Abstract
Regulation of ribonucleotide reductase (RNR) is important for cell survival and genome integrity in the face of genotoxic stress. The Mec1/Rad53/Dun1 DNA damage response kinase cascade exhibits multifaceted controls over RNR activity including the regulation of the RNR inhibitor, Sml1. After DNA damage, Sml1 is degraded leading to the up-regulation of dNTP pools by RNR. Here, we probe the requirements for Sml1 degradation and identify several sites required for in vivo phosphorylation and degradation of Sml1 in response to DNA damage. Further, in a strain containing a mutation in Rnr1, rnr1-W688G, mutation of these sites in Sml1 causes lethality. Degradation of Sml1 is dependent on the 26S proteasome. We also show that degradation of phosphorylated Sml1 is dependent on the E2 ubiquitin-conjugating enzyme, Rad6, the E3 ubiquitin ligase, Ubr2, and the E2/E3-interacting protein, Mub1, which form a complex previously only implicated in the ubiquitylation of Rpn4.
INTRODUCTION
DNA damage activates a checkpoint kinase cascade that both halts the cell cycle and concurrently activates factors that repair the damage. One consequence of checkpoint activation is to increase dNTP production, which causes about a 6- to 8-fold increase in dNTP pools after DNA damage treatment (1). Transient up-regulation of dNTP pools leads to increased resistance to DNA-damaging agents, but also increased mutation rates (1). Furthermore, constitutively high dNTP pools inhibit the entry into S phase by delaying replication initiation and also impair activation of the DNA damage checkpoint (2). Therefore, proper dNTP regulation is crucial for cell growth and DNA damage repair.
dNTP production is tightly controlled throughout the cell cycle and in response to DNA damage. This is accomplished through the regulation of ribonucleotide reductase (RNR), the enzyme that performs the rate-limiting step in de novo synthesis of dNTPs (3). In most eukaryotes, the RNR enzyme is a heterotetramer, comprised of one large homodimeric R1 subunit and one small homodimeric R2 subunit. In Saccharomyces cerevisiae, there are four genes (RNR1-4) that encode RNR polypeptides, but only RNR1, RNR2 and RNR4 are essential (4–6). The yeast enzyme is comprised of one homodimeric Rnr1 subunit, as well as a heterodimeric Rnr2/Rnr4 subunit. Although protein levels of the second large polypeptide, Rnr3, increase dramatically in response to DNA damage, there is no detectable growth or DNA repair defect for rnr3Δ (7). However, Rnr3 is important for cell survival in response to genotoxic stress when the target of rapamycin (TOR) pathway is inhibited by Rapamycin treatment (8).
The regulation of RNR is multifaceted and includes both allosteric regulation (9) and checkpoint-dependent regulation controlled by the Mec1/Rad53/Dun1 kinases. Following damage, Mec1, the ataxia telangiectasia-related (ATR) homolog in yeast, is activated and initiates a kinase cascade that controls many aspects of the DNA damage response including cell-cycle progression, expression of transcriptional targets, replication fork stability and late-replication origin firing (10). Additionally, all of the RNR genes are transcriptionally induced in a Dun1-dependent manner following checkpoint activation, varying from about 3-fold for RNR1 to more than 100-fold for RNR3 (4–6,11). The RNR proteins are also regulated by changes in their subcellular localization (12). At all stages of the cell cycle, Rnr1 and Rnr3 are found in the cytoplasm, where dNTP synthesis is thought to occur. In contrast, Rnr2 and Rnr4, the small subunits, are localized to the nucleus during G1 and are co-transported to the cytoplasm during S phase and after DNA damage treatment (13). Wtm1, a WD40-containing protein, is involved in anchoring Rnr2 and Rnr4 to the nucleus in G1 (14,15), while the cytoplasmic protein, Dif1, is required for nuclear import of Rnr2 (16,17). In response to DNA damage, Wtm1 releases the small RNR heterodimeric subunit from the nucleus and Dif1 is degraded, allowing Rnr2 and Rnr4 to remain in the cytoplasm (14–17).
In budding yeast, RNR is also regulated by the protein inhibitor Sml1, which was first identified as a suppressor of the lethality of mec1 and rad53 mutations (18). A sml1Δ mutation leads to increased levels of all four dNTPs compared to wild type (18) and Sml1 binds to Rnr1 and inhibits RNR activity in vitro (19,20). The Sml1 protein is degraded in response to DNA damage and this regulation is dependent on the Mec1, Rad53 and Dun1 checkpoint kinases, mutations of which completely stabilize Sml1 (21). This degradation correlates with the appearance of Dun1-dependent phosphorylated forms of Sml1 (21); however, it was not shown directly whether this phosphorylation is required for the degradation of the protein. Purified Dun1 from yeast directly phosphorylates recombinant Sml1 in vitro and Sml1 physically interacts with Dun1 in a two-hybrid assay (22). Additionally, three serines in the Sml1 protein (56, 58 and 60) can be phosphorylated by Dun1 in vitro (23). Recently, Sml1 degradation was shown to be a very sensitive indicator of DNA damage checkpoint activation (24) and its degradation occurs even when Rad53 phosphorylation is undetectable (25).
Ubiquitylation, an important post-translational modification, commonly targets proteins for degradation by the 26S proteasome [for review see Refs (26) and (27)]. Protein ubiquitylation is controlled by a sequence of reactions carried out by three types of conjugating enzymes: E1 (ubiquitin-activating enzyme), E2 (ubiquitin-conjugating enzyme) and E3s (ubiquitin-protein ligases) as well as by deubiquitylation enzymes. The E3 enzymes determine specificity for the target protein, and also regulate where the ubiquitin will be added (28).
An E2 that is involved in the DNA damage response, Rad6, associates with several E3 enzymes, including Ubr1, Bre1 and Rad18 (29–31) and is known to ubiquitylate the proliferating cell nuclear antigen (PCNA) and 9-1-1 clamps, among other targets (32,33). Rad6 has also been shown to associate with the E3 Ubr2, which was discovered due to its sequence homology to Ubr1 (34) and was later shown to have a role in the ubiquitylation and degradation of the proteasomal regulator Rpn4 (35). Mub1 is an additional factor required for the ubiquitylation of Rpn4 in vivo and in vitro (36).
In the present study, site-directed mutagenesis was used to identify the in vivo phospho-acceptors important for Sml1 degradation. Changing four serines (56, 58, 60 and 61) to alanines, sml1-4SA, prevents the degradation of the protein by blocking its in vivo and in vitro Dun1 phosphorylation. Endogenous expression of the sml1-4SA gene alone does not affect cell growth or DNA damage repair since other forms of RNR regulation are still intact. However, when sml-4SA is overexpressed, it slows S phase progression. Additionally, failure to degrade Sml1 is toxic when Rnr1 function is compromised (rnr1-W688G). Sml1 phosphorylation is required for its degradation in response to DNA damage. The degradation of Sml1 following DNA damage treatment also depends on the Rad6–Ubr2–Mub1 E2/E3 ubiquitin complex. Our results suggest a model whereby DNA damage-induced phosphorylation and ubiquitylation of Sml1 occur sequentially triggering degradation of Sml1 by the 26S proteasome.
MATERIALS AND METHODS
Strains and media
The strains used in this study are listed in Supplementary Table S1. Sml1 phosphorylation was detected in strains that have increased levels of Sml1 due to overexpression of RNR1, which does not affect the regulation or function of Sml1 (21). In Figure 1D and Supplementary Figure S1, strains were transformed with pWJ841, a 2-µ plasmid that carries RNR1 (21). All mutations were generated by polymerase chain reaction (PCR). Integrations at the respective chromosomal loci were done by the cloning-free PCR-based allele replacement method (37). The correct integration was verified through sequencing of an amplified segment from the respective genomic region. To distinguish between the different alleles, mutations were engineered to either introduce or delete a restriction site: sml1-4SA is detected by loss of an MboII site; rnr1-W688G has a new SfcI site. The chromosomal GAL-SML1 locus described previously (21) was used to make the GAL-sml1-4SA strain (W3332-5C). Media and growth conditions used in all experiments are standard (38) with the addition of twice the amount of leucine (60 µg/ml) in all synthetic complete (SC)-based media. Cultures were grown in Yeast extract, peptone, dextrose (YPD), SC or SC-dropout for plasmid selection. YPD or Yeast extract, peptone, galactose (YPGal) contains 1% yeast extract, 2% peptone and 2% glucose or 2% galactose, respectively. YPGly (2% peptone, 1% yeast extract, 3% glycerol and 3% d-lactic acid) or YPRaffinose (1% yeast extract, 2% peptone and 2% raffinose) medium was used in experiments where galactose induction was required. For the experiments in Figures 2 and 4D, cells were grown in YPGly medium, pH 4.5, to facilitate the induction of GAL-sml1 constructs. To synchronize cells in G1, 3.4 µg/ml α-factor was added for 2.5 h. Galactose (2%) induction was initiated in the last 30 min of this treatment and continued after the cells were released from α-mating factor by rapid filtration into YPGal medium. S-phase progression was monitored by analysis of the DNA content through flow cytometry.
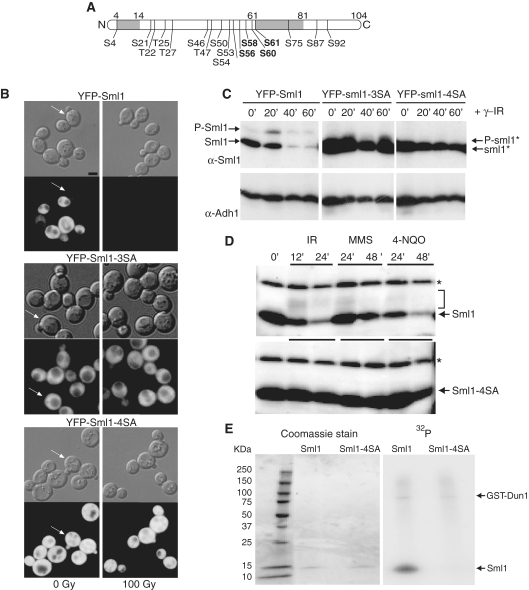
Figure 1.
Mutations that eliminate putative phosphorylation sites in Sml1 stabilize the protein after DNA damage treatment and prevent in vivo and in vitro phosphorylation. (A) Depiction of the Sml1 protein with the positions of all serine and threonine residues that are potential phosphorylation sites. The shaded regions indicate the positions of two α helices, including the C-terminal helix, which is important for Sml1 binding to Rnr1. The four serines changed in the sml1-4SA mutant are shown in bold type. (B) Mid-log phase cultures of cells expressing YFP-Sml1 (W4622-14B), YFP-sml1-3SA (W6976-4A) or YFP-sml1-4SA (W4748-4D) fusion proteins were treated with 100 Gy of γ-irradiation. Protein stability was examined by visualizing YFP fluorescence before and after treatment. White arrows indicate cells that are in S phase (small buds). The scale bar is equal to 3 µm. (C) Total yeast extracts of the strains shown in (B) were probed with anti-Sml1 antibody to examine stability and in vivo phosphorylation, as determined by mobility shift of immunoblot, of the fusion proteins in logarithmically growing cultures. To control for loading, the membrane was stripped and re-probed using anti-Adh1 antibody. (D) Total yeast extracts from wild-type (W1588-4C) and sml1-4SA (W3329-7D) strains were examined for Sml1 in vivo phosphorylation in response to treatment with γ-irradiation (300 Gy), 0.05% MMS and 4-NQO (0.25 mg/l). The arrow indicates the position of Sml1 proteins. The slower migrating bands (indicated by a bracket) are due to phosphorylation (21). Immunoblots were probed with anti-Sml1 serum. The top band, labeled with an asterisk, is a Sml1-independent cross-reacting band used as a loading control. (E) Recombinant purified Sml1 and sml1-4SA were incubated with GST-Dun1 fusion protein purified from yeast. A portion of the reaction was resolved on a 4–20% SDS–PAGE gradient gel, stained with Coomassie blue and subsequently visualized by autoradiography for 32P incorporation. Both reactions contained the same amount of recombinant protein (left) and exhibited the same level of kinase activity as observed by GST-Dun1 autophosphorylation (right).
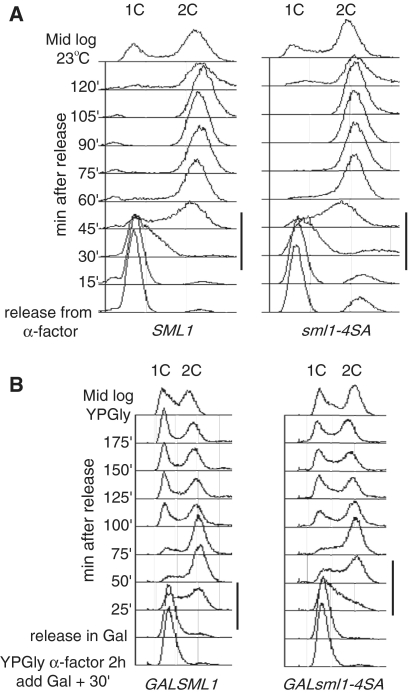
Figure 2.
Overexpression of sml1-4SA, but not SML1, slows S phase progression in wild-type cells. (A) Wild-type and sml1-4SA strains were analyzed for cell-cycle progression. There is no significant difference in the duration of S phase between the two strains when the proteins are expressed endogenously (black lines to the right of the panels). (B) Overexpression of sml1-4SA (W3332-5C), but not SML1 (W2056-8A), driven by a strong galactose promoter slows S phase progression in wild-type cells. Black lines to the right of the panels indicate S-phase.
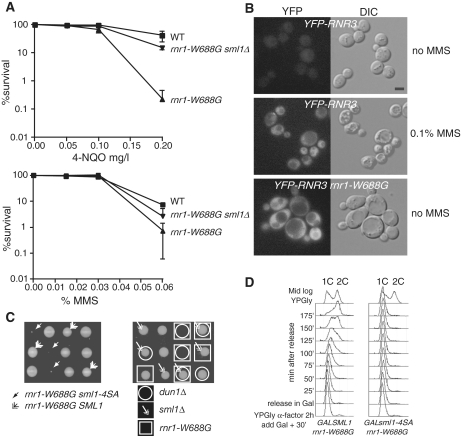
Figure 4.
sml1-4SA sensitizes cells that are defective in dNTP regulation. (A) rnr1-W688G is sensitive to genotoxic stress. The DNA damage sensitivities of the strains [wild type (W1588-4C), rnr1-W688G (W4383-1B) and rnr1-W688G sml1Δ (W4383-10C)] were examined in a quantitative survival assay used to determine the LD50 values after 4-NQO (mg/l) and MMS (%) treatments. Mid-log phase cultures were sonicated and appropriate dilutions were plated on YPD for viability, or on YPD plates containing 4NQO (top) and MMS (bottom) at various concentrations. The reported value is the mean of three experiments and the error bars represent the standard error of the mean. (B) rnr1-W688G induces Rnr3 expression in the absence of exogenous damage. The activation of the DNA damage checkpoint was monitored through the induction of a YFP-Rnr3 fusion protein (W6986-1B). Rnr3 is induced in response to DNA damage and the induction depends on checkpoint signaling (55). In the absence of DNA damage, YFP-Rnr3 is not expressed in wild-type cells (top). To induce YFP-Rnr3 expression, mid-log phase cells were treated with 0.1% MMS for 5 h (middle). The rnr1-W688G mutation causes YFP-Rnr3 to be expressed without any exogenous DNA damage treatment (bottom). (C) Dissection of heterozygous diploid SML1/sml1-4SA rnr1-W688G/RNR1 (W4383) and SML1/sml1Δ DUN1/dun1Δ RNR1/rnr1-W688G (W4384) strains shows a genetic interaction between rnr1-W688G and the sml1-4SA and dun1Δ alleles. Deletion of sml1Δ suppresses the synthetic lethality between rnr1-W688G and dun1Δ. The four spores of each tetrad are positioned in the three rows shown. (D) Yeast cells (W3755-14D: GAL-SML1 rnr1-W688G and W3756-3B: GAL-sml1-4SA rnr1-W688G) growing in YPGly at 30°C were synchronized at G1 with α-factor for 2 h. Galactose was added 30 min into the treatment. The pheromone was removed through rapid filtration and cells were released in fresh YPGal without α-factor. At each time point, samples were fixed in 70% ethanol for DNA content analysis by FACS. WT, wild-type strain.
Protein extracts and immunoblots
Several methods of protein extraction were used to detect protein levels. For the experiments in Figures 1C, 5A, C and D; Supplementary Figure S1, extracts were made by the ‘boiling method’ as described in ref. (21). For the experiment in Figure 3C, protein extracts were prepared by the trichloroacetic acid (TCA) method (39), and for the experiments in Figure 1D, a variation of this experiment using NP-40 extraction buffer (1% NP-40, 150 mM NaCl, 50 mM Tris, pH 8.0) was performed. Immediately before use, this buffer was supplemented with a protease inhibitor cocktail (Roche, as per manufacturer’s instructions), 1 µM pepstatin, 1 mM PMSF, 30 mM NaF and 1 mM DTT. Proteins were separated by SDS–PAGE, transferred to polyvinylidene fluoride membranes and blots were probed with anti-Sml1 serum (21), anti-Rad53 antibody (Santa Cruz) or anti-Adh1 (alcohol dehydrogenase) antibody (Chemicon International, AB1202). Sml1 bands were detected using ECL+ (Amersham).
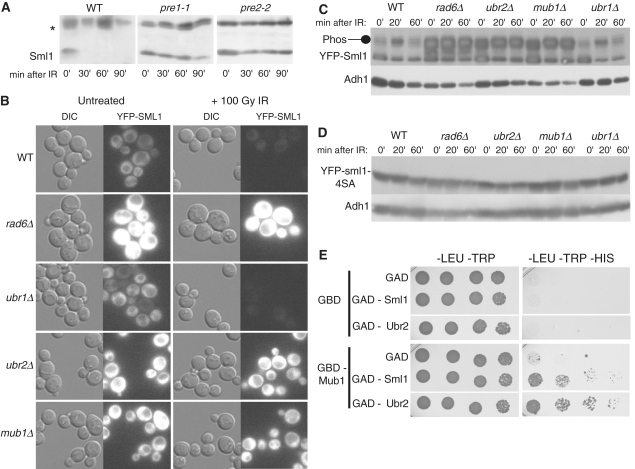
Figure 5.
Sml1 is degraded by the 26S proteasome and the degradation is dependent on Rad6, Ubr2 and Mub1. (A) Wild-type (MHY686) or mutant cells were shifted to 37°C for 3 h to inactivate the proteasome in the pre1-1 (MHY687) and pre2-2 (MHY689) mutants, and treated with 0.03% MMS. Samples were run on a 15% SDS–PAGE gel. Using anti-Sml1 serum, total yeast extracts were examined for Sml1 protein levels at the indicated time points after MMS treatment. In both immunoblots, the top band, labeled with an asterisk, is a Sml1-independent cross-reacting band used as a loading control. (B) Mid-log cultures of WT, rad6Δ, ubr1Δ, ubr2Δ and mub1 strains containing a genomic copy of YFP-Sml1 (W9174-5D, W9174-10C, W9177-8B, W9175-7C and W9176-4D, respectively) were treated with 100 Gy of γ-irradiation. Protein stability was examined by visualizing YFP fluorescence before and after treatment. (C) (Top panel) Total yeast extracts from logarithmically growing cultures of the strains shown in (B) were immunoblotted and probed with anti-Sml1 antibody to examine stability and in vivo phosphorylation. To control for loading, the membrane was stripped and re-probed using anti-Adh1 antibody. (D) Total yeast extracts of mid-log cultures of WT, rad6Δ, ubr1Δ, ubr2Δ and mub1 strains containing a genomic copy of YFP-sml1-4SA (W9261-7D, W9261-11B, W9264-11C, W9262-2D and W9263-7B, respectively) were treated with 100 Gy of γ-irradiation, immunoblotted and probed with anti-Sml1 antibody. To control for loading, the membrane was stripped and re-probed using anti-Adh1 antibody. (E) Diploids containing different combinations of GBD or GBD-Mub1 with GAD, GAD-Sml1 or GAD-Ubr2 (as indicated) were spotted in 5-fold serial dilutions onto -LEU–TRP and -LEU–TRP–HIS media. The -LEU–TRP medium selects for diploids containing a GBD and GAD plasmids. Growth on medium lacking histidine indicates a two-hybrid interaction and was observed for GBD-Mub1 and GAD-Sml1 as well as GBD-Mub1 and GAD-Ubr2. Plates were scanned after 4 days of growth. WT, wild-type strain.
Figure 3.
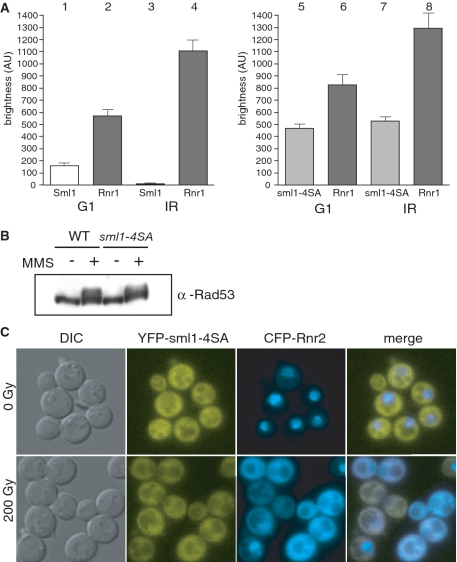
RNR regulation and checkpoint activation are normal in a sml1-4SA strain. (A) Cells expressing YFP-Sml1 (W5530-6C) or YFP-sml1-4SA (W5755-2A) along with CFP-Rnr1 were visualized by fluorescent microscopy before and an hour after 200 Gy of γ-irradiation. Live cell images were captured identically and YFP levels are depicted on the graphs. Cells without buds are G1 cells and were used for subsequent analyses. (B) Protein extracts were taken from wild-type (W1588-4C) and sml1-4SA (W3329-7D) cells before and an hour after 0.03% MMS treatment and analyzed by immunoblot using a Rad53 antibody. (C) Cells expressing both YFP-sml1-4SA and CFP-Rnr2 proteins (W5766-1D) were visualized by fluorescent microscopy before and an hour after 200 Gy of γ-irradiation. WT, wild-type strain.
In vitro kinase assays
The ORFs of SML1 and sml1-4SA were PCR amplified and cloned in the pET3a expression vector (Stratagene) to generate plasmids pWJ1265 and pWJ1266, respectively (primer sequences available on request). Recombinant proteins were expressed in BL21(DE3)pLYS(S) bacteria and purified as described previously (19). Expression of pWJ772, a GST-Dun1 fusion protein (22), was induced for 5 h with 4% galactose in a pep4Δ strain exponentially growing in SC-Ura medium with 2% raffinose. Preparation of GST-Dun1 extracts and in vitro kinase assays were carried out essentially as described previously (22). A portion of the reaction was resolved on a 4–20% SDS–PAGE gradient gel (Bio-Rad) then stained with Coomassie blue R-250 (Bio-Rad) and subsequently autoradiographed.
DNA damage sensitivity experiments
Exponential cultures were sonicated and plated at the appropriate dilutions on YPD plates with methyl methane sulfonate (MMS) or 4-nitroquinoline 1-oxide (4-NQO), prepared 24 h before the experiment. Cells were grown at 30°C and viable colonies were counted after 4 days. DNA damage sensitivity experiments were repeated at least three times and a minimum of two strains for each genotype were tested.
YFP fusions, construction and fluorescence microscopy
Yellow fluorescent protein (YFP) and cyan fluorescent protein (CFP) fusions of the proteins (Sml1 variants, Rnr1, Rnr2 and Rnr3) were made by the cloning-free PCR-based allele replacement method and are at the corresponding chromosomal loci (40). All fusions are to the N-terminal end of the proteins and are separated by an eight-alanine linker. Cells were processed for differential interference contrast (DIC) and fluorescence microscopy as described previously (41) and fluorescence was quantified using Openlab software (Improvision).
Two-hybrid construction and testing
Sml1 was cloned using primers BamHI_Sml1 and Sml1_PstI. Ubr2 was cloned using SmaI_Ubr2 and Ubr2_PstI. Mub1 was cloned using SmaI_Mub1 and Mub1_PstI. All primer sequences are available upon request. PCR products from the Sml1 PCR were cut with BamHI and PstI as inserted into pGAD-C2 and pGBD-C2 (42). Ubr2 and Mub1 PCR products were digested with SmaI and PstI and inserted into pGAD-C1 and pGBD-C1 (42). All pGAD derived plasmids, including pGAD-C1, were transformed into PJ69-4A and all pGBD-derived plasmids, including pGBD-C1, were transformed into PJ69-4α (42). Strains containing pGBD-Sml1 and pGBD-Ubr2 were not further tested due to autoactivation of the reporters. Strains containing pGBD-C1, and pGBD-Mub1 were mated with strains containing pGAD-C1, pGAD-Sml1 and pGAD-Ubr2. Diploids were grown up overnight in medium lacking LEU and TRP to select for plasmids. Strains were diluted to an optical density (OD600) = 1.0, were serially diluted 5-fold and spotted on to -LEU–TRP, and -LEU–TRP–HIS plates. Plates were scanned after 4 days of growth.
RESULTS
Multiple serine to alanine changes in Sml1 stabilize the protein after DNA damage treatment and prevent in vivo and in vitro phosphorylation
Previously, we showed that the degradation of Sml1 after DNA damage treatment depends on the MEC1/RAD53/DUN1 checkpoint pathway and correlates with the phosphorylation of Sml1 (21). In addition, Dun1, a serine/threonine kinase (43), phosphorylates Sml1 in vitro (22). Since there are 13 serines and 4 threonines in the 104 amino acids Sml1 protein (Figure 1A), we used site-directed mutagenesis to identify potential phosphorylation sites in Sml1 that would prevent Sml1 degradation following DNA damage treatment. Notably, three serines (56, 58 and 60) were found by Dealwis and colleagues (23) to be phosphorylated by Dun1 in vitro. Although mutation of these three serines to alanines, referred to as sml1-3SA, results in a stable protein as determined by fluorescent microscopy (Figure 1B), this protein still undergoes a mobility shift following DNA damage treatment, likely resulting from phosphorylation (Figure 1C). Therefore, Sml1 mutants containing different combinations of serine to alanine changes were examined by fluorescence microscopy and immunoblot for their stability and phosphorylation state following γ-irradiation (data not shown). While many of the mutants are stable, it was necessary to change four serines (56, 58, 60 and 61) to alanines to block any detectable phosphorylation of the protein, indicated by a mobility shift on the gel (Figure 1C). Henceforth, we call this mutant sml1-4SA because four serines were changed to alanines.
Protein levels and localization of Sml1 and sml1-4SA were analyzed by visualizing the fluorescence of YFP-tagged fusion proteins. In unirradiated cells, wild-type Sml1 levels are lower during S phase, while sml1-4SA levels remain unaltered throughout the cell cycle (Figure 1B, arrows). Furthermore, as shown in the right panel of Figure 1B and in Figure 1C, wild-type Sml1 protein is completely degraded within an hour after γ-irradiation, while the sml1-4SA mutant protein is stable. A similar result was observed on immunoblots of untagged proteins using anti-Sml1 serum (Figure 1D). YFP-Sml1 and YFP-sml1-4SA show the same dispersed cytoplasmic localization. Using anti-GFP antibody as well as fluorescence intensity for quantification, we find that the levels of YFP-sml1-4SA are 2- to 2.5-fold higher compared to the levels of YFP-Sml1 in exponentially growing cultures, indicating that YFP-sml1-4SA is present at higher levels in the cell.
Next, we analyzed the in vivo phosphorylation state of untagged sml1-4SA in response to a variety of DNA damaging agents in the presence of RNR1 overexpression, which facilitates detection of phosphorylated Sml1 on an immunoblot (21). In wild-type cells, DNA damage treatment results in the appearance of phosphorylated species of Sml1 observed as a band or bands with reduced electrophoretic mobility (Figure 1D, top) (21). No such bands are seen in the sml1-4SA mutant, demonstrating the absence of any detectable in vivo phosphorylation of the sml1-4SA protein (Figure 1D, bottom).
The increased stability and undetectable phosphorylated protein in the sml1-4SA strain following DNA damage is reminiscent of Sml1 behavior in a dun1Δ mutant. Therefore, we measured the phosphorylation of the sml1-4SA mutant protein by Dun1 in vitro. While GST-Dun1 purified from total yeast extracts phosphorylates wild-type Sml1 in vitro (22), it does not detectably phosphorylate sml1-4SA mutant protein (Figure 1E). The in vitro phosphorylation experiment is consistent with the in vivo phosphorylation data suggesting that the mutations in sml1-4SA prevent phosphorylation by Dun1.
Since we find no detectable growth defect in a sml1-4SA strain (Figure 2A), we investigated the effect of overexpressing the non-degradable sml1-4SA protein on the cell cycle after release from G1 arrest. We confirmed that overexpression of wild-type Sml1 does not affect entry into or progression through the cell cycle (Figure 2B, first panel and ref. (21)). In contrast, overexpression of sml1-4SA delays the start of and extends the progression of S phase (Figure 2B, second panel). At 25 min post-release, most of the cells in the GAL-SML1 strain have started DNA replication and by 50 min the majority of the DNA is replicated. Comparing the same time points in the GAL-sml1-4SA strain shows that while some DNA replication has initiated at 25 min, the majority is delayed ∼25 min and initiation occurs at the 50-min time point.
Taken together, these results show that changing the four serines identified in our analyses to alanines likely abolishes Sml1 phosphorylation by Dun1 both in vivo and in vitro. Furthermore, the stability of the non-phosphorylatable mutant sml1-4SA indicates that phosphorylation of Sml1 is essential to target the protein for degradation after DNA damage.
Other aspects of dNTP regulation and checkpoint activation are functional in sml1-4SA
While there are many facets to RNR regulation, their relationships are not well understood. Therefore, we investigated the induction of Rnr1 as well as the relocalization of the small Rnr2 subunit from the nucleus to the cytoplasm when Sml1 phosphorylation is blocked. Using fluorescently tagged proteins, Sml1 and Rnr1 levels were analyzed before and after γ-irradiation. Both Sml1 and Rnr1 levels fluctuate throughout the cell cycle in unirradiated cells. After irradiation, all of the cells are comparably arrested in G2/M. To eliminate the variability that occurs between different cell-cycle stages in the unirradiated samples, we only measured fluorescence in G1 cells. As previously shown in Figure 1B, YFP-Sml1 disappears after γ-irradiation while YFP-sml1-4SA remains stable (Figure 3A). It was previously reported that Rnr1 protein levels increase about 2-fold following DNA damage treatment (7). We also see a 2-fold increase in endogenously tagged CFP-Rnr1 levels in wild-type cells treated with γ-irradiation (Figure 3A; bars 2 and 4). Similarly, Rnr1 levels increase following DNA damage treatment in sml1-4SA cells (Figure 2A; bars 6 and 8). These results suggest that Rnr1 transcriptional regulation is still intact in the sml1-4SA strain. Furthermore, in unirradiated G1 cells, Rnr1 levels are increased in the sml1-4SA strain compared to wild type (Figure 3A; bars 2 and 6; P < 0.001). We suggest that this increase compensates for the higher Sml1 levels in this strain during unperturbed growth and after DNA damage treatment.
In response to DNA damage treatment, Rad53 is phosphorylated (44) and Rnr2 and Rnr4 move from the nucleus to the cytoplasm (12). We examined these upstream and downstream events in both wild-type and sml1-4SA cells after γ-irradiation. In contrast to untreated cells, Rad53 is phosphorylated following MMS treatment in both wild-type and sml1-4SA cells (Figure 3B). Thus, checkpoint activation appears normal even in the absence of Sml1 degradation. Additionally, when we examine a downstream event, we find that CFP-tagged Rnr2 moves from the nucleus to the cytoplasm in both wild-type and sml1-4SA cells after γ-irradiation (data not shown and Figure 3C). This result demonstrates that blocking phosphorylation and degradation of Sml1 does not affect RNR relocalization. Altogether, these experiments indicate that the relative stability of the Sml1 protein does not affect these other aspects of the DNA damage response.
Sml1 protein down-regulation is necessary for survival of mutants compromised in dNTP regulation
Unlike a dun1Δ strain, which is also defective in Sml1 degradation, a sml1-4SA strain is not sensitive to DNA damaging agents (data not shown). We suspect that the sml1-4SA mutant does not show increased DNA damage sensitivity because other aspects of the DNA damage response and of RNR regulation remain functional as shown in Figure 3. To explore this question further, sml1-4SA was combined with a mutation that is defective in dNTP regulation: rnr1-W688G. We chose the rnr1-W688G strain because it is particularly sensitive to Sml1 regulation. This allele was isolated based on its ability to physically interact with sml1 mutants that do not bind to wild-type Rnr1 (45). Since rnr1-W688G can also interact with wild-type Sml1, the mutation may cause a stronger interaction between Sml1 and Rnr1. In support to this view, rnr1-W688G causes increased sensitivity to DNA damaging agents, which can be suppressed by deleting SML1 (Figure 4A). In addition, rnr1-W688G most likely leads to endogenous DNA damage, indicated by the constitutive expression of YFP-Rnr3 in these cells (Figure 4B).
Next, we analyzed the spores from a cross between an rnr1-W688G strain and a sml1-4SA strain to test the genetic interaction between stabilized Sml1 and this sensitizing mutation. Genetic analysis of this diploid shows that spores of the genotype rnr1-W688G sml1-4SA are synthetic lethal (Figure 4C, left). This interaction is identical to that seen when the rnr1-W688G allele is combined with dun1Δ (Figure 4C, right), which is also defective for Sml1 degradation (22) as well as RNR regulation (12,43). Furthermore, deletion of SML1 suppresses the synthetic lethality between rnr1-W688G and dun1Δ (Figure 4C, right). Interestingly, the levels of YFP-Sml1 fluorescent protein in an unperturbed rnr1-W688G strain are undetectable (data not shown), also indicating that there is a constitutively active checkpoint in this strain. However, even the low levels of Sml1 in an rnr1-W688G strain must account for some dNTP inhibition, as deletion of SML1 rescues the severe petite phenotype, indicative of low dNTP pools (18) observed in an rnr1-W688G strain (data not shown). Taken together, these results show that the degradation of Sml1 becomes essential in the rnr1-W688G strain, likely due to the aberrant regulation of this sensitized RNR subunit.
To more closely inspect the effects of expressing Sml1 and sml1-4SA in the rnr1-W688G strain, both genes were placed under the control of a conditional galactose promoter. Transient overexpression of either Sml1 or sml1-4SA in the rnr1-W688G mutant has a dramatic effect on growth. For example, the rnr1-W688G strain is sensitive to increased levels of wild-type Sml1 since entry into S phase is significantly delayed (Figure 4D, first panel; Figure 2B). However, these cells do resume the cell cycle and continue to grow. On the other hand, overexpression of sml1-4SA in the rnr1-W688G strain completely blocks entry into S phase after release from G1 (Figure 4D, second panel). This result provides an explanation for the lethality of the rnr1-W688G sml1-4SA double mutant shown in Figure 4C, namely, it is unable to progress through S phase.
Sml1 degradation is dependent on the 26S proteasome as well as the RAD6–UBR2–MUB1 ubiquitin ligase complex
Phosphorylation is often a signal for substrates targeted for degradation by the 26S proteasome via ubiquitylation (46,47). To determine whether Sml1 degradation is dependent on the 26S proteasome, Sml1 stability after DNA damage treatment was examined following inactivation of two essential genes of the proteasome (48,49). Figure 5A shows that Sml1 protein degradation is impaired in the temperature-sensitive mutants, pre1-1 or pre2-2, at the restrictive temperature (50).
To determine the ubiquitin ligase(s) responsible for the ubiquitylation of Sml1, we introduced a YFP-Sml1 plasmid into all 36 of the known non-essential E3 ubiquitin ligases (Supplementary Table S2). Sml1 stability was examined by fluorescent microscopy following 100 Gy of γ-irradiation and YFP-Sml1 was only stable in the ubr2Δ strain (data not shown). Ubr2 has previously been shown to interact with the Rad6 E2 ubiquitin conjugating enzyme and with Mub1, an E2/E3 interacting protein (35,36). This complex ubiquitylates Rpn4, a transcription factor involved in the biosynthesis of proteasome components (35,36). Therefore, we introduced a YFP-Sml1 plasmid into rad6Δ and mub1Δ strains and found that YFP-Sml1 is also stable following DNA damage treatment. Notably, stabilization of YFP-Sml1 following 100 Gy of γ-irradiation is not seen in deletions of any of the other six known non-essential E2s (Supplementary Table S2) (data not shown).
To avoid potential problems associated with plasmid-based expression of YFP-Sml1, a genomic copy of YFP-Sml1 was introduced into the rad6Δ, ubr2Δ and mub1Δ genetic backgrounds. In addition, Ubr1 is a known interactor with Rad6 (30) and shows homology with Ubr2 (34). Therefore, we also introduced a genomic copy of YFP-Sml1 into a ubr1Δ strain. Fluorescent protein levels were examined in all strains before and after DNA damage treatment. As shown in Figure 5B, YFP-Sml1 is degraded within 40 min following treatment with 100 Gy of γ-irradiation in both wild-type and ubr1Δ strains, but is stable in rad6Δ, ubr2Δ, and mub1Δ strains. Furthermore, endogenous levels of Sml1 are higher in the rad6Δ, ubr2Δ, and mub1Δ mutants, even in the absence of DNA damage. Next, YFP-Sml1 protein from these strains was examined by immunoblot following γ-irradiation (Figure 5C). In wild-type cells at 20 min post damage, a slower migrating band appears, which is consistent with phosphorylation of the protein (Figure 1C) and this band is degraded at 45 min post-irradiation. Similar results are also observed in a ubr1Δ mutant. Interestingly, in rad6Δ, ubr2Δ and mub1Δ mutants, this slower migrating band is seen even in the absence of DNA damage treatment and accumulates post-DNA damage with no noticeable degradation. Next, the non-phosphorylatable YFP-sml1-4SA was introduced into the rad6Δ, ubr2Δ, mub1Δ, ubr1Δ mutants and a wild-type strain and immunoblots were performed following DNA damage treatment. As seen in Figure 5D, the slower migrating band observed in Figure 5C is absent, consistent with the notion that this is the phosphorylated form of Sml1. In addition, as expected (Figure 1D), the YFP-sml1-4SA protein is stable following DNA damage treatment in all strains. Finally, untagged Sml1 is more stable in rad6Δ, ubr2Δ and mub1Δ strains following 100 Gy of γ-irradiation compared to wild-type and ubr1Δ strains (Supplementary Figure S1).
Mub1 has been implicated in the substrate specificity of the E2/E3 complex during the ubiquitylation of Rpn4 (36). Using a two-hybrid approach, we investigated the interactions between Ubr2, Mub1 and Sml1. Unfortunately, GBD-Sml1 and GBD-Ubr2 show non-specific interactions with an empty GAD construct and could not be tested further (data not shown). On the other hand, GBD-Mub1 shows an interaction with GAD-Ubr2 confirming results found previously (36). Interestingly, GBD-Mub1 also interacts with GAD-Sml1 indicating a direct link between these two proteins (Figure 5E).
DISCUSSION
Sml1, a regulator of RNR, is phosphorylated and degraded in response to DNA damage treatment (22). Although the precise serine residue(s) that is/are phosphorylated is not known, we show that it is necessary to change four serines to alanines in the Sml1 protein to eliminate any detectable phosphorylation in vivo (Figure 1C and data not shown). There are numerous examples of phosphorylation at multiple sites to control protein stability. The detailed analysis of Sic1, a cell-cycle regulator and a substrate of the SCF Cdc4 complex, showed that phosphorylation at any six, but not five, of the nine possible phosphorylation sites, targets it for ubiquitylation and degradation (51). In another example, mutation of six phospho-acceptor residues in FANCI is necessary to abolish its phosphorylation, monoubiquitylation, focus formation and DNA repair activity (52). Thus, the number of phosphorylated residues, rather than their position, is often more important for targeting a protein for degradation. Furthermore, the amino acid sequence at the site that we identified in Sml1 contains multiple serines in close proximity (SASASS), making it unlikely that phosphorylation at a particular site is important. Recently, a small cytoplasmic protein, Dif1, was shown to be required for Rnr2 import into the nucleus (16,17). Interestingly, Dif1 shares homology with Sml1 at three of the four serines that are mutated in sml1-4SA (56, 60 and 61). Since Dif1 is also phosphorylated and degraded in a Dun1-dependent manner, perhaps it shares a similar mechanism for degradation with Sml1.
The serine to alanine mutations in sml1-4SA completely block detectable phosphorylation and degradation of the protein. However, the non-degradable protein does not alter cell survival or resistance to DNA damage unless sml1-4SA is combined with a mutation that further impairs RNR activity (Figure 3C). Our results confirm that dNTP regulation is robust and there is a discernible biological effect only when multiple components of this regulation are eliminated. For example, a dun1Δ strain is DNA damage sensitive, since it affects dNTP regulation not only by preventing Sml1 degradation, but also by affecting induction of the RNR genes, as well as the relocalization of the R2 subunit. However, in a sml1-4SA strain, only sml1-4SA stability is affected, while other aspects of RNR regulation are normal (Figure 3). In addition, there is a small but significant increase in Rnr1 levels in a sml1-4SA strain compared to wild type (Figure 3A, bars 2 and 6; P < 0.001). We suggest that cells expressing the non-degradable sml1-4SA compensate for low dNTP pools by increasing RNR1 transcription.
We show that the Sml1 protein is degraded via the 26S proteasome (Figure 5A). Screening the 36 known non-essential E3 enzymes, we found that Ubr2 is required for Sml1 degradation (Figure 5B). Ubr2 ubiquitylates the transcription factor, Rpn4, in concert with the Rad6 E2 ligase and the Mub1 proteins (35,36), and both of these proteins are also important for Sml1 degradation (Figure 5). Since Rpn4 is involved in the regulation of proteasomal genes (53), its stability in ubr2Δ, rad6Δ and/or mub1Δ mutants could potentially lead to an increase in proteasomes. Up-regulation of proteasomal subunits due to increased Rpn4 levels would in fact destabilize Sml1, making it unlikely that these mutants are having an indirect effect on Sml1 stability. Furthermore, the two-hybrid experiment demonstrates a direct interaction between Mub1 and Sml1 (Figure 5D). Therefore, Ubr2, Rad6 and Mub1 likely ubiquitylate Sml1 directly, leading to its degradation in response to DNA damage. This E2/E3 complex may also be involved in the turnover of Sml1 protein during S phase, since increased levels of phosphorylated Sml are observed in ubr2Δ, rad6Δ and mub1Δ strains even without DNA damage treatment (Figure 5C).
Although Rad6, in conjunction with several E3 ubiquitin ligases, has many targets in the DNA damage pathway including PCNA and members of the 9-1-1 complex (29–33), this is the first interaction, to our knowledge, that also requires the Ubr2/Mub1 proteins. Sml1 protein localizes to the cytoplasm where it is bound to Rnr1 (Figure 1B and (12)). Ubr2 is also found in the cytoplasm (54), perhaps facilitating Sml1 ubiquitylation. It will be important to determine whether the Rad6–Ubr2–Mub1 complex ubiquitylates other DNA damage-regulated proteins. Of particular interest is Dif1, another cytoplasmically localized protein, which shares homology with Sml1 and is involved in dNTP regulation (16,17).
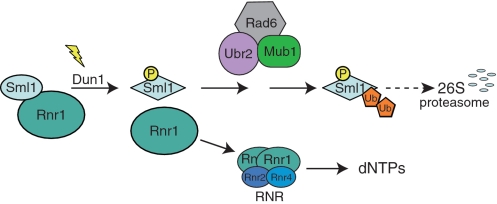
Taken together, our results can be summarized in a model describing Sml1 modifications that target the protein for degradation in response to DNA damage (Figure 6). In this model, before DNA damage, Sml1 is bound to Rnr1, inhibiting RNR activity. Following DNA damage, the Mec1/Rad53/Dun1 kinase cascade is activated and phosphorylates Sml1. This phosphorylation triggers a conformational change in Sml1 leading to its dissociation from Rnr1. Phosphorylated Sml1 is recognized by the Rad6–Ubr2–Mub1 E2/E3 ligase complex, which ubiquitylates Sml1 targeting it for degradation by the 26S proteasome. In the end, loss of Rnr1 inhibition after Sml1 degradation allows the formation of an active RNR enzyme leading to an increase in the production of dNTPs to facilitate DNA damage repair.
Figure 6.
A model for Sml1 regulation in response to DNA damage. Prior to modification, Sml1 is bound to Rnr1 and inhibits RNR activity. Following DNA damage, Dun1 is activated and phosphorylates Sml1 on serines 56, 58, 60 and/or 61. Phosphorylation promotes ubiquitylation of Sml1 by the Rad6–Ubr2–Mub1 complex, targeting Sml1 for degradation by the 26S proteasome. The released Rnr1 associates with Rnr2 and Rnr4 to form an active RNR enzyme allowing the production of dNTPs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: National Institutes of Health (GM50237).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Kara Bernstein, Rebecca Burgess, Michael Lisby, Peter Thorpe and Xiaolan Zhao for thoughtful comments on the manuscript. We also thank Mark Hochstrasser for strains.
REFERENCES
- 1.Chabes A, Georgieva B, Domkin V, Zhao X, Rothstein R, Thelander L. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell. 2003;112:391–401. doi: 10.1016/s0092-8674(03)00075-8. [DOI] [PubMed] [Google Scholar]
- 2.Chabes A, Stillman B. Constitutively high dNTP concentration inhibits cell cycle progression and the DNA damage checkpoint in yeast Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2007;104:1183–1188. doi: 10.1073/pnas.0610585104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reichard P. Interactions between deoxyribonucleotide and DNA synthesis. Annu. Rev. Biochem. 1988;57:349–374. doi: 10.1146/annurev.bi.57.070188.002025. [DOI] [PubMed] [Google Scholar]
- 4.Elledge SJ, Davis RW. Two genes differentially regulated in the cell cycle and by DNA-damaging agents encode alternative regulatory subunits of ribonucleotide reductase. Genes Dev. 1990;4:740–751. doi: 10.1101/gad.4.5.740. [DOI] [PubMed] [Google Scholar]
- 5.Elledge SJ, Davis RW. Identification and isolation of the gene encoding the small subunit of ribonucleotide reductase from Saccharomyces cerevisiae: DNA damage-inducible gene required for mitotic viability. Mol. Cell. Biol. 1987;7:2783–2793. doi: 10.1128/mcb.7.8.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang M, Elledge SJ. Identification of RNR4, encoding a second essential small subunit of ribonucleotide reductase in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997;17:6105–6113. doi: 10.1128/mcb.17.10.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Domkin V, Thelander L, Chabes A. Yeast DNA damage-inducible Rnr3 has a very low catalytic activity strongly stimulated after the formation of a cross-talking Rnr1/Rnr3 complex. J. Biol. Chem. 2002;277:18574–18578. doi: 10.1074/jbc.M201553200. [DOI] [PubMed] [Google Scholar]
- 8.Shen C, Lancaster CS, Shi B, Guo H, Thimmaiah P, Bjornsti MA. TOR signaling is a determinant of cell survival in response to DNA damage. Mol. Cell. Biol. 2007;27:7007–7017. doi: 10.1128/MCB.00290-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thelander L, Reichard P. Reduction of ribonucleotides. Annu. Rev. Biochem. 1979;48:133–158. doi: 10.1146/annurev.bi.48.070179.001025. [DOI] [PubMed] [Google Scholar]
- 10.Nyberg KA, Michelson RJ, Putnam CW, Weinert TA. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 2002;36:617–656. doi: 10.1146/annurev.genet.36.060402.113540. [DOI] [PubMed] [Google Scholar]
- 11.Wang PJ, Chabes A, Casagrande R, Tian XC, Thelander L, Huffaker TC. Rnr4p, a novel ribonucleotide reductase small-subunit protein. Mol. Cell. Biol. 1997;17:6114–6121. doi: 10.1128/mcb.17.10.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yao R, Zhang Z, An X, Bucci B, Perlstein DL, Stubbe J, Huang M. Subcellular localization of yeast ribonucleotide reductase regulated by the DNA replication and damage checkpoint pathways. Proc. Natl Acad. Sci. USA. 2003;100:6628–6633. doi: 10.1073/pnas.1131932100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.An X, Zhang Z, Yang K, Huang M. Cotransport of the heterodimeric small subunit of the Saccharomyces cerevisiae ribonucleotide reductase between the nucleus and the cytoplasm. Genetics. 2006;173:63–73. doi: 10.1534/genetics.105.055236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee YD, Elledge SJ. Control of ribonucleotide reductase localization through an anchoring mechanism involving Wtm1. Genes Dev. 2006;20:334–344. doi: 10.1101/gad.1380506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Z, An X, Yang K, Perlstein DL, Hicks L, Kelleher N, Stubbe J, Huang M. Nuclear localization of the Saccharomyces cerevisiae ribonucleotide reductase small subunit requires a karyopherin and a WD40 repeat protein. Proc. Natl Acad. Sci. USA. 2006;103:1422–1427. doi: 10.1073/pnas.0510516103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee YD, Wang J, Stubbe J, Elledge SJ. Dif1 is a DNA-damage-regulated facilitator of nuclear import for ribonucleotide reductase. Mol. Cell. 2008;32:70–80. doi: 10.1016/j.molcel.2008.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu X, Huang M. Dif1 controls subcellular localization of ribonucleotide reductase by mediating nuclear import of the R2 subunit. Mol. Cell. Biol. 2008;28:7156–7167. doi: 10.1128/MCB.01388-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- 19.Chabes A, Domkin V, Thelander L. Yeast Sml1, a protein inhibitor of ribonucleotide reductase. J. Biol. Chem. 1999;274:36679–36683. doi: 10.1074/jbc.274.51.36679. [DOI] [PubMed] [Google Scholar]
- 20.Zhao X, Georgieva B, Chabes A, Domkin V, Ippel JH, Schleucher J, Wijmenga S, Thelander L, Rothstein R. Mutational and structural analyses of the ribonucleotide reductase inhibitor Sml1 define its Rnr1 interaction domain whose inactivation allows suppression of mec1 and rad53 lethality. Mol. Cell. Biol. 2000;20:9076–9083. doi: 10.1128/mcb.20.23.9076-9083.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao X, Chabes A, Domkin V, Thelander L, Rothstein R. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J. 2001;20:3544–3553. doi: 10.1093/emboj/20.13.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao X, Rothstein R. The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc. Natl Acad. Sci. USA. 2002;99:3746–3751. doi: 10.1073/pnas.062502299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uchiki T, Dice LT, Hettich RL, Dealwis C. Identification of phosphorylation sites on the yeast ribonucleotide reductase inhibitor Sml1. J. Biol. Chem. 2004;279:11293–11303. doi: 10.1074/jbc.M309751200. [DOI] [PubMed] [Google Scholar]
- 24.Torres-Rosell J, Sunjevaric I, De Piccoli G, Sacher M, Eckert-Boulet N, Reid R, Jentsch S, Rothstein R, Aragon L, Lisby M. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat. Cell Biol. 2007;9:923–931. doi: 10.1038/ncb1619. [DOI] [PubMed] [Google Scholar]
- 25.Barlow JH, Lisby M, Rothstein R. Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol. Cell. 2008;30:73–85. doi: 10.1016/j.molcel.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hofmann K. Ubiquitin-binding domains and their role in the DNA damage response. DNA Repair. 2009;8:544–556. doi: 10.1016/j.dnarep.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 27.Xu P, Peng J. Dissecting the ubiquitin pathway by mass spectrometry. Biochim. Biophys. Acta. 2006;1764:1940–1947. doi: 10.1016/j.bbapap.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 29.Bailly V, Lamb J, Sung P, Prakash S, Prakash L. Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev. 1994;8:811–820. doi: 10.1101/gad.8.7.811. [DOI] [PubMed] [Google Scholar]
- 30.Dohmen RJ, Madura K, Bartel B, Varshavsky A. The N-end rule is mediated by the UBC2(RAD6) ubiquitin-conjugating enzyme. Proc. Natl Acad. Sci. USA. 1991;88:7351–7355. doi: 10.1073/pnas.88.16.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, Dean K, Golshani A, Zhang Y, Greenblatt JF, et al. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell. 2003;11:267–274. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- 32.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 33.Fu Y, Zhu Y, Zhang K, Yeung M, Durocher D, Xiao W. Rad6-Rad18 mediates a eukaryotic SOS response by ubiquitinating the 9-1-1 checkpoint clamp. Cell. 2008;133:601–611. doi: 10.1016/j.cell.2008.02.050. [DOI] [PubMed] [Google Scholar]
- 34.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 35.Wang L, Mao X, Ju D, Xie Y. Rpn4 is a physiological substrate of the Ubr2 ubiquitin ligase. J. Biol. Chem. 2004;279:55218–55223. doi: 10.1074/jbc.M410085200. [DOI] [PubMed] [Google Scholar]
- 36.Ju D, Wang X, Xu H, Xie Y. Genome-wide analysis identifies MYND-domain protein Mub1 as an essential factor for Rpn4 ubiquitylation. Mol. Cell. Biol. 2008;28:1404–1412. doi: 10.1128/MCB.01787-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Erdeniz N, Mortensen UH, Rothstein R. Cloning-free PCR-based allele replacement methods. Genome Res. 1997;7:1174–1183. doi: 10.1101/gr.7.12.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams A, Gottschling D, Kaiser C, Stearns T. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1997. [Google Scholar]
- 39.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Reid RJD, Lisby M, Rothstein R. Cloning-free genome alterations in Saccharomyces cerevisiae using adaptamer-mediated PCR. Meth. Enzymol. 2002;350:258–277. doi: 10.1016/s0076-6879(02)50968-x. [DOI] [PubMed] [Google Scholar]
- 41.Lisby M, Rothstein R, Mortensen UH. Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl Acad. Sci. USA. 2001;98:8276–8282. doi: 10.1073/pnas.121006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Z, Elledge SJ. DUN1 encodes a protein kinase that controls the DNA damage response in yeast. Cell. 1993;75:1119–1127. doi: 10.1016/0092-8674(93)90321-g. [DOI] [PubMed] [Google Scholar]
- 44.Pellicioli A, Lucca C, Liberi G, Marini F, Lopes M, Plevani P, Romano A, Di Fiore PP, Foiani M. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 1999;18:6561–6572. doi: 10.1093/emboj/18.22.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Georgieva B, Zhao X, Rothstein R. Damage response and dNTP regulation: the interaction between ribonucleotide reductase and its inhibitor, Sml1. Cold Spring Harb. Symp. Quant. Biol. 2000;65:343–346. doi: 10.1101/sqb.2000.65.343. [DOI] [PubMed] [Google Scholar]
- 46.Ciechanover A, Finley D, Varshavsky A. The ubiquitin-mediated proteolytic pathway and mechanisms of energy-dependent intracellular protein degradation. J. Cell. Biochem. 1984;24:27–53. doi: 10.1002/jcb.240240104. [DOI] [PubMed] [Google Scholar]
- 47.Hershko A, Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 48.Heinemeyer W, Simeon A, Hirsch HH, Schiffer HH, Teichert U, Wolf DH. Lysosomal and non-lysosomal proteolysis in the eukaryotic cell: studies on yeast. Biochem. Soc. Trans. 1991;19:724–725. doi: 10.1042/bst0190724. [DOI] [PubMed] [Google Scholar]
- 49.Heinemeyer W, Gruhler A, Mohrle V, Mahe Y, Wolf DH. PRE2, highly homologous to the human major histocompatibility complex-linked RING10 gene, codes for a yeast proteasome subunit necessary for chrymotryptic activity and degradation of ubiquitinated proteins. J. Biol. Chem. 1993;268:5115–5120. [PubMed] [Google Scholar]
- 50.Richter-Ruoff B, Wolf DH, Hochstrasser M. Degradation of the yeast MAT alpha 2 transcriptional regulator is mediated by the proteasome. FEBS Lett. 1994;354:50–52. doi: 10.1016/0014-5793(94)01085-4. [DOI] [PubMed] [Google Scholar]
- 51.Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- 52.Ishiai M, Kitao H, Smogorzewska A, Tomida J, Kinomura A, Uchida E, Saberi A, Kinoshita E, Kinoshita-Kikuta E, Koike T, et al. FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nat. Struct. Mol. Biol. 2008;15:1138–1146. doi: 10.1038/nsmb.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mannhaupt G, Schnall R, Karpov V, Vetter I, Feldmann H. Rpn4p acts as a transcription factor by binding to PACE, a nonamer box found upstream of 26S proteasomal and other genes in yeast. FEBS Lett. 1999;450:27–34. doi: 10.1016/s0014-5793(99)00467-6. [DOI] [PubMed] [Google Scholar]
- 54.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 55.Allen JB, Zhou Z, Siede W, Friedberg EC, Elledge SJ. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 1994;8:2401–2415. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.