Abstract
The recent discovery of genomic 5-hydroxymethylcytosine (hmC) and mutations affecting the respective Tet hydroxylases in leukemia raises fundamental questions about this epigenetic modification. We present a sensitive method for fast quantification of genomic hmC based on specific transfer of radiolabeled glucose to hmC by a purified glucosyltransferase. We determined hmC levels in various adult tissues and differentiating embryonic stem cells and show a correlation with differential expression of tet genes.
INTRODUCTION
DNA methylation plays a crucial role in the epigenetic regulation of gene expression during development and disease (1). The post-replicative addition of a methyl group to the carbon-5 of cytosine has long been the only known enzymatic modification to bases in mammalian genomic DNA, and due to its crucial role as an epigenetic mark it is often referred to as the fifth base. Recently, the Ten-Eleven Translocation 1 gene (tet1) was shown to encode a 2-ketoglutarate- and Fe(II)-dependant hydroxylase that converts genomic 5-methylcytosine (mC) to 5-hydroxymethylcytosine (hmC) and on the basis of sequence homology the closely related Tet2 and 3 proteins are expected to catalyze the same reaction (2,3). To date hmC has been detected in genomic DNA isolated from embryonic stem cells (ESCs) and some adult tissues and it appears to be relatively abundant in the central nervous system (2,4–6). The functional relevance of hmC in these tissues is unknown and roles as an epigenetic mark and/or an intermediate of oxidative demethylation are intriguing possibilities (2,7). In addition, translocations as well as nonsense and missense mutations of tet genes have been identified in myelodysplastic syndromes including several forms of myeloid leukemia (8–10), raising the possibility that aberrant genomic hmC patterns may be involved in these pathologies. These observations grant sustained efforts to define the role(s) of hmC in mammalian genomes.
Quantification and selective detection of genomic hmC is technically challenging due to the relatively low abundance and similarity of hmC to the more abundant mC, not only in structural terms but also with respect to lack of deamination by bisulfite treatment (11–12). We sought to exploit enzymes involved in hmC modification that evolved as part of the struggle between prokaryotes and their viruses.
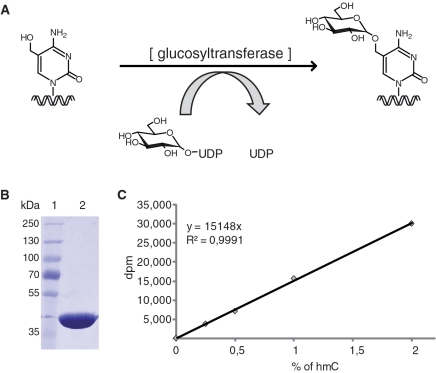
The three methods used so far to quantify global hmC content in mammalian genomes are designed to detect hmC in hydrolyzed DNA globally (HPLC/esi-ms/ms) or at subsets of CpG sites (2,4,5). As hmC may also occur at non-CpG sites, the latter type of methods may underestimate its abundance. In addition, none of these procedures is easily applicable to large sample numbers. We therefore sought to establish a highly sensitive and accurate method to detect hmC independently of sequence context and with higher throughput capacity. To this aim, we turned our attention to glucosyltransferases of T-even bacteriophages that transfer glucose from UDP-glucose donor to genomic hmC. Notably, all cytosines in the T4 genome are replaced by hmC residues that are invariably modified by α- and β-glucosyltransferases (α- and β-gt; Figure 1A). We reasoned that by using UDP-[3H]glucose the incorporation of radiolabeled glucose in DNA should reflect the abundance of hmC. We focused on β-gt rather than α-gt, as it was shown to glucosylate to completion all tested hmC-containing DNA substrates both in vivo and in vitro, including the non-glucosylated T4 genome (13,14). This indicates that β-gt can glucosylate hmC independently of DNA sequence and structural context and therefore is ideally suited for this assay.
Figure 1.
Elements of the hmC glucosylation assay. (A) Schematic representation of the hmC glucosylation reaction catalyzed by β-gt. (B) Coomassie blue stained gel showing the purified β-gt preparation. (C) Example of calibration curve using mixtures of hmC-containing and unmodified reference fragment (equal total DNA amounts). Note the linear relationship between [3H]glucose incorporation and percentage of hmC.
MATERIALS AND METHODS
Cell culture and differentiation of ESCs
Undifferentiated J1 and E14 ESCs were maintained on gelatin-coated dishes in Dulbecco's modified Eagle's medium containing 16% fetal bovine serum (PAA Laboratories GmbH), 0.1 mM β-mercaptoethanol (Invitrogen), 2 mM l-glutamine, 1 × MEM Non-essential Amino Acid Solution, 100 U/ml penicillin, 100 μg/ml streptomycin (PAA Laboratories GmbH) and 1000 U/ml recombinant mouse LIF (Millipore). To induce embryoid body (EB) formation, ESCs were resuspended in the same medium as above but without LIF and cultured in hanging drops (600 cells/20 μl drop) for 4 days. Subsequently, EBs were cultured in bacterial culture dishes and the medium was replaced every 4 days.
DNA and RNA isolation
All tissue samples were prepared from 6-week-old 129sv mice. Genomic DNA and total RNA were isolated from tissue samples using the NucleoSpin Triprep Kit (Macherey-Nagel). Genomic DNA and total RNA were isolated from ESCs and EBs using the Blood & Cell culture DNA mini kit (QIAGEN) and TRIzol reagent (Invitrogen), respectively. To avoid genomic DNA contamination, isolated RNA was digested with recombinant RNase-free DNase I (Roche) and further purified with the QIAGEN RNeasy kit. Genomic DNA samples were sheared to 500–1500 bp fragments by sonication to reduce the viscosity and improve homogeneity. The concentration of genomic DNA samples was measured by fluorometry. Fifty microliters of diluted sample were mixed with 50 µl of 2 × TNE (Tris 20 mM, pH 7.4; NaCl 400 mM and EDTA 2 mM) containing 200 ng/µl of Hoechst 33258. Fluorescence was measured in a TECAN infinite M1000 plate reader (Ex: 350/10; Em: 455/10). Serial dilutions (20–2000 ng/ml) of the hmC containing reference DNA fragment (see below) were used as standard for quantification.
Protein expression and purification
The sequence encoding bacteriophage T4 β-gt was synthesized at Mr. Gene GmbH (Regensburg) and cloned into pET28b vector (Novagen). BL21(DE3) E. coli cells carrying the expression construct were grown at 37°C until A600 = 0.6–0.7 and induced with 1 mM isopropyl β-d-thiogalactopyranoside for 16 h at 20°C. Lysates were prepared by sonication in 300 mM NaCl, 50 mM Na2HPO4, 10 mM imidazole, 1 mM β-mercaptoethanol, cleared by centrifugation and applied to nickel-nitrilotriacetic acid column (QIAGEN) pre-equilibrated with lysis buffer. Washing and elution were performed with lysis buffer containing 20 and 250 mM imidazole, respectively. Eluted proteins were applied to a Superdex S-200 preparative gel filtration column (GE Healthcare) in 150 mM NaCl, 20 mM Tris, pH 8.0, 1 mM DTT. Fractions containing the β-gt peak were pooled and applied to a ResourceQ anion exchange column (GE Healthcare) in order to eliminate residual contaminants, resulting in pure β-gt in the flowthrough.
Preparation of reference DNA fragments
Reference DNA fragments (1139 bp) containing 0 and 100% hmC were prepared by polymerase chain reaction (PCR), using dCTP and 5-hydroxymethyl-dCTP (Bioline GmbH), respectively. T4 phage DNA was used as template with primers: 5′-TGG AGA AGG AGA ATG AAG AAT AAT-3′ and 5′-GTG AAG TAA GTA ATA AAT GGA TTG-3′, Phusion HF DNA Polymerase (Finnzymes) and the following cycling profile: one cycle of 98°C for 2 min and 35 cycles of 98°C for 10 s; 58°C for 10 s; and 72°C for 30 s. Primer sequences were selected that do not contain cytosine residues. PCR products were purified by gel electrophoresis followed by silica column purification (Nucleospin, Macherey-Nagel).
Quantitative hmC glucosylation assay
Reactions contained 150 mM NaCl, 20 mM Tris, pH 8.0, 25 mM CaCl2, 1 mM DTT, 2.8 μM ‘cold’ UDP-glucose (Sigma-Aldrich), 0.86 nM UDP-[3H]glucose (glucose-6-3H; 60 Ci/mmol; Hartmann Analytic GmbH), 1 µg of DNA substrate and 36 nM recombinant β-gt in a total volume of 50 µl. Reactions were incubated for 20 min at room temperature and terminated by heating at 65°C for 10 min. Twenty microliters of each reaction were spotted in duplicate on paper filters (Whatmann) and DNA was precipitated by incubation in 5% TCA for 15 min at room temperature. Filters were washed twice with 5% TCA and once with 70% ethanol. Remaining radioactivity was measured using a Liquid Scintillation Analyzer Tri-Carb 2100TR (Packard) with quench indicating parameter set on tSIE/AEC (transformed spectral index of the external standard/automatic efficiency control) in 4 ml of Rotiszint Eco Plus scintillation liquid (Roth GmbH) in Snaptwist vials (Simport). Samples were measured for 30 min or until the 2σ value reached 2%. The percentage of hmC per total cytosine was calculated from the incorporation of [3H]glucose using a calibration curve measured with the reference fragment series for every experiment. The percentage of hmC was then corrected for the difference in C abundance between reference fragment (35%) and mouse genome (42%).
cDNA synthesis and real-time PCR
Five hundred nanograms of total RNA were used for cDNA synthesis with the High-Capacity cDNA Reverse Transcription Kit (with RNase Inhibitor; Applied Biosystems). Equal amounts of cDNA were used for real-time PCR with Power SYBR Green PCR Master Mix (Applied Biosystems) on a 7500 Fast Real-Time PCR System (Applied Biosystems) according to the manufacturer's instructions. Gene expression levels were normalized to Gapdh and calculated using the comparative CT method (ΔΔCT method).
Primers for quantitative real-time PCR were designed with the Primer Express software (Applied Biosystems) and contained the following sequences: Gapdh forward 5′-CAT GGC CTT CCG TGT TCC TA-3′, Gapdh reverse 5′-CTT CAC CAC CTT CTT GAT GTC ATC-3′, Tet1 forward 5′-CCA GGA AGA GGC GAC TAC GTT-3′, Tet1 reverse 5′-TTA GTG TTG TGT GAA CCT GAT TTA TTG T-3′, Tet2 forward 5′-ACT TCT CTG CTC ATT CCC ACA GA-3′, Tet2 reverse 5′-TTA GCT CCG ACT TCT CGA TTG TC-3′, Tet3 forward 5′-GAG CAC GCC AGA GAA GAT CAA-3′ and Tet3 reverse 5′-CAG GCT TTG CTG GGA CAA TC-3′.
RESULTS AND DISCUSSION
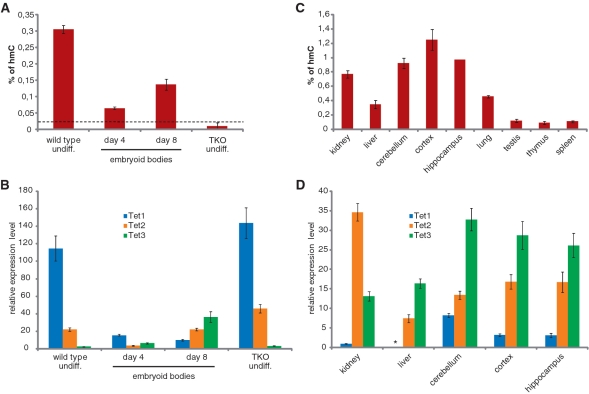
T4 β-gt was expressed in bacteria as a 6 × His tag fusion and purified to homogeneity by sequential nickel-NTA, size exclusion and ion exchange chromatography (Figure 1B). To assess whether transfer of [3H]glucose to DNA is proportional to the hmC content within the range previously reported for mammalian tissues, we prepared a series of standard DNA substrate samples with global hmC content ranging from 0.25 to 2% of total cytosine by mixing corresponding proportions of two preparations of the same 1.2 kb DNA fragment, one having all cytosine residues replaced by hmC and the other containing no hmC (Figure 1C). Using a 325-fold excess of unlabelled UDP-glucose, the incorporation of radiolabeled glucose in 1 µg of total DNA substrate was strictly linear in this range. This standard sample series was measured in every assay to generate a calibration curve for the calculation of hmC content in genomic DNA samples. We first measured genomic hmC levels in wild-type and Dnmt1, 3a and 3b triple knockout (TKO) J1 ESCs (15) (Figure 2A and B). Due to the absence of all three major DNA methyltransferases, genomic DNA from TKO ESCs is expected to contain very little, if any, cytosine methylation. Indeed, the measured level of genomic hmC in TKO ESCs was at the detection limit (0.025%) of our assay, while genomic DNA from wild-type ESCs contained 0.3% hmC relative to total cytosine. Real-time reverse transcription (RT) PCR analysis showed that Tet1–3 mRNA levels are similar in wild-type and TKO ESCs, with Tet1 transcripts largely preponderant and Tet3 mRNA the least abundant (more than 40-fold lower than Tet1). It was previously shown that differentiation of mouse ESCs by withdrawal of LIF from monolayer cultures for 5 days results in a reduction of genomic hmC and concomitant decrease in Tet1 transcripts (2). We followed genomic hmC and Tet1–3 transcript dynamics during EB differentiation of two commonly used wild-type ESC lines (Figure 2A and B and Supplementary Figure S1). In both cases, a sharp decrease of hmC content was evident after 4 days of EB culture, but a substantial recovery was observed after additional 4 days of culture (Day 8). Interestingly, the tet genes showed distinct expression dynamics during ESCs differentiation. Tet1 transcripts drastically decreased in the first 4 days and further dropped by Day 8 of EB culture. Tet2 mRNA levels also decreased in the first 4 days, but were fully restored in Day 8 EBs. In contrast, Tet3 transcripts doubled at Day 4 and increased about 20 times by Day 8 of EB culture. Thus, the relatively high hmC content in undifferentiated ESCs correlates with high levels of Tet1 and, to a lower extent, Tet2 transcripts, while the partial recovery of genomic hmC in Day 8 EBs correlates with increased Tet2 and Tet3 mRNA levels.
Figure 2.
Quantification of genomic hmC and Tet transcripts in mouse tissues, undifferentiated ESCs and EBs. (A and C) hmC glucosylation assays. The percentage of hmC per total cytosine was calculated from the incorporation of [3H]glucose using a calibration curve from the reference fragment (see Figure 1C). Shown are average values and error bars from two (A) or one (C) biological replicates, each measured in two independent assays, with the exception of hippocampus that was measured only once. In every assay, each sample was measured in duplicate. The dashed line in (A) indicates the estimated limit of detection. (B and D) Real-time RT–PCR analysis for Tet transcript levels. Expression levels are all relative to Tet1 in kidney (set to 1), so that values in b and d are directly comparable. Shown are average values and error bars from two (B) and one (D) biological replicates, each measured from two independent cDNA synthesis reactions. In every real-time PCR reaction, each sample was measured in triplicate. Genomic DNA and RNA samples used in A/C and B/C, respectively, were isolated from the very same cell and tissue lysates.
We then analyzed several adult mouse tissues (Figure 2C and D). As reported earlier, the highest levels of genomic hmC were found in brain regions, although kidney also showed relatively high levels. In all cases, the hmC content was at least four times higher than the detection limit, while in a previous report using a different assay the same non-neural tissues were either marginally above or right at the detection limit (5). Abundant hmC in brain tissues correlated with high levels of Tet3 and to a lower extent Tet2, a pattern similar to Day 8 EBs. Thus, most differentiated tissues are characterized by very low levels of Tet1 and high levels of Tet3, while undifferentiated ESCs show the opposite pattern. It will be interesting to determine whether all pluripotent cell types have this pattern and at which stages along the specification of the various somatic lineages the relative expression levels of tet genes change. Interestingly, kidney represents an exception among the adult tissues analyzed as it shows relatively high hmC content and a prevalence of Tet2 transcripts. This is consistent with a cellular defect in proximal convoluted tubules of the kidney as the only phenotype described for Tet2 null mice (16). These observations suggest that Tet proteins have redundant roles and that the lack of a specific Tet family member may result in phenotypic alterations only in tissues where high levels of that Tet enzyme cannot be compensated by the other family members. In this context, it should be noted that the assay described here could also be employed to measure the enzymatic activity of Tet proteins and their mutants identified in leukemia patients by using mC-containing DNA substrates.
In conclusion, we have established an accurate assay for the quantification of genomic hmC that: (i) is more sensitive than previously described methods; (ii) is not subject to sequence bias; (iii) allows simultaneous processing of large sample numbers; and (iv) does not require specialized and expensive equipment. It should be noted that lower concentrations of ‘cold’ UDP-glucose should allow scaling down the amount of substrate DNA without loss of signal. This assay will be highly useful to determine the global abundance of hmC in genomic DNA, especially in situations where limited amounts of tissue are available such as isolates of rare primary cell types and clinical samples.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft (Grants DFG SPP 1356, SFB 646 and 684 to H.L.); financial support by the Elite Network of Bavaria (International Doctorate Program NanoBioTechnology) and the International Max Planck Research School for Molecular and Cellular Life Sciences (IMPRS-LS to C.S.S.). S.B. is a fellow of the graduate school Life Science Munich (LSM). Funding for open access charge: Deutsche Forschungsgemeinschaft (DFG SPP 1356, SFB 646 and 684 to H.L.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Daniela Meilinger, Andrea Steiner and Andreas Brachmann (Ludwig Maximilians University Munich) for help with EB culture, dissection of mouse tissues and scintillation counting, respectively. TKO ESCs were provided by Masaki Okano (RIKEN Center for Developmental Biology, Kobe, Japan).
REFERENCES
- 1.Rottach A, Leonhardt H, Spada F. DNA methylation-mediated epigenetic control. J. Cell. Biochem. 2009;108:43–51. doi: 10.1002/jcb.22253. [DOI] [PubMed] [Google Scholar]
- 2.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iyer LM, Tahiliani M, Rao A, Aravind L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8:1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Penn NW, Suwalski R, O’Riley C, Bojanowski K, Yura R. The presence of 5-hydroxymethylcytosine in animal deoxyribonucleic acid. Biochem. J. 1972;126:781–790. doi: 10.1042/bj1260781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loenarz C, Schofield CJ. Oxygenase catalyzed 5-methylcytosine hydroxylation. Chem. Biol. 2009;16:580–583. doi: 10.1016/j.chembiol.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 8.Lorsbach RB, Moore J, Mathew S, Raimondi SC, Mukatira ST, Downing JR. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23) Leukemia. 2003;17:637–641. doi: 10.1038/sj.leu.2402834. [DOI] [PubMed] [Google Scholar]
- 9.Ono R, Taki T, Taketani T, Taniwaki M, Kobayashi H, Hayashi Y. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23) Cancer Res. 2002;62:4075–4080. [PubMed] [Google Scholar]
- 10.Langemeijer SMC, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, Stevens-Linders E, van Hoogen P, van Kessel AG, Raymakers RAP, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat. Genet. 2009;41:838–842. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- 11.Hayatsu H, Shiragami M. Reaction of bisulfite with the 5-hydroxymethyl group in pyrimidines and in phage DNAs. Biochemistry. 1979;18:632–637. doi: 10.1021/bi00571a013. [DOI] [PubMed] [Google Scholar]
- 12.Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS ONE. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Georgopoulos CP, Revel HR. Studies with glucosyl transferase mutants of the T-even bacteriophages. Virology. 1971;44:271–285. doi: 10.1016/0042-6822(71)90259-5. [DOI] [PubMed] [Google Scholar]
- 14.Kornberg SR, Zimmerman SB, Kornberg A. Glucosylation of deoxyribonucleic acid by enzymes from bacteriophage-infected Escherichia coli. J. Biol. Chem. 1961;236:1487–1493. [PubMed] [Google Scholar]
- 15.Tsumura A, Hayakawa T, Kumaki Y, Takebayashi S, Sakaue M, Matsuoka C, Shimotohno K, Ishikawa F, Li E, Ueda HR, et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells. 2006;11:805–814. doi: 10.1111/j.1365-2443.2006.00984.x. [DOI] [PubMed] [Google Scholar]
- 16.Tang H, Araki K, Li Z, Yamamura K. Characterization of Ayu17-449 gene expression and resultant kidney pathology in a knockout mouse model. Transgenic Res. 2008;17:599–608. doi: 10.1007/s11248-008-9170-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.