Abstract
The helical tension of chromosomal DNA is one of the epigenetic landmarks most difficult to examine experimentally. The occurrence of DNA crosslinks mediated by psoralen photobinding (PB) stands as the only suitable probe for assessing this problem. PB is affected by chromatin structure when is done to saturation; but it is mainly determined by DNA helical tension when it is done to very low hit conditions. Hence, we developed a method for genome-wide analysis of DNA helical tension based on PB. We adjusted in vitro PB conditions that discern DNA helical tension and applied them to Saccharomyces cerevisiae cells. We selected the in vivo cross-linked DNA sequences and identified them on DNA arrays. The entire procedure was robust. Comparison of PB obtained in vivo with that obtained in vitro with naked DNA revealed that numerous chromosomal regions had deviated PB values. Similar analyses in yeast topoisomerase mutants uncovered further PB alterations across specific chromosomal domains. These results suggest that distinct chromosome compartments might confine different levels of DNA helical tension in yeast. Genome-wide analysis of psoralen–DNA PB can be, therefore, a useful approach to uncover a trait of the chromosome architecture not amenable to other techniques.
INTRODUCTION
In eukaryotic cells, multiple molecular interactions drive DNA to fold into nucleosomes and chromatin fibers (1). Protein modifications and other molecular ensembles organize chromatin fibers into domains of different conformational and functional capabilities (2). In recent years, the development of genome-wide analyses began to uncover the complex landscape of eukaryotic chromatin structure and its role on epigenetic regulation (3,4). In Saccharomyces cerevisiae, the distribution of several DNA-binding proteins (5–7), DNA-acting enzymes (8–10), histone modifications (11,12), and the bulk of nucleosome positions (13,14) had been mapped genome wide. Whereas these analyses provide valuable information about what is interacting with DNA in a given experimental condition, not as much is known about the conformational state of DNA.
One conformational aspect of DNA with deep implications in the genome transactions is the helical tension of the duplex. This condition of altered twist facilitates or hinders the melting of DNA strands, as well as its interactions with structural and regulatory factors (15). DNA helical tension also promotes the formation of supercoils that contribute to the juxtaposition of distant DNA sites and to the global DNA compaction (16). In bacteria, several lines of evidence indicate that chromosomes are organized into supercoiled DNA domains (17–19), in which DNA helical tension regulates DNA transcription, replication and recombination (20). In eukaryotic cells, the issues of whether DNA helical tension configures chromatin domains and regulates genome transactions are less clear and remain controversial (21–23). A conceptual difficulty for the above questions is to dissect the multiplicity of factors that determine the generation, transmission, and dissipation of DNA twisting forces in vivo. Thus far, besides the in vivo evidence of transcription-driven supercoiling of DNA (24) and the known capability of different topoisomerases to relax DNA (25), our knowledge about DNA helical tension in eukaryotes is very limited. A main hurdle for these studies is the lack of techniques to examine the topology of chromosomal DNA.
To date, many studies assessing the helical tension in chromosomal DNA had relied on the use of psoralens. These compounds have a planar aromatic structure that allows them to cross cell membranes and to randomly intercalate into DNA (26). Upon exposure to ‘UV’ light (360 nm), intercalated psoralens photobind to DNA and crosslink its complementary strands (Figure 1A). The most favorable contacts for crosslink formation occur at 5′-TA dinucleotides, where the adjacent thymines on the opposite strand become covalently bonded at each end of the psoralen (26,27). Due to the intercalation requirement, the probability of psoralen binding and crosslink formation increases with DNA negative helical tension, since it facilitates the unwinding of the duplex (28). Following this correlation, in vivo measurements of global psoralen–DNA photobinding (PB) in Escherichia coli had indicated that the bacterial chromosome has, in average, significant levels of unconstrained negative helical tension (28,29). In contrast, analogous studies conducted in eukaryotic cells did not detect significant DNA torsional stress in their chromosomal DNA (28,29). Yet, local analyses of psoralen PB denoted the presence of negative helical tension in particular gene loci in yeast (30,31), Drosophila (32,33) and human cells (34–36).
Figure 1.
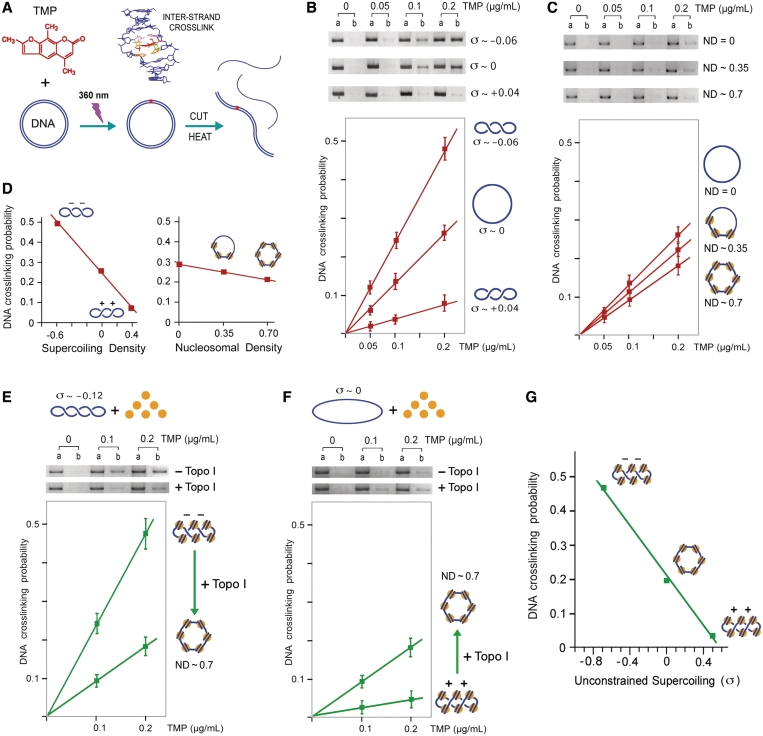
Effect of DNA helical tension and DNA–protein interactions on DNA-crosslinking probability mediated by psoralen PB. (A) Incubation of circular DNA with TMP followed by ‘UV’ irradiation produces TMP-mediated DNA inter-strand crosslinks. The fraction of linearized DNA molecules resistant to thermal denaturation indicates DNA-crosslinking probability. (B) Negatively supercoiled (σ ∼ −0.06), relaxed (σ ∼ 0), and positively supercoiled (σ ∼ +0.04) forms of YCp50 (an 8-kb plasmid) were dissolved in TE (100 µg/ml) and incubated with TMP (0, 0.05, 0.1, 0.2 µg/ml). Following irradiation with 360 nm light at a dose of 1.2 kJ/m2/min during 120 s, DNA was purified and linearized with EcoRI endonuclease. One half of each DNA sample was directly loaded on an agarose gel (lane A). The other half was boiled for 1 min and quickly chilled on ice prior inspection by the gel electrophoresis (lane B). DNA-crosslinking probability was calculated from the un-denatured molecular fraction as described in the ‘Materials and Methods’ section. The graph averages results from three experiments. (C) Chromatin was assembled in the YCp50 plasmid as described in the ‘Materials and Methods’ section and Supplementary Figure S1 to obtain nucleosome densities of 0, 0.35 and 0.7. TMP PB and evaluation of DNA-crosslinking probability was conducted as in B. (D) Comparison of the effect of DNA supercoiling density and DNA ND on the probability to produce one DNA crosslink per plasmid molecule. (E) Highly negatively supercoiled (σ ∼ −0.12) YCp50 was used to assembly chromatin (ND of 0.7) in the absence of topoisomerase activity. TMP PB (0, 0.1, 0.2 µg/ml) was examined both before and after relaxation of the chromatin with topoisomerase I. PB and evaluation of DNA-crosslinking probability were conducted as in B. (F) Experiment identical to E, but using relaxed (σ ∼ 0) YCp50. (G) Plot of TMP–DNA PB probability and unconstrained DNA helical tension on chromatinized DNA. Values of unrestrained DNA supercoiling on chromatin were estimated after experiments shown in (E) and (F), and given that ND of 0.7 corresponds to ∼40 nucleosomes per 8-kb plasmid molecule and that each nucleosome is able to constrain about one negative supercoil.
The above in vivo psoralen PB studies focused either in whole cell measurements or in the analysis of specific loci. Therefore, relative differences of psoralen PB among different chromosomal regions had not been systematically inspected. Thus far, one genome-wide study had been carried out in polytene chromosomes of Drosophila, in which the distribution of biotinylated psoralen was visualized with fluorescent streptavidin. The relative intensity of the signals denoted substantial differences of DNA helical tension between many chromosomal interbands and puffs (37). In the present study, we pursued to develop a general method for a genome-wide analysis of psoralen PB, as it stands the only suitable biochemical probe for assessing the helical state of chromosomal DNA.
To do so, we show first how to calibrate in vitro psoralen-mediated DNA-crosslinking conditions that discern differences of DNA helical tension. Next, we show how to reproduce these conditions in vivo, select the psoralen-mediated crosslinked DNA sequences, and convert them into array signals. In order to minimize the contribution of DNA base pair sequence in the crosslinking probability, we show how to calculate for each array signal the ratio of PB in vivo to that obtained in vitro with naked DNA. We applied the procedure to S. cerevisiae cells and topoisomerase mutants. The technique readily uncovered that distinct chromosome compartments might confine different levels of DNA helical tension in yeast.
MATERIALS AND METHODS
DNA substrates for in vitro TMP–DNA PB
TMP (4,5′, 8-trimethylpsoralen from Sigma) was kept in stock solution (200 µg/ml in ethanol). Relaxed and supercoiled DNA forms of the yeast plasmid YCp50 (8 kb) with specific DNA linking number differences (σ) +0.04, 0, −0.06 and −0.12 were prepared as described previously (38). Chromatin was assembled in vitro by incubating during 4 h at 35°C relaxed plasmid YCp50 with a yeast extract supplemented with an ATP-regenerating system, vaccinia virus topoisomerase I and increasing amounts of core histones (purified from yeast chromatin) as described previously (39). Nucleosome assembly was monitored by the reduction of DNA linking number, given that topoisomerase activity removed the compensatory supercoils generated during the wrapping of DNA around core histones (Supplementary Figure S1A). Nucleosomal density was calculated as nucleosomal DNA length/total DNA length, assuming that each assembled nucleosome constrains dLk ∼ -1 (Supplementary Figure S1). Micrococcal nuclease digestion of the assembled minichromosomes was done to confirm the typical phasing of loaded nucleosomes (Supplementary Figure S1B). Minichromosomes with unconstrained DNA supercoils were obtained by the same experimental procedure, but using yeast extracts depleted of topoisomerase activity (from Δtop1 top2tscells).
Calculation of DNA-crosslinking probability
Following TMP–DNA PB reactions, one half of each DNA sample was boiled during 1 min and quickly chilled on ice prior to examination by agarose gel electrophoresis. Gels were stained with Ethidium Bromide or SYBR Green, and DNA bands quantified using Kodak Molecular Imaging Software. The mean number of crosslinks per unit length (nX) was calculated assuming a Poisson distribution of hits with nX = −ln(1 – Fx), in which Fx is the fraction of crosslinked DNA molecules produced in each reaction. Fx is determined by dividing amount of DNA resistant to denaturation by the total amount of input DNA.
In vivo TMP–DNA PB and selection of DNA-crosslinked fragments
Saccharomyces cerevisiae strain FY251 (MATa his3-Δ200 leu2-Δ1 trp1-Δ63 ura3–52) and its mutant JCW28 (Δtop1 top2ts FY251), that harbors a deletion of the TOP1 gene and a thermo-sensitive mutation in the TOP2 gene, had been described previously (40). Cells were grown at 30°C in YPD media up to exponential phase. When indicated, thermal inactivation of topoisomerase II was carried out by the addition of hot media to equilibrate the cultures at 37°C during 10 min. Cultures were then split to conduct psoralen–DNA PB and transcription run-on analyses in independent flasks. An aliquot of TMP was added to a 20 ml of cell culture (OD ∼0.6) to yield a final TMP concentration of 4 µg/ml. Incubation continued during 10 min in a dark environment. Yeast cells were then poured into a Petri dish, exposed for 120s to 360 nm light at a dose of 1.2 kJ/m2/min and immediately washed by three rounds of sedimentation in cold water. Around 5 × 108 cells were then resuspended in 500 µl of buffer L (Tris–HCl pH 8, 50 mM, EDTA 1 mM, NaCl 150 mM, BME 7 mM, SDS 1%) plus 300 µl glass beads (425 µm from Sigma). Cell disruption was done at room temperature in 1.5 ml centrifuge tubes using the Fast-Prep Bio101 apparatus (2 × 30 s pulses at setting 5.5). Cell lysates were diluted by addition of 1 ml of TE and chromosomal DNA was then fragmented down to a 2 kb average size by using a Branson Sonifier (3 × 6s pulses at setting 5). Following an overnight incubation at 37°C with proteinase K and RNAse I, the lysates were extracted twice with phenol and once with phenol–chloroform. DNA was precipitated with EtOH, washed with 70% EtOH and resuspended in 100 µl of water. To select the crosslinked fragments, DNA samples were boiled for 2 min, chilled on ice and then digested with excess Exonuclease I (NE BioLabs) during 4 h. Undigested DNA was recovered with a GFX filtration kit (Amersham) in 50 µl of water and further digested with excess λ Exonuclease (NE BioLabs) during 6 h. The reaction terminated by inactivation of the enzyme at 80°C during 10 min. To examine TMP PB to purified yeast DNA, unreacted cells were disrupted using the Fast-Prep apparatus and total DNA was fragmented down to a 2 kb average size by using a Branson Sonifier, as above. Following proteinase K and RNAse I incubations, DNA fragments were extracted twice with phenol and once with phenol–chloroform, precipitated with EtOH, washed with 70% EtOH and resuspended in 100 µl of water. Genomic DNA fragments were incubated with TMP (0.4 µg/ml) and irradiated (360 nm, at a dose of 1.2 kJ/m2/min during 120s) to produce an amount of crosslinks comparable with that obtained in vivo (1 crosslink/10 kb). Crosslinked fragments were selected by exonuclease digestions as above.
DNA array hybridization
DNA arrays were provided by the DNA Chip Laboratory at Universitat de Valencia (Spain) and consist of nylon membranes printed with 6020 PCR-amplified ORFs segments of S. cerevisiae (41). Exonuclease resistant DNA chains were subjected to random priming with the Strip-EZ DNA kit (Ambion), which incorporated 33P-dATP and a modified dCTP nucleotide that produces easily strippable chains. The arrays were pre-hybridized with 10 ml UltraHyb solution (Ambion) in a roller oven during 1 h at 46°C. 33P-dATP labeled DNA (5 × 106 dpm/ml) was then added and the hybridization conducted during 16 h at 46°C. Arrays were washed at 65°C, once in 2 × SSC plus 0.1% SDS for 20 min and twice in 0.2 × SSC plus 0.1% SDS for 30 min. Arrays were then exposed for 1–3 days to an imaging plate (BAS-MP, FujiFilm). Signal intensities were read at 50 nm resolution in a phosphor-imager scanner (FLA-3000, FujiFilm). The degradation buffer of Strip-EZ DNA (Ambion) was used to remove the signals from the arrays, so allowing up to 6–8 re-hybridization rounds (41).
Quantification of array signals and data analysis
Three biological replicates of each experiment were conducted by using independent yeast colonies and by swapping array membranes among them. Spot intensities were analyzed by using ArrayVision 7.0 software (Imaging Research, Inc.), taking the sARM density (with subtracted local background) as signal. Intensity values were normalized within each experiment by the global mean procedure. Reproducibility of the replicates was tested by the ArrayStat software (Imaging Research, Inc.) considering the data as independent and allowing the program to take a minimum of two valid replicates to calculate a mean value for every spot signal (ORF). Normalized data sets were used to calculate for each array spot the ratio (PB ratio) of TMP PB of the in vivo to the in vitro conditions. In order to determine whether subsets of array spots had a mean PB ratio statistically different from that of the global distribution of PB ratios, the two-tailed Student’s t-test was used to detect significant variations of mean PB ratios among different chromosomal arms and different chromosomal regions spanning 20 genes.
Genome-wide transcription run-on
Yeast transcription run-on, array hybridization and data analyses were done as described by García-Martínez et al. (42) with minor modifications. Around 5 × 108 cells were washed in cold water and permeabilized with 0.5% N-lauryl sarcosine sodium sulfate. In vivo transcription was conducted by incubating cells during 5 min at 30°C in 250 µl of buffer T (20 mM Tris–HCl pH 7.7, 200 mM KCl, 50 mM MgCl2, 2 mM DTT) plus 4 mM each of CTP, ATP, GTP and 12 µl of 33P-UTP (3000 Ci/mmol, 10 µCi/µl). In vivo labeled RNA was purified and hybridized (0.2–2 × 107 dpm/5 ml) on array membranes of the same lot used for TMP PB analyses. Genomic DNA was random priming labeled and hybridized on the same arrays. These data were used for array and probe normalization as described (42).
RESULTS AND DISCUSSION
Effect of negative and positive DNA helical tension on TMP–DNA PB probability
Before carrying out in vivo experiments, we examined in vitro how different levels of DNA helical tension would affect psoralen–DNA PB probability. Earlier studies by Sinden et al. (28) had shown that psoralen PB probability increases over 2-fold in negatively supercoiled plasmids (superhelical density ∼−0.06, a DNA linking number reduction of ∼6% relative to relaxed DNA). Since both negative and positive helical tension might occur at intracellular DNA (25), we extended their in vitro analysis to positively supercoiled plasmids. We prepared an 8 kb plasmid (YCp50) with three physiological degrees of superhelical density (σ): negatively supercoiled (σ ∼ −0.06), relaxed (σ ∼ 0), and positively supercoiled (σ ∼ +0.04). We used TMP by its good solubility and DNA-crosslinking efficiency (28,43). Each plasmid form was incubated with low concentrations of TMP and briefly irradiated with ‘UV’ light (360 nm), in order to produce no more than one TMP-mediated DNA inter-strand crosslink per plasmid molecule. After the reactions, plasmids were cut into a linear form, submitted to thermal denaturation and examined by agarose gel electrophoresis. DNA-crosslinking probability in each sample was determined from the fraction of DNA resistant to denaturation (Figure 1A). In the three topological forms, a linear dose response for DNA crosslinking was observed with TMP concentrations up to 0.2 µg/ml. In this range, DNA-crosslinking probability in the negatively supercoiled molecules was ∼2-fold higher than in the relaxed, and 5-fold higher than the positively supercoiled ones (Figure 1B). Therefore, with this low hit condition, the probability of DNA crosslinking mediated by TMP PB nearly followed a linear correlation with the examined range of DNA supercoiling density (Figure 1D).
Effect of DNA–protein interactions on TMP–DNA PB probability
In addition to DNA helical tension, psoralen–DNA PB can be affected by DNA–protein interactions. Accordingly, this technique has been used to examine chromatin organization (44,45), local changes of chromatin structure in RNA polymerase I transcribed rDNA (46,47) and in RNA polymerase II transcribed genes (48,49), and to determine nucleosome positioning (50). These analyses required a density of PB hits much higher (20–1000 hits/10 kb) than that we calibrated here to discern different levels of positive and negative DNA helical tension (<1 hit/10 kb). Yet, we need to examine if DNA–protein interactions would significantly affect PB probability in these very low hit conditions. To do so, we used cell-free chromatin assembly system derived from the yeast S. cerevisiae, which efficiently packages DNA into minichromosomes (39). Nucleosome assembly was monitored as topoisomerase activity relaxed the compensatory supercoils generated during the wrapping of DNA around histone octamers (Supplementary Figure S1). We obtained plasmids with nucleosomal densities of 0, 0.35 and 0.7 (nucleosomal DNA length/total DNA length); and photoreacted them with low concentrations of TMP to produce no more than one DNA inter-strand crosslink per plasmid molecule. A linear dose response for crosslink formation was observed with TMP concentrations up to 0.2 µg/ml. Yet, relative to naked DNA, a physiological nucleosomal density of 0.7 reduced only by 0.3-fold the probability of producing one DNA crosslink per plasmid molecule (Figure 1C). Therefore, in comparison to the larger variation produced by the helical state of DNA, PB probability at low hit conditions was minimally affected by DNA–protein interactions (Figure 1D).
TMP–DNA PB probability on chromatin domains with unconstrained DNA helical tension
Finally, in order to reproduce more closely the state of intracellular DNA, we examined how TMP PB probability at low hit conditions would be affected in chromatinized DNA with unrestrained negative and positive DNA supercoils. To do so, a highly negative superhelical form (σ ∼ −0.12) and the relaxed form (σ ∼ −0) of the YCp50 plasmid were incubated with an excess of core histones (to reach a physiological nucleosomal density of 0.7) in the cell-free chromatin assembly system, as above, but lacking DNA topoisomerase activity. Consequently, most of the negative helical tension in the highly superhelical form of the plasmid (σ ∼ −0.12) would remain unrestrained because wrapping of DNA around histones could only constrain a supercoiling density of ∼−0.5. On the other hand, in the relaxed form of the plasmid (σ ∼ −0), positive DNA helical tension would be generated to compensate the negative supercoils constrained by the wrapping of DNA around histones. TMP PB was then examined in both topological forms of chromatinized DNA before and after their relaxation with topoisomerase I. The results showed that, when the initial plasmid was highly negatively supercoiled, PB probability was 2.5-fold higher than after its relaxation (Figure 1E), thus confirming that most negative helical tension was not constrained by nucleosome assembly. Conversely, when the initial plasmid was relaxed, PB probability was 3.5-fold lower than after its relaxation (Figure 1F), thus confirming that compensatory positive helical tension was generated by nucleosome assembly. These results indicated that, in chromatinized DNA with unrestrained negative and positive supercoils, PB probability changes by several fold (Figure 1G) within a magnitude and slope comparable to that observed with naked DNA (Figure 1D). Therefore, by using low hit conditions in vivo (1 hit/10 kb), regional differences of DNA helical tension should translate in substantial changes of psoralen PB probability, whereas differences in the extent of DNA–protein interactions should barely affect it. Namely, only a large depletion of nucleosomes, leaving intracellular DNA naked along several kilobites, would appreciably increase the probability to produce one DNA crosslink in a given chromosomal region.
Calibration of the extent of DNA crosslinks produced in vivo by TMP PB
In order to detect genome-wide differences of DNA helical tension in S. cerevisiae cells, we first determined the conditions to produce one TMP-mediated crosslink per about 10 kb of intracellular DNA. These low hit conditions would be also adequate to minimize plausible interferences with physiological DNA transactions during the PB reaction. To do so, exponentially growing cells were incubated with low concentrations TMP and briefly irradiated as described in the ‘Materials and Methods’ section. Next, total yeast DNA was purified and randomly fragmented to an average size of ∼2 kb. The extent of DNA crosslinks produced was then calculated from the portion of fragments resistant to thermal denaturation.
For our reference strain FY251, we found a lineal increase of DNA-crosslinked fragments by incubating the cells with increasing TMP concentrations up to 16 µg/ml during 10 min, followed by irradiation with 360 nm light during 2 min, at a dose of 1.2 kJ/m2/min (Supplementary Figure S2A). In this range, a TMP concentration of 4 µg/ml produced ∼20% DNA fragments containing one crosslink (that is ∼1 DNA crosslink/10 kb). Genomic DNA samples obtained with this condition were thus selected to carry out the genome-wide analyses.
Selection of DNA-crosslinked fragments produced by TMP PB
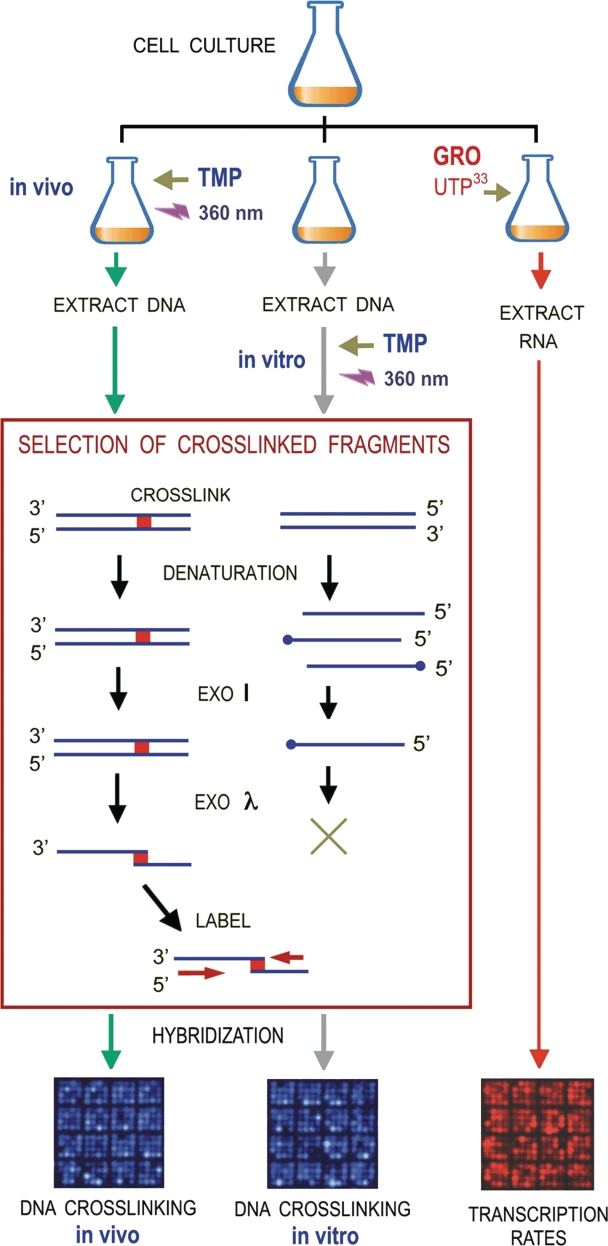
In order to identify the in vivo crosslinked sequences, un-crosslinked DNA fragments had to be efficiently removed from the genomic DNA sample. To do so, we carried out a sequential digestion with Exonuclease I and Exonuclease λ as illustrated in Figure 2. Quality enzymes provided by most manufacturers (free of other detectable activities) were appropriate for this step. First, DNA was thermally denatured, such that un-crosslinked fragments were converted into single DNA strands. Exonuclease I degraded then these single DNA strands by from their 3′-end. The remaining crosslinked duplexes and plausible DNA strands with a blocked 3′-end with a topoisomerase I cleaved-DNA intermediate (51) were then digested by Exonuclease λ from their 5′-ends. As a result, all DNA was degraded except crosslinked fragments, which were converted into a pair of single DNA chains covalently linked by a TMP molecule near their 5′-ends. This selection process proved to be efficient and specific (Supplementary Figure S2B). First, the final fraction of DNA resistant to the exonucleases was ∼10% (w/w) of the input DNA, an amount consistent with the presence of approximately one DNA crosslink per 10 kb. Second, when DNA from un-reacted cells (TMP omitted) was subjected to the same treatment, the final fraction of DNA resistant to the exonuclease treatment was negligible (<1%).
Figure 2.
Experimental setting for genome-wide analysis of DNA-crosslinking probability mediated by TMP PB. In a typical experiment, exponentially growing yeast cells were split in three identical fractions. In the first fraction, cells were incubated with TMP and irradiated to produce a limited amount of intracellular DNA crosslinks (one crosslink per 10 kb). Cellular DNA was extracted and fragmented (2-kb average length). In the second fraction, cellular DNA was extracted, fragmented (2-kb average length) and then incubated with TMP and irradiated to produce one crosslink per 10 kb. Genomic DNA fragments from the above in vivo and in vitro experiments were thermally denatured to convert un-crosslinked segments into ssDNA. Exonuclease I degraded then ssDNA chains from their 3′-end, such that only crosslinked duplexes (dsDNA) and DNA chains with a blocked 3′-end remained. Exonuclease λ digested then ssDNA and dsDNA and from their 5′-ends. As a result, un-crosslinked fragments were degraded and crosslinked fragments were converted into a pair of ssDNA chains bridged by a TMP linkage. Random priming along these chains produced radiolabeled sequences that were hybridized on DNA arrays. The third fraction of the yeast culture was used to conduct a genome-wide analysis of ongoing transcription (GRO). In vivo radiolabeled RNA was purified and hybridized on arrays of the same lot used to analyze the TMP-mediated DNA crosslinks.
Genomic analysis of TMP-induced DNA crosslinks by means of array hybridization
The in vivo crosslinked sequences selected above (single-stranded DNA chains covalently linked by a TMP molecule) were suitable for direct labeling and hybridization on DNA arrays. This possibility avoids PCR amplification steps, which could potentially distort the distribution of crosslinked DNA sequences. Radiolabeled sequences were hybridized on DNA arrays displaying 6020 ORFs of S. cerevisiae, which are currently used in our laboratory to conduct transcription run-on analyses. It should be stressed then, that the resolution provided by the previous DNA-fragmentation step and these arrays to allocate the DNA-crosslink sites was >2 kb, a length that surpasses the average intergenic distance of S. cerevisiae (∼1.5 kb). Therefore, the distribution of array signals obtained in this method study was only suitable to compare genomic regions spanning several genes. Yet, using tiling arrays or massively parallel DNA sequencing should attain a higher resolution. In such a case, genomic data sets should allow comparisons of psoralen PB probability between inter and intragenic regions and shorter DNA segment motifs.
Following array hybridization, quantification of the spot signals was done as detailed in the ‘Materials and Methods’ section. Three biological replicates of the experiment were examined to obtain independent sets of spot intensities. No >2% of the spot signals analyzed were discarded by the ArrayStat software because of their inconsistency among replicates. Pair-wise comparisons between the independent sets of intensities yield high Pearson coefficients (r > 0.95). Therefore, our procedure for in vivo TMP-PB, selection of crosslinked fragments, and array hybridization proved to be robust.
Correction of TMP-PB preferences due to DNA base pair sequence
The above datasets of DNA-crosslinking probability should inform on relative differences of DNA helical tension genome wide, but could reflect also the preference of some genomic DNA base pair sequences to interact with TMP. For instance, since most favorable contacts for TMP PB occur at 5′-TA dinucleotides, genomic regions enriched in this sequence could bias the distribution DNA crosslinks. To correct this bias, we determined also the TMP-mediated DNA-crosslinking probability on purified genomic DNA fragments extracted from the same yeast cells (Figure 2). As expected, these in vitro data sets (reflecting only the effect of genomic DNA base pair sequence) showed a reduced correlation (r ∼ 0.65) with those obtained in vivo. This r-value denoted yet a significant contribution of DNA sequence during in vivo PB, which could be also attributed to signals generated from distinct nearby sequences within a given array spot. We calculated then the quotient of the in vivo to the in vitro PB values for each array spot to obtain a PB ratio. These ratios better reflected the contribution of intracellular DNA topology on the probability of DNA crosslinking mediated by TMP PB. Finally, since in vitro PB was conducted on genomic DNA extracted at the same time and from the same the cells being examined in vivo, PB ratios also corrected plausible variations due to amplification of genomic regions.
PB ratios exposed domains of different DNA helical tension along the yeast chromosomes
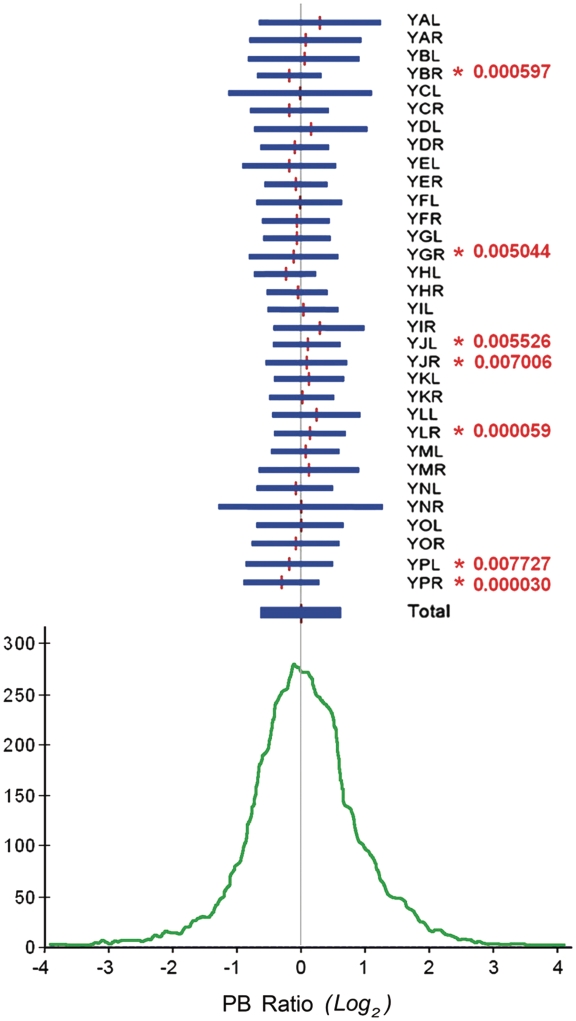
The distribution of PB ratios obtained in yeast cells revealed that 68% of array-spots changed no >2-fold in their in vivo to their in vitro DNA-crosslinking probability, while 27% changed 2–10-fold. The rest of the spots, ∼200, increased or decreased their relative DNA-crosslinking probability by over a factor of 10 (Figure 3). We examined then if the distribution of PB ratios was comparable in all 32 yeast chromosomal arms. Surprisingly, we found that the mean PB ratio of few chromosomal arms statistically diverged from the global mean. In all the biological replicates, mean PB ratios in YBR, YGR, YPL and YPR were slightly below genomic average, whereas in YJL, YJR and YLR they were above (Figure 3). This observation anticipated that DNA helical tension was not homogeneously distributed across the yeast chromosomes. We examined then the topographic distribution of PB ratios across individual chromosomal arms. To do so, we plotted PB ratios according to the ORF position in all yeast chromosomes (Figure 4 and Supplementary Figure S3). These plots denoted numerous regions where long runs of PB ratios appeared either above or below the global genomic average. We evaluated the significance of these regional deviations by conducting a sliding Student’s t-test in windows of 20 or more consecutive ORFs, moving one ORF at each step. As expected, several windows were found to be different from the global distribution (Supplementary Table S1). For instance, the mean PB ratio in a run of 35 ORFs (from YMR145C to YMR183 C) was 0.6 log2 units above the global mean (P-value < 3.10−6), whereas in a run of 25 ORFs (from YGR004W to YGR028W) that was near 0.5 log2 units below (P-value < 7.10−5). These deviations were observed in all the biological replicates and their statistical significance was further confirmed by doing a similar sliding analysis on randomized data sets (i.e. the same distribution of PB ratios but randomizing the ORFs position). In such simulations, the number of deviated runs (P-value < 10−3) dropped by 10-fold. Therefore, considering that the difference of mean PB ratios between some runs was > 2-fold, those runs with mean PB increased were likely reflecting chromosomal domains in which DNA tended to be more un-wound than the global average. Conversely, those runs with mean PB reduced likely reflected chromosomal domains where DNA tended to be more over-wound.
Figure 3.
Global distribution of PB ratios. The PB ratio for each array spot was calculated as the quotient of the in vivo to the in vitro relative DNA-crosslinking probability. The histogram (bottom) shows the frequency distribution of PB ratios (log2 scale being 0 the global mean). Horizontal segments (top) graphically compare the mean and variance of PB ratios for each of the 32 yeast chromosome arms. Student’s t-test (P-values in red) exposed chromosomal arms with mean PB ratios statistically deviated from the global distribution.
Figure 4.
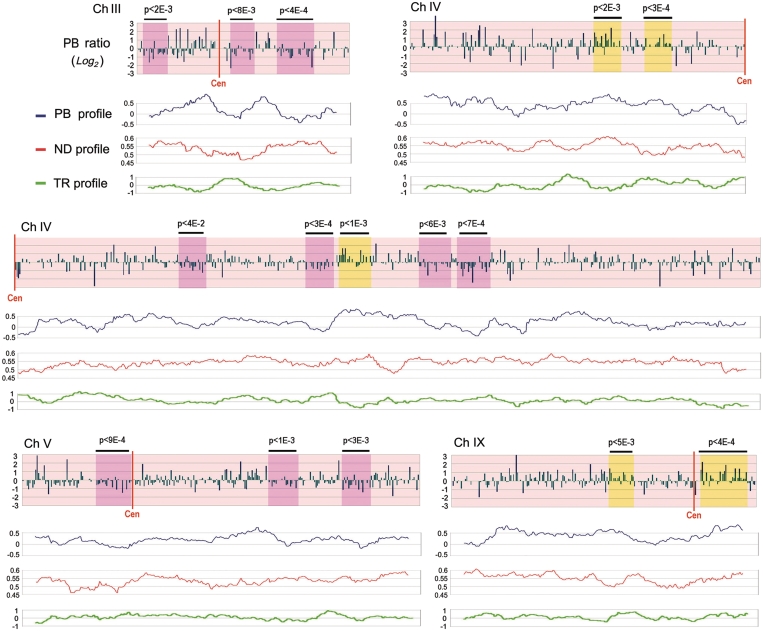
Topography of PB ratios along individual yeast chromosomes. The genomic distribution of PB ratios is plotted (log2 scale being 0 the global mean) by their corresponding ORF position ordering along chromosomes III, IV, V and IX of S. cerevisiae (the full set of chromosomes is depicted in Supplementary Figure S1). Student’s t-test determined whether runs of consecutive ORFs had a mean PB equal or statistically distinct from that of the global distribution. Runs with mean PB statistically increased and reduced are shadowed by yellow or pink boxes, respectively, with indication of their P-value on top. Below each chromosome, regional profiles of PB ratios (blue, log2 scale), of ND (in red) and of TR (in green, log2 scale) are compared. ND data was derived from Mavrich et al. (50). The three profiles were obtained by calculating the mean value of the corresponding parameter in sliding windows of 20 ORFs, moving one ORF at each step. Resulting means were then plotted in the centre of each sliding window.
Chromosomal profiles of PB ratios do not correlate to nucleosome density and transcription probability
According to our in vitro calibrations, deviations in the genomic profiles of PB ratios should mostly reflect differences in the helical state of DNA, rather than other properties of chromatin structure such as nucleosome density (ND). We corroborated this extreme by comparing genomic distributions of PB and ND. ND has been well characterized along the yeast genome and found to range between 0.4 and 0.8 for most yeast genes (52). As seen in Figure 1D, this ND variation should produce little impact on TMP PB probability. Accordingly, when we ordered the genomic distribution of PB ratios by the ND of the corresponding genes, a small increase of PB values was only discernible for the subset of genes with lowest ND (Supplementary Figure S4). We compared then regional means of ND and PB ratios across the yeast chromosomes and, as expected, no correlation was found (r = −0.04) between both parameters. Visual inspection of both ND and PB profiles did not expose also any consistent parallelism between them (Figure 4). This lack of correlation further sustained that the observed PB deviations were mainly produced by differences in the helical state of DNA.
Since DNA transcription markedly alters the helical state of DNA (24), we asked if the genomic profiles of PB ratios would correlate to the local levels of gene activity. This test was particularly suitable in our experimental setting because we could analyze ongoing transcription in one aliquot of the same yeast cultures used to examine TMP PB (Figure 2). To do so, elongating RNA molecules were radiolabeled in vivo and hybridized on DNA arrays alike those used to determine PB values. Normalized array signals indicated then the relative transcription rate (TR) of each gene at the time that TMP PB was conducted. Pair-wise comparison of the global TR and PB data sets showed no global correlation (r = −0.02). Regional means of PB and TR did not correlate either (r = −0.03). Furthermore, both un-wound and over-wound DNA domains did not overlap with regions of high or low TR (Figure 4). We observed also that PB ratios corresponding to the highly transcribed rDNA genes (represented by 18S and 25S array spots) were only slightly above the genomic average (Supplementary Figure S3). Therefore, it was clear that DNA conformational differences exposed by PB ratios did not anticipate the transcription probability of a particular genomic region. We reasoned this lack of correlation in several ways. First, since PB ratios provide information on the average conformation of DNA, concurrent positive and negative DNA helical tension produced during transcription might not produce a net difference on TMP PB probability along a broad region. Second, since DNA torsion waves generated in one gene might diffuse to neighboring genes, the transcription process and its associated structural changes do not have necessarily the same boundaries. Finally, since the duration of on going transcription in any given gene is generally very small compared to its resting state, PB ratios mostly reflect the basal conformation state rather than the brief dynamic state. Therefore, the regional differences of DNA topology uncovered with our method likely reflected some basal state of the yeast chromosome architecture.
Topoisomerase inactivation markedly alters PB ratios along specific chromosomal regions
In order to examine whether the chromosomal PB profiles reported above could change in different experimental conditions or genetic backgrounds, we applied our method in a yeast mutant defective in topoisomerase I and II activities. The strain FY251 (TOP1 TOP2) and its derivative JCW28 (Δtop1 top2ts) were grown at 30°C and both shifted to 37°C during 10 min to inactivate the thermo-sensitive topoisomerase II allele. In vivo TMP–DNA PB and transcription run-on analyses were conducted as above in three biological replicates of both strains. The amount of crosslinked DNA fragments recovered in these conditions was comparable to that of the parental strain FY251 at 30°C. Pair-wise comparison of the global distribution of PB ratios did not anticipate major alterations between the TOP1 TOP2 strain before and after the thermal shift (r > 0.96); and between the TOP1 TOP2 and the Δtop1 top2ts strain after the thermal shift (r >0.92). However, when we plotted then PB ratios along each individual chromosome and calculated regional means, the resulting profiles revealed that inactivation of topoisomerase activities markedly altered PB ratios across few specific chromosomal regions (Supplementary Figure S5).
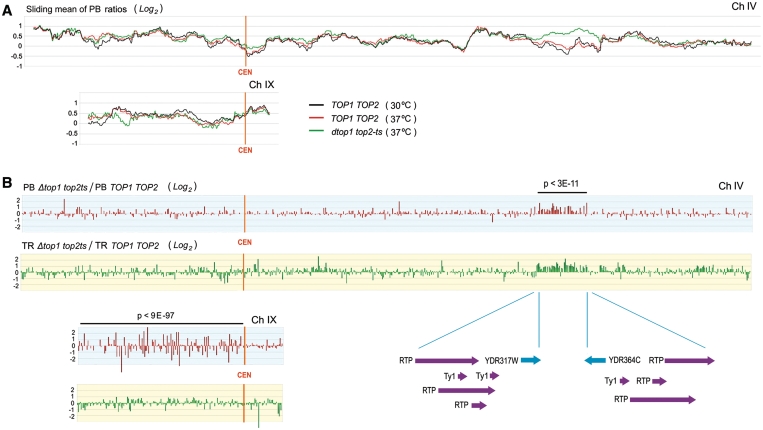
Two of the above alterations became very evident by plotting the PB ratios and TR values for each array spot as a quotient of the Δtop1 top2ts (at 37°C) to the TOP1 TOP2 (at 37°C) data sets (Figure 5 and Supplementary Figures S6 and S7). First, PB ratios increased on average by >2-fold along a run of 50 ORFs (Student’s t-test, P-value < 3.10−11) in the right arm of chromosome IV (from YDR317C to YDR364C). Interestingly, this sharp alteration correlated with a boost of TR along the same domain (Figure 5). Second, PB ratios underwent large fluctuations all along the left arm of chromosome IX (f-test, P-value < 9.10−97). The increased dispersion produced in this arm did not occur in any other chromosomal region and was not paralleled to significant changes of transcription activity (Figure 5). It is remarkable that these two vast alterations spread along chromosomal regions apparently flanked by putative topological boundaries. In one case, DNA transposons had been identified as functional insulators (53,54) and the domain affected in chromosome IV is precisely flanked by an unusual concentration of transposable elements (Figure 5). In the other case, yeast centromeres had been identified as main anchoring points of interphase chromosomes to the nuclear structure (55) and the sharp transition between the left and right arm of chromosome IX clearly identifies the CEN region as the boundary. To the question of why the large alterations caused by topoisomerase inactivation are restricted to few chromosome domains, it is possible that, as soon as those domains with a weaker structural threshold are altered, stress responses are triggered to avoid further disruptions of the DNA topology.
Figure 5.
Alteration of PB ratios after the inactivation of topoisomerase activity. (A) Regional profiles of PB ratios (obtained as in Figure 4, log2 scale being 0 the global mean) in TOP1 TOP2 (at 30 and 37°C) and in Δtop1 top2ts cells (at 37°C) are compared for chromosomes IV and IX. (B) Alterations of PB ratios and of TRs are illustrated (PB in red; TR in green) along chromosomes IV and IX as the quotient of values of the Δtop1 top2ts to the TOP1 TOP2 cells (log2 scales, being 0 the global means). In the Δtop1 top2ts strain, PB ratios statistically increased (P < 3.10−11 by t-test) in a run of 50 ORFs of chromosome IV (from YDR317C to YDR364C); and the distribution PB ratios underwent also a large dispersion (P < 3.10−97 by f-test) all along the left arm of chromosome IX.
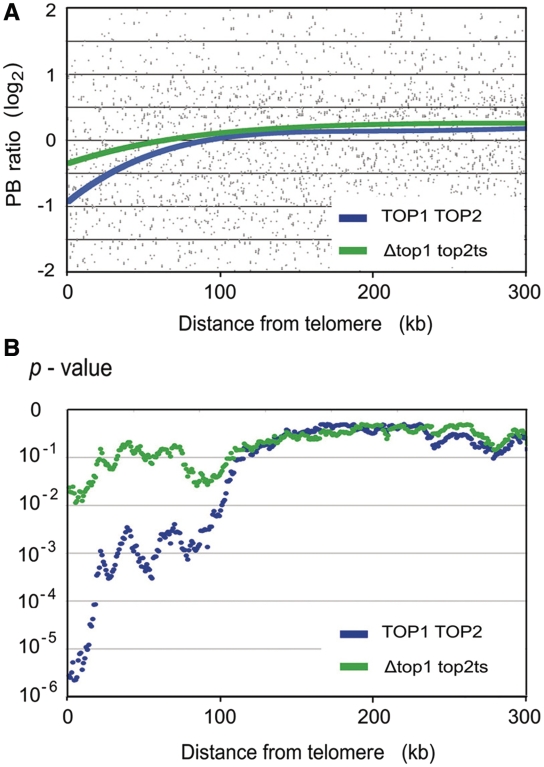
Finally, another noteworthy deviation between the Δtop1 top2ts and TOP1 TOP2 strains was observed when plotting the genomic distribution of PB ratios according to the gene distances to their corresponding telomere. As illustrated in Figure 6, as genes become proximal to the chromosomal extremity (<100 kb from the telomere), the average PB ratio tends to gradually decrease with respect to the global mean. This deviation was patent in the TOP1 TOP2 cells, but it was much less pronounced in the Δtop1 top2ts mutant. Analogous analyses with respect to gene distances to the centromere did not show any alteration. We believe that this positional effect on PB values is reflecting the dissipation of DNA helical tension that occurs at yeast chromosomal ends (56). Since DNA tends to be more relaxed near the telomeres, the terminal drop of PB values would then indicate that the average helical tension of DNA in the yeast genome is slightly negative. The different effect observed after the inhibition of topoisomerase activities further support that this deviation of PB ratios is related to DNA helical state.
Figure 6.
Deviation of PB ratios at the yeast chromosome extremities. (A) The genomic distribution of PB ratios (log2) obtained for TOP1 TOP2 cells and the Δtop1 top2ts mutant are plotted against the distance (kb) of the corresponding gene to its telomere. Plot trendlines (order 4 polynomial) are shown. (B) The significance of the deviations observed in A is examined by conducting a Student’s t-tests that compares the global PB distribution and that in sliding windows of 200 genes, moving one gene at each step (starting from the chromosomal end). Obtained P-values are plotted against the distance from the telomere of the first gene of each window.
CONCLUSION
Our procedure for genome-wide analysis of inter-strand DNA crosslinks mediated by psoralen PB has proved to be robust tool to assess relative differences of helical tension across chromosomal DNA. The distribution of PB ratios obtained in S. cerevisiae revealed for the first time different levels of DNA helical tension might be constituted in separated domains along eukaryotic chromosomes. Further implementation of this procedure by the use of tiling arrays of massive-parallel sequencing should strongly increase the resolution to allocate the psoralen photobinding sites. This information might reveal how the helical state of DNA changes at intra- and inter-genic regions, as well as the precise position of twist diffusion boundaries that separate topological compartments. The analysis of psoralen photobinding can be thus a valuable complement to other genome-wide structural and functional studies, and help to uncover the often forgotten relevance of DNA topology.
ACCESSION NUMBER
Genomic data sets (biological triplicates of all the reported experiments) are stored in the GEO databases. GEO series accession number is GSE16673.
SUPPLEMENTARY DATA
Supplementary Table S1 and Supplementary Figures S1–S7 are available at NAR Online.
FUNDING
Spanish National Plan I+D+I (Grant numbers BFU2007-67575-CO3-01/BMC to J.E.P-O. and BFU2008-00366 to J.R.). Funding for open access charge: Spanish National Plan I+D+I (Grant number BFU2008-00366 to J.R.).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 2.Luger K, Hansen JC. Nucleosome and chromatin fiber dynamics. Curr. Opin. Struct. Biol. 2005;15:188–196. doi: 10.1016/j.sbi.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 3.Mendenhall EM, Bernstein BE. Chromatin state maps, new technologies, new insights. Curr. Opin. Genet. Dev. 2008;18:109–115. doi: 10.1016/j.gde.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schones DE, Zhao K. Genome-wide approaches to studying chromatin modifications. Nat. Rev. Genet. 2008;9:179–191. doi: 10.1038/nrg2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borneman AR, Gianoulis TA, Zhang ZD, Yu H, Rozowsky J, Seringhaus MR, Wang LY, Gerstein M, Snyder M. Divergence of transcription factor binding sites across related yeast species. Science. 2007;317:815–819. doi: 10.1126/science.1140748. [DOI] [PubMed] [Google Scholar]
- 7.Schäfer G, McEvoy CR, Patterton HG. The Saccharomyces cerevisiae linker histone Hho1 p is essential for chromatin compaction in stationary phase and is displaced by transcription. Proc. Natl Acad. Sci. USA. 2008;105:14838–14843. doi: 10.1073/pnas.0806337105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steinmetz EJ, Warren CL, Kuehner JN, Panbehi B, Ansari AZ, Brow DA. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol. Cell. 2006;24:735–746. doi: 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 9.Lindroos HB, Ström L, Itoh T, Katou Y, Shirahige K, Sjögren C. Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol. Cell. 2006;22:755–767. doi: 10.1016/j.molcel.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 10.Bermejo R, Doksani Y, Capra T, Katou YM, Tanaka H, Shirahige K, Foiani M. Top1- and Top2-mediated topological transitions at replication forks ensure fork progression and stability and prevent DNA damage checkpoint activation. Genes Dev. 2007;21:1921–1936. doi: 10.1101/gad.432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 12.Kurdistani SK, Tavazoie S, Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117:721–733. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 13.Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 14.Jiang C, Pugh BF. A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol. 2009;10:R109. doi: 10.1186/gb-2009-10-10-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vologodskii AV, Cozzarelli NR. Conformational and thermodynamic properties of supercoiled DNA. Annu. Rev. Biophys. Biomol. Struct. 1994;23:609–643. doi: 10.1146/annurev.bb.23.060194.003141. [DOI] [PubMed] [Google Scholar]
- 16.Vologodskii A, Cozzarelli NR. Effect of supercoiling on the juxtaposition and relative orientation of DNA sites. Biophys. J. 1996;70:2548–2556. doi: 10.1016/S0006-3495(96)79826-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delius H, Worcel A. Electron microscopic visualization of the folded chromosome of Escherichia coli. J. Mol. Biol. 1974;82:107–109. doi: 10.1016/0022-2836(74)90577-4. [DOI] [PubMed] [Google Scholar]
- 18.Sinden RR, Pettijohn DE. Chromosomes in living Escherichia coli cells are segregated into domains of supercoiling. Proc. Natl Acad. Sci. USA. 1981;78:224–228. doi: 10.1073/pnas.78.1.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Postow L, Hardy CD, Arsuaga J, Cozzarelli NR. Topological domain structure of the Escherichia coli chromosome. Genes Dev. 2004;18:1766–1779. doi: 10.1101/gad.1207504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pruss GJ, Drlica K. DNA supercoiling and prokaryotic transcription. Cell. 1989;56:521–523. doi: 10.1016/0092-8674(89)90574-6. [DOI] [PubMed] [Google Scholar]
- 21.Eissenberg JC, Cartwright IL, Thomas GH, Elgin SC. Selected topics in chromatin structure. Annu. Rev. Genet. 1985;19:485–536. doi: 10.1146/annurev.ge.19.120185.002413. [DOI] [PubMed] [Google Scholar]
- 22.Esposito F, Sinden RR. DNA supercoiling and eukaryotic gene expression. Oxf. Surv. Eukaryot. Genes. 1988;5:1–50. [PubMed] [Google Scholar]
- 23.Freeman LA, Garrard WT. DNA supercoiling in chromatin structure and gene expression. Crit. Rev. Eukaryot. Gene Expr. 1992;2:165–209. [PubMed] [Google Scholar]
- 24.Giaever GN, Wang JC. Supercoiling of intracellular DNA can occur in eukaryotic cells. Cell. 1988;55:849–856. doi: 10.1016/0092-8674(88)90140-7. [DOI] [PubMed] [Google Scholar]
- 25.Salceda J, Fernández X, Roca J. Topoisomerase II, not topoisomerase I, is the proficient relaxase of nucleosomal DNA. EMBO J. 2006;25:2575–2583. doi: 10.1038/sj.emboj.7601142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cimino GD, Gamper HB, Isaacs ST, Hearst JE. Psoralens as photoactive probes of nucleic acid structure and function, organic chemistry, photochemistry, and biochemistry. Annu. Rev. Biochem. 1985;54:1151–1193. doi: 10.1146/annurev.bi.54.070185.005443. [DOI] [PubMed] [Google Scholar]
- 27.Sinden RR, Hagerman PJ. Interstrand psoralen cross-links do not introduce appreciable bends in DNA. Biochemistry. 1984;23:6299–6303. doi: 10.1021/bi00321a002. [DOI] [PubMed] [Google Scholar]
- 28.Sinden RR, Carlson JO, Pettijohn DE. Torsional tension in the DNA double helix measured with trimethylpsoralen in living E. coli cells, analogous measurements in insect and human cells. Cell. 1980;21:773–783. doi: 10.1016/0092-8674(80)90440-7. [DOI] [PubMed] [Google Scholar]
- 29.Sinden RR, Ussery DW. Analysis of DNA structure in vivo using psoralen photobinding, measurement of supercoiling, topological domains, and DNA-protein interactions. Methods Enzymol. 1992;212:319–335. doi: 10.1016/0076-6879(92)12020-q. [DOI] [PubMed] [Google Scholar]
- 30.Schultz MC, Brill SJ, Ju Q, Sternglanz R, Reeder RH. Topoisomerases and yeast rRNA transcription, negative supercoiling stimulates initiation and topoisomerase activity is required for elongation. Genes Dev. 1992;6:1332–1341. doi: 10.1101/gad.6.7.1332. [DOI] [PubMed] [Google Scholar]
- 31.Liang CP, Garrard WT. Template topology and transcription, chromatin templates relaxed by localized linearization are transcriptionally active in yeast. Mol. Cell Biol. 1997;17:2825–2834. doi: 10.1128/mcb.17.5.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jupe ER, Sinden RR, Cartwright IL. Stably maintained microdomain of localized unrestrained supercoiling at a Drosophila heat shock gene locus. EMBO J. 1993;12:1067–1075. doi: 10.1002/j.1460-2075.1993.tb05748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jupe ER, Sinden RR, Cartwright IL. Specialized chromatin structure domain boundary elements flanking a Drosophila heat shock gene locus are under torsional strain in vivo. Biochemistry. 1995;34:2628–2633. doi: 10.1021/bi00008a029. [DOI] [PubMed] [Google Scholar]
- 34.Ljungman M, Hanawalt PC. Localized torsional tension in the DNA of human cells. Proc. Natl Acad. Sci. USA. 1992;89:6055–6059. doi: 10.1073/pnas.89.13.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ljungman M, Hanawalt PC. Presence of negative torsional tension in the promoter region of the transcriptionally poised dihydrofolate reductase gene in vivo. Nucleic Acids Res. 1995;23:1782–1789. doi: 10.1093/nar/23.10.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kramer PR, Sinden RR. Measurement of unrestrained negative supercoiling and topological domain size in living human cells. Biochemistry. 1997;36:3151–3158. doi: 10.1021/bi962396q. [DOI] [PubMed] [Google Scholar]
- 37.Matsumoto K, Hirose S. Visualization of unconstrained negative supercoils of DNA on polytene chromosomes of Drosophila. J. Cell Sci. 2004;117:3797–3805. doi: 10.1242/jcs.01225. [DOI] [PubMed] [Google Scholar]
- 38.Roca J. Varying levels of positive and negative supercoiling differently affect the efficiency with which topoisomerase II catenates and decatenates DNA. J. Mol. Biol. 2001;305:441–450. doi: 10.1006/jmbi.2000.4307. [DOI] [PubMed] [Google Scholar]
- 39.Rodríguez-Campos A, Koop R, Faraudo S, Beato M. Transcriptionally competent chromatin assembled with exogenous histones in a yeast whole cell extract. Nucleic Acids Res. 2004;32:e111. doi: 10.1093/nar/gnh107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roca J, Gartenberg M, Oshima Y, Wang JC. A hit-and-run system for targeted genetic manipulations in yeast. Nucleic Acids Res. 1992;20:4671–4672. doi: 10.1093/nar/20.17.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alberola TM, García-Martínez J, Antúnez O, Viladevall L, Barceló A, Ariño J, Pérez-Ortín JE. A new set of DNA macrochips for the yeast Saccharomyces cerevisiae, features and uses. Int. Microbiol. 2004;7:199–206. [PubMed] [Google Scholar]
- 42.García-Martínez J, Aranda A, Pérez-Ortín JE. Genomic run-on evaluates transcription rates for all yeast genes and identifies gene regulatory mechanisms. Mol. Cell. 2004;15:303–313. doi: 10.1016/j.molcel.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 43.Cech T, Pardue ML. Crosslinking of DNA with trimethylpsoralen is a probe for chromatin structure. Cell. 1977;11:631–640. doi: 10.1016/0092-8674(77)90080-0. [DOI] [PubMed] [Google Scholar]
- 44.Carlson JO, Pfenninger O, Sinden R, Lehman JM, Pettijohn DE. New procedure using a psoralen derivative for analysis of nucleosome associated DNA sequences in chromatin of living cells. Nucleic Acids Res. 1982;10:2043–2063. doi: 10.1093/nar/10.6.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ostrander EA, Kurkinen NA, Hallick LM. DNA structural alterations in the SV40 enhancer region are retained in vivo. Virology. 1988;165:274–277. doi: 10.1016/0042-6822(88)90682-4. [DOI] [PubMed] [Google Scholar]
- 46.Conconi A, Widmer RM, Koller T, Sogo JM. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell. 1989;57:753–761. doi: 10.1016/0092-8674(89)90790-3. [DOI] [PubMed] [Google Scholar]
- 47.Toussaint M, Levasseur G, Tremblay M, Paquette M, Conconi A. Psoralen photocrosslinking, a tool to study the chromatin structure of RNA polymerase I-transcribed ribosomal genes. Biochem Cell Biol. 2005;83:449–459. doi: 10.1139/o05-141. [DOI] [PubMed] [Google Scholar]
- 48.De Bernardin W, Koller T, Sogo JM. Structure of in-vivo transcribing chromatin as studied in simian virus 40 minichromosomes. J. Mol. Biol. 1986;191:469–482. doi: 10.1016/0022-2836(86)90142-7. [DOI] [PubMed] [Google Scholar]
- 49.Cavalli G, Thoma F. Chromatin transitions during activation and repression of galactose-regulated genes in yeast. EMBO J. 1993;12:4603–4613. doi: 10.1002/j.1460-2075.1993.tb06149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wellinger RE, Sogo JM. In vivo mapping of nucleosomes using psoralen-DNA crosslinking and primer extension. Nucleic Acids Res. 1998;26:1544–1545. doi: 10.1093/nar/26.6.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Champoux JJ. DNA topoisomerases: structure, function and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 52.Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 54.Valenzuela L, Kamakaka RT. Chromatin insulators. Annu. Rev. Genet. 2006;40:107–138. doi: 10.1146/annurev.genet.39.073003.113546. [DOI] [PubMed] [Google Scholar]
- 55.Perrod S, Gasser SM. Long-range silencing and position effects at telomeres and centromeres: parallels and differences. Cell Mol. Life Sci. 2003;60:2303–2318. doi: 10.1007/s00018-003-3246-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Joshi RS, Piña B, Roca J. Positional dependence of transcriptional inhibition by DNA torsional stress in yeast chromosomes. EMBO J. 2010;29:740–748. doi: 10.1038/emboj.2009.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.