Abstract
HDM2 is a p53-specific E3 ubiquitin ligase. Its overexpression leads to excessive inactivation of tumor protein p53, diminishing its tumor suppressor function. HDM2 also affects the cell cycle, apoptosis and tumorigenesis through interacting with other molecules, including several ribosomal proteins. To identify novel HDM2 regulators, we performed a yeast two-hybrid screening using HDM2 as bait. Among the candidates, ribosomal protein L26 (RPL26) was characterized as a novel HDM2-interactor. The interaction between HDM2 and RPL26 was further validated by in vivo and in vitro assays. RPL26 modulates the HDM2–p53 interaction by forming a ternary complex among RPL26, HDM2 and p53, which stabilize p53 through inhibiting the ubiquitin ligase activity of HDM2. The ribosomal stress caused by a low dose of Act D enhances RPL26–HDM2 interaction and activates p53. Overexpression of RPL26 results in activating of p53, inhibits cell proliferation and induces a p53-dependent cell cycle arrest. These results provide a novel regulatory mechanism of RPL26 to activate p53 by inhibiting HDM2.
INTRODUCTION
Tumor suppressor p53 is a transcription factor that acts by stopping cell cycle progression or promoting apoptosis when cells encounter stress stimuli such as oncogene activation or DNA damage (1). The importance of p53 in cancer development is illustrated by the fact that p53 is highly mutated in many different cancers (2) and is probably rendered inactive by a range of indirect mechanisms (for example, HDM2 amplification or loss of ARF) in most other cancer types.
Having a short half-life, p53 is normally maintained at low levels in unstressed mammalian cells by continuous ubiquitination and subsequent degradation by the 26S proteasome. This is primarily due to the interaction of p53 with the RING-finger ubiquitin E3 ligase MDM2 (also known as HDM2; 3). The mdm2 gene is one of the target genes the transcription of which is activated by the p53 proteins, thus forming a tight auto-regulatory feedback loop (4,5).
The ability of HDM2 to keep p53 in check is essential for normal cell function. The repression operates via three mechanisms. First, HDM2 interacts with the N-terminal transactivation domain of p53, which is the primary binding site for HDM2. Through binding to p53 at its transactivation domain, HDM2 inhibits p53 transcriptional activity (4). Second, HDM2 labels p53 with ubiquitin for degradation (6). Lastly, HDM2 is responsible for the export of p53 from nucleus to cytoplasm to abrogate its transcriptional activity (7). Many cellular stresses such as DNA damage stabilize p53 protein by blocking the HDM2–p53 feedback loop (8). One prominent example is that, in response to oncogene activation, p14ARF activates p53 by inhibiting the ubiquitin ligase activity of HDM2 and relieving HDM2-dependent inhibition of p53 (9).
Besides ARF, a number of factors that alter the p53–HDM2 feedback loop have been identified, including the retinoblastoma protein (Rb) and the transcription factor Yin Yang 1 (YY1). Nucleolar proteins are also prominent among this group, including the ribosomal proteins L5, L11, L23, S7 (10–14), PML (15) and nucleophosmin (also called B23). Ribosomal proteins L5, L11, L23 and S7 interacted with HDM2 and inhibited the HDM2–p53 feedback loop in response to ribosomal stress, such as treatment with low dose actinomycin D (Act D), serum starvation (13), 5-fluorouracil (16) and mycophenolic acid treatment (17). Thus, releasing small protein molecules such as the ribosomal proteins from the nucleolus leads to p53 activation in response to ribosomal stress.
In this study, we designed to search for novel HDM2-binding proteins. We conducted stringent yeast two-hybrid (Y2H) screening using full-length HDM2 to screen human liver cDNA library. RPL26 was a novel ribosomal protein that can directly interact with HDM2, and its interaction with HDM2 was confirmed in vivo and in vitro. Furthermore, RPL26 modulates the HDM2-p53 interaction by forming a ternary complex among RPL26, HDM2 and p53, which leads to the stabilization of p53 and HDM2 by inhibiting the ubiquitin ligase activity of HDM2. RPL26 activates p53 by overcoming HDM2-mediated p53 degradation through the proteasome. The interaction of RPL26 and HDM2 was increased and activated p53. Overexpression of RPL26 results in activating of p53, inhibits cell proliferation and induces a p53-dependent cell cycle arrest. From previous report, RPL26 was found to bind to the 5′ untranslated region (UTR) of p53 mRNA and control p53 translation and induction after DNA damage (18). Thus, RPL26 is the only identified ribosomal protein that activates p53 by simultaneously potentiating its translation and attenuating its degradation till now. These observations provide an additional regulatory mechanism associated with RPL26 in regulating p53 function.
MATERIALS AND METHODS
Cell culture
Human embryonic kidney HEK293, human osteosarcoma U2OS cells, mouse p53−/−/mdm2−/− MEF cells and HCT116 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10%(v/v) fetal bovine serum, and Human lung small cell adenocarcinoma H1299 cells were maintained in RPMI medium 1640 with 10% FBS.
Plasmids and reagents
The pCMV-p53, pCMV-HDM2, and pcDNA3/poly-HA-tagged ubiquitin were kindly provided by Dr Y. Xiong, (University of North Carolina, Chapel Hill, NC, USA), and pG13-Luc were obtained from Dr B. Vogelstein (Johns Hopkins Medical Institutions, Baltimore, MD, USA). Various constructs of RPL26 were generated according to standard molecular techniques. All deletion mutants were created by polymerase chain reaction (PCR) to obtain DNA fragments and subjected to sequencing verification. The details of the primer sequences used for deletion mutations are available upon request. The proteasome inhibitor MG132 and protein synthesis inhibitor cycloheximide (CHX) were purchased from Sigma (St Louis, MO, USA). Anti-FLAG and RPL26 antibody were purchased from Sigma (St Louis), anti-HA antibody from Roche (Basel, Switzerland), anti-Myc, anti-Flag-HRP, anti-GFP and anti-HDM2 antibody were from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
Yeast two-hybrid screening
The two-hybrid screening was performed with the ProQuest™ two-hybrid system (Invitrogen, CA, USA). Full length human HDM2 was cloned in-frame with the GAL4 DNA binding domain in the vector pDBLeu to create pDBLeu-HDM2. MaV203 yeast cells were successively transformed with pDBLeu-HDM2 and human liver cDNA library. A total of ∼1 × 106 independent transformants were analyzed and clones were selected for positive interactions based on screening for expression of reporter genes His3, LacZ and URA3. To eliminate interactions that originated from nonspecific promoter activation, we only considered DB-HDM2-AD-Prey pairs if they activated at least two out of three reporter genes. Positives were subsequently retested in fresh yeast cells, and their AD-Prey identities were determined with interaction sequence tags (ISTs) obtained by sequencing. The AD-Prey reading frame was verified for each IST to avoid the recovery of out-of-frame peptides.
Transfection, immunobloting and immunoprecipitation analyses
H1299, U2OS, MEF, HCT116 or HEK293 cells were transfected with plasmids as indicated in each figure legend using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Cells were harvested at 24 h post-transfection and lysed in HEPES lysis buffer [50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1% (v/v) Tween 20, 0.2% NP-40, 10% glycerol] supplemented with protease inhibitor cocktail (Roche, Basel, Switzerland) and phosphatase inhibitors (10 mM NaF and 1 mM Na3VO4). Immunoprecipitation and immunoblotting analysis were performed as described previously (19).
In vitro GST pull-down assays
Bacteria-expressed GST or GST–HDM2 proteins were immobilized on glutathione-Sepharose 4B beads (GE, UK) and washed, and then beads were incubated with His-RPL26 expressed in Escherichia coli BL21 and purified with Ni-nitrilotriacetate-agarose beads (GE, UK) for 3 h at 4°C. Beads were washed with GST binding buffer (100 mM NaCl, 50 mM NaF, 2 mM EDTA, 1% Nonidet P40 and protease inhibitor cocktail) and proteins were eluted, followed by western blotting.
Immunofluorescent staining
U2OS cells were transfected with GFP-RPL26 and HDM2 expression plasmid. Twenty-four hours after transfection, cells were fixed with 5% paraformaldehyde for 30 min and permeabilized in 0.2% Triton X-100 (PBS). Then cells were incubated with mouse anti-HDM2 (dilution 1:50; Santa Cruz) for 1 h, followed by incubation with TRITC-conjugated secondary antibody 1h at room temperature. Nuclei were stained with 100 mg/ml 4,6-diamidino-2-phenylindole. Images were visualized with a confocal microscope.
Reporter gene assays
Luciferase reporter plasmids pG13-Luc and pRL-CMV (Promega) were cotransfected with plasmids indicated in figures. After 24 h, cells were lysed and measured for luciferase activity with the Luciferase Assay System (Promega) according to the manufacturer’s protocol.
In vivo ubiquitination assays
H1299 cells (60% confluence, 100-mm diameter plate) were transfected with plasmids expressing HA-ubiquitin (2 μg), Flag-p53 (2 μg), Myc-RPL26 (2 μg) or HDM2 (2 μg) in various combinations. At 48 h after transfection, cells from each plate were harvested and split into two aliquots, one for western blotting and the other for ubiquitination assays. Cell pellets were lysed in HEPES lysis buffer (0.02 M HEPES, 0.05 M NaCl,1 mM Triton-100, 1 mM NaF, 1 mM DTT, 1 mM Na3VO5) and incubated with HA antibody at 4°C for 2 h, then incubated with protein A/G-sepharose (Santa Cruz) for 8 h. Sepharoses were washed three times with lysis buffer. And they were analyzed by western blotting with indicated antibodies.
siRNA experiments
Cells were transfected with 100 nmol RPL26 siRNA complex or a scrambled control (UUCUCCGAACGUGUCACGUTT) and other plasmids showed in figures. The RPL26 siRNA complex contained three siRNA sequences. They are CCGAAAGGAUGAUGAAGUUTT(18), CACAUUCGAAGGAAGAUUATT and AGGUUGUACGUGGACACUAUATT. After 24 h, cells were washed with phosphate-buffered saline, lysed directly into sample buffer and resolved by 12% SDS–polyacrylamide gel electrophoresis.
Cell proliferation assays
For assessing the effect of RPL26 expression on growth rate, cells were cultured into 96-well plates in triplicate for 24 h. Then U2OS or H1299 cells were transiently transfected with indicated plasmid or siRNA. After 24 h post-transfection, cell proliferation was measured by the MTS-based cell proliferation assay. The experiment was performed on three independent occasions in triplicate.
Cell cycle analysis
Cells were transfected with plasmids encoding GFP, GFP-RPL26. At 24 h post-transfection, cells were harvested by trypsinization, fixed in 70% ethanol and stained with propidium iodide (50 mg/ml) containing 1 mg/ml of RNase A at 37°C for 30 min and then analyzed for DNA content. At least 10 000 GFP-positive cells were gated for cell cycle analysis.
RESULTS
RPL26 interacts with HDM2 in vitro and in vivo
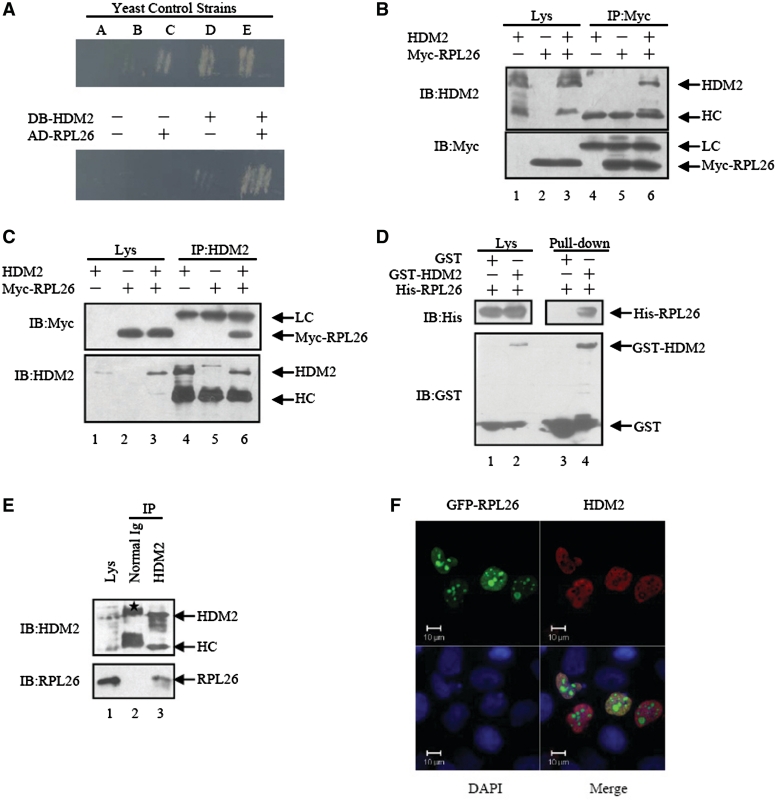
To search for novel binding partners of HDM2, we conducted stringent Y2H screening using full-length HDM2 as bait. Three ribosomal proteins L11, S7 and L26 were identified as candidate HDM2 interacting proteins. RPL11 (20) and RPS7 (14) have previously been shown to bind to HDM2, while RPL26 has been found to bind to the 5′UTR of p53 mRNA and augment its translation and induction after DNA damage (18). We retested the interaction between HDM2 and RPL26 by yeast retransformation assay. The yeast colonies that contained DNA binding (DB)-HDM2 and activating domain (AD)-RPL26 simultaneously activated the two reporter genes (HIS3 and URA3; Figure 1A).
Figure 1.
RPL26 binds to HDM2. (A) HDM2 binds RPL26 in yeast two hybrid assays. A collection of yeast control strains, from A to E, have been developed that contain plasmid pairs expressing fusion proteins with a spectrum of interaction strengths from none, weak, moderately strong, strong to very strong. Transformants with indicated plasmids were cultured on SC-Trp-Leu-His-Ura plates and incubated at 30°C for 3–4 days. (B and C) Exogenous HDM2 and RPL26 interact with each other in HEK293 cells. HDM2 (3 μg), Myc-RPL26(3 μg) or both vectors (3 μg) were used for transfection. Whole-cell lysates were subjected to immunoprecipitation with anti-HDM2 or anti-Myc antibodies followed by immunoblotting with anti-Myc or anti-HDM2 antibodies. IP, immunoprecipitation; IB, immunoblotting; Lys, lysate; HC, heavy chain. LC, light chain. (D) Direct interaction between RPL26 and HDM2 revealed by GST pull-down assays. Both input and pull-down samples were subjected to immunoblotting with anti-His and anti-GST antibodies. (E) Binding between endogenous HDM2 and RPL26 in MCF7 cells. The lysates of MCF7 cells were immunoprecipitated with nonimmune mouse IgG or an antibody against HDM2. The immunoprecipitates were further separated by SDS–PAGE, and target proteins were detected with RPL26 and HDM2 antibodies. Ig, immunoglobulin. Asterisk indicates non-specific band. (F) HDM2 colocalized with RPL26 in the nucleoplasm. U2OS cells were transfected with GFP-RPL26 and HDM2. Scale bar, 10 μm.
To further confirm the association of HDM2 and RPL26 in mammalian cells, we expressed Myc-tagged RPL26 and untagged HDM2 individually or together in HEK293 cells followed by co-immunoprecipitation (CoIP) analysis with either anti-Myc or anti-HDM2 antibodies. Indeed, HDM2 specifically immunoprecipitated with RPL26 by anti-Myc antibody when expressed with HDM2 (Figure 1B, lane 6). Conversely, RPL26 also specifically co-immunoprecipitated with HDM2 by anti-HDM2 antibody (Figure 1C, lane 6). In contrast, there was no HDM2 or RPL26 detected in Myc or HDM2 antibody immunoprecipitates from cells transfected with Myc-RPL26 alone or HDM2 alone. Consistently, the direct physical binding between HDM2 and RPL26 was demonstrated by in vitro pull-down assay. As shown, GST-HDM2, but not GST, bound to His-RPL26 expressed in E. coli BL21 and purified with Ni-nitrilotriacetate agarose beads (Figure 1D, lane 4). Moreover, the binding of endogenous HDM2 and RPL26 was confirmed in MCF7 cells with anti-HDM2 antibody. Western blotting analysis showed that the HDM2 immunoprecipitate contained RPL26 (Figure 1E, lane 3). To determine the subcellular position of the RPL26-HDM2′s binding in cells, the U2OS cells were transfected with GFP-RPL26 and HDM2. As shown in Figure 1F, HDM2 and RPL26 were co-localized in nucleoplasm. All these results indicate that HDM2 direct binds to RPL26 under normal cellular conditions.
Determination of mutual interaction regions in HDM2 and RPL26
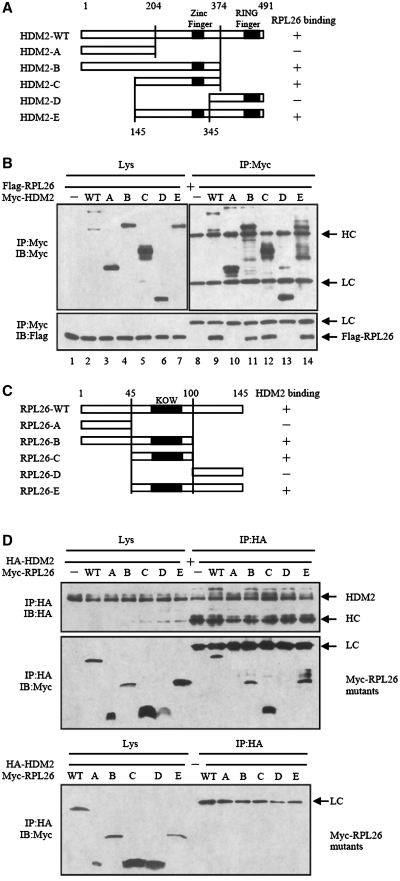
To determine the interacted region of HDM2 and RPL26, series deletion mutants were constructed, as indicated schematically (Figure 2A and 2C). When co-transfected with Myc-HDM2, the full-length RPL26 was co-immunoprecipitated with Myc-HDM2 (Figure 2B, lower panel, lane 9), while RPL26 was not observed in the control lane (Figure 2B, lower panel, lane 8). HDM2 deletion mutants B, C and E retained their ability to form a complex with RPL26 (Figure 2B). However, HDM2 deletion mutants A and D lost their ability to interact with RPL26 (Figure 2B), indicating that the region around amino acids (aa) 204–345 in HDM2 is critical for RPL26 interaction.
Figure 2.
Determination of mutual interaction regions in HDM2 and RPL26. (A) A diagram for the deletion mutants of HDM2 is shown. (B) Mapping of the HDM2 domain for RPL26 binding. Extracts from HEK293 cells transfected with the indicated plasmid DNA encoding deletion mutants of HDM2 and Flag-RPL26 were immunoprecipitated with Myc antibody, and followed by immunoblotting with Flag or Myc antibodies. (C) A diagram for the deletion mutants of RPL26 is shown. (D) Mapping of the RPL26 domain for HDM2 binding. Extracts from HEK293 cells transfected with the indicated plasmid DNA encoding deletion mutants of RPL26 and HA-HDM2 were immunoprecipitated with HA antibody, and followed by immunoblotting with HA and Myc antibodies.
We co-transfected different deletion mutants of RPL26 with the full-length HDM2 to HEK293 cells to define the binding region of RPL26 with HDM2. As shown in Figure 2D, all the RPL26 mutants, with the exception of A and D, interacted with HDM2, indicating that the region spanning aa 45–100 of RPL26 is critical for HDM2 interaction (Figure 2D). While the central domain of RPL26 contains aa 63–90 is also particularly important in regulation of p53 translation (18).
RPL26 enhances p53′s stability and activity by overcoming HDM2-mediated suppression
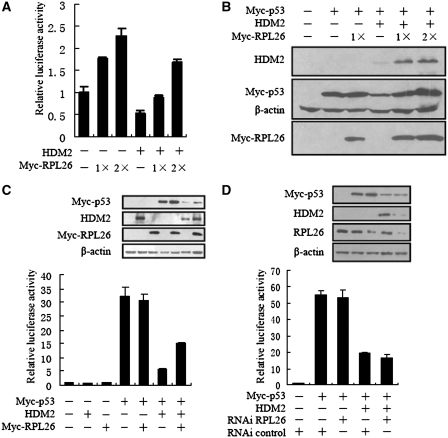
HDM2, a p53-specific E3 ubiquitin ligase, is the principal cellular antagonist of p53. Disruption of the p53–HDM2 interaction by multiple regulators is the pivot event for p53 activation, leading to p53 induction and its biological response. To investigate the functional consequences of RPL26–HDM2 interaction, we examined whether RPL26 could affect the function of HDM2 in degrading p53 and p53-dependent transcription. The p53-posivite U2OS cells were transfected with Myc-RPL26 or untagged-HDM2 together with a luciferase reporter plasmid which containing a consensus p53 target sequence. Ectopic expression of full-length RPL26 markedly stimulated endogenous p53 transactivation activity, as presented as luciferase activity, in a dose-dependent manner (Figure 3A).
Figure 3.
RPL26 enhances p53′s stability and activity by overcoming HDM2-mediated suppression. (A) Ectopic expression of RPL26 increases p53-dependent transactivation from the pG13-Luc reporter in p53-positive U2OS cells. U2OS cells were transfected with increasing amounts of Myc-RPL26 [0.4 μg (1×) and 0.8 μg (2×)] in the presence or absence of ectopically expressed HDM2 (0.2 μg). After 28 h, the luciferase assay was performed. Data are plotted as mean ± standard errors for three independent experiments. (B) Ectopic expression of RPL26 reverses HDM2-mediated p53 degradation. H1299 cells were transfected with Myc-RPL26 (0.2 μg or 0.4 μg) in the presence of p53 (0.1 μg) with (−) or without (+) HDM2 (0.4 μg) as indicated. Cell lysates were immunoblotted with anti-HDM2, anti-Myc, anti-Flag or anti-actin antibodies. (C) Expression of RPL26 alleviated exogenous HDM2-mediated p53 suppression in the MEF (p53−/−/mdm2−/−) cells. MEF (p53−/−/mdm2−/−) cells were transfected with (−) or without (+) Myc-RPL26 (0.4 μg) in the presence or absence of ectopically expressed HDM2 (0.2 μg). After 24 h, the luciferase assay was performed. Data are plotted as mean ± standard errors for three independent experiments. Inset shows similar expression levels of relevant transfected proteins. (D) Knockdown of RPL26 did not inhibit p53 in cooperation with HDM2 in MEF (p53−/−/mdm2−/−) cells. MEF (p53−/−/mdm2−/−) cells were transfected with or without RPL26 siRNA (100 nM) in the presence or absence of ectopically expressed p53 (0.1 μg) or HDM2 (0.4 μg). After 24 h, the luciferase assay was performed. Data are plotted as mean ± standard errors for three independent experiments.
All of the reported ribosomal proteins that interact with HDM2 can activate p53 through inhibiting HDM2. RPL26 might adopt similar mechanisms. As RPL26 binds to the 5′UTR of p53 mRNA and effects the translation of p53 (18), we examined overexpression of RPL26′s effect on HDM2-mediated p53 degradation by introducing exogenous proteins into p53-deficient human non-small-cell carcinoma H1299 cells. Plasmids expressing HDM2, Flag-tagged p53, and Myc-tagged RPL26 were transfected to H1299 cells, and the expression of p53 was under the control of CMV promoter that did not contain the 5′UTR of p53. As expected, overexpression of HDM2 remarkably reduced p53 levels (Figure 3B). In contrast, overexpression of RPL26 rescued HDM2-mediated p53 degradation in a dose-dependent fashion (Figure 3B).
That the RPL26 activates p53 by inhibiting HDM2 was further validated in MEF (p53−/−/mdm2−/−) cells. The p53-dependent reporter was cotransfected in combinations with HDM2, Myc-tagged p53, Myc-tagged RPL26 and siRNA as indicated. HDM2 and RPL26 did not affect p53 transcription activity without p53 in MEF (p53−/−/mdm2−/−) cells (Figure 3C, lane 1 to 3). RPL26 did not directly affect the exogenous p53 transcription activity in MEF (p53−/−/mdm2−/−) cells (Figure 3C, lane 4 and 5). In contrast, expression of HDM2 inhibited the exogenous p53 transcriptional activity, and co-expression of RPL26 alleviated exogenous HDM2-mediated p53 suppression in the MEF (p53−/−/mdm2−/−) cells (Figure 3C, lane 6 and 7). Conversely, knockdown of RPL26 had no measurable effect on the exogenous p53 transcriptional activity while lacking HDM2 (Figure 3D, lane 2 and 3) in the MEF (p53−/−/mdm2−/−) cells. But knockdown of RPL26 did not give notable effect on the exogenous p53 transcriptional activity with HDM2 in the MEF (p53−/−/mdm2−/−) cells (Figure 3D, lane 4 and 5). Knockdown of RPL26 reduced HDM2′s expression level. These results demonstrated that RPL26 stimulates p53-dependent transcription by inhibiting the activity of HDM2.
RPL26 attenuates HDM2-mediated ubiquitination and degradation
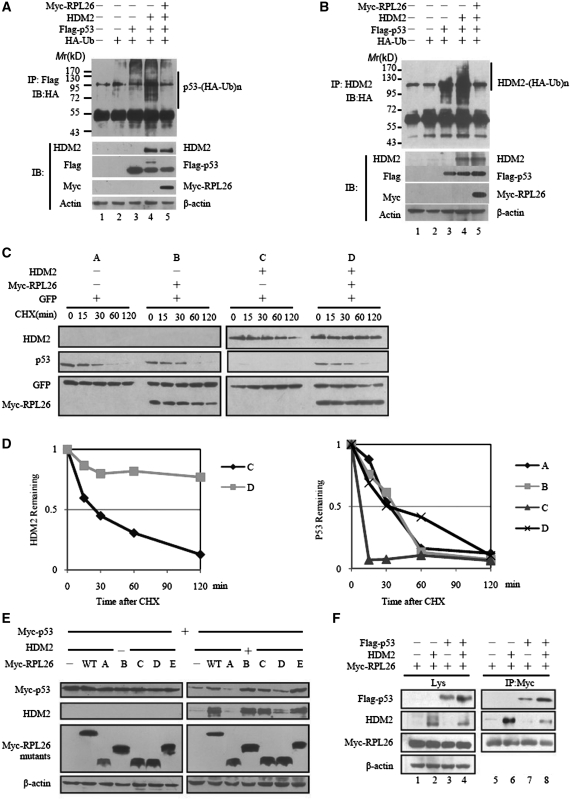
As we know, HDM2 is a RING-finger ubiquitin E3 ligase (3). HDM2-dependent suppression of p53 may be related with its E3 ligase activity on p53 (6). To determine whether RPL26 stabilize p53 by affecting HDM2-mediated ubiquitination and degradation, we did an in vivo ubiquitination assay. Overexpression of RPL26 not only led to marked inhibition of p53 ubiquitination (Figure 4A, lane 4 and 5) but also stabilized HDM2 (Figure 4B, lane 4 and 5). The ability of RPL26 to inhibit HDM2-induced p53 degradation could be explained by its inhibiting of HDM2-mediated p53 polyubiquitination.
Figure 4.
RPL26 attenuates HDM2-mediated ubiquitination and degradation. (A and B) RPL26 inhibits HDM2-mediated p53 and HDM2 ubiquitination in H1299 cells. H1299 cells were transfected with combinations of RPL26 (2 μg)-, p53 (1 μg)- or HDM2 (2 μg)-encoding plasmids in the presence of the HA-ubiquitin (HA-Ub) (2 μg) plasmid as indicated. The cells were treated with MG132 (20 μM) for 8 h before harvesting. The cell extracts was immunoprecipited by Flag antibody (A) or HDM2 antibody (B). Ubiquitinated proteins were detected by immunoblotting with the anti-HA antibodies. Ubiquitinated p53s [p53-(HA-Ub)n] (A) or Ubiquitinated HDM2s [HDM2-(HA-Ub)n] (B) is indicated to the upper panels. The expression levels of HDM2, p53 and L26 are shown in the lower panels. (C) Half-life analysis of p53 and HDM2 in the presence or absence of overexpressed RPL26. U2OS cells were transfected with 0.2 μg of Myc-RPL26 in the presence of GFP (50 ng) with (−) or without (+) HDM2 (0.4 μg) as indicated. After 24 h transfection, cells were exposed to the protein synthesis inhibitor CHX (100 μg/ml) for different times. Target proteins were detected by immunoblotting. (D) Plot of the p53 and HDM2-expression levels following CHX treatment. The value is normalized to the levels of GFP. (E) RPL26 modulates p53 by interacting with HDM2. Wild-type and mutant RPL26 were overexpressed in MEF (p53−/−/mdm2−/−) cells with or without HDM2 in the presence of p53, and the target proteins in whole-cell lysates were detected by immunoblotting. (F) RPL26 modulates the HDM2–p53 interaction by forming a ternary complex among RPL26, HDM2 and p53. HEK293 cells were transfected with combinations of plasmids as indicated. Cell lysates were immunoprecipitated by Myc antibody followed by immunoblotting with Myc, Flag-HRP, HDM2 and β-actin antibodies for the immunoprecipitates or the whole-cell lysates.
To further determine if RPL26 acts on HDM2 and p53 at the post-translational level, plasmids expressing HDM2, GFP and RPL26 were co-expressed in U2OS cells as indicated, which were then exposed to the protein synthesis inhibitor, cycloheximide, for different times (Figure 4C). The p53 protein did not show apparent difference in RPL26 transfected cells. The level of p53 protein decreased when transfected with HDM2, but it can be rescued by the expression of RPL26. Compared to the control, the degradation of HDM2 protein was much more slowly in RPL26 transfected cells. The results suggest that RPL26 affect HDM2 and p53 at the post-translational level. The RPL26 might inhibit HDM2′s E3 ligase function, while HDM2 harbors a self- and p53-specific E3 ubiquitin ligase activity (6).
Furthermore, the binding of RPL26 and HDM2 seems to be crucial for this process. In MEF (p53−/−/mdm2−/−) cells which were overexpressed with different mutants of RPL26 plasmids, RPL26 mutants did not affect the p53 level without HDM2 (Figure 4E, left panels). But RPL26 mutants retaining the capacity for binding to HDM2 (B, C and E) stabilized p53 and HDM2, while those without binding capacity, A and D, did not increase the p53 and HDM2 levels in MEF (p53−/−/mdm2−/−) cells which were co-transfected with HDM2 (Figure 4E, right panels). It demonstrated that the HDM2 binding capacity of RPL26 was pivot for the regulation of HDM2-dependent degradation.
To explore the relationship of RPL26, HDM2 and p53, HEK293 cells were transiently transfected with various combinations of plasmids (Figure 4F). As shown, Flag-p53 was detected in Myc-RPL26 immunoprecipitates of lysates from cells only transfected with Myc-RPL26 and Flag-p53 (Figure 4F, lane 7), albeit with lower affinity compared to the presence of HDM2 (Figure 4F, lane 8). The result illustrates the formation of a complex among RPL26, HDM2 and p53, while p53 and RPL26 can immunoprecipitated in a complex without HDM2. Together, RPL26 decreases HDM2-mediated ubiquitination and degradation of p53 and HDM2 by binding to HDM2.
Act D enhances RPL26-HDM2 interaction and activates p53
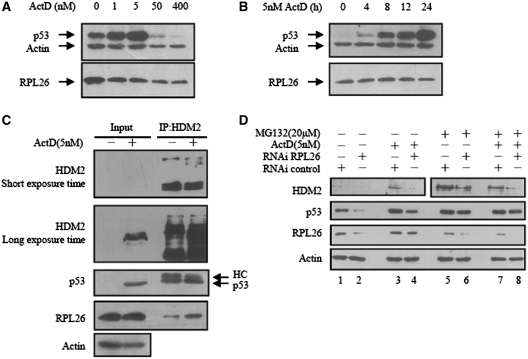
It has been demonstrated that, a low dose of Act D (5 nM) specifically inhibits RNA polymerase I and consequently reduces rRNA synthesis, leading to perturbation of ribosomal biogenesis (21). Some ribosomal proteins, including L5, L11, L23 and S7, are released and modulate the HDM2–p53 interaction. We examined the level of RPL26 with different dose of Act D. As shown in Figure 5A, the level of RPL26 was decreased a little by treatment with high dose of Act D. With the low dose of Act D, the level of RPL26 did not apparently change, but the level of p53 enhanced time-dependently (Figure 5B).
Figure 5.
Act D enhances RPL26-HDM2 interaction and activates p53. (A) Low doses of Act D induce p53, whereas high doses of Act D inhibit p53. U2OS cells were treated with increasing amounts of Act D (Act D) as indicated at the top. Cell lysates were used for an immunoblot analysis with antibodies indicated to the left. (B) Time-dependent effect of Act D on p53 and RPL26 levels in U2OS cells. U2OS cells were treated with 5 nM Act D and harvested at different time points as indicated at the top. Cell lysates were used for an immunoblot analysis with antibodies indicated to the left of each panel. (C) Low dose of Act D enhances HDM2–RPL26 interaction. U2OS cells were treated with (−) or without (+) 5 nM Act D for 24 h before harvesting. (D) Inhibition of endogenous RPL26 by siRNA inhibits the Act D-induced p53 levels but did not effect the protein level of p53 with the treatment of MG132. U2OS cells were transfected with or without RPL26 siRNA (100 nM). Cells were then incubated with (−) or without (+) 5 nM Act D for 12 h before harvesting. Cells were incubated with (−) or without (+) MG132 (20 μM) for 8 h before harvesting. Cell lysates were then immunoblotted with anti-HDM2, anti-p53, anti-RPL26 or anti-actin antibodies.
We examined the binding of HDM2, RPL26 and p53, with treatment of 5 nM Act D. The level of p53 and HDM2 enhanced a lot (Figure 5C, left panel) and the endogenous binding between RPL26 and HDM2 was increased (Figure 5C, right panel). It might due to the formation of a complex among RPL26, HDM2 and p53 following the release of RPL26 in response to ribosomal stress.
To determine if RPL26 is necessary for p53 induction caused by Act D, U2OS cells were transfected transiently with RPL26 siRNA or control siRNA prior to treatment with 5 nM of Act D. As shown in Figure 5D, the expression level of RPL26 was decreased after RNAi. While the protein level of p53 and HDM2 also drastically decreased. The results suggest that knockdown of RPL26 inhibit Act D induced p53 levels. Moreover, knockdown of RPL26 inhibited the expression level of HDM2 with the treatment of 20 μM MG132, but did not affect the protein level of p53 (Figure 5D, lane 6 and 8). These results suggest that RPL26 might affect the stabilization of HDM2 and p53 by inhibiting the ubiquitin ligase activity of HDM2.
RPL26 inhibits cell proliferation and induces cell cycle arrest by p53
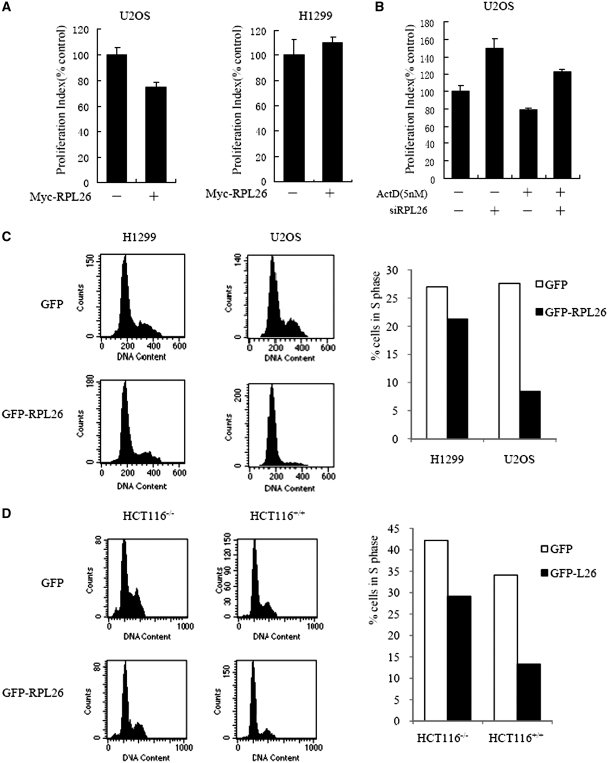
To investigate the cellular consequences of overexpresion RPL26, we transfected Myc-RPL26 or control vector into U2OS or H1299 cells. Such overexpression of Myc-RPL26 resulted in the inhibition of cell proliferation in U2OS cells which contains functional p53 (Figure 6A). But RPL26 did not affect the cell proliferation in H1299 cells which is deficient of p53 (Figure 6A). So RPL26 can regulate cell proliferation by mediating p53 activity.
Figure 6.
RPL26 enhances the activities of p53. (A) RPL26 inhibits a p53-dependent cell’s proliferation. U2OS and H1299 cells were transiently transfected with Myc-RPL26 or Myc cells. After 24 h post-transfection, cell proliferation was measured by the MTS-based cell proliferation assay. Data are plotted as mean ± standard errors for three independent experiments. (B) siRNA knockdown of RPL26 increases U2OS cell’s proliferation. U2OS cells were transfected transiently with RPL26 siRNA or control siRNA. After 12 h post-transfection, cell was treated with 5 nM of Act D for 12 h. Then cell proliferation was measured by the MTS-based cell proliferation assay. Data are plotted as mean ± standard errors for three independent experiments. (C) RPL26 induces a p53-dependent cell cycle arrest. U2OS or H1299 cells were transfected with the indicated plasmid expressing a GFP or GFP-RPL26 proteins. Transfected cells were sorted, and their cell cycle distribution characteristics were determined by flow cytometry at 24 h after transfection. The proportions of cells in S phase in each transfected cell population were compared using bar graphs. A minimum of 10 000 GFP-positive cells were analyzed for each transfection. (D) RPL26 induces a p53-dependent cell cycle arrest. HCT116+/+(p53-positive) and HCT116−/−(p53-negative) cells were transfected with the indicated plasmid expressing a GFP or GFP-RPL26 proteins. Transfected cells were sorted, and their cell cycle distribution characteristics were determined by flow cytometry at 24 h after transfection. The proportions of cells in S phase in each transfected cell population were compared using bar graphs. A minimum of 10 000 GFP-positive cells were analyzed for each transfection.
To test the effect of siRNA knockdown of RPL26 on cell proliferation, U2OS cells were transfected transiently with RPL26 siRNA or control siRNA prior to treatment with 5 nM of Act D. As shown in Figure 6B, Act D decreased the cell proliferation, while knockdown of RPL26 increased the cell proliferation with or without Act D.
Next, we examined whether activation of p53 by RPL26 could result in cell cycle distribution change. The p53-proficient U2OS or p53-deficient H1299 cells were transiently transfected with either the GFP-RPL26 or the GFP. GFP-positive cells were then gated for cell cycle analysis. As shown in Figure 6C, overexpression of GFP-RPL26 induced cell cycle arrest only in U2OS, but not in H1299 cells. To role out the influence of the genetic alterations in cell types, the experiments were repeated in the HCT116+/+ (p53 positive) and HCT116−/− (p53 negative) cells. Overexpression of RPL26 induced significantly cell cycle arrest in p53-positive cells than in negative cells (Figure 6D).
DISCUSSION
RPL26 is one of the ribosomal proteins in 60S ribosome. Previous report suggest that RPL26 may bind to 5′UTR of p53 mRNA and control p53 translation and induction after DNA damage (18). In this study, we identify that RPL26 regulate p53 protein level by physical interacting with HDM2. Unlike reported HDM2 interactors, RPL26 is the first ribosomal proteins that can regulate p53 activity at both translational and post-translational level.
As far as we know, several ribosomal proteins L5, L11, L23 and S7 have been shown to regulate p53 activity via binding to HDM2 and blocking HDM2-mediated p53 ubiquitination and degradation (10–14). Further studies suggest that ribosomal proteins that can bind to HDM2 may regulate HDM2–p53 pathway in a cooperative way by forming a ribosomal proteins–HDM2–p53 complex (10,22). Our data demonstrated that RPL26 also can interact with HDM2 and form a complex with HDM2–p53 (Figures 1 and 4F), but further studies will be needed to illustrate the interrelationship of RPL26 and other ribosomal proteins in regulating p53 activity. Although these ribosomal proteins can block HDM2-p53 loop in response to ribosomal stress, the expression level of them changed in a different way in response to stress, such as 5-fluorouracil (5-FU) or low dose Act D treatment (13,16). We also detected the RPL26 expression level in response to low-dose Act D treatment (Figure 5), and it changed differently from other ribosomal proteins. The expression level of RPL26 by Act D is dosage dependent. It was decreased sharply after treatment of U2OS cells with 50 nM Act D. But the expression level of RPL26 by a low dose of Act D is not timely dependent. While the change of p53 level is not only effected by the dose of Act D but also the working time of low dose of Act D. These data suggest that exist of different mechanisms in regulating free ribosomal proteins response to perturbation.
Moreover, our data show that the HDM2 protein level increased in U2OS cells when RPL26 overexpressed (Figure 4C). It suggests that the increase of HDM2 is related to the decreasing of HDM2′s E3 ubiquitin ligase activity, not to the p53 transactivation of hdm2 gene. This phenomenon must be induced by the interaction of RPL26′s binding to the central region of HDM2 which contains nuclear localization, export sequences and a zinc-finger domain. In the context of cancer-associated mutations in the HDM2 zinc-finger domain disrupt ribosomal protein interaction and attenuate HDM2-induced p53 degradation (23), ribosomal proteins’ binding to HDM2 is an important mechanism in regulating p53.
RPL26 induces p53 through direct binding to HDM2 and decreasing HDM2′s ubiquitin E3 ligase activity. This mode is different from the previous reported mechanism that RPL26 enhances p53 mRNA’s translation and expression level. But both mechanisms could improve p53′s level and activity and involve the middle domain of RPL26 which can bind to HDM2 and 5′UTR of p53 mRNA (18). This suggests that the two mechanisms might operate together in regulating p53 by RPL26. Recently, HDM2 has been reported to attenuate the association of RPL26 with p53 mRNA and repress RPL26-mediated augmentation of p53 protein synthesis by promoting RPL26′s proteasome-mediated degradation and binding of RPL26 (24). Together, the interaction of RPL26 and HDM2 seems to have mutual action that adjusts HDM2 and RPL26′s effecting on p53′s expression level and activity. These suggest that RPL26 and p53 may competitively bind to HDM2. In this regard, the cancer-associated mutations in RPL26 gene that inhibit its interaction with 5′UTR of p53 mRNA or HDM2 are strong evidence of RPL26′s importance in cancer. Moreover, previous study identified mutations in RPL26 in two murine tumor cell lines resulting in a unique tumor-associated antigen and a more aggressive tumor phenotype (25). Therefore, RPL26 might be a tumor suppressor that works by regulation HDM2–p53 loop.
To sum up, we establish that RPL26 has the capacity to stabilize p53 by binding to HDM2. Different from other HDM2 binding ribosomal proteins, RPL26 is the only identified ribosomal protein that can activate p53 by simultaneously potentiating its translation and attenuating its degradation till now. It would be important to further investigate the roles of RPL26 in p53-related cancer.
FUNDING
National High-tec Research Developing Programme (2006AA02A310); Special Funds for Major State Basic Research of China (2006CB910802); Chinese National Natural Science Foundation Projects (Innovation group project) (30621063). Funding for open access charge: National High-tec Research Developing Programme (863).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Drs Moshe Oren and Bert Vogelstein for p53 reporter gene plasmids; Dr Yue Xiong for p53, Hdm2 and ubiquitin expression constructs; Dr Mian Wu for MEF (p53−/−/mdm2−/−) cell; Dr Qimin Zhan for p53+/+ HCT116 and p53−/− HCT116 cells.
REFERENCES
- 1.Braithwaite AW, Prives CL. p53: more research and more questions. Cell Death Differ. 2006;13:877–880. doi: 10.1038/sj.cdd.4401938. [DOI] [PubMed] [Google Scholar]
- 2.Olivier M, Hussain SP, Caron de Fromentel C, Hainaut P, Harris CC. TP53 mutation spectra and load: a tool for generating hypotheses on the etiology of cancer. IARC Sci. Publ. 2004;157:247–270. [PubMed] [Google Scholar]
- 3.Brooks CL, Gu W. Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr. Opin. Cell Biol. 2003;15:164–171. doi: 10.1016/s0955-0674(03)00003-6. [DOI] [PubMed] [Google Scholar]
- 4.Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 5.Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 7.Geyer RK, Yu ZK, Maki CG. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat. Cell Biol. 2000;2:569–573. doi: 10.1038/35023507. [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez MS, Desterro JM, Lain S, Lane DP, Hay RT. Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol. Cell Biol. 2000;20:8458–8467. doi: 10.1128/mcb.20.22.8458-8467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barlev NA, Liu L, Chehab NH, Mansfield K, Harris KG, Halazonetis TD, Berger SL. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell. 2001;8:1243–1254. doi: 10.1016/s1097-2765(01)00414-2. [DOI] [PubMed] [Google Scholar]
- 10.Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J. Biol. Chem. 2004;279:44475–44482. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- 11.Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3:577–587. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- 12.Jin A, Itahana K, O'Keefe K, Zhang Y. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol. Cell Biol. 2004;24:7669–7680. doi: 10.1128/MCB.24.17.7669-7680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol. Cell Biol. 2004;24:7654–7668. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen D, Zhang Z, Li M, Wang W, Li Y, Rayburn ER, Hill DL, Wang H, Zhang R. Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene. 2007;26:5029–5037. doi: 10.1038/sj.onc.1210327. [DOI] [PubMed] [Google Scholar]
- 15.Bernardi R, Scaglioni PP, Bergmann S, Horn HF, Vousden KH, Pandolfi PP. PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat. Cell Biol. 2004;6:665–672. doi: 10.1038/ncb1147. [DOI] [PubMed] [Google Scholar]
- 16.Sun XX, Dai MS, Lu H. 5-fluorouracil activation of p53 involves an MDM2-ribosomal protein interaction. J. Biol. Chem. 2007;282:8052–8059. doi: 10.1074/jbc.M610621200. [DOI] [PubMed] [Google Scholar]
- 17.Sun XX, Dai MS, Lu H. Mycophenolic acid activation of p53 requires ribosomal proteins L5 and L11. J. Biol. Chem. 2008;283:12387–12392. doi: 10.1074/jbc.M801387200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 19.Zhang L, Xing G, Tie Y, Tang Y, Tian C, Li L, Sun L, Wei H, Zhu Y, He F. Role for the pleckstrin homology domain-containing protein CKIP-1 in AP-1 regulation and apoptosis. EMBO J. 2005;24:766–778. doi: 10.1038/sj.emboj.7600532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, Xiong Y. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol. Cell Biol. 2003;23:8902–8912. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashcroft M, Taya Y, Vousden KH. Stress signals utilize multiple pathways to stabilize p53. Mol. Cell Biol. 2000;20:3224–3233. doi: 10.1128/mcb.20.9.3224-3233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horn HF, Vousden KH. Cooperation between the ribosomal proteins L5 and L11 in the p53 pathway. Oncogene. 2008;27:5774–5784. doi: 10.1038/onc.2008.189. [DOI] [PubMed] [Google Scholar]
- 23.Lindstrom MS, Jin A, Deisenroth C, White Wolf G, Zhang Y. Cancer-associated mutations in the MDM2 zinc finger domain disrupt ribosomal protein interaction and attenuate MDM2-induced p53 degradation. Mol. Cell Biol. 2007;27:1056–1068. doi: 10.1128/MCB.01307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol. Cell. 2008;32:180–189. doi: 10.1016/j.molcel.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beck-Engeser GB, Monach PA, Mumberg D, Yang F, Wanderling S, Schreiber K, Espinosa R, III, Le Beau MM, Meredith SC, Schreiber H. Point mutation in essential genes with loss or mutation of the second allele: relevance to the retention of tumor-specific antigens. J. Exp. Med. 2001;194:285–300. doi: 10.1084/jem.194.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]