Abstract
Multiple KH-domain proteins, collectively known as vigilins, are evolutionarily highly conserved proteins that are present in eukaryotic organisms from yeast to metazoa. Proposed roles for vigilins include chromosome segregation, messenger RNA (mRNA) metabolism, translation and tRNA transport. As a step toward understanding its biological function, we have identified the fission yeast vigilin, designated Vgl1, and have investigated its role in cellular response to environmental stress. Unlike its counterpart in Saccharomyces cerevisiae, we found no indication that Vgl1 is required for the maintenance of cell ploidy in Schizosaccharomyces pombe. Instead, Vgl1 is required for cell survival under thermal stress, and vgl1Δ mutants lose their viability more rapidly than wild-type cells when incubated at high temperature. As for Scp160 in S. cerevisiae, Vgl1 bound polysomes accumulated at endoplasmic reticulum (ER) but in a microtubule-independent manner. Under thermal stress, Vgl1 is rapidly relocalized from the ER to cytoplasmic foci that are distinct from P-bodies but contain stress granule markers such as poly(A)-binding protein and components of the translation initiation factor eIF3. Together, these observations demonstrated in S. pombe the presence of RNA granules with similar composition as mammalian stress granules and identified Vgl1 as a novel component that required for cell survival under thermal stress.
INTRODUCTION
RNA localization and spatial restriction of translation are important processes for gene regulation (1), and defects in these processes can contribute to diseases (2). Many of these processes appear to be regulated by RNA-binding proteins containing K homology (KH) domains. The KH motif was first biochemically characterized in the major pre-mRNA-binding protein K (heterogeneous nuclear ribonucleoprotein K, hnRNP K) and described as a 45-amino-acid repeat detected by sequence similarity in a number of RNA-binding proteins (3). Clinically significant KH-domain proteins include the FMR protein (2), which is involved in fragile X syndrome, the major cause of heritable human mental retardation, and Nova-1, which is important in the motor control disorder paraneoplastic opsoclonus-ataxia (4).
KH motifs can occur in multiple copies. In Saccharomyces cerevisiae Scp160, a multi KH-domain protein, has been identified (5). Scp160 has been shown to be associated with polysomes (6,7) and is in close proximity to translation elongation factor 1A and the WD-repeat protein Asc1 (8). Subcellular localization studies, using both immunofluorescence and green fluorescence protein-tagged alleles, have demonstrated that the majority of Scp160 is cytoplasmic, with significant enrichment around the nuclear envelope and rough endoplasmic reticulum (ER) (5,6), consistent with the distribution of polysomes in yeast. Although Scp160 is dispensable for growth, scp160Δ mutants display defects in cell morphology and nuclear segregation, resulting in cells with increased size and DNA content (5). In addition, loss or mutation of EAP1, which encodes an eIF4E binding protein involved in both translation initiation and spindle pole body functions, is synthetically lethal with loss of SCP160 (9). In line with these data, Scp160 was identified as a part of the Smy2–Eap1–Scp160–Asc1 (SESA) network of proteins that link duplication of the yeast centrosome with the protein translation machinery (10). Finally, Guo et al. (11) recently identified Scp160 as a potential effector of Gα-mediated signal transduction in yeast, although the mechanism and the extent of this function remain unclear.
In Drosophila, a functional homolog of Scp160 has also been identified. This protein, DDP1, binds dodeca satellite repeat regions of centromeric heterochromatin in embryonic and larval cell nuclei, and contributes to centromeric silencing and chromosome segregation (12,13). Overexpression of the DDP1 protein complements the cell morphology and nuclear segregation defects in scp160Δ mutants (12) and, consistent with the function of the Drosophila ortholog, the contribution of Scp160 to silencing at telomeres and the mating-type locus has been described (14). Similar multi-KH-domain proteins are found ubiquitously in all eukaryotic cells. In vertebrate species this protein is known as vigilin (15–17). A clear picture of the cellular function and of specific RNA targets of these proteins has not yet emerged. In Xenopus, vigilin binds specifically to the 3′-untranslated region of vitellogenin mRNA, stabilizing the RNA by blocking cleavage by an endonuclease (18). In contrast to these results, human vigilin was proposed to be involved in binding and transport of tRNA (19).
In this study, we describe the characterization of fission yeast vigilin, designated Vgl1, and investigate its role in the cellular response to environmental stress. We demonstrated that Vgl1 is a novel component of the fission yeast stress granules that required for cell survival under thermal stress.
MATERIALS AND METHODS
Fission yeast strains and methods
Conditions for growth, maintenance, and genetic manipulation of fission yeast were as described previously (20). A complete list of the strains used in this study is given in Table 1. One-step gene disruption or modification via homologous recombination was performed following polymerase chain reaction (PCR)-mediated generation of ura4+ or KanMX selectable cassettes flanked by 80-bp segments from appropriate regions of vgl1+ using oligonucleotides described in Table 2. The original hht2-GFP, GFP-scp160 (6) and sec72-GFP-HA (21) strains were gifts from Drs MK Balasubramanian, M. Seedorf and Yeast Genetic Resource Center Japan (YGRC/NBRP), respectively. Except where otherwise stated, strains were grown at 30°C in yeast extract (YE) or Edinburgh Minimal Medium (EMM2) with appropriate supplements.
Table 1.
Schizosaccharomyces pombe strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| 972 | h− | Lab. stock |
| pop1Δ | h− pop1::kanr | (25) |
| SG01 | h− vgl1::ura4+ ura4 | This study |
| SG02 | h− vgl1-tdTomato::kanr | This study |
| SG03 | h− vgl1-GFP::kanr leu1 ura4 | This study |
| SG04 | h− vgl1-TAP::kanr ade6 leu1-32 ura4 | This study |
| SG05 | h− vgl1-tdTomato::kanr hht2-GFP::ura4+ ura4 | This study |
| SG06 | h− vgl1-tdTomato::kanr Sec72-GFP-HA:: kanr | This study |
| SG07 | h− vgl1-tdTomato::kanr adf1::[adh-GFP-adf1-ura4+] leu1-32 ura4 | This study |
| SG08 | h− vgl1-tdTomato::kanr kanMx6-p81-GFP-atb2+ leu1-32 | This study |
| SG09 | h− vgl1-TAP::kanr Sec72-GFP-HA:: kanr | This study |
| SG10 | h− dcp2-tdTomato::kanr vgl1-GFP:: kanr | This study |
| SG11 | h− pabp-tdTomato::kanr vgl1-GFP:: kanr | This study |
| SG12 | h− pabp-GFP:: kanr vgl1::ura4+ ura4 | This study |
| SG13 | h− vgl1-GFP:: kanr pabp::ura4+ ura4 | This study |
| SG14 | h− vgl1-GFP:: kanr eif3b-tdTomato::kanr | This study |
| SG15 | h− hri2:: leu1+gcn2:: ura4+ vgl1-GFP:: kanr leu1 ura4 | This study |
| SG16 | h− hri2:: leu1+gcn2:: ura4+ pabp-GFP:: kanr leu1 ura4 | This study |
| SG17 | h− hri2:: leu1+gcn2:: ura4+ eif3b-tdTomato::kanr ade6 leu1 ura4 | This study |
| SFY5 | MATa GFP-scp160::ura3+ ura3 trp1 leu2 | (6) |
Table 2.
Oligonucleotides used in this study
| Name | Sequence (5′-3′) |
|---|---|
| Vgl1delF | ATGGAACATCTCTCTAATCTTGAGCAACCAACAACGATGGACTCATATGATTTTCAGAAACTGACTAACGAAATCCCACTGGCTATATGT |
| Vgl1delR | AATGTAAGTACTAAATAAAGGCTCCAAAATTTTACCTTACTCTTGATTTTGGTTTTCTTGAAGTCTCTCGAATTCTAAATGCCTTCTGAC |
| Vgl1tagF | TAGTCCAAGGAAGTCGAGCTGGCGTTGTCAAGGCTAAAGATCTGATTTTCGAGAGACTTCAAGAAAACCAAAATCAAGAGCGGATCCCCGGGTTAATTAA |
| Vgl1tagR | TACACATAATTTTCAAATACACTCTCAAAAAAATTTGAGAAATGAATGTAAGTACTAAATAAAGGCTCCAAAATTTTACCGAATTCGAGCTCGTTTAAAC |
| Dcp2tagF | AATCCGATTTCAAAGGGTCTGATGATCATTTTCTGTCATATTTACAATCTGTCGTGTCTTCTAACTCCAATGGTCTTCATCGGATCCCCGGGTTAATTAA |
| Dcp2tagR | ATAACTTTAGAAGAGAATACCATATGTGTATTACACAGTCGTTCATTAGTTTCTAATGCAATGCCAATTAGAATAACAAAGAATTCGAGCTCGTTTAAAC |
| PABPdelF | GCAAACCACAAAACCTATAAATTTAAAAAAATCTCGAAAAATATCATAAAAGCCCAAGTTTTTAAAATTCTGAAAACGCCAAATCCCACTGGCTATATGT |
| PABPdelR | TAAATAAACTATACTAAATTTCCAACAGGCTTAACCCTTATTTCTTCCAATAGTGAAAACTTTCATAGAAATGTCATTCAAATTCTAAATGCCTTCTGAC |
| PABPtagF | ACTCCGCTCTTAATGAAAGAGTCAACGAAGCTATTGGGGTTTTGCAAGAATTCGTTGATCAAGAGCCTGGCTTCACTGAGCGGATCCCCGGGTTAATTAA |
| PABPtagR | TAAATAAACTATACTAAATTTCCAACAGGCTTAACCCTTATTTCTTCCAATAGTGAAAACTTTCATAGAAATGTCATTCAGAATTCGAGCTCGTTTAAAC |
| eIF3btagF | CTGTTCCTGCTGAAGAAGAAGAAATAATTCAGGAAACGGTTGAGGAAGTAATCTCTGAAGAGATAGAACCCGTTGAAGATCGGATCCCCGGGTTAATTAA |
| eIF3btagR | AAGATCATGATTTTAATCAAGTAACATCCGATTTCGAAGAAGGCCCCAATTTTTTTATGACAAAAGGTAGCGTAAAGCCCGAATTCGAGCTCGTTTAAAC |
Microscopy and flow cytometry
Living cells growing in EMM2 were stained by the addition of 5 µg/ml bis-benzimide (Hoechst 33342, Sigma) before examination by fluorescence microscopy. Visualization of tdTomato red and green fluorescent protein (GFP)-tagged protein was performed at room temperature. The SYTO RNASelect (1 µM, Invitrogen) stain of cells after methanol-fixation was used to reveal RNA. Images were acquired using a Leica DM RA2 microscope equipped with a Leica DC 350F camera, and were assembled using Adobe PhotoShop. For flow cytometry, cells fixed with 70% ethanol were re-hydrated in 10 mM EDTA, pH 8.0, 0.1 mg/ml RNase A, 1 µM sytox green and incubated at 37°C for 2 h. Cells were analyzed using a Coulter Epics XL-MCL (Fullerton, CA, USA).
Cell fractionation
Total lysate from cycloheximide-treated cells (100 µg/ml) by glass bead lysis in ice-cold low-salt buffer (20 mM HEPES-KOH, pH 7.6, 100 mM potassium acetate, 5 mM magnesium acetate, 1 mM EDTA, 2 mM dithiothreitol, 100 µg/ml cycloheximide, 0.1 mM phenylmethylsulfonyl fluoride, complete protease inhibitor mix) was obtained by centrifugation for 2 min at 1200g. These lysates were fractionated by consecutive centrifugation at 6000g and 18 000g for 30 min at 4°C, and subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) gels and immunoblotting analysis.
Polysome profile analysis
Polysomes were obtained using a protocol modified from Dunand-Sauthier et al. (22). Cycloheximide (100 µg/ml) was added to 200 ml of cultures at OD600 of 0.5 grown at 30°C or after a 1-h incubation at 42°C. The cultures were harvested and processed for polysome profiling on 7–47% sucose gradients. Proteins in the polysome profile fractions were concentrated by TCA precipitation, and samples were analyzed by SDS–PAGE and immunoblotting.
Antibodies and immunoblotting
Whole-cell protein extracts were prepared by alkaline extraction with the following modification; 5 × 107 cells were harvested and resuspended by vortexing in 1 ml of 0.3 M NaOH. One-hundred and fifty microliters of 55% (w/v) trichloroacetic acid was added before vortexing and incubation on ice for 10 min. The cells were pelleted at 4°C for 10 min at 14 000 r.p.m. The supernatant was removed by aspiration, and the cells were spun briefly for a second time to remove remaining trichloroacetic acid. The pellet was resuspended in 500 µl of SDS gel-loading buffer [50 mM Tris–HCl (pH 8.8), 2% SDS, 0.05% bromophenol blue, 10% glycerol, 4% 2-mercaptethanol] before denaturing at 100°C for 10 min. Samples were centrifuged briefly, before loading onto SDS–PAGE gels and subjected to immunoblotting using anti-HSP70 polyclonal antibody (SPA-757, Stressgen), which cross-reacted with yeast proteins, and anti-PAP antibody (Peroxidase-Anti-Peroxidase soluble complex, Sigma) to reveal Vgl1-TAP proteins. Antibodies that specifically recognize eIF2α phosphorylated at serine 51 and total eIF2α were from Invitrogen. Antibody against α-tubulin (Sigma) was used as controls.
Microarray experiments and data evaluation
The stress time course experiments with wild-type and vgl1Δ mutants were performed as two independent biological repeats. Thermal stress: exponentially growing cultures of yeast cells in YE medium were quickly transferred from 30°C to a large prewarmed flask in a 42°C water bath, reaching temperature equilibrium after 2 min. Cells were harvested immediately before as well as 15 and 60 min after heat stress from the same culture. Total RNA was isolated by hot phenol extraction and purified using RNeasy (Qiagen). Twenty micrograms of total RNA was labeled by directly incorporating Cy3- and Cy5-dCTP through reverse transcription and the resulting cDNA was hybridized onto DNA microarrays containing probes for >99% of all known and predicted genes of Schizosaccharomyces pombe spotted in duplicate onto glass slides. Performing hybridization and initial data processing and normalization were as previously described (23). Labeled samples from each time point of the wild-type and mutant experiments were hybridized with a labeled reference pool, containing an equal amount of all the RNA samples from the wild-type and mutant time points. For duplicate experiments, the Cy dyes were swapped for the experimental and reference samples. After data acquisition and within-array normalization, the ratios of each gene (time point/reference pool) were divided by the corresponding ratios of untreated wild-type cells (0 h wild-type/reference pool). Thus, the reported ratios represent the expression levels at each time point relative to the expression levels of the untreated wild-type cells from the same stress experiment (24). Expression ratios of biological repeat experiments (wild-type and vgl1Δ strains) were averaged. Data were analyzed using GeneSpring (Agilent) and Excel (Microsoft). Hierarchical clustering was performed in GeneSpring using Pearson correlation with genes containing no data in ≥50% of the conditions being discarded. The entire processed data set will be available at http://www.bahlerlab.info/projects/collaborators.
RESULTS
Fission yeast Vgl1 is not required for the maintenance of cell ploidy
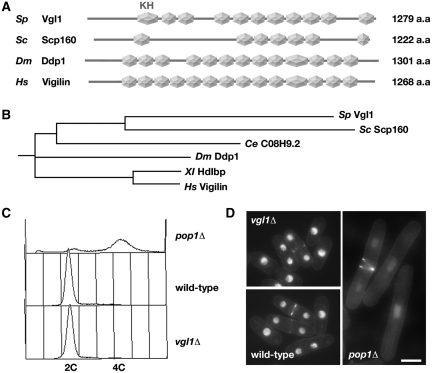
BLAST searches of the complete S. pombe genome using the S. cerevisiae Scp160 amino acid sequence as a query identified a single S. pombe gene SPCC550.14 that is significantly similar to SCP160. ProfileScan analysis of the 1279-amino-acid sequence revealed multiple KH domains over its entire length (Figure 1A). While only seven of the 14 KH domains are classical (containing noninterrupting Gly-X-X-Gly sequences) in S. cerevisiae Scp160, the predicted S. pombe protein sequence contains 11 classical KH domain structures and is more closely related to human Vigilin (Figure 1B). In recognition of this level of sequence conservation, and in deference of the existing gene name in S. pombe GeneDB, we refer to the S. pombe vigilin as Vgl1·
Figure 1.
Fission yeast Vgl1 is not required for the maintenance of cell ploidy. (A) Schematic representation of the domain structures of S. pombe Vgl1, S. cerevisiae Scp160, Drosophila Ddp1 and human Vigilin. Note that only the classical KH domain structures (hexagon) containing non-interrupting Gly-X-X-Gly sequences are shown. (B) Cladogram showing the relationship between S. pombe Vgl1 and multi-KH domain proteins from S. cerevisiae Scp160 to human Vigilin. The length of each pair of branches represents the distance between sequence pairs. (C) Flow cytometric analysis of the DNA content of ethanol-fixed, Sytox Green-stained samples from exponentially growing cultures of wild-type, vgl1Δ and pop1Δ cells. (D) Fluorescence micrographs of Hoechst 33342-stained living wild-type, vgl1Δ and pop1Δ cells. Bar: 5 µm.
As a first step toward determining the function of vgl1+ in S. pombe, the one-step gene disruption method was used in a ura4−/ura4− diploid S. pombe strain to replace one copy of the entire vgl1+ open reading frame with the ura4+ selectable marker, and after induction of meiosis, tetrads were dissected. Haploid ura4+ progeny segregated 2:2, indicating that the vgl1+gene is not essential for growth. Microscopic examination of exponentially growing cells with the vgl1::ura4+ genotype (vgl1Δ) showed that there were no gross abnormalities in cell length or division associated with vgl1 deletion (Figure 1D). First identified in 1995, Scp160 was originally hypothesized to function in the maintenance of cell ploidy in yeast, due in large part to the null phenotype of abnormal cell size and shape, increased DNA content and aberrant segregation of genetic markers through meiosis (5). However, biochemical studies from others quickly challenged this hypothesis (7,9). To test whether or not Vgl1 is required for the maintenance of cell ploidy in fission yeast, flow cytometric analysis was performed to determine the DNA content of cells without functional Vgl1. As shown in Figure 1C, the asynchronous population of vgl1Δ cells consists mainly of G2 cells with 2C DNA content, as expected of a wild-type exponential population of haploid cells (the G2 phase occupies most of the cell cycle and cells replicate their DNA close to the time of septation). In contrast, deletion of the pop1+ gene, which is important for the maintenance of cell ploidy (25), leads to mutants with a higher amount of 4C DNA content and doubling in cell size on average as compared with wild-type cells (Figure 1D). Taking together, we conclude that fission yeast Vgl1 is not required for the maintenance of cell ploidy.
Vgl1 is predominantly localized to the ER in a microtubule-independent manner
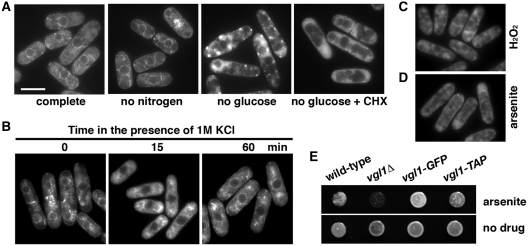
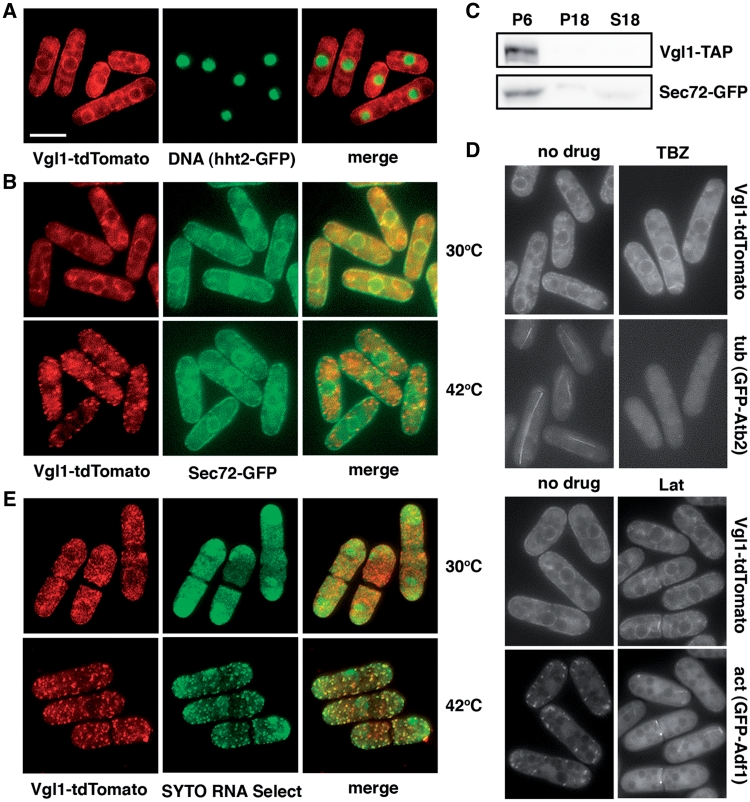
To characterize Vgl1 in S. pombe, targeted recombination was used to add various epitope tag sequences to the 3′ end of the vgl1 open reading frame in its normal chromosomal context. The tagged proteins appeared to be functional as judged by the lack of hypersensitivity to arsenite of the vgl1-GFP and vgl1-TAP strains compared with vgl1Δ (Figure 5E). Examination of living cells by fluorescence microscopy showed that Vgl1-tdTomato was concentrated around the nucleus and in patches at the periphery of the cell (Figure 2A), reminiscent of ER staining. To correlate this more directly, the fluorescent ER membrane protein Sec72 (21) was used to reveal the ER structures. The overlapping localization of Vgl1-tdTomato with the Sec72-GFP ring-like ER structures (Figure 2B) as well as in cell fractions generated by consecutive centrifugation (Figure 2C) suggest that Vgl1 is localized in the ER membranes. Furthermore, using sucrose gradient fractionation we demonstrated that Vgl1 co-migrated with polysomes (Figure 3A), as found in S. cerevisiae that Scp160 bound polysomes accumulated at ER (5,6). However, unlike Scp160, the accumulation of fission yeast Vgl1 at the ER does not require the function of microtubules or the actin cytoskeleton (6), as treatment with thiabendazole (TBZ) or Latrunculin A (Lat A) did not change the localization of tdTomato-tagged Vgl1 (Figure 2D).
Figure 5.
Analysis of Vgl1-GFP localization under stress. (A) Localization of Vgl1-GFP in living cells grown at 30°C (complete) and after deprivation of glucose or nitrogen for 1 and 3 h, respectively. One-hundred micrograms per milliliter of cycloheximide (CHX) were added 1 min before depletion of glucose. Bar: 5 µm. (B) Localization of Vgl1-GFP after exposure to 1 M KCl for the times indicated. (C and D) Localization of Vgl1-GFP in living cells, after exposure for 60 min to 1 mM H2O2 or 2 mM arsenite, respectively. (E) 104 cells of the indicated strains were spotted onto YES agar containing 0.6 mM arsenite or no drug. Plates were photographed after 2 days of incubation at 30°C.
Figure 2.
Fission yeast Vgl1 is predominantly localized to the endoplasmic reticulum (ER) in a microtubule-independent manner and is rapidly relocalized to cytoplasmic granules stained with an RNA-selective dye under thermal stress. (A) Merged images of fluorescence micrographs showing Vgl1-tdTomato and DNA (hht2-GFP) localization in living cells. Bar: 5 µm. (B) Merged images of fluorescence micrographs showing Vgl1-tdTomato and Sec72-GFP localization in living cells grown at 30°C and after a 15-min incubation at 42°C. (C) Total lysate from cells co-expressing Vgl1-TAP and Sec72-GFP were fractionated by consecutive centrifugation, resulting in 6000g and 18 000g pellets (P6 and P18) and a supernatant (S18), before separated by SDS–PAGE and subjected to immunoblotting using anti-PAP (Peroxidase-Anti-Peroxidase soluble complex) antibodies to reveal Vgl1 proteins and anti-GFP antibodies for Sec72 proteins. (D) Localization of Vgl1-tdTomato in cells after a 120-min incubation with 20 µg/ml thiabendazole (TBZ) or a 60-min incubation with 10 µM Latrunculin A (Lat A). The fluorescent Atb2 and Adf1 proteins were used to reveal the microtubule (tub) and actin (act) structures. (E) Merged images of fluorescence micrographs showing Vgl1-tdTomato localization in methanol-fixed cells stained with SYTO RNA Select dye (green). Samples were taken from exponentially growing cultures grown at 30°C or after a 15-min incubation at 42°C.
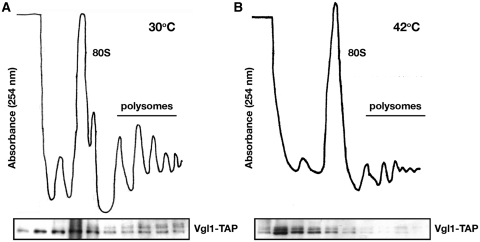
Figure 3.
Analysis of Vgl1-containing complexes using sucrose gradient fractionation. Cytosolic extracts from cycloheximide-treated yeast cells grown at 30°C (A) and after a 60-min incubation at 42°C (B) were separated on 7–47% sucrose gradients. The distribution of the rRNA is shown by absorption profiles at 254 nm and the distribution of Vgl1 by Western blotting.
Vgl1 is rapidly relocalized to cytoplasmic granules under thermal stress
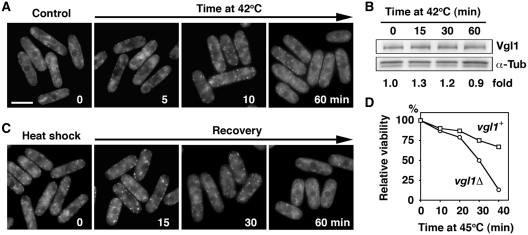
Intriguingly, during the characterization of Vgl1 in S. pombe, we found that under thermal stress, Vgl1 is rapidly relocalized to cytoplasmic granules without changes of ER structures (Figure 2B). To investigate the dynamic distribution of Vgl1, again we monitored the Vgl1-containing complexes using sucrose gradient fractionation. In line with the relocalization of Vgl1, we found that disruption of the polysomes by the treatment of cells under thermal stress shifted the Vgl1 signal to the top of the gradient (Figure 3B). Consistent with a function in RNA metabolism, these Vgl1 granules were overlapping with SYTO nucleic acid stains specific for RNA (Figure 2E), suggesting that Vgl1 might escort RNA from ER-associated polyribosomes to the cytosol under thermal stress. Detailed analysis revealed that this process is highly dynamic and reversible. Upon thermal stress, the accumulation of Vgl1 at the ER rapidly disappeared within 2 min and small patches of granule-like structures occurred in the cytoplasm at around 5 min (Figure 4A). By 10 min these granule structures become evident and remain stable for up to 60 min. Western blotting demonstrated that these changes in cellular localization occurred in the absence of any effect on Vgl1 protein level (Figure 4B). Following removal of the thermal stress, these granule-like structures rapidly dispersed, and Vgl1 accumulated again at the ER (Figure 4C). In line with these data, we found that vgl1Δ mutants were more susceptible to thermal stress and, when incubated at high temperature, lost their viability more rapidly than wild-type cells (Figure 4D). We conclude that Vgl1 mediates cell survival under thermal stress.
Figure 4.
Assembly and disassembly of heat shock-induced Vgl1 granules. (A) Vgl1-GFP was visualized after growth to mid-logarithmic phase at 30°C (time 0) or after a shift to 42°C for the time indicated. Bar: 5 µm. (B) Whole-cell protein extracts from cells expressing Vgl1-TAP incubated at 42°C for the times indicated were prepared by alkaline extraction followed by trichloroacetic acid precipitation. The extracts were separated by SDS–PAGE and subjected to immunoblotting using anti-PAP (Peroxidase-Anti-Peroxidase soluble complex) antibodies to reveal Vgl1 proteins. Antibodies against α-tubulin were used as controls. The relative level of Vgl1 is indicated beneath each lane (average of two independent experiments). (C) Localization of Vgl1-GFP after a 15-min incubation at 42°C (time 0) followed by a shift back to 30°C for the times indicated. (D) The indicated strains were grown in liquid culture to mid-logarithmic phase at 30°C and shifted to 45°C. Samples of 500 cells taken at the indicated times after the shift to 45°C were plated in duplicate onto YES agar and incubated at 30°C. After 3 days of growth, viability was scored as a percentage of the number of colonies formed by the sample taken at time zero.
Localization of Vgl1-GFP under different stress conditions
We next examined the localization of Vgl1-GFP under additional types of stress. As shown in Figure 5A, nitrogen depletion did not change the localization of Vgl1-GFP. In contrast, glucose starvation caused a rapid relocalization of Vgl1-GFP from the ER to the cytosol with distinct granules evident in some cells that can be inhibited by the addition of cycloheximide (Figure 5A +CHX). Similarly, after exposure of cells to 1 M KCl, Vgl1-GFP rapidly relocalized to the cytosol, although the granules were less conspicuous than in heat stress. The accumulation of Vgl1-GFP at the ER was resumed after extended periods of exposure (60 min), indicating that cells adapted to osmotic stress (Figure 5B). Oxidative stress (1 mM H2O2) had little or no effect on the localization of Vgl1-GFP (Figure 5C). Intriguingly, exposure of cells to arsenite, a chemical used to induce stress granules (SGs) in mammalian cells, also caused some relocalization of Vgl1-GFP to the cytosol (Figure 5D). Moreover, vgl1Δ mutants were hypersensitive to the treatment of arsenite (Figure 5E). Taken together, these results raise the possibility that the Vgl1 granules reflect cytoplasmic aggregates of ribonucleoprotein complexes that correspond to SGs emerging in response to cellular stress.
Catalog of the protein composition of Vgl1 granules
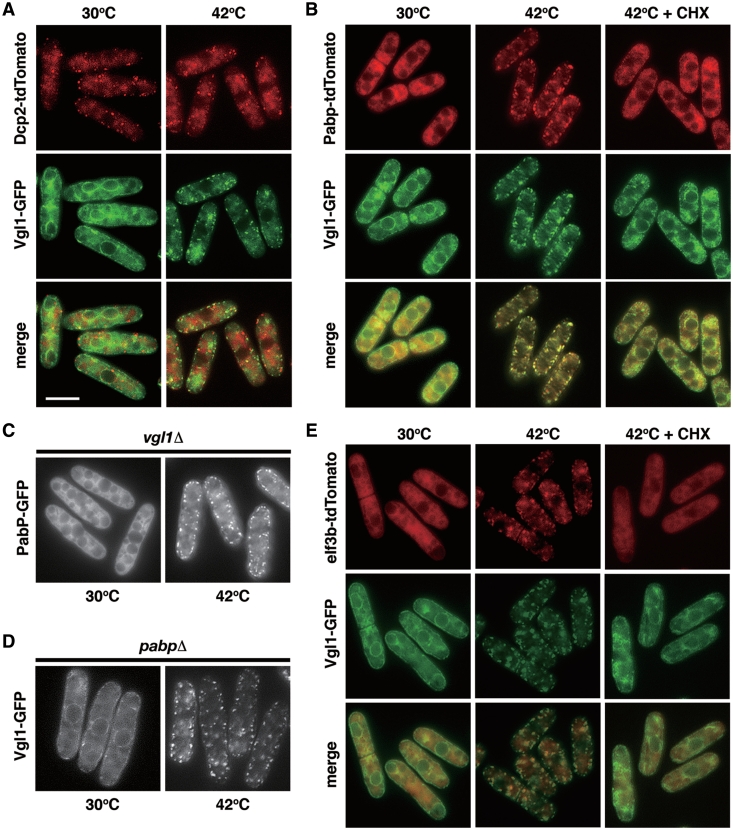
Several different types of RNA granules have been described (26). Cytoplasmic processing bodies, also known as P-bodies (PBs), are observed in actively growing unstressed cells, that contain untranslating mRNA in conjunction with enzymes involved in translation repression and mRNA decapping and degradation. In contrast, SGs are cytoplasmic phase-dense structures that are not seen in cells growing under favorable conditions but are rapidly induced in response to environmental stress. Although SGs and PBs share some protein and mRNA components, they also contain a number of unique markers specific to each structure. To catalog the Vgl1 granules, we used fluorescence microscopy to determine the protein composition of Vgl1 granules. The signature constituents of PBs are components of the decay machinery that removes the m7G (7-methylguanosine) cap and degrades mRNA from the 5′ ends, for example, the Dcp1–Dcp2 complex. To determine if these proteins were components of a single granule type, we examined their localization with a Dcp2-tdTomato fusion protein. As shown in Figure 6A, Dcp2 formed cytoplasmic foci in actively growing unstressed cells that we refer to as fission yeast PBs. Thermal stress has little effect on these structures that are largely distinct from the Vgl1 cytoplasmic granules. In contrast, we observed that, under thermal stress, poly(A) binding protein Pabp-tdTomato (the signature constituents of SGs) colocalized almost completely with GFP fusions of Vgl1 (Figure 6B). This colocalization indicated that these proteins are components of a single granule with similar composition to mammalian SGs. This is further supported by the fact that, as in mammalian cells, drugs such as cycloheximide (that stabilize polysomes by freezing ribosomes on translating mRNA) inhibited the assembly of these granules (Figure 6B + CHX). We refer to these granules as fission yeast SGs and note that they are almost certainly the same eIF3-positive granules identified earlier (22).
Figure 6.
Colocalization of Vgl1 with poly(A)-binding protein (PABP) and eIF3b, a component of the translation initiation complex, but not with the decapping enzyme Dcp2 under thermal stress. (A) Merged images of fluorescence micrographs showing Dcp2-tdTomato (red) and Vgl1-GFP (green) localization in living cells grown at 30°C and after a 15-min incubation at 42°C. Bar: 5 µm. (B) Colocalization of Pabp-tdTomato (red) and Vgl1-GFP (green) in living cells under thermal stress (15 min at 42°C) that is inhibited by the addition of 100 µg/ml cycloheximide (CHX) 1 min before temperature shift. (C) Fluorescence micrographs of the vgl1Δ mutants expressing Pabp-GFP grown at 30°C and after a 15-min incubation at 42°C. (D) Fluorescence micrographs of pabpΔ mutants expressing Vgl1-GFP grown at 30°C and after a 15-min incubation at 42°C. (E) Merged images of fluorescence micrographs showing Vgl1-GFP (green) and eIF3b-tdTomato (red) localization in living cells grown at 30°C and after a 15-min incubation at 42°C that is inhibited by the addition of 100 µg/ml cycloheximide (CHX) 1 min before temperature shift.
To determine the role of Vgl1 in SG assembly, we examined how the absence of Vgl1 affected the assembly of SGs under thermal stress. We observed that, under thermal stress, the accumulation of cytoplasmic Pabp granules (and, by inference SGs) still occurred in vgl1Δ mutants (Figure 6C). Similar results were observed in the pabpΔ mutants (Figure 6D), suggesting that both proteins are not the effectors of SGs; instead, they seem to act as a chaperone to escort mRNA from ER-associated polyribosome to the cytosol under thermal stress.
As part of our analysis, we also assessed the colocalization of Vgl1 with translation initiation factors eIF3b, another SG marker in mammalian cells (Figure 6E). Although to a less degree, under thermal stress, eIF3b-tdTomato rapidly relocalized to cytoplasmic foci overlapping with Vgl1-GFP, suggesting that these proteins are components of a single granule, which can be inhibited by the addition of cycloheximide (Figure 6E +CHX).
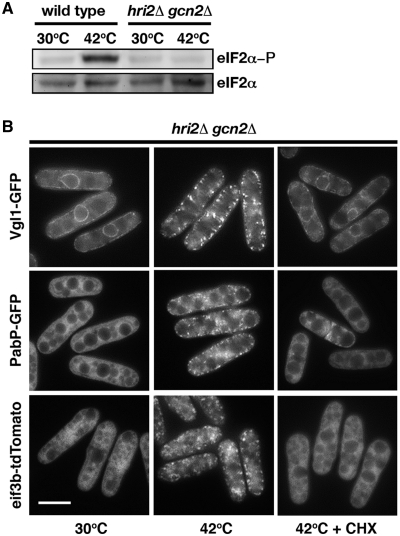
Although SG formation can occur as a consequence of impaired translation initiation independently of eIF2α phosphorylation (27), SG assembly usually requires the stress-induced phosphorylation of eIF2α (28). To determine how fission yeast SGs assemble, we examine how the absence of eIF2α phosphorylation affected the assembly of SGs under thermal stress. It has been previously demonstrated that Hri2 and Gcn2 are the primary and secondary eIF2 kinases in response to heat shock in S. pombe (29), and the double-knockout strain contained no phosphorylated eIF2α (Figure 7A). We found that the relocalization of Vgl1, Pabp and eIF3b still occurred in the absence of Hri2 and Gcn2, which can be inhibited by the addition of cycloheximide (Figure 7B). We conclude that assembly of SGs in S. pombe is independent of eIF2α phosphorylation.
Figure 7.
Assembly of SGs in S. pombe is independent of eIF2α phosphorylation. (A) Whole-cell protein extracts from wild-type cells and hri2Δ gcn2Δ mutants grown at 30°C and after a 15-min incubation at 42°C were prepared by alkaline extraction followed by trichloroacetic acid precipitation. The extracts were separated by SDS–PAGE and subjected to immunoblotting using antibodies that specifically recognize eIF2α phosphorylated at serine 51 and total eIF2α· (B) Fluorescence micrographs of the hri2Δgcn2Δ mutants expressing Vgl1-GFP (top), Pabp-GFP (middle) and eIF3b-tdTomato (lower) grown at 30°C and after a 15-min incubation at 42°C. One-hundred micrograms per milliliter of cycloheximide (CHX) were added 1 min before temperature shift. Bar: 5 µm.
Gene expression patterns in vgl1Δ mutants
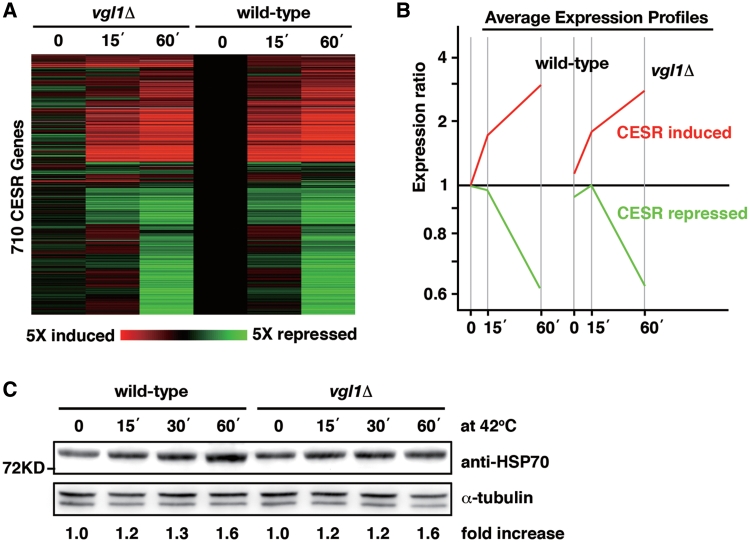
Eukaryotic cells have developed response mechanisms to combat the harmful effect of a variety of stress conditions. Such a response involves changes in cellular gene expression, leading to increased levels and activities of proteins that have stress protective function. To understand further the role of Vgl1 in cell survival under thermal stress, we used microarrays to determine the gene expression signature in vgl1Δ mutants under thermal stress. Figure 8A shows a cluster analysis of the expression of induced and repressed core environmental stress response (CESR) genes (24) under thermal stress in wild-type and vgl1Δ mutants. Despite the changes of a small number of genes before thermal stress, the overall profiles of upregulated (red) and downregulated transcripts (green) in vgl1Δ mutants are similar to that in wild-type cells (Figure 8A and B). We conclude that Vgl1 has no global effect on changes in transcript levels under thermal stress. Next, we asked whether or not Vgl1 might work through regulation of the translational activities of mRNAs functioning in stress protection by measuring protein levels of heat shock proteins. However, the accumulation of heat shock proteins under thermal stress still occurred in the vgl1Δ mutants (Figure 8C).
Figure 8.
Expression profiles of the core environmental stress response (CESR) genes and the accumulation of Hsp70 proteins under thermal stress are not affected by the deletion of vgl1+. (A) CESR genes (24) were hierarchically clustered based on their expression patterns. Horizontal strips represent genes, and columns represent experimental time points. The fold changes in expression, relative to the untreated wild-type sample (time point 0), are color-coded as shown in the bar. The labels on the bottom indicate CESR induced genes (red) and repressed genes (green). (B) Average expression patterns of the CESR-induced (red) and -repressed (green) genes as labeled in A. (C) Whole-cell protein extracts from wild-type and vgl1Δ cells during a 60-min incubation at 42°C were prepared by alkaline extraction followed by trichloroacetic acid precipitation. The extracts were separated by SDS–PAGE and subjected to immunoblotting using anti-HSP70 antibody. Antibody against α-tubulin was used as controls. The relative increase in the level of the Hsp70 is indicated beneath each lane (average of two independent experiments).
Localization of S. cerevisiae Scp160-GFP under thermal stress
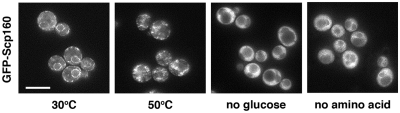
As part of our analysis, we also assessed the localization of S. cerevisiae Scp160-GFP under thermal stress. As shown in Figure 9, we found that Scp160 also rapidly relocalized from the ER to cytoplasmic granules under thermal stress. Similarly, relocalization of Scp160-GFP was observed in cells after deprivation of glucose or amino acids. However, no granule-like structures were observed in these cells.
Figure 9.
Localization of S. cerevisiae GFP-Scp160 under thermal stress and after deprivation of glucose or amino acids. Bar: 5 µm.
DISCUSSION
Characterization of fission yeast vigilin
The large family of multiple KH-domain proteins, collectively known as vigilins, are evolutionarily highly conserved proteins from the yeast S. cerevisiae (Scp160) to Drosophila (DDP1), and vertebrates (Vigilin). Although all vigilins reported to date appear to bind nucleic acids, in most cases both the type of nucleic acid bound and the biological relevance of this interaction remain barely understood. As an independent approach toward understanding the biological function of these proteins, we have characterized the fission yeast vigilin Vgl1 (Figure 1A), and have investigated its role in cellular response to environmental stress. We found that, as its counterpart in S. cerevisiae, S. pombe vgl1 is not essential for normal cell growth (Figure 1D). Scp160 was originally hypothesized to function in the maintenance of cell ploidy in S. cerevisiae (5), but we found no indication that Vgl1 is required for the maintenance of cell ploidy in S. pombe (Figure 1C). Instead, Vgl1 is required for cell survival under thermal stress, and vgl1Δ mutants loose their viability more rapidly than wild-type cells when incubated at high temperature (Figure 4D).
Vgl1 in stress granules
Subcellular localization studies have demonstrated that, as its counterpart in S. cerevisiae, the majority of Vgl1 is cytoplasmic, with significant enrichment at the ER (Figure 2A). In keeping with a function relevant to promoting cell survival under thermal stress, Vgl1 is rapidly relocalized from the ER to cytoplasmic granules upon heat treatment (Figure 2B). These granules appear to be the equivalent of mammalian SGs, as suggested by the observation that, under thermal stress, Vgl1 aggregated into foci that can be separate from P-bodies (Figure 6A), and which contain RNA (Figure 2E). Moreover, these granules contain proteins analogous to those seen in mammalian SGs including Pabp, G3BP and components of the eIF3 translation initiation complex (Figure 6B and E, and data not shown). Like mammalian SGs, assembly of these proteins into granule-like structures is blocked by trapping mRNAs in polysomes with cycloheximide (Figure 7). Unlike mammalian cells and similar to trypanosomes and S. cerevisiae (30,31), however, the assembly of SGs in S. pombe is independent of eIF2α phosphorylation (Figure 7) and not induced by oxidative stress (Figure 5C). Experiments are underway to identify additional components and proteins that affected the assembly of Vgl1 granules.
Role of Vgl1 in cell response to thermal stress
Our results argue that Vgl1 does not function in global transcription or regulation of mRNA turnover during stress (Figure 8A and B). An alternative possibility is that Vgl1 might function to regulate the translational activities of mRNAs functioning in stress protection. However, no such effect was seen on the accumulation of heat shock protein 70 after thermal stress, which still occurred normally in the vgl1Δ mutants (Figure 8C). Furthermore, Vgl1 is not required for the assembly of SGs despite its dynamic translocation (Figure 6C). Taking together, these results lead us to suggest that Vgl1 acts as an escort moving with mRNA not required for stress protection from ER-associated polysomes to the cytosol for storage during thermal stress, thus inhibiting their translation to provide space for stress proteins. The colocalization with translation initiation factors could serve to enhance translation initiation once stress is removed (Figure 6E). Experiments are underway to test this hypothesis. A similar system may be operating in other organisms as we found that S. cerevisiae Scp160 also rapidly relocalized from the ER to cytoplasmic granules under thermal stress (Figure 9), suggesting the function in stress granules might be the general role of vigilin with the exception of nuclear protein DDP1 in Drosophila (12,13). Further experiments are required to determine the composition and property of these structures in budding yeast. Recently, ribonucleoprotein granules referred to as EGP-bodies containing eIF4E, eIF4G and Pab1, which are components of mammalian SGs, have been described to form in S. cerevisiae during glucose deprivation and to both overlap and be distinct from PBs (32,33). It will be interesting to determine if all these proteins are components of a single granule type. Yeast is an ideal system for studying the relationship between SGs and PBs, as PB assembly can be prevented or modified in various mutant strains (34). Yeast genetics will continue to point the way toward understanding SG dynamics and regulation, but because many relevant components (e.g. OGT and HBP enzymes) (35) are not found in yeast, studies of higher eukaryotes are also essential. A deeper understanding of the smallest and most ancient RNA granules in these unicellular eukaryotes may reveal principles that are applicable to other granules in general.
FUNDING
Funding for open access charge: The National Health Research Institute, Taiwan (MG-099-PP-10).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Drs Mohan K. Balasubramanian, Yasushi Hiraoka, Matthias Seedorf, and Takashi Toda for yeast strains.
REFERENCES
- 1.Lipshitz HD, Smibert CA. Mechanisms of RNA localization and translational regulation. Curr. Opin. Genet. Dev. 2000;10:476–488. doi: 10.1016/s0959-437x(00)00116-7. [DOI] [PubMed] [Google Scholar]
- 2.Siomi H, Choi M, Siomi MC, Nussbaum RL, Dreyfuss G. Essential role for KH domains in RNA binding: impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome. Cell. 1994;77:33–39. doi: 10.1016/0092-8674(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 3.Siomi H, Matunis MJ, Michael WM, Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993;21:1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckanovich RJ, Darnell RB. The neuronal RNA binding protein Nova-1 recognizes specific RNA targets in vitro and in vivo. Mol. Cell. Biol. 1997;17:3194–3201. doi: 10.1128/mcb.17.6.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wintersberger U, Kuhne C, Karwan A. Scp160p, a new yeast protein associated with the nuclear membrane and the endoplasmic reticulum, is necessary for maintenance of exact ploidy. Yeast. 1995;11:929–944. doi: 10.1002/yea.320111004. [DOI] [PubMed] [Google Scholar]
- 6.Frey S, Pool M, Seedorf M. Scp160p, an RNA-binding, polysome-associated protein, localizes to the endoplasmic reticulum of Saccharomyces cerevisiae in a microtubule-dependent manner. J. Biol. Chem. 2001;276:15905–15912. doi: 10.1074/jbc.M009430200. [DOI] [PubMed] [Google Scholar]
- 7.Lang BD, Fridovich-Keil JL. Scp160p, a multiple KH-domain protein, is a component of mRNP complexes in yeast. Nucleic Acids Res. 2000;28:1576–1584. doi: 10.1093/nar/28.7.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baum S, Bittins M, Frey S, Seedorf M. Asc1p, a WD40-domain containing adaptor protein, is required for the interaction of the RNA-binding protein Scp160p with polysomes. Biochem. J. 2004;380:823–830. doi: 10.1042/BJ20031962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mendelsohn BA, Li AM, Vargas CA, Riehman K, Watson A, Fridovich-Keil JL. Genetic and biochemical interactions between SCP160 and EAP1 in yeast. Nucleic Acids Res. 2003;31:5838–5847. doi: 10.1093/nar/gkg810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sezen B, Seedorf M, Schiebel E. The SESA network links duplication of the yeast centrosome with the protein translation machinery. Genes Dev. 2009;23:1559–1570. doi: 10.1101/gad.524209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo M, Aston C, Burchett SA, Dyke C, Fields S, Rajarao SJ, Uetz P, Wang Y, Young K, Dohlman HG. The yeast G protein alpha subunit Gpa1 transmits a signal through an RNA binding effector protein Scp160. Mol. Cell. 2003;12:517–524. doi: 10.1016/s1097-2765(03)00307-1. [DOI] [PubMed] [Google Scholar]
- 12.Cortes A, Huertas D, Fanti L, Pimpinelli S, Marsellach FX, Pina B, Azorin F. DDP1, a single-stranded nucleic acid-binding protein of Drosophila, associates with pericentric heterochromatin and is functionally homologous to the yeast Scp160p, which is involved in the control of cell ploidy. EMBO J. 1999;18:3820–3833. doi: 10.1093/emboj/18.13.3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cortes A, Azorin F. DDP1, a heterochromatin-associated multi-KH-domain protein of Drosophila melanogaster, interacts specifically with centromeric satellite DNA sequences. Mol. Cell. Biol. 2000;20:3860–3869. doi: 10.1128/mcb.20.11.3860-3869.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marsellach FX, Huertas D, Azorin F. The multi-KH domain protein of Saccharomyces cerevisiae Scp160p contributes to the regulation of telomeric silencing. J. Biol. Chem. 2006;281:18227–18235. doi: 10.1074/jbc.M601671200. [DOI] [PubMed] [Google Scholar]
- 15.Schmidt C, Henkel B, Poschl E, Zorbas H, Purschke WG, Gloe TR, Muller PK. Complete cDNA sequence of chicken vigilin, a novel protein with amplified and evolutionary conserved domains. Eur. J. Biochem. 1992;206:625–634. doi: 10.1111/j.1432-1033.1992.tb16967.x. [DOI] [PubMed] [Google Scholar]
- 16.Plenz G, Kugler S, Schnittger S, Rieder H, Fonatsch C, Muller PK. The human vigilin gene: identification, chromosomal localization and expression pattern. Hum. Genet. 1994;93:575–582. doi: 10.1007/BF00202827. [DOI] [PubMed] [Google Scholar]
- 17.Dodson RE, Shapiro DJ. Vigilin, a ubiquitous protein with 14 K homology domains, is the estrogen-inducible vitellogenin mRNA 3′-untranslated region-binding protein. J. Biol. Chem. 1997;272:12249–12252. doi: 10.1074/jbc.272.19.12249. [DOI] [PubMed] [Google Scholar]
- 18.Cunningham KS, Dodson RE, Nagel MA, Shapiro DJ, Schoenberg DR. Vigilin binding selectively inhibits cleavage of the vitellogenin mRNA 3′-untranslated region by the mRNA endonuclease polysomal ribonuclease 1. Proc. Natl Acad. Sci. USA. 2000;97:12498–12502. doi: 10.1073/pnas.220425497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kruse C, Willkomm DK, Grunweller A, Vollbrandt T, Sommer S, Busch S, Pfeiffer T, Brinkmann J, Hartmann RK, Muller PK. Export and transport of tRNA are coupled to a multi-protein complex. Biochem. J. 2000;346(Pt 1):107–115. [PMC free article] [PubMed] [Google Scholar]
- 20.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 21.Hayashi A, Ding DQ, Tsutsumi C, Chikashige Y, Masuda H, Haraguchi T, Hiraoka Y. Localization of gene products using a chromosomally tagged GFP-fusion library in the fission yeast Schizosaccharomyces pombe. Genes Cells. 2009;14:217–225. doi: 10.1111/j.1365-2443.2008.01264.x. [DOI] [PubMed] [Google Scholar]
- 22.Dunand-Sauthier I, Walker C, Wilkinson C, Gordon C, Crane R, Norbury C, Humphrey T. Sum1, a component of the fission yeast eIF3 translation initiation complex, is rapidly relocalized during environmental stress and interacts with components of the 26S proteasome. Mol. Biol. Cell. 2002;13:1626–1640. doi: 10.1091/mbc.01-06-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyne R, Burns G, Mata J, Penkett CJ, Rustici G, Chen D, Langford C, Vetrie D, Bahler J. Whole-genome microarrays of fission yeast: characteristics, accuracy, reproducibility, and processing of array data. BMC Genomics. 2003;4:27. doi: 10.1186/1471-2164-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen D, Toone WM, Mata J, Lyne R, Burns G, Kivinen K, Brazma A, Jones N, Bahler J. Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell. 2003;14:214–229. doi: 10.1091/mbc.E02-08-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kominami K, Toda T. Fission yeast WD-repeat protein pop1 regulates genome ploidy through ubiquitin-proteasome-mediated degradation of the CDK inhibitor Rum1 and the S-phase initiator Cdc18. Genes Dev. 1997;11:1548–1560. doi: 10.1101/gad.11.12.1548. [DOI] [PubMed] [Google Scholar]
- 26.Anderson P, Kedersha N. RNA granules. J. Cell. Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mazroui R, Sukarieh R, Bordeleau ME, Kaufman RJ, Northcote P, Tanaka J, Gallouzi I, Pelletier J. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2alpha phosphorylation. Mol. Biol. Cell. 2006;17:4212–4219. doi: 10.1091/mbc.E06-04-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 29.Zhan K, Narasimhan J, Wek RC. Differential activation of eIF2 kinases in response to cellular stresses in Schizosaccharomyces pombe. Genetics. 2004;168:1867–1875. doi: 10.1534/genetics.104.031443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kramer S, Queiroz R, Ellis L, Webb H, Hoheisel JD, Clayton C, Carrington M. Heat shock causes a decrease in polysomes and the appearance of stress granules in trypanosomes independently of eIF2(alpha) phosphorylation at Thr169. J. Cell. Sci. 2008;121:3002–3014. doi: 10.1242/jcs.031823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grousl T, Ivanov P, Frydlova I, Vasicova P, Janda F, Vojtova J, Malinska K, Malcova I, Novakova L, Janoskova D, et al. Robust heat shock induces eIF2{alpha}-phosphorylation-independent assembly of stress granules containing eIF3 and 40S ribosomal subunits in budding yeast, Saccharomyces cerevisiae. J. Cell. Sci. 2009;122:2078–2088. doi: 10.1242/jcs.045104. [DOI] [PubMed] [Google Scholar]
- 32.Brengues M, Parker R. Accumulation of polyadenylated mRNA, Pab1p, eIF4E, and eIF4G with P-bodies in Saccharomyces cerevisiae. Mol. Biol. Cell. 2007;18:2592–2602. doi: 10.1091/mbc.E06-12-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoyle NP, Castelli LM, Campbell SG, Holmes LE, Ashe MP. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J. Cell. Biol. 2007;179:65–74. doi: 10.1083/jcb.200707010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell. Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohn T, Kedersha N, Hickman T, Tisdale S, Anderson P. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat. Cell. Biol. 2008;10:1224–1231. doi: 10.1038/ncb1783. [DOI] [PMC free article] [PubMed] [Google Scholar]