA hybridization and SDLS. Additionally, the hybridized arrays were exposed from 10 min to 5 h to optimize the hybridization intensities for gene copy number estimation. The entire MA experiment, from array preparation to gene copy number estimation, was replicated for 4–8 times. Part of the MA results were further verified by the SDLS method using 7 of the 187 lines hybridized with the probes used in the MA hybridization and the WSBA method using the sequenced genomes of rice cv. Nipponbare and cv. 93-11 (19–22).
Statistical analyses
The data set of the numbers of genes in the NBS and RLK families estimated by the MA hybridization were first log10 transformed to normalize their distribution and then subjected to analysis of variance (ANOVA), Pearson’s correlation and t-test at two-tailed significance levels using SPSS (Statistical Package for the Social Sciences).
RESULTS
Intra- and inter-specific variations in numbers of genes in the NBS and RLK families
We initiated this study based on the results from screening bacterial artificial chromosome (BAC) libraries of two rice (O. sativa) cultivars, Nipponbare and Teqing, using 10 genes representing the rice NBS family (32) (Supplementary Table S3). Although the libraries were constructed with the same restriction enzymes and a similar genome coverage of clones screened (33–35), surprisingly, for every probe we obtained multiple-fold more positive clones from the Teqing library than from the Nipponbare library (Supplementary Table S6). While the difference could be partly attributed to the different insert sizes of the libraries (151 versus133 kb) and distribution of the NBS genes in the genomes (21), we could not exclude the possibility that the two cultivars have different numbers of NBS genes. Independently, we were also screening the BAC libraries of two soybean (G. max) cultivars, Williams 82 (36) and Forrest (37), both of which were constructed with EcoRI and have similar insert sizes (151 versus 157 kb) with the same genome coverage (5.5×), using an NBS gene representing a subfamily of the soybean NBS family (38) (Supplementary Table S3). As seen in rice, we obtained multiple-fold more positive clones from the Forrest library than from the Williams 82 library (Supplementary Table S6). Together, these results led us to hypothesize that the number of genes in a gene family may vary significantly among cultivars of a species.
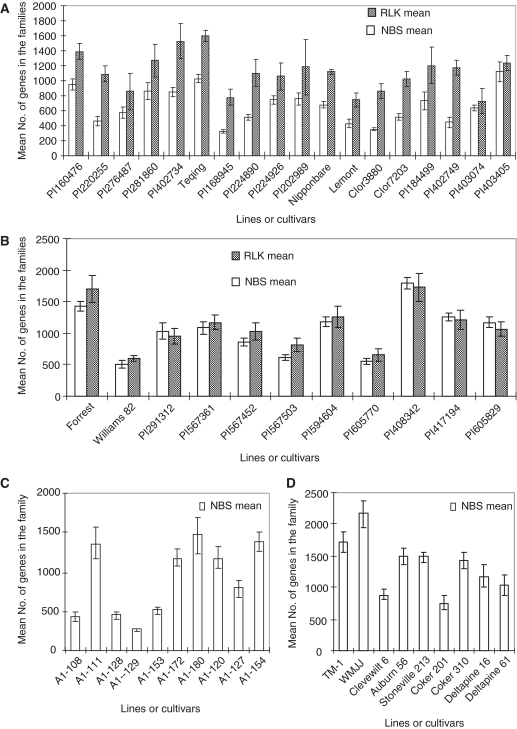
Therefore, we decided to conduct further research in the variation and evolution of gene family sizes among lines within a species and among congeneric species using Oryza, Glycine and Gossypium, and the NBS and RLK families for verifying the results from one to another. As described in ‘Materials and Methods’ section, we first evaluated the methods, WSBA, MA, M, RGCS, qrtPCR and SDLS, that have been used in estimation of the copy number or number of genes in a family (2,3,27–31) (Supplementary Table S2). Then, we conducted pilot experiments to test the three methods selected by the analysis for this study (‘Materials and Methods’ section in Supplementary Data). According to the results (Table 1; Materials and Methods in Supplementary Data, Supplementary Tables S3–S5), the MA method was selected to measure the numbers of genes in the NBS and RLK families in the genomes of the 187 lines. Figure 1 shows examples of the mean numbers of genes in the NBS and RLK families and their variations among cultivars within a species of the three genera (for more information, see Supplementary Table S1). The numbers of genes in the NBS and RLK families varied from 328–1120 (3.4-fold) and 747–1604 (2.1-fold), respectively, among the 18 O. sativa lines analyzed (Supplementary Table S1A), and 501–1801 (3.6-fold) and 597–1704 (2.8-fold), respectively, among the 11 G. max lines (Supplementary Table S1B). The numbers of NBS and RLK genes in O. sativa are in great agreement with those (508–597 NBS genes in Nipponbare and 1147 RLK genes in 93-11) estimated from their genome sequences by the WSBA method (15,19–22) and those by the SDLS method in this study (Supplementary Table S5). The numbers of genes in the NBS family varied from 268–1465 (5.4-fold) and 758–2161 (2.8-fold) among the 10 G. herbaceum (2x) lines and 9 G. hirsutum (4x) lines analyzed, respectively (Figure 1; Supplementary Table S1C), which was also well agreed with those obtained by the SDLS method in this study (Supplementary Table S5). The variation in number of NBS genes reached 19.4-fold, ranging from 88–1710, among the 30 Gossypium diploid species (Supplementary Table S1C).
Figure 1.
Intra-specific variation of the NBS and RLK family sizes within (A) O. sativa (2x), (B) G. max (2x), (C) G. herbaceum (2x) and (D) G. hirsutum (4x). The mean number of genes in each accession or cultivar was calculated from four to eight replicates for each NBS or RLK family and the vertical bars indicate SDs (±SE). Up to 5.4-fold variation in number of genes in the gene families was detected among the lines of the species. The intra-specific variations were verified by ANOVA (Table 1).
To further confirm the intra- and inter-specific variations in the number of NBS and RLK genes, we conducted ANOVA or t-test for the species having two or more lines analyzed, by which experimental errors, if any, could be excluded from the analysis. The intra-specific variations in number of NBS genes were significant for 13 of the 25 species analyzed, whereas those in number of RLK genes were significant (P ≤ 0.05, 0.01 or 0.001) for 8 of the 13 species analyzed (Table 1). The inter-specific variations within a genus in numbers of both NBS and RLK genes were significant (P ≤ 0.05, 0.01 or 0.001) for all three genera at both diploid and polyploid levels (Table 1). In contrast, no significant variation was detected among different plants of either cv. Nipponbare or cv. Teqing (Table 1). These results indicate that the variations in numbers of genes in both NBS and RLK families exist, not only among congeneric species, but also among conspecific lines.
Variation correlation in number of genes between the NBS and RLK families
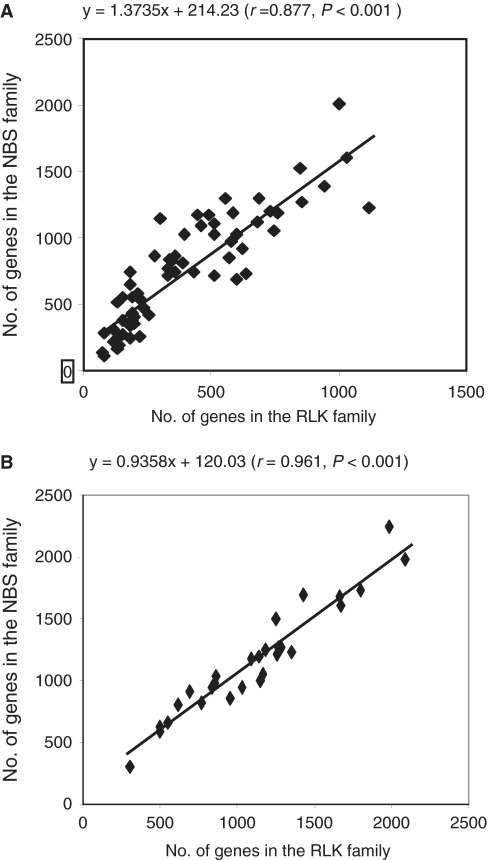
Although the NBS and RLK families represent two different families, both play a significant role in plant defense (23–26). Our previous study showed that the genes having related functions are interacted or correlated (39). Therefore, we hypothesized that the variation in number of genes in the NBS family may correlate with that in the RLK family because they might have been subjected to similar selection pressures. To test this hypothesis, we calculated the correlation coefficients between the variations in the two gene family sizes in Oryza and Glycine diploid species (Figure 2; Supplementary Table S1A and B). In both genera, variation correlation in number of genes was detected between the two families (r = 0.877, P ≤ 0.001 for Oryza and r = 0.961, P ≤ 0.001 for Glycine). This result agreed with the previous finding between Arabidopsis (22,40) and rice (22) that rice having more NBS genes also contains more RLK genes than Arabidopsis. Together, both our result and that of Arabidopsis versus rice support our hypothesis that the number of genes in the NBS family varies correlatively with that in the RLK family.
Figure 2.
Correlation of family size variation between NBS and RLK families in (A) Oryza and (B) Glycine.
Roles of natural selection, artificial selection, genome size variation and polyploidization in the variation and evolution of NBS or RLK family size
The question now is what drives the fate of a gene family. Several hypotheses, including polyploidization, genome size variation and natural selection, have been proposed to answer the question, but few lines of evidences have been reported. Therefore, we tested the roles of each of these factors and artificial selection in the variation and evolution of the NBS and RLK family sizes using the data set.
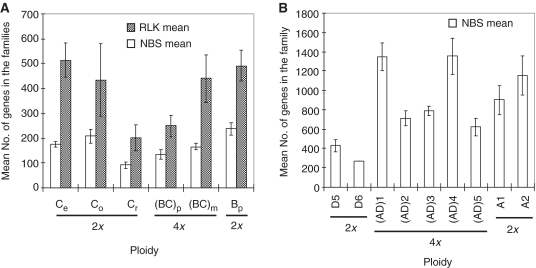
Polyploidization is a prominent process in flowering plant evolution; it is estimated that ∼70% of these species are polyploids. A significant effect of the process is chromosome doubling or combining two genomes in a single cell; thus, the number of genes in the resultant polyploid is expected to be doubled if autopolyploidization occurred, or increased by an additional set of genes from its donor diploid species if allopolyploidization occurred. To estimate the effect of polyploidization on the variation and evolution of the NBS and RLK families, we examined the gene family size variations in the complexes of Oryza BBCC (Supplementary Table S1A) and Gossypium AADD (41,42) (Supplementary Table S1C). Surprisingly, all seven polyploid species studied, O. punctata (BBCC), O. minuta (BBCC), G. hirsutum (AADD)1, G. barbadense (AADD)2, G. tomentosum (AADD)3, G. mustelinum (AADD)4 and G. darwinii (AADD)5, contained similar (P > 0.05) numbers of genes to those of one of their putative diploid donor species for both NBS and RLK families (Figure 3). This result is in contrast to the variation and evolution of 5S rRNA gene family size found in the same species that the numbers of 5S rRNA genes in the polyploids are nearly two-fold higher than the sum of their two diploid donors (M. Zhang et al., in preparation). These results suggest that although polyploidization might lead to an instant expansion of the families, nearly half of the combined number of genes were lost rapidly during the postpolyploidization process. While the underlying mechanism of the process remains to be studied, the result suggests that organisms tend to not maintain ‘surplus’ genes in their genomes. Therefore, in a long run it seems that polyploidization has played little roles in the expansion of the gene families in the Oryza and Gossypuim polyploid species.
Figure 3.
Roles of polyploidization on the variation and evolution of the NBS and RLK family sizes in the (A) Oryza BBCC complex and the (B) Gossypium AADD complex. The mean number of genes in each species (genome) was calculated from 4 to 6 technical replicates per line and 2 to 12 line biological replicates for each NBS or RLK family, and the vertical bars indicate SDs (±SE). Note that the polyploid species have the numbers of NBS or RLK genes similar to that of one of their donor diploid species (P > 0.05). Ce, O. eichinggeri; Co, O. officinalis; Cr, O. rhizomatis; (BC)p, O. punctata (4x); (BC)m, O. minuta; Bp, O. punctata (2x); D5, G. raimondii; D6, G. gossypioides; (AD)1, G. hirsutum; (AD)2, G. barbadense; (AD)3, G. tomentosum; (AD)4, G. mustelinum; (AD)5, G. darwinii; A1, G. herbaceum; and A2, G. arboreum.
Genome sizes can vary by million-fold in living organisms. It was reported in bacteria that gene family sizes increase with the increase of their genome sizes (1,6), but no study has yet been reported in the relationship in plants and other higher organisms. It was found from genome sequence analysis by the WSBA method that A. thaliana ecotype Columbia that has a genome size of ∼125 Mb/1C contains approximately 150 NBS genes and 629 RLK genes (22,40), and O. sativa cv. Nipponbare and cv. 93-11 that have a genome size of ∼440 Mb/1C contain 500–600 NBS genes (15,19–21) and 1147 RLK genes (22), respectively. It appears that the species with larger genomes tend to have more genes in the NBS or RLK family than those with smaller genomes. To confirm this observation and determine the role of genome size variation in the variation and evolution of gene family sizes, we calculated the correlation coefficients between genome size (43–45) and log10-transformed gene family size (Materials and Methods in Supplementary Data) at the diploid species levels of Oryza, Glycine, Gossypium and their combinations (Table 2). The correlation coefficients for both NBS and RLK families were positive for Gossypium, Oryza + Glycine, and Oryza + Glycine + Gossypium species and negative for Oryza and Glycine species, but only those for Oryza species were significant (r = −0.851, P = 0.007; r = −0.859, P = 0.006), even though their genome sizes vary from 0.37–2.84 pg/1C (7.7-fold) (Supplementary Table S1). This result neither agreed with that observed in bacteria (1,6) nor supported the above-observed relationship between genome size and gene family size in Arabidosis and rice, but was consistent with the numbers of NBS genes observed in maize, sorghum, Brachypodium and rice (15). Interestingly, the numbers of genes in the NBS or RLK family did not increase, but decreased with the increase of their genome sizes among the Oryza species, while they were not affected by genome size variation for the Glycine and Gossypium species.
Table 2.
Correlation between genome size (pg/1C) and log10-transformed number of genes in the NBS and RLK families (Supplementary Table S1)
| Species analyzed | N | NBS |
RLK |
||
|---|---|---|---|---|---|
| Coefficient (r) | P-value | Coefficient (r) | P-value | ||
| Oryza | 8 | −0.851** | 0.007 | −0.859** | 0.006 |
| Glycine | 10 | −0.372 | 0.290 | −0.260 | 0.468 |
| Gossypium | 26 | 0.198 | 0.333 | Not studied | |
| Oryza + Glycine | 18 | 0.312 | 0.208 | 0.038 | 0.881 |
| Oryza + Glycine + Gossipium | 44 | 0.062 | 0.691 | Not studied | |
The variation is significant in two-tailed test at **P ≤ 0.01.
There has been no doubt on the role of natural selection in organismal speciation and evolution; nevertheless, no research has yet been reported about its role in the fate of a gene family. For this research purpose, we included in this study a few pairs of diploid lines that have the same genomes, but are known to be different in ecotypes so that the effects of polyploidization, genome size variation and artificial selection on the analysis could be minimized and ecological effect could be estimated (Table 3). Comparative analysis of the NBS and RLK family sizes between the ecotype pairs showed that O. sativa ssp. indica, native to subtropic, had more NBS and RLK genes than its sister subspecies, japonica, native to temperate (P < 0.001), and the G. max US southern ecotype Forrest had more NBS and RLK genes than its northern ecotype Williams 82 (P < 0.001), though no significant difference in NBS and RLK family sizes was detected between Asian wild rice, O. rufipogon and African wild rice, O. barthii. Moreover, of the O. sativa species, southern ecotypes, such as Teqing that was adapted to Southern China (subtropic), had many more NBS and RLK genes than northern ecotypes, such as Nipponbare that was adapted to Japan (temperate). These results agreed with the BAC library and SDLS results in which many more NBS positive clones were identified from the Teqing libraries than from the Nipponbare libraries (Supplementary Tables S4 and S6). Therefore, ecological environments or natural selection may play an important role in the variation and evolution of NBS and RLK gene family sizes.
Table 3.
Comparison by t-test in the log10-transformed number of genes in the NBS and RLK families between species of different geographical origins and ecotypes in Oryza and Glycine
| Species | N | NBS |
RLK |
||||
|---|---|---|---|---|---|---|---|
| Mean ± SD | Mean difference | P-value | Mean ± SD | Mean difference | P-value | ||
| Oryza | |||||||
| O. barthii (Africa) versus | 24 | 2.7401 ± 0.20969 | −0.03599 | 0.532 | 3.0626 ± 0.19297 | 0.12677 | 0.189 |
| O. rufipogon (Asia) | 18 | 2.7761 ± 0.13964 | 2.9358 ± 0.2178 | ||||
| O. sativa ssp. indica (subtropic) versus | 54 | 2.9032 ± 0.17977 | 0.19380*** | 0.000 | 3.1268 ± 0.13808 | 0.16683*** | 0.000 |
| O. sativa ssp. japonica (temperate) | 30 | 2.7094 ± 0.17976 | 2.9599 ± 0.13638 | ||||
| Glycine | |||||||
| G. max cv. Forrest (S. ecotype) versus | 8 | 3.1492 ± 0.07208 | 0.47036*** | 0.000 | 3.2030 ± 0.17654 | 0.43739*** | 0.000 |
| G. max cv. Williams 82 (N. ecotype) | 8 | 2.6789 ± 0.14579 | 2.7656 ± 0.10607 | ||||
The variation is significant in two-tailed test at ***P ≤ 0.001.
Artificial selections, including domestication, breeding and cultivation, have played a significant role in the perceived variation and evolution of crop plants since men have been involved, on purpose, in the process; however, research in its effects on genome evolution remains. To determine the effect of artificial selection on the fate of a gene family, we included a few pairs of cultivated and wild donor species that have the same genome and geographical origin so that the background of the effects of genome size variation, polyploidization and natural selection, if any, on the analysis could be minimized (Table 4). Of the six pairs of cultivated/wild species compared, O. sativa ssp. indica versus O. rufipogon, O. glaberrina versus O. barthii and G. max versus G. soja were different in number of genes in the NBS and RLK families (P < 0.05–0.001). Interestingly, the cultivated species O. sativa ssp. indica had more NBS and RLK genes than its wild donor species, O. rufipogon, while the cultivated species, O. glaberrina and G. max, had fewer NBS and RLK genes than their wild donor species, O. barthii and G. soja. Therefore, the role of artificial selection in the variation and evolution of the NBS and RLK families has been confirmed; this activity could lead to either expansion or contraction of the gene families, depending on the objectives and strengths of the selection. Furthermore, given that humans domesticated and bred these crops between 3000 and 7000 years ago (46), their activities since then have led to a gain of 156 (24.9%) NBS and 412 (41.1%) RLK genes for O. sativa ssp. indica, a loss of 147 (31.6%) NBS and 250 (25.5%) RLK genes for O. glaberrina and a loss of 274 (26.3%) NBS and 189 (17.1%) RLK genes for G. max. This implies a gain or loss at a rate of 2–6 genes per 100 years. As did Sakai and Itoh (16), we also found that cultivated O. sativa ssp. japonica (575) had fewer NBS genes than the wild rice, O. rufipogon (629), but the difference was not statistically significant (P = 0.184).
Table 4.
Comparison by t-test in the log10-tranformated number of genes in the NBS and RLK families between cultivated and wild species of Oryza and Glycine
| Species | N | NBS |
RLK |
||||
|---|---|---|---|---|---|---|---|
| Mean ± SD | Mean difference | P-value | Mean ± SD | Mean difference | P-value | ||
| Oryza (Asian rice): cultivated versus wild | |||||||
| O. sativa ssp. indica versus | 54 | 2.9032 ± 0.17977 | 0.12703** | 0.008 | 3.1268 ± 0.13808 | 0.19095** | 0.006 |
| O. rufipogon | 18 | 2.7761 ± 0.13964 | 2.9358 ± 0.21780 | ||||
| O. sativa ssp. japonica versus | 30 | 2.7094 ± 0.17976 | −0.06677 | 0.184 | 2.9599 ± 0.13638 | 0.02412 | 0.748 |
| O. rufipogon | 18 | 2.7761 ± 0.13964 | 2.9358 ± 0.21780 | ||||
| O. sativa ssp. javanica versus | 36 | 2.7564 ± 0.19568 | −0.01971 | 0.705 | 2.9956 ± 0.13904 | 0.05978 | 0.401 |
| O. rufipogon | 18 | 2.7761 ± 0.13964 | 2.9358 ± 0.21780 | ||||
| O. sativa versus | 120 | 2.8107 ± 0.20221 | 0.03456 | 0.486 | 3.0446 ± 0.15494 | 0.10877 | 0.094 |
| O. rufipogon | 18 | 2.7761 ± 0.13964 | 2.9358 ± 0.21780 | ||||
| Oryza (African rice): cultivated versus wild | |||||||
| O. glaberrina versus | 42 | 2.6399 ± 0.17400 | −0.10026* | 0.041 | 3.0017 ± 0.18582 | −0.06084 | 0.380 |
| O. barthii | 24 | 2.7401 ± 0.20969 | 3.0626 ± 0.19297 | ||||
| Glycine: cultivated versus wild | |||||||
| G. max versus | 88 | 2.9777 ± 0.19760 | −0.11342*** | 0.000 | 2.9935 ± 0.21865 | −0.07224* | 0.037 |
| G. soja | 72 | 3.0911 ± 0.16170 | 3.0657 ± 0.21465 | ||||
The variation is significant in two-tailed test at *P ≤ 0.05, **P ≤ 0.01 and ***P ≤ 0.001.
Association of the size variations of NBS and RLK families with their host organismal phylogeny
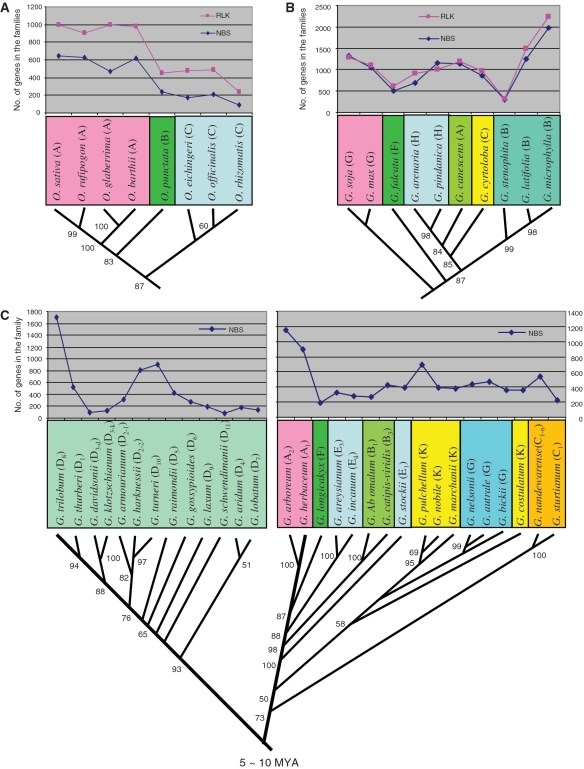
It has been hypothesized that the contraction and expansion of a gene family may play a role in the observed difference between organisms (1–3,8–10,17,18), but research in this regard is limited. To test this hypothesis, we compared the size variations of the NBS and RLK families with the phylogenetic relationships among the species shown by a phylogenetic tree (Figure 4). There seemed an association between gene family size and species relationships. To confirm this inference, we calculated pairwisely the differences in family size between lines or species of the genera, retrieved the phylogenetic distances between them and determined their variation correlation (Supplementary Table S7). For the Oryza and Gossypium species, the size variations of NBS and RLK families were all positively correlated with those of phylogenetic distances (r = 0.692–0.792, P < 0.01; r = 0.241, P < 0.001), whereas for the Glycine species the variation of the RLK family size was negatively correlated with that of phylogenetic distances (r = −0.526, P < 0.05). Therefore, our inference was confirmed that the variation of the NBS or RLK family size is associated with organismal speciation, thus likely providing an additional source of perceived genetic variation.
Figure 4.
Association of gene family size variation with organismal phylogentic relationships indicated by phylogenetic distance. The phylogenetic trees and corresponding phylogenetic distances of (A) Oryza, (B) Glyine and (C) Gossypium species were from previous studies (52,55–57). The pairwised difference in number of genes in the families between two species was calculated by subtracting the mean number of the genes in one species from that of the other (Supplementary Table S1), and the phylogenetic distance between them was retrieved from the genetic distance matrix of their phylogenetic tree (Supplementary Table S7). The numbers near each branch of the trees indicate its percentage confidence estimated by bootstrap replications. Note that the size variations of the NBS and RLK families in the Oryza and Gossypium species positively (r = 0.692–0.792, P ≤ 0.004; r = 0.241, P = 0.000) and those in the Glycine species negatively (r=−0.526, P = 0.044) correlate with the species phylogenetic distances.
DISCUSSION
We uncovered in this study that the numbers of genes in the NBS and RLK families vary by multiple fold (P ≤ 0.001), not only among congeneric species (up to 19.4-fold), surprisingly, also among conspecific cultivars or lines (up to 5.4-fold) (Table 1; Figure 1; Supplementary Table S1). The inter-specific variation was observed among the species of all three genera studied, Oryza, Glycine and Gossypium, and the intra-specific variation observed in 13 of the 25 Oryza, Glycine and Gossypium species analyzed for the NBS family and in 8 of the 13 Oryza and Glycine species analyzed for the RLK family. This suggests that the gene family size variation is a common phenomenon in the plant genera, despite that only a limited number of lines have been studied for each species. Furthermore, these results have been confirmed by statistical analysis and verified with several independent experiments. First, as described above, the difference in number of NBS genes was discovered in an occasion by BAC library screening, then confirmed by the MA and SDLS methods and verified by blast analysis of the rice cv. Nipponbare and cv. 93-11 genome sequences (47). The discrepancy (13.7%) in the numbers of NBS genes obtained between the MA (679) and WSBA (597) methods for Nipponbare was close to the 17.5% (508–597) variation in numbers of NBS genes estimated with the WSBA method by different researchers (15,19–21) and probably due to the incomplete genome coverage of the rice sequence (47) and/or improper assembly of identical copies of the genes that often leads to underestimation of gene numbers (3). Second, the data of Gossypium, Glycine and Oryza were obtained independently; so, systematic artificial errors, if any, could have been minimized. Third, previous research (30) and this study both showed that the MA method, though it cannot count the change of individual copy numbers of genes as do the WSBA, RGCS and SDLS methods, has a similar sensitivity to these and other methods for our research purposes (Supplementary Table S2). As described in Materials and Methods in Supplementary Data, the hybridization signal data used to estimate the number of genes could be directly used for the statistical analyses, without the calculation of gene copy number, from which the same results were obtained. Nevertheless, it should be pointed out that although the expansion or contraction of a gene family could be contributed by both functional and nonfunctional (e.g. pseudogenes) gene members (2,9,10,13–16), the MA method itself could not distinguish them. To estimate the contribution of each type of the genes to the gene family size variation, sequence- and expression-based studies of the gene families will be needed. Fourth, if artificial experimental errors existed, they could be excluded from the ANOVA; a larger artificial error would lead to a smaller F-value, thus a lower probability of statistically significant variation. Finally, the gene number variation is further confirmed by the fact obtained in the study that the degree of variation among congeneric species was much larger than that among conspecific lines and there was no significant difference in the family sizes among different plants of a self-crossing rice line, Nipponbare or Teqing.
The inter-specific variation of the NBS and RLK family sizes observed in the plant species, as expected, agrees with those of other gene families previously observed among species of archaea (6), bacteria (1,6), Drosophila (3) and mammals (2,9,10); however, the intra-specific variation of the gene family sizes is striking, which has not been reported previously. Nevertheless, the intra-specific variation is supported by the local intra-specific violation of genetic colinearity found between maize inbred lines (48,49) and copy number variations discovered in human (50,51). Moreover, the rapid and massive gain or loss of NBS genes has been observed recently among related plant species (14,16). The variation of gene family size could be attributed to gene duplication, deletion, pseudogenization and/or functional diversification (2,3,5,11,12), but may be subjected to several factors discussed below. Since both NBS and RLK families are crucial to plant defense, thus plant adaptation and fitness, this rapid variation in the number of their genes may be necessary to allow plants to meet the need of defending themselves from rapidly varying populations, including races and types, of pathogens in an environment. Therefore, the observed variation of the gene family sizes, especially the intra-specific variation, provides novel insights into the roles of the gene family size variation in plant genetic variation and evolution.
The variations of the NBS and RLK family sizes could be driven by a number of factors, including genome size variation, polyploidization, natural selection, artificial selection and/or gene interaction, depending on their host organisms and living environments. Variation in number of genes in a gene family was previously shown to be positively correlated with genome size among bacterial species (1,6). This study shows that the numbers of genes in both NBS and RLK families are negatively correlated with the genome sizes in the Oryza species and that there is no correlation between them in the species of Gossypium, Glycine, Oryza + Glycine and Oryza + Glycine + Gossypium. Therefore, a gene family may expand, contract or be consistent in size as its host genome size changes. This conclusion is consistent with a recent study in grasses that has shown that the species with larger genomes need not necessarily have more gene members for a gene family (15).
It appears from this study that the size variations of the NBS and RLK families have not been affected much by polyploidization. It is surprising that the polyploids of Gossypium and Oryza have similar numbers of NBS and RLK genes (P > 0.05) to those of one of their putative diploid donor species, even though the process combines the two sets of genes contained in the two donor species when the polyploids originated. This implies that the ‘surplus’ genes in a polyploid may disappear rapidly during postpolyploidization, as suggested by earlier studies in Arabidopsis (11,12). In the case of the Gossypium polyploid species, this process must have occurred within the past 1–2 million years after they originated, with an average rate of 242–484 genes per million years (Supplementary Table S1C) if G. raimondii and the ancestor of G. herbaceum and G. arboreum are the diploid donors of the AD-genome polyploids (41,52). A previous study (53) showing that polyploid Gossypium species have many more pseudogenes of the NBS family than diploids seems to suggest that the pseudogenization mechanism has played an important role in this regard. Therefore, we conclude that while polyploidization may lead to an immediate expansion of the NBS and RLK families, it plays little roles in their size variation and evolution in a long run.
It is apparent from this study that the expansion and contraction of the NBS and RLK families are significantly regulated by natural and artificial selection. The significant difference in the NBS and RLK family sizes between the species native to different geographical regions or different ecotypes has provided a line of evidence in the role of the variation of ecological environments or natural selection in the variation and evolution in number of genes in the two gene families. The gene members that are favorable for fitness are selected and accumulated in the genomes, but those that are not favorable for fitness are lost in natural selection. However, it is surprising that the number of genes in the families is so different (P = 0.041–0.000) between the cultivated and wild species that have been diverged only thousands of years ago. As both NBS and RLK families are extensively involved in plant defenses (23–26), it is expected that wild species have more NBS and RLK genes than cultivated ones due to the genetic bottleneck of domestication (54). Although the expectation was observed for the African wild rice, O. barthii, and the wild soybean, G. soja, it was not for the Asian wild rice, O. rufipogon. The fact that the number of genes in the families in the cultivated indica rice is larger (P ≤ 0.001) than that in its wild donor species (O. rufipogon) indicates that plant breeding, especially for disease resistance, likely allows accumulation of NBS and RLK genes that potentially confer resistance to pathogens. Therefore, plant breeders, in fact, select for not only favorable alleles and their combinations, as expected, but also the number of genes. If man is assumed to have participated in crop domestication, breeding and cultivation some 7000 years ago, his contribution to the size variations of the NBS and RLK families could be from 156 to 413 genes in expansion in O. sativa ssp. indica or contraction in the cultivated soybean and African rice, with an average rate of 2–6 genes per 100 years. This number is larger by million-fold than the 0.09 gene/million years calculated in the mammalian species (2), suggesting that the role of artificial selection is much larger than that of natural selection.
Moreover, this study also shows that the size variation of the NBS family is correlated with that of the RLK family. This indicates that the expansion or contraction of a gene family may be regulated or correlated by those of other gene families as well. Although only NBS and RLK families were investigated in this study, it is likely that as individual genes, different gene families may correlate with each other in number of members. This result reveals that the expansion or contraction of a gene family is a complicated process. Study of the size variations and relationships of many more gene families will be needed to understand the underlying molecular basis of their expansion and contraction.
Furthermore, the variations of both NBS and RLK family sizes correlate with the variation of the host plant phylogenetic distances. The correlation could be positive, for instance in Oryza and Gossypium, or negative, for instance in Glycine (Supplementary Table S8). The correlation suggests that the expansion or contraction of the NBS and RLK families may play a role in their host plant speciation and evolution. Furthermore, since the process of speciation and evolution is considered to result from the organism’s genetic variation, the expansion and contraction of the gene families may provide a source of genetic variation essential for plant speciation and evolution. However, further studies remain to decipher how the variation of the NBS and RLK family sizes influence the plant speciation and evolution.
Finally, it should be pointed out that since this study represents the first report in the size variation and evolution of a gene family, particularly among different cultivars or lines of a species, many studies remain to understand the underlying molecular mechanisms of the gene family size variation and evolution. These include, but are not limited to, the contribution of functional and nonfunctional gene members to gene family size variation, contribution of each diploid donor species to the gene family size variation of resultant polyploids, size variation of other gene families, correlation of gene families, relationships of variation between gene family size and trait including morphology, biology and complexity, and genetics of gene family size. It is believed that these studies will greatly promote our understanding of the molecular mechanisms of gene family size variation in organism’s genetics, variation and evolution.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Research grant (203232-85360 to H.-B.Z.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank R. Fan and C. W. Smith for their suggestions and discussion in the data statistical analysis, and S. Ge for kindly providing the phylogenetic distance matrices of Oryza species.
REFERENCES
- 1.Pushker R, Mira A, Rodríguez-Valera F. Comparative genomics of gene-family size in closely related bacteria. Genome Biol. 2004;5:R27. doi: 10.1186/gb-2004-5-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Demuth JP, De Bie T, Stajich JE, Cristianini N, Hahn MW. The evolution of mammalian gene families. PLoS ONE. 2006;1:e85. doi: 10.1371/journal.pone.0000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hahn MW, Han MV, Han SG. Gene family evolution across 12 Drosophila genomes. PLoS Genet. 2007;3:e197. doi: 10.1371/journal.pgen.0030197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 5.Demuth JP, Hahn MW. The life and death of gene families. Bioessays. 2009;31:29–39. doi: 10.1002/bies.080085. [DOI] [PubMed] [Google Scholar]
- 6.Jordan IK, Makarova KS, Spouge JL, Wolf YI, Koonin EV. Lineage-specific gene expansions in bacterial and archaeal genomes. Genome Res. 2001;11:555–565. doi: 10.1101/gr.166001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karlsson M, Stenlid J. Comparative evolutionary histories of the fungal chitinase gene family reveal non-random size expansions and contractions due to adaptive natural selection. Evol. Bioinfo. 2008;4:47–60. doi: 10.4137/ebo.s604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soanes DM, Alam I, Cornell M, Wong HM, Hedeler C, Paton NW, Rattray M, Hubbard SJ, Oliver SG, Talbot NJ. Comparative genome analysis of filamentous fungi reveals gene family expansions associated with fungal pathogenesis. PLoS ONE. 2008;3:e2300. doi: 10.1371/journal.pone.0002300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grus WE, Shi P, Zhang Y-P, Zhang J. Dramatic variation of the vomeronasal pheromone receptor gene repertoire among five orders of placental and marsupial mammals. Proc. Natl Acad. Sci. USA. 2005;102:5767–5772. doi: 10.1073/pnas.0501589102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prachumwat A, Li W-H. Gene number expansion and contraction in vertebrate genomes with respect to invertebrate genomes. Genome Res. 2008;18:221–232. doi: 10.1101/gr.7046608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seoighe C, Gehring C. Genome duplication led to highly selective expansion of the Arabidopsis thaliana proteome. Trends Genet. 2004;20:461–464. doi: 10.1016/j.tig.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 12.Maere S, DeBodt S, Raes J, Casneuf T, Van Montagu M, Kuiper M, VandePeer Y. Modeling gene and genome duplications in eukaryotes. Proc. Natl Acad. Sci. USA. 2005;102:5454–5459. doi: 10.1073/pnas.0501102102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tarr DE, Alexander HM. TIR-NBS-LRR genes are rare in monocots: evidence from diverse monocot orders. BMC Res. Notes. 2009;2:197. doi: 10.1186/1756-0500-2-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Q, Han Z, Jiang H, Tian D, Yang S. Strong positive selection drives rapid diversification of R-genes in Arabidopsis relatives. J. Mol. Evol. 2010;70:137–148. doi: 10.1007/s00239-009-9316-4. [DOI] [PubMed] [Google Scholar]
- 15.Li J, Ding J, Zhang W, Zhang Y, Tang P, Chen J-Q, Tian D, Yang S. Unique evolutionary pattern of numbers of gramineous NBS–LRR genes. Mol. Genet. Genomics. 2010;283:427–438. doi: 10.1007/s00438-010-0527-6. [DOI] [PubMed] [Google Scholar]
- 16.Sakai H, Itoh T. Massive gene losses in Asian cultivated rice unveiled by comparative genome analysis. BMC Genomics. 2010;11:121. doi: 10.1186/1471-2164-11-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sheps JA, Ralph S, Zhao Z, Baillie DL, Ling V. The ABC transporter gene family of Caenorhabditis elegans has implications for the evolutionary dynamics of multidrug resistance in eukaryotes. Genome Biol. 2004;5:R15. doi: 10.1186/gb-2004-5-3-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi P, Zhang J. Comparative genomic analysis identifies an evolutionary shift of vomeronasal receptor gene repertoires in the vertebrate transition from water to land. Genome Res. 2007;17:166–174. doi: 10.1101/gr.6040007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koczyk G, Chełkowski J. An assessment of the resistance gene analogues of Oryza sativa ssp. japonica: their presence and structure. Cell. Mol. Biol. Lett. 2003;8:963–972. [PubMed] [Google Scholar]
- 20.Monosi B, Wisser RJ, Pennill L, Hulbert SH. Full-genome analysis of resistance gene homologues in rice. Theor. Appl. Genet. 2004;109:1434–1447. doi: 10.1007/s00122-004-1758-x. [DOI] [PubMed] [Google Scholar]
- 21.Zhou T, Wang Y, Chen J-Q, Araki H, Jing Z, Jiang K, Shen J, Tian D. Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes. Mol. Genet. Genomics. 2004;271:402–415. doi: 10.1007/s00438-004-0990-z. [DOI] [PubMed] [Google Scholar]
- 22.Shiu S-H, Karlowski WM, Pan R, Tzeng Y-H, Mayer KFX, Li W-H. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell. 2004;16:1220–1234. doi: 10.1105/tpc.020834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takken FLW, Joosten MHAJ. Plant resistance genes: their structure, function and evolution. Eur. J. Plant Pathol. 2000;106:699–713. [Google Scholar]
- 24.McHale Ll, Tan X, Koehl P, Michelmore RW. Plant NBS-LRR proteins: adaptable guards. Genome Biol. 2006;7:212. doi: 10.1186/gb-2006-7-4-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shiu S-H, Bleecker AB. Plant receptor-like kinase gene family: diversity, function, and signaling. Sci. STKE. 2001;113:re22. doi: 10.1126/stke.2001.113.re22. [DOI] [PubMed] [Google Scholar]
- 26.Afzal AJ, Wood AJ, Lightfoot DA. Plant receptor-like serine threonine kinases: roles in signaling and plant defense. Mol. Plant Microbe Interact. 2008;21:507–517. doi: 10.1094/MPMI-21-5-0507. [DOI] [PubMed] [Google Scholar]
- 27.Chung Y-J, Jonkers J, Kitson H, Fiegler H, Humphray S, Scott C, Hunt S, Yu Y, Nishijima I, Velds A, et al. A whole-genome mouse BAC microarray with 1-Mb resolution for analysis of DNA copy number changes by array comparative genomic hybridization. Genome Res. 2004;14:188–196. doi: 10.1101/gr.1878804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hawkins JS, Kim H, Nason JD, Wing RA, Wendel JF. Differential lineage-specific amplification of transposable elements is responsible for genome size variation in Gossypium. Genome Res. 2006;16:1252–1261. doi: 10.1101/gr.5282906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferreira ID, do Rosário VE, Cravo PVL. Real-time quantitative PCR with SYBR Green I detection for estimating copy numbers of nine drug resistance candidate genes in Plasmodium falciparum. Malar. J. 2006;5:1. doi: 10.1186/1475-2875-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diaz MGQ, Ryba M, Leung H, Nelson R, Leach JE. Detection of deletion mutants in rice via overgo hybridization onto membrane spotted arrays. Plant Mol. Biol. Rep. 2007;25:17–26. [Google Scholar]
- 31.Yi CX, Zhang J, Chan KM, Liu XK, Hong Y. Quantitative real-time PCR assay to detect transgene copy number in cotton (Gossypium hirsutum) Anal. Biochem. 2008;375:150–152. doi: 10.1016/j.ab.2007.11.022. [DOI] [PubMed] [Google Scholar]
- 32.Leister D, Kurth J, Laurie DA, Yano M, Sasaki T, Devos K, Graner A, Schulze-Lefert P. Rapid reorganization of resistance gene homologues in cereal genomes. Proc. Natl Acad. Sci. USA. 1998;95:370–375. doi: 10.1073/pnas.95.1.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang H-B, Choi S-D, Woo S-S, Li Z-K, Wing RA. Construction and characterization of two rice bacterial artificial chromosome libraries from the parents of a permanent recombinant inbred mapping population. Mol. Breed. 1996;2:11–24. [Google Scholar]
- 34.Tao Q, Chang Y-L, Wang J, Chen H, Schuering C, Islam-Faridi MN, Wang B, Stelly DM, Zhang H-B. Bacterial artificial chromosome-based physical map of the rice genome constructed by restriction fingerprint analysis. Genetics. 2001;158:1711–1724. doi: 10.1093/genetics/158.4.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tao Q, Wang A, Zhang H-B. One large-insert plant-transformation-competent BIBAC library and three BAC libraries of japonica rice for genome research in rice and other grasses. Theor. Appl. Genet. 2002;105:1058–1066. doi: 10.1007/s00122-002-1057-3. [DOI] [PubMed] [Google Scholar]
- 36.Marek LF, Shoemaker RC. BAC contig development by fingerprint analysis in soybean. Genome. 1997;40:420–427. doi: 10.1139/g97-056. [DOI] [PubMed] [Google Scholar]
- 37.Wu C, Sun S, Nimmakayala P, Santos FA, Springman R, Meksem K, Ding K, Lightfoot D, Zhang H-B. Construction and characterization of a soybean bacterial artificial chromosome library and use of multiple complementary libraries for genome physical mapping. Theor. Appl. Genet. 2004;109:1041–1050. doi: 10.1007/s00122-004-1712-y. [DOI] [PubMed] [Google Scholar]
- 38.Kanazin V, Marek LF, Shoemaker RC. Resistance gene analogs are conserved and clustered in soybean. Proc. Natl Acad. Sci. USA. 1996;93:11746–11750. doi: 10.1073/pnas.93.21.11746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu C, Wang S, Zhang H-B. Interactions among genomic structure, function and evolution revealed by comprehensive analysis of the Arabidopsis genome. Genomics. 2006;88:394–406. doi: 10.1016/j.ygeno.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 40.Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. Genome-wide analysis of NBS-LRR–encoding genes in Arabidopsis. Plant Cell. 2003;15:809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wendel JF, Cronn RC. Polyploidy and the evolutionary history of cotton. Adv. Agron. 2003;78:139–186. [Google Scholar]
- 42.Zhang H-B, Li Y, Wang B, Chee P. Recent advances in cotton genomics. Int. J. Plant Genomics. 2008;2008:742304. doi: 10.1155/2008/742304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bennett MD, Leitch IJ. Nuclear DNA amounts in angiosperms. Ann. Bot. 1995;76:113–176. [Google Scholar]
- 44.Hendrix B, Stewart JMcD. Estimation of the nuclear DNA content of Gossypium species. Ann. Bot. 2005;95:789–797. doi: 10.1093/aob/mci078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyabayashi T, Nonomura K-I, Morishima H, Kurata N. Genome size of twenty wild species of Oryza determined by flow cytometric and chromosome analyses. Breed. Sci. 2007;57:73–78. [Google Scholar]
- 46.Carter TE, Nelson RL, Sneller CH, Cui Z. Genetic diversity in soybean. In: Boerma HR, Specht JE, editors. Soybeans: Improvement, Production and Uses (Agronomy) 3rd edn. Madison, WI: American Society of Agronomy, Crop Science Society of America, Soil Science Society of America; 2004. pp. 303–416. [Google Scholar]
- 47.International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 48.Fu H, Dooner HK. Intraspecific violation of genetic colinearity and its implications in maize. Proc. Natl Acad. Sci. USA. 2002;99:9573–9578. doi: 10.1073/pnas.132259199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brunner S, Fengler K, Morgante M, Tingey S, Rafalski A. Evolution of DNA sequence nonhomologies among maize inbreds. Plant Cell. 2005;17:343–360. doi: 10.1105/tpc.104.025627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 51.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Månér S, Massa H, Walker M, Chi M, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 52.Rong Y. Ph.D Thesis. TX: Texas A&M University, College Station; 2004. Phylogeny of the genus Gossypium and genome origin of its polyploid species inferred from variation in nuclear repetitive DNA sequences. [Google Scholar]
- 53.He L, Du CG, Covaleda L, Robinson AF, Yu JZ, Kohel RJ, Zhang H-B. Cloning, characterization, and evolution of the NBS-LRR-encoding resistance gene analogue family in polyploid cotton (Gossypium hirsutum L.) Mol. Plant Microbe Interact. 2004;17:1234–1241. doi: 10.1094/MPMI.2004.17.11.1234. [DOI] [PubMed] [Google Scholar]
- 54.Hyten DL, Song Q, Zhu Y, Choi I-Y, Nelson RL, Costa JM, Specht JE, Shoemaker RC, Cregan PB. Impacts of genetic bottlenecks on soybean genome diversity. Proc. Natl Acad. Sci. USA. 2006;103:16666–16671. doi: 10.1073/pnas.0604379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hui D, Chen S, Zhuang B. Phylogeny of 12 species of genus Glycine Willd. reconstructed with internal transcribed region in nuclear ribosomal DNA. Sci. China C Life Sci. 1997;40:137–144. doi: 10.1007/BF02882041. [DOI] [PubMed] [Google Scholar]
- 56.Zou X-H, Zhang F-M, Zhang J-G, Zang L-L, Tang L, Wang J, Sang T, Ge S. Analysis of 142 genes resolves the rapid diversification of the rice genus. Genome Biol. 2008;9:R49. doi: 10.1186/gb-2008-9-3-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang L, Zou X-H, Achoundong G, Potgieter C, Second G, Zhang D-Y, Ge S. Phylogeny and biogeography of the rice tribe (Oryzeae): evidence from combined analysis of 20 chloroplast fragments. Mol. Phylogenet. Evol. 2009;54:266–277. doi: 10.1016/j.ympev.2009.08.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.