Abstract
The basal mitochondrial transcription machinery is essential for biogenesis of the respiratory chain and consists of mitochondrial RNA polymerase, mitochondrial transcription factor A (TFAM) and mitochondrial transcription factor B2. This triad of proteins is sufficient and necessary for mtDNA transcription initiation. Abolished mtDNA transcription caused by tissue-specific knockout of TFAM in the mouse heart leads to early onset of a severe mitochondrial cardiomyopathy with lethality within the first post-natal weeks. Here, we describe a mouse model expressing human TFAM instead of the endogenous mouse TFAM in heart. These rescue mice have severe reduction in mtDNA transcription initiation, but, surprisingly, are healthy at the age of 52 weeks with near-normal steady-state levels of transcripts. In addition, we demonstrate that heavy-strand mtDNA transcription normally terminates at the termination-associated sequence in the control region. This termination is abolished in rescue animals resulting in heavy (H)-strand transcription of the entire control region. In conclusion, we demonstrate here the existence of an unexpected mtDNA transcript stabilization mechanism that almost completely compensates for the severely reduced transcription initiation in rescue hearts. Future elucidation of the underlying molecular mechanism may provide a novel pathway to treat mitochondrial dysfunction in human pathology.
INTRODUCTION
Mitochondrial diseases are among the most common forms of human genetic disease. In addition, mitochondrial dysfunction is implicated in many different types of human age-associated diseases, e.g. diabetes mellitus type 2, heart failure, neurodegeneration and the ageing process itself (1,2). A molecular understanding of the regulation of oxidative phosphorylation capacity is therefore not only of basic scientific interest but might provide insights into different types of human diseases and ageing.
The mitochondrial network is responsible for a variety of metabolic processes including oxidative phosphorylation, which provides the bulk part of the cellular energy in the form of ATP (3). The oxidative phosphorylation system is under bipartite genetic control requiring both nuclear and mtDNA-encoded genes for its biogenesis. The two strands of mtDNA contain different mitochondrial genes and are transcribed from dedicated promoters; designated the light- and heavy-strand promoters (LSP and HSP). Processing of long primary polycistronic transcripts releases 13 mRNAs, which all encode respiratory chain subunits, and 2 rRNAs and 22 tRNAs, which are needed for mitochondrial translation.
We have previously defined the basal mammalian mitochondrial transcription machinery as consisting of three nuclear-encoded proteins; mitochondrial transcription factor A (TFAM), mitochondrial RNA polymerase and mitochondrial transcription factor B2 (TFB2M). This triad of proteins is sufficient and necessary for promoter-specific initiation of transcription in a pure recombinant system (3–6). The TFAM protein is not only an indispensable component of the mtDNA transcription machinery (7), but there is also a wealth of data supporting a direct role for TFAM and its homologues in packaging and stabilizing mtDNA in a variety of organisms such as yeast (8), chicken (9), mouse (10) and humans (11,12). This latter function is consistent with the fact that TFAM belongs to the high mobility group-box-domain protein family, which has the capacity to bind, bend and wrap DNA in a largely sequence-independent manner (13,14). Furthermore, the yeast homologue of TFAM, denoted ABF2, is dispensable for mtDNA transcription but is necessary for mtDNA maintenance (15). In vivo data from the mouse have demonstrated that disruption of Tfam leads to lack of mtDNA, which, in turn, causes a profound respiratory chain deficiency and embryonic lethality in mid-gestation (16). The human TFAM (hTFAM) protein has a similar capacity to bind DNA as the mouse TFAM (mTFAM) protein, but hTFAM is a poor activator of mtDNA transcription in the mouse recombinant transcription system (7,10). Consistent with these in vitro results, we have demonstrated that hTFAM cannot rescue the embryonic lethality of mTFAM knockout mice (10). Mouse embryos lacking mTFAM and expressing hTFAM can maintain near-normal levels of mtDNA but have very low levels of mitochondrial transcripts and can therefore not sustain normal oxidative phosphorylation (10). Expression of hTFAM in mice with normal endogenous mTFAM expression results in increased mtDNA levels without a significant increase of mtDNA expression, further demonstrating that it is possible to dissociate mtDNA copy number regulation from mtDNA transcription (10).
We have previously created a series of tissue-specific knockout mice with disruption of Tfam in cardiomyocytes and demonstrated that such animals develop a progressive respiratory chain deficiency in heart during post-natal life (17–19). These knockout mice develop a progressive dilated cardiomyopathy with heart conduction blocks and die within the first months of life. These conditional knockout mice have provided valuable models for severe mitochondrial cardiomyopathy, but have not been suited to model the more moderate decline of respiratory chain function associated with age-associated heart disease. In the present study, we therefore used a genetic strategy to express hTFAM in mice lacking mTFAM expression in heart; based on the assumption that hTFAM would partially rescue the cardiomyopathy phenotype. Surprisingly, we report here that hTFAM almost fully complements the absence of mTFAM in the mouse heart. These rescued animals have severely decreased de novo transcription of mtDNA in heart but contain normal levels of mtDNA and near-normal steady-state levels of the mtDNA-encoded transcripts. The animals survive and develop normally with only a minor respiratory chain deficiency in the heart. In addition, we observe high levels of a novel transcript spanning the non-coding region of mtDNA, indicative of altered RNA processing. This study thus presents experimental evidence that a highly specialized and energy-demanding tissue such as heart can activate compensatory mechanisms to increase the stability of mtDNA-encoded transcripts if de novo transcription of mtDNA is severely reduced. This compensation does not only include regulation of transcript stability but may also involve alteration of transcription termination events.
MATERIALS AND METHODS
Mouse breeding and genotyping
Previously engineered TfamloxP/loxP mice of mixed genetic background (16) were crossed to transgenic animals expressing hTFAM from a genomic phage artificial chromosome (hTFAMPAC-tg) transgene (10), generating TfamloxP/loxP; hTFAMPAC-tg animals [clone PAC19 in reference (10)]. Mice expressing the P1 phage recombinase, cre, with a nuclear localization signal (NLS-cre) from the muscle creatine kinase promoter (Ckmm-NLS-cre) were crossed to these transgenic TfamloxP/loxP; hTFAMPAC-tg animals (Supplementary Figure S1). Ear clip DNA from offspring was genotyped for the PAC transgene, the floxed mTfam gene and the Ckmm-NLS-cre gene, as previously reported (10,16,17).
Mitochondrial preparation
Hearts were collected from 11 to 12-week-old animals and crude mitochondrial preparations were obtained as described (20). In brief, the hearts were homogenized in ice-cold 75 mM sucrose, 225 mM sorbitol, 1 mM EGTA, 0.1% bovine serum albumin (BSA) and 10 mM Tris–HCl, pH 7.4, using a Teflon pestle homogenizer (six strokes at 1000 r.p.m.). Cell membranes and debris were pelleted twice for 5 min at 1000g. The mitochondrial supernatants were washed three times and pelleted at 12000g.
DNA isolation and Southern blot analysis
Genomic or mitochondrial DNA was isolated by standard Tris/EDTA/SDS and proteinase K treatment. DNA was purified by phenol–chloroform extraction, precipitated with sodium acetate (NaOAc) and ethanol, and dissolved in 10 mM Tris–HCl pH 8.5 or dH2O. Total DNA (3μg) from heart tissue of either 11- (±1) or 52 (±1)-week-old animals were digested over night with 20 U PstI (New England Biolabs), precipitated by NaOAc–ethanol precipitation and used for Southern blotting.
RNA isolation and Northern blot analysis
Genomic or mitochondrial RNA from heart or kidney preparations was isolated with Trizol® reagent (Invitrogen) or ToTALLY RNA (Ambion), following the manufacturer’s instructions. RNA quality was analysed on a BioAnalyser 2100 (Agilent) and an appropriate amount of genomic or mtRNA was used for standard Northern blot analysis. Mt-tRNAs were separated on a 10% 7 M UREA/polyacrylamide gel and blotted onto a nylon membrane (Amersham) by electrical transfer.
Probes and labelled oligonucleotides
For double-stranded DNA and single-stranded RNA probes, appropriate PCR products were cloned into pCR-II using the pCR-II TOPO cloning kit (Invitrogen). DNA probes were excised and purified by gel extraction (Qiagen), followed by random-labelling using [α-32P]-dCTP (Amersham) with the prime-it II kit (Stratagene). For RNA probes, the appropriate vectors were linearized and in vitro transcribed using Sp6 or T7 polymerase (both New England Biolabs) in the presence of [α-32P]-UTP (Amersham). End-labelled oligonucleotide probes were generated by polynucleotide kinase treatment (New England Biolabs) in the presence of [γ-32P] ATP (Amersham). Probe hybridization was performed in RapidHyb buffer (Amersham) at 65°C (for probes) or 42°C (for oligonucleotides) for at least 90 min. After washing, the membranes were exposed to an autoradiography Hyperfilm (Amersham), and/or analysed by phosphor imager analysis (BioRad). A list of primers and oligonucleotides used is shown in the Supplementary Table S1.
Isolation of proteins and Western blot analysis
Total protein extracts were generated as previously described (10). Briefly, tissue homogenates were suspended in equal parts of suspension buffer (0.1 M NaCl, 0.01 M Tris–HCl, pH 7.6, 10 mM EDTA, 1% Aprotinin, 1% PMSF), and loading buffer (0.1 M Tris–HCl, pH 6.8, 4% SDS, 20% glycerol, 200 mM DTT). Samples were boiled for 10 min, sonicated and centrifuged for 10 min at 10000g. The supernatants were collected and protein concentration determined with a total protein quantification kit (Sigma). Western blot analysis was performed as previously described (10).
Biochemical evaluation of respiratory chain function and glycolysis
Heart tissue samples of rescued animals (n = 5 per age group) and control animals (n = 5 at 12 weeks, and 4 at 52 weeks) were collected at 12 and 52 weeks of age and mitochondria were isolated. The respiratory chain enzyme activities and mitochondrial ATP production rate (MAPR) were measured blind as described (21,22). For determination of glycolytic enzyme activities, a frozen sample of heart tissue (10 mg) was thawed and analysed as previously described (17).
Histochemistry was performed on 10-μm sections from fresh-frozen hearts of 12- and 28-week-old animals as previously described (22).
In organello transcription assay
De novo transcription was measured in isolated mitochondria as described previously (23–25). Mitochondrial preparations were suspended in transcription buffer, containing 25 mM sucrose, 75 mM sorbitol, 100 mM KCl, 10 mM K2HPO4, 50 mM EDTA, 5 mM MgCl2, 1 mM ADP, 10 mM glutamate, 2.5 mM malate and 10 mM Tris–HCl (pH 7.4), with 1 mg BSA per millilitre. Mitochondria containing 200 μg protein, as determined by Bradford method (BioRad), were incubated for 45 min in 500 μl of transcription buffer containing 30 μCi [α-32P]-UTP. After incubation, samples were pelleted and washed twice in 1 ml 10 mM Tris–HCl (pH 6.8), 0.15 mM MgCl2, containing 10% glycerol. Mitochondrial RNA was isolated from the final pellet using the ToTALLY RNA isolation kit (Ambion), followed by Northern blot analysis.
S1 protection assay
S1 nuclease protection was performed essentially as described (19) following the ribonuclease protection assay kit III (Ambion) protocol, except that an S1 nuclease (Promega) was used for digestion instead of the supplied RNases, since DNA oligonucleotides were used instead of riboprobes. The end-labelled oligonucleotide probes, against 7S RNA 5′-GAC ATA TAA TAT TAA CTA TCA AAC CCT ATG TCC TGA TCA ATT CTA GTA GT-3′ (mismatch to 7S RNA underlined) and ND6 5′-AAT AAA AAT ACC CGC AAA CAA-3′, were used to hybridize to mitochondrial RNA, followed by S1 nuclease digestion and separation on an 8% 7M UREA/polyacrylamide gel.
Mapping the 3′-end of the D-loop transcript
A ligation-mediated reverse transcription protocol was used to sequence the 3′-ends of transcripts. In a 100 μl reaction, 3 μg of RNeasy purification kit (Qiagen)-purified total heart RNA was ligated to 100 pmol of linker: 5′-Phosphate-GAA TTC ACT GGC CGT CGT TTT ACA-spacer C3-3′ (antisense to standard M13 forward sequencing primer) using 20 U T4 RNA ligase (New England Biolabs), followed by NaOAc–ethanol precipitation. First strand reverse transcription was performed using M13 forward sequencing primer and the SuperScript™ II Reverse Transcriptase kit (Invitrogen). Second strand amplification was performed using the M13 forward sequencing primer and the mtDNA-specific primer 5′-TCA AAC CCT ATG TCC TGA TC-3′ (position 16164). The resulting PCR products were cloned and sequenced in-house on a 3130XL cycle sequencer (Applied Biosystems), using 0.5× big dye v1.1 (Applied Biosystems) in a 20 μl reaction.
Ligation-mediated quantitative real-time PCR
Quantification of nascent replication strands was performed using a ligation-mediated quantitative real-time PCR method, as described previously (26). Two independent extension primers complement to mtDNA sequence either within the terminated replication strand (primer W), or within the non-terminated strand (primer E) were annealed independently to total DNA and extended using Vent polymerase (New England Biolabs), followed by ligation of a double-stranded linker/adaptor to the ends of these extensions. Nested primers to the linker/adaptor and to sequence complement to both extended strands were then used to quantify the amount of extensions in the reaction, using quantitative real-time PCR on a LightCycler 2.0 (Roche) with FastStart DNA master SyBr Green 1 mix (Roche). Extension primers E: 5′-TAC ATA AAT TTA CAT AGT AC-3′; and W: 5′-GGT CAT AAA ATA ATC ATC AAC-3′; linker sequences L1: 5′-GTG ACC CGG GAG ATC TGT ATTC-3′; L2: 5′-GAA TAC AGA TC-3′; and the common internal primer L3: 5′-ATC CTC CGT GAA ACC AAC AA-3′. Annealing and extension was controlled for, using cloned mouse mtDNA (pAM1), linearized by endonuclease Bsu36 I (position 16027 bp in mtDNA). Previous amplifications were pooled, gel purified (Qiagen gel purification kit), and quantified on a Hoefner DNynaliser, to be used as standards to evaluate PCR efficiency.
RESULTS
hTFAM rescues the cardiac hypertrophy of tissue-specific Tfam knockout mice
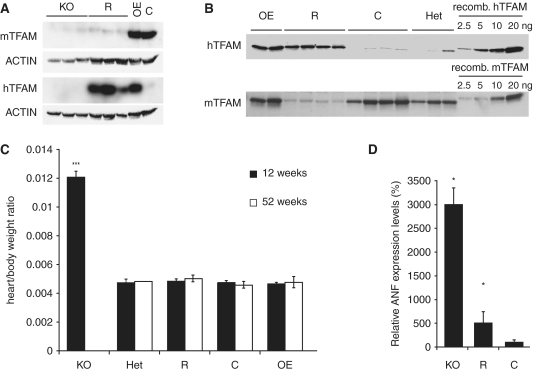
We have previously generated mice with disruption of Tfam in heart (17,19); herein referred to as ‘cardiomyopathy mice’. In the present study, we investigated whether the phenotype of such cardiomyopathy mice could be rescued by expression of the hTFAM protein. Mice containing a human phage P1 artificial chromosome (PAC) transgene, which ubiquitously expresses hTFAM (10), were crossed into the cardiomyopathy mice background (Supplementary Figure S1), to generate mice of the genotype TfamloxP/loxP, Ckmm-NLS-cre; +/hTFAM PAC-tg, hereafter denoted ‘rescue mice’. Rescue mice were born at the expected mendelian ratio and appeared normal at birth. Western blot analyses of hearts showed very low levels of the mTFAM protein both in cardiomyopathy and rescue mice, whereas the hTFAM protein only was detected in rescue hearts (Figure 1A). The expression of hTFAM was similar in hearts of rescue mice and hearts of mice expressing both hTFAM and mTFAM (hereafter denoted TFAM over-expression mice, OE). The results show that expression of hTFAM in the mouse heart is not dependent on endogenous mTFAM levels, and that endogenous mTFAM levels are not altered if hTFAM is expressed (Figure 1B). Thus, there seems to be no feedback mechanism to sense total TFAM levels in cardiomyocytes.
Figure 1.
hTFAM can compensate for loss of mTFAM in the heart. (A) Western blot analysis of total heart protein extracts using polyclonal antibodies against mouse and hTFAM, demonstrating severe reduction of the mTFAM protein in both cardiomyopathy (KO) and rescue (R) samples and the presence of hTFAM in R and over-expressor (OE) samples at 12 weeks of age. (B) Quantification of protein levels of both mTFAM and hTFAM in R and control (C) animals in comparison with increasing amounts of recombinant protein. Rescue hearts contain similar amounts of TFAM as control samples. (C) Heart weight to body weight ratios from animals at 12 (dark bars) and 52 weeks (white bars) of age. KO mice have increased heart to body weight ratios, while heterozygous knockouts (HET), R or OE mice show heart to body weight ratios comparable with C. (N ≥ 10 except Het and OE at 52 weeks, where N = 1 and 7, respectively). (D) KO and R animals (N = 3) present with increased expression levels of the ANF, as determined by Northern blot analysis. Values are given as percentage of controls. Data are presented as means ± SEM; *P < 0.05; ***P < 0.001, respectively.
We have previously shown that the cardiomyopathy mice develop a dilated cardiomyopathy with heart conduction blocks and die by 10–14 weeks of age (17,19). Consistent with previous results (17), we found that the heart weight to body weight ratio was massively increased in end-stage cardiomyopathy mice at the age of 12 weeks (Figure 1C). In contrast, rescue mice had normal heart weight to body weight ratios at all studied ages (Figure 1C) and survived without obvious disease symptoms until the age of 62 weeks, when we stopped observing the animals. Northern blot analysis of atrial natriuretic factor (ANF) expression, a commonly used molecular marker for heart failure, showed dramatically increased ANF transcript levels in the myocardium of cardiomyopathy mice, whereas only a mild induction of ANF expression was found in the hearts of rescue mice (Figure 1D). These results thus demonstrate that hTFAM expression is sufficient to almost completely revert the cardiomyopathy phenotype of tissue-specific TFAM knockout mice.
Rescue hearts have decreased de novo transcription of mtDNA but near-normal steady-state levels of transcripts
We assessed de novo transcription of mtDNA in rescue heart mitochondria with in organello transcription assays and found a decrease of at least 45% in comparison with control heart mitochondria (Figure 2A). Levels of transcription initiation at LSP were also assessed by S1 mapping of the 7S RNA, which is a good indicator of de novo transcription of mtDNA (25). There was a profound decrease of 7S RNA transcripts in rescue hearts in comparison with control hearts (Figure 2B and C). The finding of very low levels of transcription initiation of mtDNA in hearts of rescue mice is consistent with the previous biochemical observation that hTFAM is poor at promoting mtDNA transcription initiation in the mouse system (10).
Figure 2.
Measurement of de novo transcription. (A) In organello transcription of 16-week-old rescue and control hearts, followed by normalization by Northern blotting for the mitochondrial transcripts of ND6. (B) Schematic diagram of the murine mtDNA control region containing tRNAT (T), tRNAP (P), TAS 1 and 2, 7S RNA transcript, 7S DNA fragment, conserved sequence blocks 1–3 (CSB), LSP, HSP and tRNAF (F). (C) S1 protection assay of LSP proximal transcription (7S RNA) and control transcript (ND6) of control (C) and rescue (R) mitochondrial RNA extracts. (D and E) Relative transcript levels normalized to the nuclear 18S rRNA transcript. Levels of the mitochondrial transcripts of COX1 (light grey bars) and ND6 (dark grey bars) and the large mitochondrial ribosomal subunit 16S rRNA (white bars) are shown at 12 (D) and 52 weeks (E). Data are presented as means ± SEM percentage of controls; *P < 0.05; ***P < 0.001, respectively.
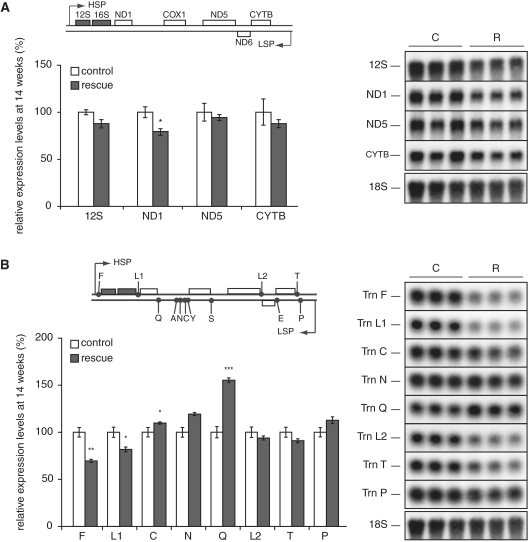
The reduced levels of mitochondrial transcription initiation in hearts of rescue animals led us to investigate the steady-state levels of several transcripts, i.e. cytochrome c oxidase subunit 1 (COX1), NADH dehydrogenase subunit 6 (ND6) and the large ribosomal RNA (16S rRNA) in rescue animals. No significant changes were observed in hearts of 12-week-old rescue animals (Figure 2D) and a moderate decrease of steady-state levels of the COX1 mRNA and the 16S rRNA was present in 52-week-old rescue animals (Figure 2E). Additional characterization of transcription patterns in heart from 14-week-old rescue animals showed normal levels of 12S rRNA, NADH dehydrogenase subunit 5 (ND5) mRNA and cytochrome b (cytb) mRNA, whereas the levels of NADH dehydrogenase subunit 1 (ND1) mRNA were slightly reduced (Figure 3A). A comprehensive survey of mitochondrial tRNAs, transcribed from either LSP or HSP, showed both increased (tRNAC and tRNAQ) and decreased (tRNAF and tRNAL1) steady state-levels (Figure 3B). The genes whose expression was significantly decreased (16S rRNA, COXI, ND1, tRNAF, tRNAL1) are all transcribed from HSP. It is interesting to note that the steady-state levels of transcripts generated from LSP were normal (ND6) or increased (tRNAC and tRNAQ) despite evidence of reduced transcription initiation at this promoter (low 7S RNA levels). This finding suggests that levels of transcripts are regulated at a stage following transcription initiation at LSP, e.g. by reduced transcription termination downstream of LSP or by increased stability of mature transcripts.
Figure 3.
Altered steady-state transcript levels. Relative mitochondrial transcript steady-state levels were measured in 12-week-old rescue (white bars) and control (black bars) hearts by Northern blot analysis. (A) 12S rRNA and the mitochondrial mRNAs from ND1, ND5 and cytb. (B) Mitochondrial tRNA steady-state levels of tRNAF (F), tRNAL1 (L1), tRNAC (C), tRNAN (N), tRNAQ (Q), tRNAL2 (L2), tRNAT (T) and tRNAP (P). Data are presented as means ± SEM percentage of controls; *P < 0.05; **P < 0.01 and ***P < 0.001, respectively.
Rescue mice have reduced mtDNA replication, but near-normal steady-state levels of mtDNA
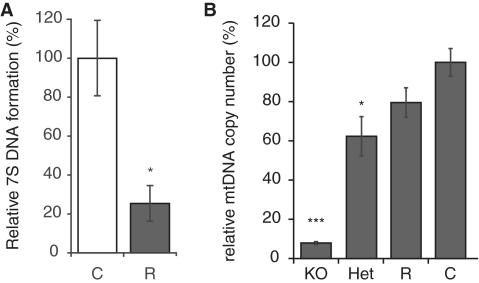
Transcription is a prerequisite for initiation of mtDNA replication (3) and we therefore investigated mtDNA replication by assaying levels of the nascent 7S DNA, which is continuously generated de novo by an abortive mtDNA replication process in the non-coding displacement (D) loop region. We used a ligation-mediated PCR technique to quantify 7S DNA formation in relation to leading-strand replication (26). Heart mitochondria isolated from rescue mice had ∼75% reduction of 7S DNA molecules (Figure 4A) indicating a decrease in mtDNA replication initiation consistent with the observed reduction in transcription initiation (Figure 2A and B).
Figure 4.
mtDNA replication and maintenance (A) The normalized ratios of steady-state levels of replication termination versus replication elongation events of control (C; N = 7) and rescue (R; N = 5) heart samples, as determined by quantitative real-time PCR are shown. Data are presented as means ± SEM percentage of controls; *P < 0.05. (B) Southern blot analysis of mtDNA copy number in comparison with 18S DNA demonstrate a significant reduction of mtDNA levels in KO (8%; N = 7) and heterozygous KO (HET) (62%; N = 3), and a modest reduction in R (80%; N = 8; P = 0.075) in comparison with control samples (N = 10).
Loss of TFAM protein leads to severe mtDNA depletion in cardiomyopathy mice at 12 weeks of age, whereas Tfam heterozygous knockout hearts have ∼60% of normal mtDNA levels. The rescue mice have near-normal levels of mtDNA in their hearts at 12 (Figure 4B) and 52 weeks of age (data not shown). These findings demonstrate that hTFAM is a good substitute for mTFAM as an mtDNA maintenance factor.
Rescue mice have decreased COX activity and decreased ATP production
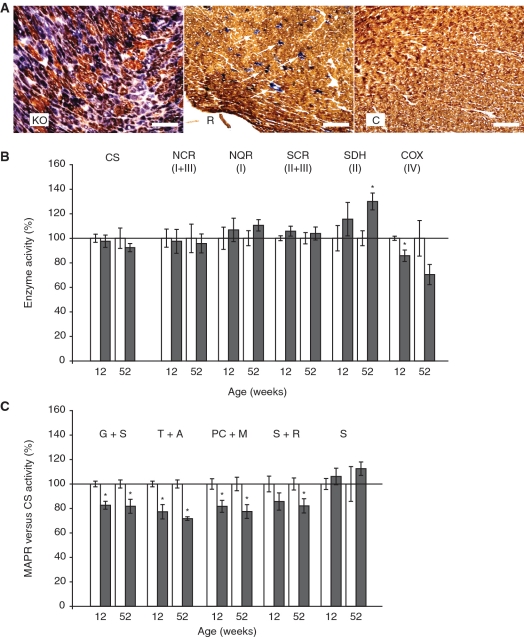
We performed COX-succinate dehydrogenase (COX-SDH) double staining on fresh-frozen sections of hearts from 10-week-old cardiomyopathy, rescue and control animals (Figure 5A). The cardiomyopathy hearts displayed many COX negative cells, which stained strongly for SDH, consistent with previous reports (18,19). Surprisingly, the rescue hearts also contained COX negative cells, albeit much less abundant than in the cardiomyopathy hearts. No COX negative cells were observed in control hearts. In order to determine whether the presence of COX negative cells was a temporary phenomenon associated with the switch between mTFAM and hTFAM at 10 weeks, we also stained sections from rescue and control mice at 28 weeks of age. No COX negative cells could be observed in control hearts, while rescue hearts contained some COX negative cells, but at lower frequency than at 10 weeks of age (Supplementary Figure S2).
Figure 5.
Biochemical measurements of the respiratory chain function in heart tissue. (A) Enzyme histochemical double staining for COX and SDH activities on heart sections of cardiomyopathy, rescue and control mice at 10 weeks of age. Size bar is 100 μm. (B and C) The results from biochemical measurements in hearts from rescue (dark bars) and control (white bars) mice at 12 and 52 weeks of age are shown. Data are presented as means ± SEM percentage of controls with *P < 0.05. (B) Relative enzyme activities of respiratory chain enzyme complexes I and III (NADH cytochrome c reductase, NCR), complex I (NADH coenzyme Q reductase, NQR), complexes II and III (SCR), complex II (SDH) and complex IV (COX). (C) Results from measurements of the MAPR per unit of CS activity in the presence of various substrates: Glutamate + succinate (G + S); TMPD + ascorbate (T + A); palmitoyl-L-carnitine + malate (PC + M); succinate + rotenone (S + R), and succinate (S).
The respiratory chain dysfunction in rescue hearts, as detected by the COX-SDH double staining, was further investigated by biochemical measurements of mitochondrial enzyme activities at both 12 and 52 weeks of age. No changes in citrate synthase activity (CS) were found, indicating that the transgenic animals had similar mitochondrial mass as control animals (Figure 5B). Biochemical measurements demonstrated an isolated COX deficiency at 12 weeks and tendency to reduction at 52 weeks in relation to CS activity (Figure 5B). In contrast, the activity of the exclusively nuclear-encoded complex II (SDH) was significantly increased at 52 weeks, but normal at 12 weeks. We measured the MAPR in freshly isolated heart mitochondria, both in relation to CS activity (Figure 5C) and in relation to heart tissue mass (Supplementary Figure S3). The MAPR/CS in heart tissue of rescued mice was reduced with substrates entering at all levels of the respiratory chain, with the exception of succinate, which enters the respiratory chain at complex II. The general decrease in ATP production rate in the heart tissue of rescue animals is consistent with an inhibition at the level of complex IV (COX).
Rescue mice express high levels of a transcript encompassing the non-coding region
We performed Northern blot analyses and found that the rescue hearts contained an abundant transcript of ∼1 kb in length encompassing the D-loop region. Characterization with strand-specific RNA probes and RNase I treatment revealed that this transcript corresponded to a transcript complementary to the heavy strand (Figure 6A and B) and we therefore termed this transcript anti-control region (ACR) transcript. This transcript has the opposite sense as the LSP transcript providing primers for initiation of mtDNA replication. The ACR transcript was not detected in the kidney of heart rescue animals or in hearts from animals expressing both mTFAM and hTFAM (Figure 6B). The presence of this transcript is thus dependent on both the presence of hTFAM and the absence of mTFAM.
Figure 6.
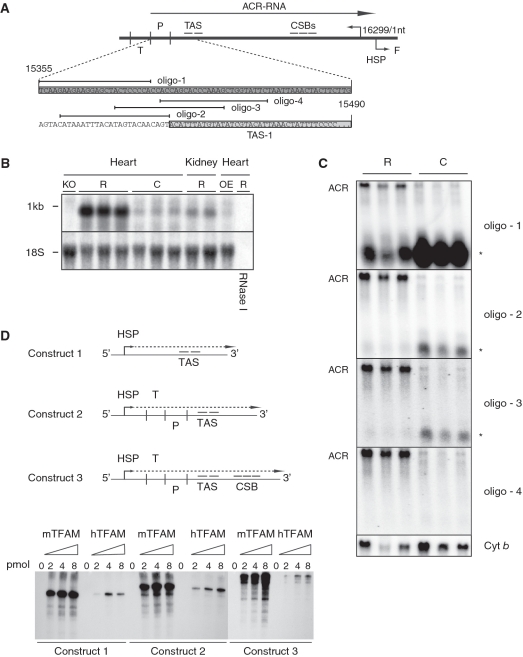
Altered transcription of the mtDNA control region. (A) Schematic diagram of the murine mtDNA control region containing tRNAT (T), tRNAP (P) (dark grey), TAS1 (light grey) and 2, the ACR transcript (ACR-RNA), conserved sequence blocks 1–3 (CSB), tLSP, HSP and tRNAF (F). (B) Northern blot analysis of total heart RNA samples from ≥12-week-old animals. The membrane was first hybridized with a single-stranded ribo-probe complement to the mtDNA light strand control region, followed by hybridization for the 18S rRNA gene product as loading control. Samples used were from cardiomyopathy (KO), rescue (R) and TFAM over-expressing hearts and from rescue kidney samples. One rescue sample was RNase 1 treated prior to loading. (C) The ACR transcript was mapped by Northern blot analysis, using end-labelled oligonucleotides complement to the mtDNA light strand. Additionally, a short RNA species (*) was observed for some oligonucleotides. (D) In vitro transcription run off assay using three artificial constructs, containing HSP and the TAS region (construct 1), HSP, tRNAT, tRNAP and TAS (construct 2) and HSP, tRNAT tRNAP, TAS, and CSBs (construct 3).
The 5′-terminus of the ACR transcript was mapped to immediately upstream tRNAP by primer extension (data not shown) and by Northern blot analysis with radiolabelled oligonucleotide probes corresponding to the heavy strand of the control region (Figure 6A and C). The 3′-terminus of the ACR transcript was identified by ligation-mediated reverse transcription PCR followed by cloning and sequencing. The 3′-terminus could be precisely aligned immediately upstream of tRNAF (position 16297–16299) and contained a polyadenylation tail of at least 50 nt (Supplementary Figure S4). The exact nucleotide position is uncertain as the two most proximal bases in the mouse mtDNA genome are adenine nucleotides and thus could be generated either by polyadenylation or transcription. On the other hand, oligonucleotide probes complementary to control region light strand transcripts failed to show any difference between the rescue and control samples (data not shown). In summary, mitochondrial transcription in rescue hearts generates high levels of a polyadenylated transcript, which extends from tRNAP at the 5′-end to tRNAF at its 3′-end and thus has a length of 943 nt (excluding the poly A tail).
The presence of the ACR transcript correlated with the loss of a short RNA species (* in Figure 6C) present in control animals. This shorter transcript extends between the tRNAT and the onset of the termination-associated sequence (TAS) region. This is consistent with the suggestion that a normally occurring transcription termination event close to the TAS region is lost in rescue hearts. We performed in vitro transcription run off assays using both mouse and hTFAM in a reconstituted mouse transcription assay. However, no transcription termination due to the presence of TFAM could be observed under these conditions (Figure 6D). We also failed to identify any specific differences in hTFAM and mTFAM binding at or near the TAS region by in vitro footprinting analysis (data not shown). In order to investigate whether transcription proceeds into tRNAF, we performed qRT-PCR experiments across HSP. No increase in transcript could be identified in this region (data not shown).
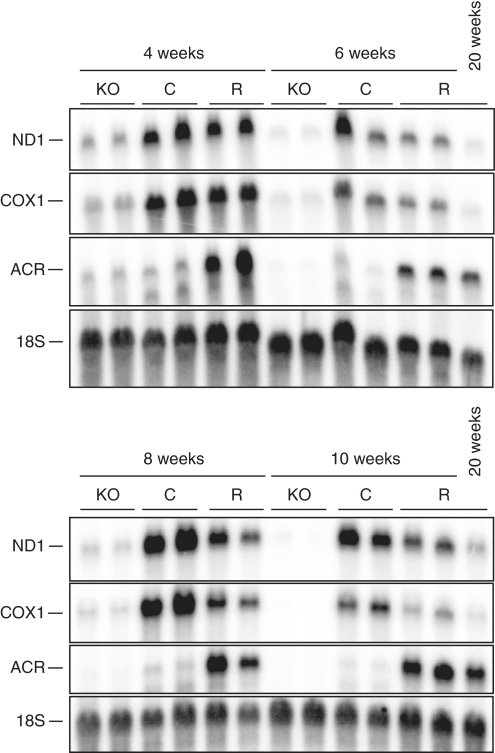
We wondered if the increased expression of the ACR transcript was a late secondary response to the increased physiological stress on heart mitochondria lacking mTFAM. We thus investigated the steady-state levels of ACR, COX1 and ND1 transcripts in heart mitochondria from 4- to 10-week-old animals. As expected, the cardiomyopathy mice have decreased mitochondrial transcripts already at 4 weeks of age (Figure 7). However, the rescue animals retain close to normal transcript levels and express the ACR transcript from at least 4 weeks of age.
Figure 7.
Altered transcription is an early event in mitochondria from rescue hearts. Northern blot analysis of steady-state transcript levels in hearts of 4-, 6-, 8-, 10- and 20-week-old control (C) and rescue (R) mice. The membranes were hybridized with double-stranded probes against, the mitochondrial transcripts of ND1, cytochrome c oxidase subunit 1 (COX1), ACR and the nuclear transcript of 18S rRNA as loading control. Increased transcript levels of ACR can already be seen at 4 weeks of age.
DISCUSSION
We demonstrate here that severely reduced transcription of mtDNA can be unexpectedly well tolerated in cardiomyocytes, a highly specialized cell type with a continuous high demand for energy. This finding is surprising and points towards the existence of substantial post-transcriptional regulatory mechanisms. Expression of hTFAM in mTFAM knockout hearts almost fully rescues the cardiomyopathy phenotype despite the fact that hTFAM is a very poor transcriptional activator in the mouse. The rescue mice have close to normal mtDNA levels but severely decreased de novo transcription of mtDNA. Thus, we were able to demonstrate a clear dissociation of mtDNA transcription and mtDNA maintenance. These findings also show that a severe reduction in transcription initiation allows maintenance of near-normal mtDNA levels. However, the decrease in transcription initiation has only a minor impact on steady-state levels of mitochondrial transcripts and there is only a slight decrease of respiratory chain function. Biochemical measurements of the activities of individual respiratory chain complexes revealed a decrease of COX activity, whereas the activities of the other complexes were normal or, in case of SDH even increased. Measurement of MAPR with substrates entering the respiratory chain at different complexes also gave a pattern consistent with a moderate COX deficiency. Enzyme histochemical staining of heart tissue sections supported the biochemical results and showed the presence of COX-deficient cells. These findings indicate that COX is particularly sensitive to a moderate reduction in mtDNA expression.
Cardiomyopathy mice lacking mTFAM in heart have increased heart weight and a massively increased expression (30-fold) of ANF, a commonly used marker for heart failure (27). However, the rescue animals have normal heart weight and only a moderate induction of ANF expression (5-fold), indicating that these animals only suffer from minor heart failure. It is possible that the reduced cardiac function observed during ageing, at least in part, stem from a reduced oxidative phosphorylation capacity of cardiac mitochondria. Microarray studies have shown an age-related decrease in the expression of genes involved in metabolism and respiratory chain function in tissues such as heart, skeletal muscle and brain (28–31). Observations made in hearts of senescent rats and flies show a reduced rate of mitochondrial transcription without affecting mtDNA copy number (32,33). Also, reduced 12S rRNA and COX1 transcript levels were found in hearts, cerebral hemispheres and cerebellum, but not in the liver, of senescent rats (34). However, a causal role of decreased mitochondrial transcription capacity has not been established in age-associated cardiovascular disease or in the ageing process itself.
We were able to show here that rescue hearts have severely reduced transcription initiation at LSP, but surprisingly retain normal ND6 and normal to increased tRNA levels. Total de novo transcription was also reduced and manifested itself with the reduction of the HSP-derived ND1, COX1 and 16S transcripts. How rescued mitochondria are able to maintain normal ND6 transcript levels is unclear, but a significant increase in transcript stability would be a plausible mechanism. Increased stability is supported by the observations that tissue-specific TFAM KO mice rapidly loose TFAM protein levels and mtDNA levels, but can retain mitochondrial transcripts for weeks and months after disappearance of TFAM in the targeted tissue (22,35).
Interestingly, we found a novel heavy strand transcript covering the D-loop region in hearts of rescue animals that is expressed from at least 4 weeks of age. Several groups have previously reported heavy strand transcripts of the D-loop region. For instance, Aloni and Attardi (36) reported that almost the entire heavy strand of HeLa cells was transcribed and several transcription termination events have been reported between tRNAP and TAS (37). In this latter study, the authors identified transcripts of up to 800 nt in length, corresponding to almost the entire D-loop region. Later it was suggested that mitochondrial HSP transcription passes through the entire length of the control region, and is terminated downstream of HSP, due to the presence of a putative polyadenylation signal (38), and an unidentified termination factor (39). Avadhani and colleagues (39) suggested that this mode of transcription termination is the normal mode of mitochondrial HSP transcription termination, and the D-loop transcript is rapidly degraded under normal physiological conditions. Here, we report the up-regulation of a transcript with identical 3′ end as observed by Avadhani and colleagues. The up-regulation of this transcript could be explained by several mechanisms.
This transcript could be the result of either specific or global transcription stabilization events in heart rescue mice. The ACR transcript has previously been reported in HepG2 cells, in which mitochondrial protein synthesis had been blocked by the addition of thiamphenicol (40). Thiamphenicol treatment leads to increased mitochondrial transcription, with a maximal induction 3 days after drug treatment. Interestingly, this D-loop transcript—termed RNA DL-L by the authors—was induced only 7 days after initiating drug treatment and was thus not due to increased transcription. One plausible explanation for this is that mitochondria responding to high demands of gene expression undergo a switch in the regulation of transcription, enabling transcription through the regulatory D-loop region. Unfortunately, we were unable to detect transcripts containing both control region and downstream RNA sequence, suggesting either the absence of such a product or very rapid processing at this site.
Instead, we observed a short RNA species, ranging from the region complementary to the tRNAP gene to the TAS region in control animals but not in rescue animals, which instead express the larger ACR transcript. The termination-associated region is comprised in the mouse of two blocks spanning 107 nt (41). We propose that the TAS region, previously identified as a putative termination site for heavy strand replication, is also the site for HSP transcription termination under normal physiological conditions. The absence of transcription termination in the present mouse model could either be due to the failure of hTFAM to promote termination, or caused by a switch in transcription regulation. It is noteworthy that there are different effects on LSP-dependent transcripts upstream and downstream of this postulated termination event. Levels of 7S RNA, which is proximal to the promoter, are decreased, whereas the levels of tRNAC and tRNAQ are increased in the heart rescue animals. This observation could be explained by increased transcript stability of tRNAC and tRNAQ, but two observations argue against this interpretation. First, increased stability would imply that there is a difference in RNA stability regulation between LSP- and HSP-initiated transcripts, which we find unlikely. Second, we have previously reported that tRNA levels reflect overall transcription rates and are not regulated by RNA stability to the same extent as are mitochondrial mRNA molecules (24,25). It is therefore possible that a decrease of transcription termination in the TAS region is compensating for a decrease in transcription initiation at LSP. This termination event apparently regulates transcription termination in a bidirectional way, since we see an up-regulation of the ACR transcript. It is possible that the ACR transcript has a specific functional role of its own, which remains to be established, but it is also possible that it simply is a secondary consequence of decreased termination of LSP-dependent transcripts in the TAS region.
Additional factors are required to promote transcription termination, since both hTFAM and mTFAM failed to promote transcription termination in a pure recombinant in vitro transcription system. Isolating the factors required for transcription termination in vivo, and whether alterations in the mechanism of transcription termination are associated with various disease states or developmental stages, will be an important step in understanding mitochondrial transcription. We previously demonstrated that hearts of TFAM KO mice undergo a genetic switch, reinitiating a fetal gene expression programme and activating mitochondrial biogenesis (17). Potentially, such a switch in transcription regulation might also occur, under milder mitochondrial stress.
In conclusion, we were able to demonstrate that in vivo mechanisms exist compensating for reduced mtDNA transcription in highly energy demanding tissues, such as the heart. Further, we report that heavy-strand transcription of mtDNA regularly terminates at TAS.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swedish Research Council; Swedish Cancer Society; European Commission (fp6 EUMITOCOMBAT); Swedish Strategic Foundation; Göran Gustafsson Foundation; Knut and Alice Wallenberg Foundation. Funding for open access charge: Max Planck Gesellschaft.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Larsson NG. Somatic mitochondrial DNA mutations in mammalian aging. Annu. Rev. Biochem. 2010 doi: 10.1146/annurev-biochem-060408-093701. doi: 10.1146/annurev-biochem-060408-093701i. [DOI] [PubMed] [Google Scholar]
- 2.Trifunovic A, Larsson NG. Mitochondrial dysfunction as a cause of ageing. J. Intern. Med. 2008;263:167–178. doi: 10.1111/j.1365-2796.2007.01905.x. [DOI] [PubMed] [Google Scholar]
- 3.Falkenberg M, Larsson N-G, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu. Rev. Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 4.Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson N-G, Gustafsson CM. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 5.Litonin D, Sologub M, Shi Y, Savkina M, Anikin M, Falkenberg M, Gustafsson C, Temiakov D. Human mitochondrial transcription revisited: only TFAM and TFB2M are required for transcription of the mitochondrial genes in vitro. J. Biol. Chem. 2010 doi: 10.1074/jbc.C110.128918. doi: 10.1074/jbc.C110.128918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sologub M, Litonin D, Anikin M, Mustaev A, Temiakov D. TFB2 is a transient component of the catalytic site of the human mitochondrial RNA polymerase. Cell. 2009;139:934–944. doi: 10.1016/j.cell.2009.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaspari M, Falkenberg M, Larsson N-G, Gustafsson CM. The mitochondrial RNA polymerase contributes critically to promoter specificity in mammalian cells. EMBO J. 2004;23:4606–4614. doi: 10.1038/sj.emboj.7600465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parisi MA, Xu B, Clayton DA. A human mitochondrial transcriptional activator can functionally replace a yeast mitochondrial HMG-box protein both in vivo and in vitro. Mol. Cell. Biol. 1993;13:1951–1961. doi: 10.1128/mcb.13.3.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsushima Y, Matsumura K, Ishii S, Inagaki H, Suzuki T, Matsuda Y, Beck K, Kitagawa Y. Functional domains of chicken mitochondrial transcription factor A for the maintenance of mitochondrial DNA copy number in lymphoma cell line DT40. J. Biol. Chem. 2003;278:31149–31158. doi: 10.1074/jbc.M303842200. [DOI] [PubMed] [Google Scholar]
- 10.Ekstrand MI, Falkenberg M, Rantanen A, Park CB, Gaspari M, Hultenby K, Rustin P, Gustafsson CM, Larsson N-G. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum. Mol. Genet. 2004;13:935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- 11.Alam TI, Kanki T, Muta T, Ukaji K, Abe Y, Nakayama H, Takio K, Hamasaki N, Kang D. Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res. 2003;31:1640–1645. doi: 10.1093/nar/gkg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takamatsu C, Umeda S, Ohsato T, Ohno T, Abe Y, Fukuoh A, Shinagawa H, Hamasaki N, Kang D. Regulation of mitochondrial D-loops by transcription factor A and single-stranded DNA-binding protein. EMBO Rep. 2002;3:451–456. doi: 10.1093/embo-reports/kvf099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisher RP, Lisowsky T, Parisi MA, Clayton DA. DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J. Biol. Chem. 1992;267:3358–3367. [PubMed] [Google Scholar]
- 14.Kaufman BA, Durisic N, Mativetsky JM, Costantino S, Hancock MA, Grutter P, Shoubridge EA. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol. Biol. Cell. 2007;18:3225–3236. doi: 10.1091/mbc.E07-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dairaghi DJ, Shadel GS, Clayton DA. Addition of a 29 residue carboxyl-terminal tail converts a simple HMG box-containing protein into a transcriptional activator. J. Mol. Biol. 1995;249:11–28. doi: 10.1006/jmbi.1995.9889. [DOI] [PubMed] [Google Scholar]
- 16.Larsson N-G, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 17.Hansson A, Hance N, Dufour E, Rantanen A, Hultenby K, Clayton DA, Wibom R, Larsson N-G. A switch in metabolism precedes increased mitochondrial biogenesis in respiratory chain-deficient mouse hearts. Proc. Natl Acad. Sci. USA. 2004;101:3136–3141. doi: 10.1073/pnas.0308710100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Wang J, Wilhelmsson H, Hansson A, Thoren P, Duffy J, Rustin P, Larsson N-G. Genetic modification of survival in tissue-specific knockout mice with mitochondrial cardiomyopathy. Proc. Natl Acad. Sci. USA. 2000;97:3467–3472. doi: 10.1073/pnas.97.7.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Wilhelmsson H, Graff C, Li H, Oldfors A, Rustin P, Brüning JC, Kahn CR, Clayton DA, Barsh GS, et al. Dilated cardiomyopathy and atrioventricular conduction blocks induced by heart-specific inactivation of mitochondrial DNA gene expression. Nat. Genet. 1999;21:133–137. doi: 10.1038/5089. [DOI] [PubMed] [Google Scholar]
- 20.Fernández-Vizarra E, López-Pérez MJ, Enriquez JA. Isolation of biogenetically competent mitochondria from mammalian tissues and cultured cells. Methods. 2002;26:292–297. doi: 10.1016/S1046-2023(02)00034-8. [DOI] [PubMed] [Google Scholar]
- 21.Wibom R, Hagenfeldt L, Döbeln UV. Measurement of ATP production and respiratory chain enzyme activities in mitochondria isolated from small muscle biopsy samples. Anal. Biochem. 2002;311:139–151. doi: 10.1016/s0003-2697(02)00424-4. [DOI] [PubMed] [Google Scholar]
- 22.Wredenberg A, Wibom R, Wilhelmsson H, Graff C, Wiener HH, Burden SJ, Oldfors A, Westerblad H, Larsson N-G. Increased mitochondrial mass in mitochondrial myopathy mice. Proc. Natl Acad. Sci. USA. 2002;99:15066–15071. doi: 10.1073/pnas.232591499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Enriquez JA, Pérez-Martos A, López-Pérez MJ, Montoya J. In organello RNA synthesis system from mammalian liver and brain. Meth. Enzymol. 1996;264:50–57. doi: 10.1016/s0076-6879(96)64008-7. [DOI] [PubMed] [Google Scholar]
- 24.Metodiev MD, Lesko N, Park CB, Cámara Y, Shi Y, Wibom R, Hultenby K, Gustafsson CM, Larsson NG. Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 2009;9:386–397. doi: 10.1016/j.cmet.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 25.Park CB, Asin-Cayuela J, Cámara Y, Shi Y, Pellegrini M, Gaspari M, Wibom R, Hultenby K, Erdjument-Bromage H, Tempst P, et al. MTERF3 is a negative regulator of mammalian mtDNA transcription. Cell. 2007;130:273–285. doi: 10.1016/j.cell.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 26.Brown TA, Tkachuk AN, Clayton DA. Native R-loops persist throughout the mouse mitochondrial DNA genome. J. Biol. Chem. 2008;283:36743–36751. doi: 10.1074/jbc.M806174200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGrath MF, de Bold MLK, de Bold AJ. The endocrine function of the heart. Trends Endocrinol. Metab. 2005;16:469–477. doi: 10.1016/j.tem.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 28.Linford NJ, Beyer RP, Gollahon K, Krajcik RA, Malloy VL, Demas V, Burmer GC, Rabinovitch PS. Transcriptional response to aging and caloric restriction in heart and adipose tissue. Aging Cell. 2007;6:673–688. doi: 10.1111/j.1474-9726.2007.00319.x. [DOI] [PubMed] [Google Scholar]
- 29.McCarroll SA, Murphy CT, Zou S, Pletcher SD, Chin C-S, Jan YN, Kenyon C, Bargmann CI, Li H. Comparing genomic expression patterns across species identifies shared transcriptional profile in aging. Nat. Genet. 2004;36:197–204. doi: 10.1038/ng1291. [DOI] [PubMed] [Google Scholar]
- 30.Zahn JM, Sonu R, Vogel H, Crane E, Mazan-Mamczarz K, Rabkin R, Davis RW, Becker KG, Owen AB, Kim SK. Transcriptional profiling of aging in human muscle reveals a common aging signature. PLoS Genet. 2006;2:e115. doi: 10.1371/journal.pgen.0020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zahn JM, Kim SK. Systems biology of aging in four species. Curr. Opin. Biotechnol. 2007;18:355–359. doi: 10.1016/j.copbio.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andreu AL, Arbos MA, Perez-Martos A, Lopez-Perez MJ, Asin J, Lopez N, Montoya J, Schwartz S. Reduced mitochondrial DNA transcription in senescent rat heart. Biochem. Biophys. Res. Commun. 1998;252:577–581. doi: 10.1006/bbrc.1998.9703. [DOI] [PubMed] [Google Scholar]
- 33.Dubessay P, Garreau-Balandier I, Jarrousse A-S, Fleuriet A, Sion B, Debise R, Alziari S. Aging impact on biochemical activities and gene expression of Drosophila melanogaster mitochondria. Biochimie. 2007;89:988–1001. doi: 10.1016/j.biochi.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 34.Gadaleta MN, Petruzzella V, Renis M, Fracasso F, Cantatore P. Reduced transcription of mitochondrial DNA in the senescent rat. Tissue dependence and effect of L-carnitine. Eur. J. Biochem. 1990;187:501–506. doi: 10.1111/j.1432-1033.1990.tb15331.x. [DOI] [PubMed] [Google Scholar]
- 35.Sörensen L, Ekstrand MI, Silva JP, Lindqvist E, Xu B, Rustin P, Olson L, Larsson N-G. Late-onset corticohippocampal neurodepletion attributable to catastrophic failure of oxidative phosphorylation in MILON mice. J. Neurosci. 2001;21:8082–8090. doi: 10.1523/JNEUROSCI.21-20-08082.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aloni Y, Attardi G. Symmetrical in vivo transcription of mitochondrial DNA in HeLa cells. Proc. Natl Acad. Sci. USA. 1971;68:1757–1761. doi: 10.1073/pnas.68.8.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sbisà E, Nardelli M, Tanzariello F, Tullo A, Saccone C. The complete and symmetric transcription of the main non coding region of rat mitochondrial genome: in vivo mapping of heavy and light transcripts. Curr. Genet. 1990;17:247–253. doi: 10.1007/BF00312616. [DOI] [PubMed] [Google Scholar]
- 38.Vijayasarathy C, Zheng YM, Mullick J, Basu A, Avadhani NG. Identification of a stable RNA encoded by the H-strand of the mouse mitochondrial D-loop region and a conserved sequence motif immediately upstream of its polyadenylation site. Gene Expr. 1995;4:125–141. [PMC free article] [PubMed] [Google Scholar]
- 39.Camasamudram V, Fang J-K, Avadhani NG. Transcription termination at the mouse mitochondrial H-strand promoter distal site requires an A/T rich sequence motif and sequence specific DNA binding proteins. Eur. J. Biochem. 2003;270:1128–1140. doi: 10.1046/j.1432-1033.2003.03461.x. [DOI] [PubMed] [Google Scholar]
- 40.Selwood SP, McGregor A, Lightowlers RN, Chrzanowska-Lightowlers ZMA. Inhibition of mitochondrial protein synthesis promotes autonomous regulation of mtDNA expression and generation of a new mitochondrial RNA species. FEBS Lett. 2001;494:186–191. doi: 10.1016/s0014-5793(01)02345-6. [DOI] [PubMed] [Google Scholar]
- 41.Sbisà E, Tanzariello F, Reyes A, Pesole G, Saccone C. Mammalian mitochondrial D-loop region structural analysis: identification of new conserved sequences and their functional and evolutionary implications. Gene. 1997;205:125–140. doi: 10.1016/s0378-1119(97)00404-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.