Abstract
The autocatalytic group II intron ai5γ from Saccharomyces cerevisiae self-splices under high-salt conditions in vitro, but requires the assistance of the DEAD-box protein Mss116 in vivo and under near-physiological conditions in vitro. Here, we show that Mss116 influences the folding mechanism in several ways. By comparing intron precursor RNAs with long (∼300 nt) and short (∼20 nt) exons, we observe that long exon sequences are a major obstacle for self-splicing in vitro. Kinetic analysis indicates that Mss116 not only mitigates the inhibitory effects of long exons, but also assists folding of the intron core. Moreover, a mutation in conserved Motif III that impairs unwinding activity (SAT → AAA) only affects the construct with long exons, suggesting helicase unwinding during exon unfolding, but not in intron folding. Strong parallels between Mss116 and the related protein Cyt-19 from Neurospora crassa suggest that these proteins form a subclass of DEAD-box proteins that possess a versatile repertoire of diverse activities for resolving the folding problems of large RNAs.
INTRODUCTION
DEAD-box proteins are members of a protein family (Helicase superfamily 2, or SF2) that participates in diverse processes such as ribosome assembly, transcription, translation and RNA processing. A common feature of these proteins is that they bind nucleic acids and hydrolyze ATP. Some members of the DEAD-box subgroup have been shown to display helicase activity. However, not all of these proteins behave as motor enzymes; some exhibit additional activities like strand annealing (1) or protein displacement activities (2,3), and one family member, eIF4-AIII, acts as an anchor for the exon junction complex (4).
Mss116, a DEAD-box protein from Saccharomyces cerevisiae, was identified in a genetic screen as affecting mitochondrial function (5). It is involved in mRNA translation, RNA end-processing and, notably, splicing of mitochondrial group I and group II introns (6). All of these functions can be rescued by overexpression of a related DEAD-box protein from Neurospora crassa, Cyt-19 (6). Indeed, either protein is necessary and sufficient for splicing of a variety of group I and group II introns in vitro under near-physiological conditions (7–9). The specific mechanism of action by Mss116 and Cyt-19 remains unclear. DEAD-box proteins are often implicated in nucleic acid remodeling, and they may facilitate proper RNA folding. However, given the diverse obstacles that can be encountered during folding, Mss116 may stimulate remodeling and splicing in several ways: for example, there are formidable electrostatic barriers to RNA collapse, and many of these can be alleviated by basic proteins. RNA molecules also have a strong tendency to adopt stable, incorrect structures that can be slow to resolve (they often form kinetically trapped species). Compounding these issues, large RNAs must sample a large amount of conformational space in order to form correct long-range interactions, potentially increasing the time for folding [they have a high contact order (10,11)]. Many of these barriers to proper folding can be overcome in vitro by employing conditions of elevated temperature and ionic strength. RNA folding in vivo, however, is generally assisted by proteins that promote RNA collapse, resolve kinetic traps [chaperones (12)], stabilize folding intermediates or stabilize the native RNA structure (13,14).
To investigate these issues, we have employed an in vitro splicing system using intron ai5γ, a group II intron in the COX1-gene of S. cerevisiae, together with its natural splicing factor, Mss116 (8,9). Intron ai5γ has been extensively characterized structurally, biophysically and enzymatically (15). It adopts the typical secondary structure of group IIB introns and self-splices efficiently under high-salt conditions via two parallel pathways, forming a lariat or a linear free intron (16) (Supplementary Figure S1). Despite its size (0.9 kb) and complexity, the ai5γ intron appears to have a relatively simple folding pathway. Under conditions of high magnesium ion concentration, the intron folds slowly to the native state via a series of on-pathway intermediates (17–21).
While the ai5γ intron is capable of efficient self-splicing in vitro under conditions of high magnesium ion concentration (∼100 mM) and high temperature (42°C), efficient splicing in vivo and in vitro requires the presence of Mss116 and ATP. Given that there are few systems in which a DEAD-box protein can be studied in the context of its natural substrate, it has been of great interest to determine the molecular mechanism by which Mss116 stimulates ai5γ splicing.
Some insights into the molecular mechanisms of DEAD-box proteins have been provided by genetic and biochemical studies of protein mutants (6,22). For example, an ATPase and helicase-deficient mutant of Mss116 (the ‘SAT-mutant’, in which the conserved SAT sequence in helicase Motif III has been mutated to AAA) displays only minor defects in splicing efficiency both in vitro and in vivo (8,23). This finding led to the suggestion that Mss116, which is a highly basic protein, might facilitate splicing without unwinding misfolded structures that constitute kinetic traps. Rather, Mss116 may facilitate the productive formation of native intermediates or structures (8,24). However, studies by Del Campo et al. (25) have shown that the Mss116 SAT mutant is still capable of unwinding very short, unstable duplexes. Based on these findings, together with the contention that helices in naturally occurring RNAs are often short, it was suggested that Mss116 might be required for unwinding a kinetic trap along the ai5γ folding pathway (25). Thus, there are different interpretations for the likely mechanistic role of Mss116 during splicing.
Although the folding pathway of ai5γ has been studied extensively and shown to be direct, previous studies were conducted on constructs that lack exons. In contrast, previous work on Mss116- and Cyt-19-facilitated splicing was conducted on ai5γ constructs that contain very long 5′ and 3′ exons (∼300 nt). These long exons may form stable secondary structures within themselves or with the intron, potentially resulting in misfolded structures that are not observed when monitoring folding of the intron alone. To differentiate between a role for DEAD-box proteins during intron folding and exon resolution, we examined the effect of proteins Mss116 and CYT-19 on splicing of ai5γ precursors with long (LE) and short (SE) exons. It was previously shown that the self-splicing kinetics of LE in vitro is biphasic, indicating a slow and a fast population of precursor molecules (16). Interestingly, if the 5′-exon is shortened, splicing becomes monophasic, suggesting that long exons interfere with splicing (26). Here, we show that a group II intron with shortened exons readily self-splices under near-physiological conditions. However, the splicing reaction can be further enhanced by adding the proteins Mss116, CYT-19 or their SAT/AAA mutant forms. These results indicate that DEAD-box proteins have more than one mechanistically distinct role in splicing: while they may resolve kinetic traps that obstruct utilization of long exons in vitro, they also accelerate the direct folding pathway of the intron.
MATERIALS AND METHODS
Plasmids and RNA transcription
LE (the ai5γ intron flanked by a 293nt 5′-exon and a 321 nt 3′-exon) was transcribed from plasmid pJD20 (16,34). SE (the ai5γ intron flanked by a 28 nt 5′-exon and a 15 nt 3′-exon) was transcribed from pAS10, a plasmid derived by PCR amplification of the region of interest from pJD20. The precursor RNAs were internally labeled with [α-32P]-UTP (Perkin-Elmer). The transcripts were purified on a 5% denaturing polyacrylamide gel and stored in a buffer of 10 mM MOPS pH 6.0, 1 mM EDTA.
Protein purification
Mss116, the Mss116 SAT/AAA mutant and Cyt-19 were purified as described (8). The Cyt-19 SAT/AAA mutant was created by site directed mutagenesis and purified in the same manner as the wild-type protein. Before diluting the proteins to working concentrations, they were briefly incubated with RNase Inhibitor (Protector, Roche) to inactivate RNases.
Splicing reactions
The full-length, body-labeled ai5γ precursor RNA was incubated in 40 mM MOPS, pH 7.5 at 90°C for 1 min and incubated at 30°C for 3 min. For Mss116 reactions, the reaction mixture contained the indicated amount of protein, 40 mM MOPS·KOH, pH 7.5, 100 mM KCl, 11 mM MgCl2 and 2 mM ATP (9 mM free Mg2+), while Cyt-19 reactions contained 100 nM protein, 40 mM MOPS·KOH, pH 7.5, 70 mM KCl, 13 mM MgCl2 and 5 mM ATP (8 mM free Mg2+). After incubation for 5 min at 30°C, the splicing reaction was initiated with the addition of the RNA to the reaction mixture at 1 nM final concentration. Reactions were quenched and resolved by PAGE (16). Self-splicing reactions in the absence of protein under high salt and temperature conditions were performed as previously described (16).
Data fitting
The precursor and product bands were quantified using a phosphorimager and the program ImageQuant TL. The relative fractions of precursor RNA, lariat and linear intron were calculated after correcting for the uridine-content of each species. The timecourse data were fit using the program KaleidaGraph 3.5 (Synergy Software). In order to quantify the length of the lag, the kobs, and the amplitude, we fit the data to the equation A· e(−kobs (t−tlag)) + y, with A: amplitude, tlag: length of the lag, y: endpoint. The reported values are averages of at least three independent timecourses.
RESULTS
Long exons interfere with ai5γ self-splicing under high-salt conditions
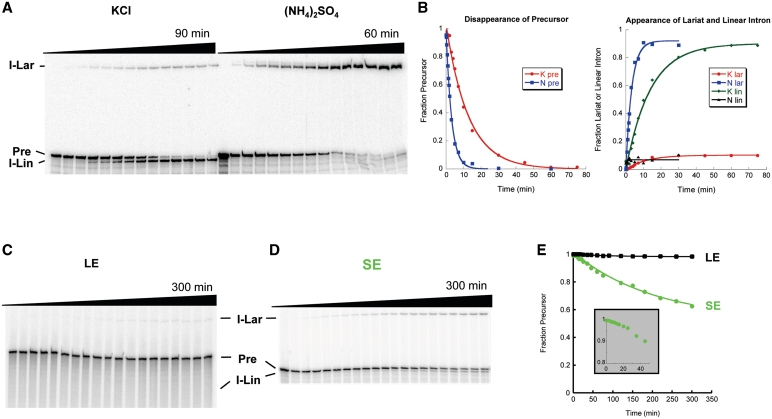
In order to understand how DEAD-box proteins influence ai5γ splicing, we designed a SE precursor RNA (SE, 28 nt 5′-exon and 15 nt 3′-exon). This construct is advantageous in that it preserves the IBS-EBS interactions, it contains a naturally occurring G for T7 transcription, and the exons are long enough to resolve the precursor from the linear spliced intron on a gel. SE spliced efficiently under high-salt self-splicing conditions (100 mM Mg2+, 500 mM monovalent cation, 42°C; Figure 1A). Like the thoroughly characterized pJD20 transcript (LE precursor RNA; 16), it reacted predominantly through the hydrolytic pathway in KCl, and the lariat pathway in (NH4)2SO4. Consistent with a previous report on a similar short exon precursor (26), shortening the exons changed the kinetic behavior of intron splicing; while LE shows marked biphasic behavior indicating a fast and a slow population, SE reacts as a single, fast population (Figure 1B) with a rate constant that is comparable to the rate constant of the fast LE population [0.065 ± 0.015 min−1 versus 0.028 ± 0.0031 min−1 (16)]. The exons may interfere directly with intron folding or form secondary structures that obstruct proper exon function (27,28). Alternatively, the length of the exons could create a steric or topological problem during exon docking.
Figure 1.
Self-splicing of SE. (A) SE self-splicing under high-salt conditions. Reactions were performed in 40 mM MOPS, pH 7.5, 100 mM MgCl2, 500 mM KCl or (NH4)2SO4 at 42°C. (B) Quantification of (A). The splicing reactions were fit to a single exponential, yielding a rate constant of 0.065 ± 0.015 min−1 in KCl and 0.36 ± 0.15 min−1 in (NH4)2SO4. (C) LE self-splicing under near-physiological conditions. (D) SE self-splicing under near-physiological conditions. (E) Quantification of (C and D). Inset: note the pronounced lag of ∼10 min for SE splicing. I-Lar, intron lariat, P, precursor, I-Lin, a population containing linear and/or broken lariat intron RNA.
The ai5γ core can self-splice under near-physiological conditions without the influence of a protein
We then investigated SE self-splicing under the near-physiological conditions that have been used previously for studies of protein-facilitated splicing (100 mM KCl, 9 mM free MgCl2, pH 7.5, 30 °C; 8,9). Although LE shows little to no reaction under these conditions (Figure 1C), SE readily self-splices (Figure 1D). This reinforces the observation made under high salt conditions that shortening the exons promotes formation of a reactive species. It is also the first observation of robust ai5γ self-splicing under near-physiological conditions without the help of a protein or a polyamine (7,16,29). The data demonstrate that ai5γ is able to fold to its native conformation, dock the exons and self-splice efficiently if surrounded by appropriate sequences. However, self-splicing under near-physiological conditions is significantly slower (0.0061 ± 0.0016 min−1) than under high-salt conditions and 62 ± 6% of the molecules either do not react at all or react very slowly (Figure 1E). This could be an effect of the remaining SEs or it may reflect an intron-specific barrier to splicing that is alleviated under high-salt conditions.
Mss116 stimulates both LE and SE splicing
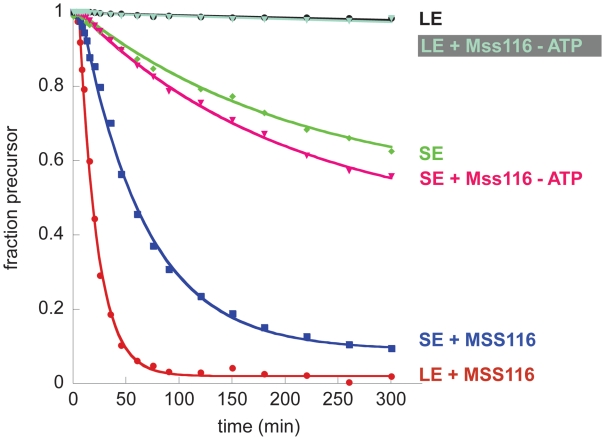
Since Mss116 was shown to promote LE splicing under near-physiological conditions (8,9), it was of interest to monitor its effects on the self-splicing of SE. Prior to initiating this comparative analysis, the reaction conditions and protocols for initiating the self-splicing reaction were extensively analyzed. For example, to reduce the kinetic influence of protein binding effects, precursor RNA concentration was reduced from 20 to 1 nM, resulting in pseudo-first order kinetics. In addition, previously reported rate constants for Mss116-catalyzed ATP hydrolysis (8,9) suggested that ATP concentration might become limiting during long timecourses with high concentrations of protein. We therefore varied ATP concentration from 1 to 10 mM, finding that 2 mM ATP is optimal for the Mss116/ai5γ-system under our experimental conditions. Finally, we varied the protein concentration in order to determine the optimal protein concentration for the two different substrates. Interestingly, Mss116 has a narrow range of optimal protein concentrations (∼10–20 nM under these conditions); higher protein concentrations are inhibitory to splicing (Supplementary Figure S2A and B). This may be an effect of protein aggregation at higher concentrations. Alternatively, high concentrations of protein may disrupt important structures throughout the intron either by binding or unwinding the RNA.
As expected from previous experiments, Mss116 greatly increases the rate constant for LE splicing under optimized near-physiological conditions. However, Mss116 also enhances the splicing of SE RNA (Figure 2). As previously shown for LE splicing, protein-mediated SE splicing is ATP-dependent (Figure 2). Given that Mss116 has a particularly strong effect on LE splicing, it seems likely that one role for the protein (at least in vitro) is to mitigate the inhibitory effects of long exons. However, the significant effects on SE suggest that Mss116 plays an additional role during folding of the intron itself and/or docking the 5′-exon into the active site, a folding step that has been demonstrated to stabilize the native state (21). Notably, in all kinetic time courses, lariat formation is preceded by a short lag, which is indicative of a sequential reaction. The lag phase was accounted for during determination of the rate constants (see ‘Materials and Methods’ section). The presence of the lag phase is consistent with previous observations indicating that splicing is preceded by formation of compact folding intermediates (21,24,30).
Figure 2.
Mss116 wt protein promotes splicing of both LE and SE in an ATP-dependent manner. Representative datasets are shown. The data were fit to a single exponential with lag and endpoint correction. Rate constants obtained from at least three independent experiments are 0.0083 ± 0.0032 min−1 for LE self-splicing, 0.0061 ± 0.0016 min−1 for SE self-splicing, 0.052 ± 0.0059 min−1 for Mss116-mediated LE splicing (15 nM Mss116), 0.011 ± 0.0024 min−1 for Mss116-mediated LE splicing without ATP, 0.016 ± 0.0016 min−1 for Mss116-mediated SE splicing (15 nM Mss116) and 0.0054 ± 0.0008 min−1 for Mss116-mediated SE splicing without ATP.
Protein-mediated splicing of LE is slightly (∼3×), but consistently, faster than SE and reaches a higher amplitude (Figure 2). Thus, the same exon sequences that are inhibitory to self-splicing seem to enhance protein-facilitated splicing. Perhaps long exons provide a structure that efficiently recruits Mss116 to a site within the intron or they may enhance oligomerization of the protein. Alternatively, the protein may promote different folding pathways in LE and SE, thus giving rise to different rate constants.
A clue to the role of Mss116 and ATP in the mechanism of splicing is provided by the observation that Mss116 increases both the amplitude and the rate constant for splicing of the LE and SE constructs. If Mss116 functioned exclusively through a kinetic redistribution mechanism, whereby it simply decreased the population of molecules caught in a kinetic trap, then one would expect an influence on the amplitude, but not the rate constant of reaction (31). The fact that marked increases in rate constant are observed for the splicing of both constructs suggest that Mss116 and ATP alter the reaction mechanism, most likely by stimulating formation of an on-pathway folding intermediate.
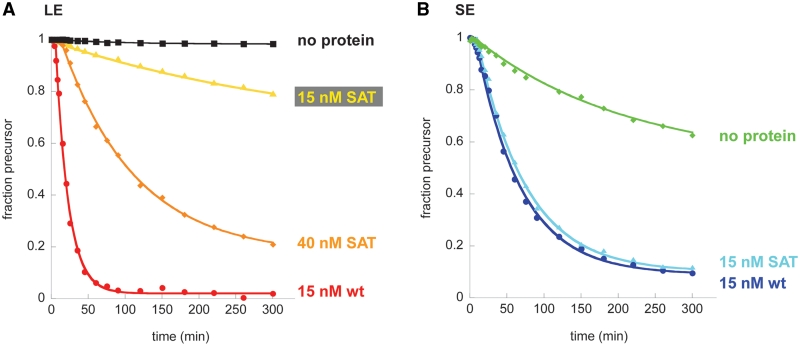
The SAT mutant shows no significant splicing defect on SE
Given that Mss116 appears to function differently during the splicing of LE and SE constructs, we decided to dissect these respective roles by employing the SAT mutant of Mss116 (Mss116SAT) and examining its effects on both types of precursor RNA. When comparing activity of the mutant on LE and SE, we observed that higher protein concentrations are required to achieve maximal kobs (Supplementary Figure S2), particularly for the LE construct. This is probably attributable to a higher Kd for RNA binding by Mss116SAT (25). While 15 nM protein promotes efficient splicing in three of the four combinations of intron precursors and Mss116 variants (Supplementary Figure S2A, B and D), we also determined kinetic data for LE splicing with 40 nM of Mss116SAT in order to measure the maximal catalytic activity of all variants (Figure 3A and Supplementary Figure S2C). Notably, the maximum kobs does not necessarily represent vmax since higher protein concentrations of Mss116SAT on LE very quickly become inhibitory. The actual catalytic activity may be even higher than reported.
Figure 3.
Mss116 SAT mutant promotes splicing of both LE and SE. Timecourses with wild-type protein and without protein from Figure 2 are included for comparison. (A) Splicing of LE. Rate constants obtained from at least three independent experiments are 0.0039 ± 0.0014 min−1 for LE splicing with 15 nM Mss116 SAT/AAA mutant and 0.010 ± 0.00062 min−1 for LE splicing with 40 nM Mss116 SAT/AAA mutant. (B) Splicing of SE. The rate constant from at least three independent experiments is 0.014 ± 0.00086 min−1 for SE splicing with 15 nM Mss116 SAT/AAA mutant.
Time courses for protein-mediated LE-splicing showed that the Mss116SAT mutant stimulates LE splicing with a rate constant that is 5- to 13-fold lower than wild-type protein, depending on whether it is used at the optimal Mss116SAT concentration or the same concentration as wild-type (Figure 3A). While protein-mediated splicing with wild type Mss116 reaches an amplitude of almost 100 % (97.8 ± 0.1 %), the reaction with Mss116SAT proceeds to only 31.2 ± 6.3 % (15 nM protein) and 82.4 ± 4.4 % (40 nM protein), respectively. In striking contrast, Mss116SAT behaves in a manner that is indistinguishable from wild-type protein during splicing of the SE RNA (Figure 3B). Assuming that Mss116 performs two functions, one on the exons and one on the intron core, the Mss116SAT mutant appears to be specifically defective in resolving problems associated with long exons.
Mss116 and Cyt-19 utilize similar mechanisms during ai5γ splicing
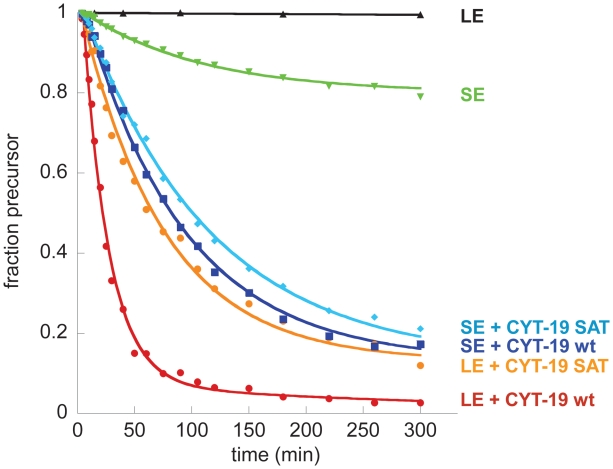
The N. crassa DEAD-box protein Cyt-19 can functionally substitute for Mss116 during ai5γ splicing in vivo (6) and Cyt-19 can also promote ai5γ splicing in vitro (7). However, assays with different model substrates [e.g. reverse branching and reverse splicing of the group II intron bI1 (9)] have suggested differences in specificity and/or catalytic mechanism between these two proteins. In order to investigate the functional similarity between Mss116 and Cyt-19, we repeated the Mss116 experiments described above with Cyt-19 and its Motif III SAT/AAA mutant (Cyt-19SAT).
Initial splicing experiments with Cyt-19 indicated that this protein has a narrower range of permissive reaction conditions than Mss116. Extensive optimization experiments varying ATP, potassium and protein concentrations (Supplementary Figure S3) have identified an uncommonly high sensitivity to monovalent salt concentrations. Nonetheless, we were able to identify optimal reaction conditions for Cyt-19 (1 nM precursor RNA, 100 nM protein, 40 mM MOPS·KOH, pH 7.5, 70 mM KCl, 13 mM MgCl2 and 5 mM ATP). Under these conditions, wild type Cyt-19 mediates LE splicing almost as efficiently as Mss116 (kobs = 0.042 ± 0.011 min−1; Figure 4). Cyt-19SAT shows ∼3-fold reduced activity (0.016 ± 0.006 min−1). Cyt-19 is also able to promote SE splicing. Under the buffer conditions used for the Cyt-19 reaction, only ∼20 % of the SE molecules self-splice in the absence of protein, with a rate constant of 0.006 ± 0.002 min−1. Analogous to Mss116, both wild type Cyt-19 and Cyt-19SAT stimulate SE splicing to the same degree (0.013 ± 0.004 min−1 for wild-type Cyt-19, 0.011 ± 0.004 min−1 for Cyt-19SAT) and raise the amplitude of the splicing reaction from 20% to 80% (Figure 4). Thus, the reactivity of Cyt-19 and Cyt-19SAT closely parallel the behavior of Mss116 and Mss116SAT under their respective optimized reaction conditions.
Figure 4.
Cyt19-mediated splicing of SE and LE. All reactions are analogous to the reactions shown in Figures 2 and 3, but they were performed in Cyt-19 splicing buffer and with 100 nM Cyt-19 or the Cyt-19 SAT→AAA mutant, where indicated. Rate constants obtained from at least three independent experiments are 0.006 ± 0.002 min−1 for SE self-splicing, 0.042 ± 0.011 min−1 for Cyt-19-mediated LE splicing, and 0.009 ± 0.002 min−1 for Cyt-19-mediated SE splicing. The Cyt-19 SAT→AAA mutant yields rate constants of 0.016 ± 0.006 min−1 for LE splicing, and 0.009 ± 0.001 min−1 for SE splicing.
DISCUSSION
Here, we have shown that the Mss116 protein, which is a DEAD-box splicing factor, stimulates group II intron self-splicing in vitro through two distinct mechanisms that can be differentiated by varying the length of precursor exons and examining the function of protein mutants.
Mss116 resolves a defect imposed by long exons
Mss116 clearly plays a role in overcoming the inhibitory effects of the LE sequences that have historically been employed in studies of ai5γ splicing. The classical pJD20 construct that has been used in previous studies of self-splicing and protein-assisted splicing contains exons of 293 (5′) and 321 (3′) nt. Several lines of evidence provide clear indication of a strong exon effect in this construct and they suggest that Mss116 serves to resolve misfolded structures within one or both exons.
Early studies of ai5γ self-splicing kinetics consistently revealed biphasic kinetic behavior under high salt conditions and it was well established that the precursor molecules existed in one of two populations (16), which exist in about equal proportions: one population is fast (0.028 min−1) and one is slow (0.0042 min−1). It had been hypothesized that the slow population represents a kinetically trapped state of the precursor. That the misfolded structure lies within one of the exons was supported by the findings of Nolte et al. (26), who showed that ai5γ self-splicing proceeds through fast, monoexponential kinetics when the exons are considerably shortened. This construct (equivalent to our SE construct) reacts uniformly as a single population with a rate constant (0.19 min−1) that matches that of the fast state for the LE construct (0.22 min−1). Here we have confirmed the Nolte result, and extended it by performing a comparative analysis of SE and LE splicing at low salt. We observe, as reported previously, that the LE construct is incapable of splicing at low salt. However, the SE construct readily self-splices under near-physiological conditions. Thus, the long exons are inhibitory to the splicing mechanism, and this effect becomes particularly pronounced under low salt conditions.
The likely mechanistic effect of the LEs only becomes apparent upon addition of the Mss116 co-factor. While Mss116 and ATP stimulate splicing by the SE construct, the effect is more pronounced for the LE construct, as if the protein alleviates multiple defects in the latter case. Most importantly, the Motif III SAT mutant (Mss116SAT→AAA), which is defective in ATPase activity (22) and RNA unwinding (8), has a greatly reduced ability to stimulate LE splicing, while it affects the behavior of the SE construct like WT Mss116. Thus, Mss116-stimulated splicing of SE clearly proceeds by a different mechanism (see below). Taken together, these results indicate that Mss116 and ATP have a role in mitigating problems imposed by the artificial long exons that have been traditionally used in studies of ai5γ self-splicing, and it is likely that these problems involve exon misfolding. Based on the experiments by Nolte et al. (26), who observed that 3′-exons of any length did not interfere with splicing, we expect the 5′-exon to be mainly responsible for the effect. Based on studies of group I splicing constructs (27), a possible mechanism of interference is the blocking of proper IBS/EBS pairing through interactions between IBS sequences and the upstream exon.
Mss116 assists folding of the ai5γ intron during splicing
In addition to its influence on the exons, and distinct from this function, we observe that Mss116 contributes to reactivity of the intron itself. The results are consistent with a role for Mss116 in promoting the productive folding pathway of the intron, by stabilizing the known series of on-pathway folding intermediates (8,24). The data is not consistent with a kinetic trap within the ai5γ intron itself. Several lines of evidence support this model.
Prior to this study, it was assumed that the ai5γ intron strictly required protein cofactors for splicing in vivo or for splicing in vitro under conditions that approximate the physiological environment. However, here we show that the SE construct can readily splice under conditions of low salt and physiological temperature. This indicates that the intron is fully capable of adopting the native fold without assistance from a chaperone, which is consistent with the many biophysical studies that have recently been conducted on the ai5γ intron RNA.
A second piece of evidence is that the Mss116SAT→AAA mutant stimulates SE splicing to the same extent as that observed for WT Mss116. Thus, whatever stimulatory role that Mss116 plays during intron function, it is unlikely to involve RNA unwinding or the resolution of kinetically trapped species. This is also corroborated by a recent study showing that ATP hydrolysis and Mss116 turnover, but not robust unwinding, are required for the splicing function of Mss116 in vivo (32).
Third, Mss116 increases both the amplitude and the rate constant for SE splicing. The fact that Mss116 increases the rate constant is inconsistent with the previously-proposed kinetic redistribution model for chaperone resolution of kinetic traps (31). In other words, if Mss116 simply served to increase the population of misfolded molecules that are capable of reaction, we would only observe an influence on amplitude. The fact that Mss116 increases the rate constant suggests that it has a positive influence on the actual splicing mechanism, increasing the rate constant at which productive intermediates actually form, as observed in studies of direct folding.
Biological relevance of Mss116-stimulated splicing in vitro
While the exon effect of Mss116 in vitro is significant, it is unlikely to be biologically relevant. There are several reasons for this. First of all, splicing precursors in vivo are not flanked by long, naked pieces of RNA. The exons would normally be coated with proteins and their behavior coordinated with splicing. The problem in this case is compounded by the artificial exons of the pJD20 construct, which contain vector sequence at the distal ends (24 nt 5′ and 33 nt 3′, Supplementary Figure 1) and are unlikely to reflect the behavior of natural flanking exon sequence. The misfolded ‘exon-effect’ that is typically observed with denatured and subsequently refolded RNA precursors (26) may never pose a problem for RNA folding in the cell. Co-transcriptional folding and ubiquitous single-stranded nucleic acid binding proteins are likely to prevent exon sequences from interfering with intron folding. Recent in vivo experiments with the Tetrahymena group I intron corroborate this idea. They suggest that after initial folding of the RNA, refolding of kinetically trapped molecules does not happen to a large extent and the misfolded RNAs are generally degraded (33). If the same is true for ai5γ, resolution of kinetic traps is unlikely to be the major role of proteins like Mss116 or Cyt-19. Instead of unfolding misfolded RNAs, they might just accelerate folding to increase the amount of functional RNA.
Previous studies of Mss116 mutant proteins and their effect on ai5γ self-splicing in vivo are not consistent with a role in resolving misfolded exons. It has been clearly shown that the Mss116SAT→AAA mutant has minimal effects on the apparent splicing activity or resultant yeast Cox2p gene expression in vivo. Side-by-side analyses of Mss116 and Mss116SAT→AAA reveal Cox2p gene expression levels and splicing efficiencies that are indistinguishable from WT in the experimental data that is publicly available (Figure 29 and 30 in ref. 23). The behavior of Mss116SAT→AAA in vivo therefore parallels the behavior of Mss116SAT→AAA behavior during splicing of the SE construct in vitro, in which exon effects are not operative and a role in stabilization of folding intermediates is likely to be predominant. This finding underscores the striking utility of DEAD-box proteins and their mechanistic flexibility, but it is most consistent with a direct and productive role for Mss116 in the folding of the ai5γ intron, as suggested by direct biophysical analyses of the folding pathway.
DEAD-box proteins are general folding factors
Comparison of the activities of the S. cerevisiae protein Mss116 (the natural interaction partner of ai5γ) and the N. crassa protein CYT-19 showed that both proteins catalyze RNA folding in a very similar manner. They require different salt conditions and protein concentrations for optimal activity, but under optimal conditions, they behave similarly. Most notably, the Motif III mutants of both proteins show a ∼5-fold defect in LE splicing, but are not significantly compromised relative to wild-type protein in SE splicing. This indicates that both proteins assist RNA folding in at least two different ways. Importantly, these proteins increase the activity of both group I and group II introns, suggesting that they have not evolved to specifically interact with a single large RNA or a single misfolded structure (6). Furthermore, CYT-19 can also act on RNAs from completely different organisms (6,7). Therefore, these proteins are versatile enzymes that can stimulate RNA folding of many different substrates. It is even possible, and perhaps likely, that these proteins can perform roles in RNA folding in vitro that are provided by other proteins in vivo.
We therefore propose that a group of DEAD-box proteins (including Ded1, which was previously shown to also facilitate ai5γ splicing; 8,9) can act as RNA chaperones, but not exclusively by disrupting misfolded structures. Rather than improving their unwinding function, they have evolved a diverse repertoire of different activities such as strand annealing, conformational switching and charge neutralization in order to facilitate collapse and structural rearrangements in large RNAs.
FUNDING
National Institutes of Health (R01GM50313 to A.M.P.); Heyl Fellowship (to A.S.); A.M.P. is a Howard Hughes Medical Institute investigator. Funding for open access charge: Howard Hughes Medical Institute.
Conflict of interest statement. None declared.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We would like to thank P. Perlman and A. Lambowitz for their generous gifts of plasmids. We are also thankful to Michael Roitzsch, Olga Fedorova, Wenxiang Cao and Aditya Paul for their help and critical discussions.
REFERENCES
- 1.Yang Q, Jankowsky E. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry. 2005;44:13591–13601. doi: 10.1021/bi0508946. [DOI] [PubMed] [Google Scholar]
- 2.Fairman ME, Maroney PA, Wang W, Bowers HA, Gollnick P, Nilsen TW, Jankowsky E. Protein displacement by DExH/D “RNA helicases” without duplex unwinding. Science. 2004;304:730–734. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- 3.Bowers HA, Maroney PA, Fairman ME, Kastner B, Luhrmann R, Nilsen TW, Jankowsky E. Discriminatory RNP remodeling by the DEAD-box protein DED1. RNA. 2006;12:903–912. doi: 10.1261/rna.2323406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shibuya T, Tange TO, Sonenberg N, Moore MJ. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat. Struct. Mol. Biol. 2004;11:346–351. doi: 10.1038/nsmb750. [DOI] [PubMed] [Google Scholar]
- 5.Faye G, Simon M. In: Mitochondria 1983: Nucleo-mitochondrial Iinteractions. Schweyen RJ, Wolf K, Kaudewitz F, editors. New York: de Gruyter, Berlin; 1983. pp. 433–439. [Google Scholar]
- 6.Huang HR, Rowe CE, Mohr S, Jiang Y, Lambowitz AM, Perlman PS. The splicing of yeast mitochondrial group I and group II introns requires a DEAD-box protein with RNA chaperone function. Proc. Natl Acad. Sci. USA. 2005;102:163–168. doi: 10.1073/pnas.0407896101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohr S, Matsuura M, Perlman PS, Lambowitz AM. A DEAD-box protein alone promotes group II intron splicing and reverse splicing by acting as an RNA chaperone. Proc. Natl Acad. Sci. USA. 2006;103:3569–3574. doi: 10.1073/pnas.0600332103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Solem A, Zingler N, Pyle AM. A DEAD protein that activates intron self-splicing without unwinding RNA. Mol. Cell. 2006;24:611–617. doi: 10.1016/j.molcel.2006.10.032. [DOI] [PubMed] [Google Scholar]
- 9.Halls C, Mohr S, Del Campo M, Yang Q, Jankowsky E, Lambowitz AM. Involvement of DEAD-box proteins in group I and group II intron splicing. Biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity. J. Mol. Biol. 2007;365:835–855. doi: 10.1016/j.jmb.2006.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swisher JF, Su LJ, Brenowitz M, Anderson VE, Pyle AM. Productive folding to the native state by a group II intron ribozyme. J. Mol. Biol. 2002;315:297–310. doi: 10.1006/jmbi.2001.5233. [DOI] [PubMed] [Google Scholar]
- 11.Woodson SA. Folding mechanisms of group I ribozymes: role of stability and contact order. Biochem. Soc. Trans. 2002;30:1166–1169. doi: 10.1042/bst0301166. [DOI] [PubMed] [Google Scholar]
- 12.Herschlag D. RNA chaperones and the RNA folding problem. J. Biol. Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 13.Weeks KM, Cech TR. Assembly of a ribonucleoprotein catalyst by tertiary structure capture. Science. 1996;271:345–348. doi: 10.1126/science.271.5247.345. [DOI] [PubMed] [Google Scholar]
- 14.Caprara M, Mohr G, Lambowitz A. A tyrosyl-tRNA synthetase protein induces tertiary folding of the group I intron catalytic core. J. Mol. Biol. 1996;257:512–531. doi: 10.1006/jmbi.1996.0182. [DOI] [PubMed] [Google Scholar]
- 15.Fedorova O, Zingler N. Group II introns: structure, folding and splicing mechanism. Biol. Chem. 2007;388:665–678. doi: 10.1515/BC.2007.090. [DOI] [PubMed] [Google Scholar]
- 16.Daniels D, Michels WJ, Pyle AM. Two competing pathways for self-splicing by group II introns; a quantitative analysis of in-vitro reaction rates and products. J. Mol. Biol. 1996;256:31–49. doi: 10.1006/jmbi.1996.0066. [DOI] [PubMed] [Google Scholar]
- 17.Su LJ, Waldsich C, Pyle AM. An obligate intermediate along the slow folding pathway of a group II intron ribozyme. Nucleic Acids Res. 2005;33:6674–6687. doi: 10.1093/nar/gki973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fedorova O, Waldsich C, Pyle AM. Group II intron folding under near-physiological conditions: collapsing to the near-native state. J. Mol. Biol. 2007;366:1099–1114. doi: 10.1016/j.jmb.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waldsich C, Pyle AM. A folding control element for tertiary collapse of a group II intron ribozyme. Nat. Struct. Mol. Biol. 2007;14:37–44. doi: 10.1038/nsmb1181. [DOI] [PubMed] [Google Scholar]
- 20.Waldsich C, Pyle AM. A kinetic intermediate that regulates proper folding of a group II intron RNA. J. Mol. Biol. 2008;375:572–580. doi: 10.1016/j.jmb.2007.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steiner M, Karunatilaka KS, Sigel RK, Rueda D. Single-molecule studies of group II intron ribozymes. Proc. Natl Acad. Sci. USA. 2008;105:13853–13858. doi: 10.1073/pnas.0804034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banroques J, Doere M, Dreyfus M, Linder P, Tanner NK. Motif III in superfamily 2 “helicases” helps convert the binding energy of ATP into a high-affinity RNA binding site in the yeast DEAD-box protein Ded1. J. Mol. Biol. 2010;396:949–966. doi: 10.1016/j.jmb.2009.12.025. [DOI] [PubMed] [Google Scholar]
- 23.Huang HR. Dallas, TX: University of Texas Southwestern Medical Center; 2004. Functional studies of intron- and nuclear-encoded splicing factors in the mitochondria of Saccharomyces cerevisiae. [Google Scholar]
- 24.Fedorova O, Solem A, Pyle AM. Protein-facilitated folding of group II intron ribozymes. J. Mol. Biol. 2010;397:799–813. doi: 10.1016/j.jmb.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Del Campo M, Tijerina P, Bhaskaran H, Mohr S, Yang Q, Jankowsky E, Russell R, Lambowitz AM. Do DEAD-box proteins promote group II intron splicing without unwinding RNA? Mol. Cell. 2007;28:159–166. doi: 10.1016/j.molcel.2007.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nolte A, Chanfreau G, Jacquier A. Influence of substrate structure on in vitro ribozyme activity of a group II intron. RNA. 1998;4:694–708. doi: 10.1017/s1355838298980165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao Y, Woodson SA. Destabilizing effect of an rRNA stem-loop on an attenuator hairpin in the 5′ exon of the Tetrahymena pre-rRNA. RNA. 1998;4:901–914. doi: 10.1017/s1355838298980621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woodson SA. Exon sequences distant from the splice junction are required for efficient self-splicing of the Tetrahymena IVS. Nucleic Acids Res. 1992;20:4027–4032. doi: 10.1093/nar/20.15.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peebles CL, Perlman PS, Mecklenburg KL, Petrillo ML, Tabor JH, Jarrell KA, Cheng H-L. A self-splicing RNA excises an intron lariat. Cell. 1986;44:213–223. doi: 10.1016/0092-8674(86)90755-5. [DOI] [PubMed] [Google Scholar]
- 30.Erat MC, Sigel RK. Divalent metal ions tune the self-splicing reaction of the yeast mitochondrial group II intron Sc.ai5gamma. J. Biol. Inorg. Chem. 2008;13:1025–1036. doi: 10.1007/s00775-008-0390-7. [DOI] [PubMed] [Google Scholar]
- 31.Bhaskaran H, Russell R. Kinetic redistribution of native and misfolded RNAs by a DEAD-box chaperone. Nature. 2007;449:1014–1018. doi: 10.1038/nature06235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bifano AL, Turk EM, Caprara MG. Structure-guided mutational analysis of a yeast DEAD-box protein involved in mitochondrial RNA splicing. J. Mol. Biol. 2010;398:429–443. doi: 10.1016/j.jmb.2010.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jackson SA, Koduvayur S, Woodson SA. Self-splicing of a group I intron reveals partitioning of native and misfolded RNA populations in yeast. RNA. 2006;12:2149–2159. doi: 10.1261/rna.184206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jarrell KA, Peebles CL, Dietrich RC, Romiti SL, Perlman PS. Group II intron self-splicing: alternative reaction conditions yield novel products. J. Biol. Chem. 1988;263:3432–3439. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.