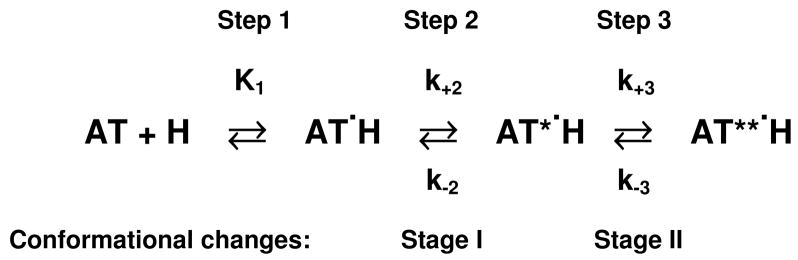
Abstract
The serpin, antithrombin, requires allosteric activation by a sequence-specific pentasaccharide unit of heparin or heparan sulfate glycosaminoglycans to function as an anticoagulant regulator of blood clotting proteases. Surprisingly, X-ray structures have shown that the pentasaccharide produces similar induced-fit changes in the heparin binding site of native and latent antithrombin despite large differences in the heparin affinity and global conformation of these two forms. Here we present kinetic evidence for similar induced-fit mechanisms of pentasaccharide binding to native and latent antithrombins and kinetic simulations which together support a three-step mechanism of allosteric activation of native antithrombin involving two successive conformational changes. Equilibrium binding studies of pentasaccharide interactions with native and latent antithrombins and the salt dependence of these interactions suggest that each conformational change is associated with distinct spectroscopic changes and is driven by a progressively better fit of the pentasaccharide in the binding site. The observation that variant antithrombins that cannot undergo the second conformational change bind the pentasaccharide like latent antithrombin and are partially activated suggests that both conformational changes contribute to allosteric activation, in agreement with a recently proposed model of allosteric activation.
Keywords: heparin, antithrombin, serpin, blood clotting, protease, allostery
Introduction
Antithrombin, a member of the serpin superfamily of protein protease inhibitors, performs a key anticoagulant function in vertebrates by regulating the activity of blood coagulation cascade proteases [1, 2]. Antithrombin inhibits clotting proteases like other serpins by an unusual branched pathway suicide substrate mechanism. In this mechanism the serpin initially binds the protease through an exposed reactive center loop (RCL) as a regular substrate and proceeds to cleave the RCL to form the acylintermediate [3]. This cleavage triggers the suicide inactivation by inducing the metastable serpin to undergo a massive conformational change that deforms the protease catalytic machinery and kinetically traps the acylintermediate complex [4, 5].
Antithrombin circulates in blood in a repressed reactivity state, but is activated by heparin and heparan sulfate glycosaminoglycans on the luminal and subluminal sides of blood vessels to control and localize the activity of blood coagulation proteases [6, 7]. The glycosaminoglycans bind to antithrombin through a specific pentasaccharide sequence and induce allosteric activating changes which enable the serpin to inhibit coagulation proteases at a rapid, physiologically relevant rate [8, 9]. The activating conformational changes overcome the repressed reactivity state by relieving repulsive interactions that diminish favorable RCL and exosite interactions of the serpin with two target proteases, factor Xa and factor IXa [10–15]. Heparin and heparan sulfate additionally enhance antithrombin reactivity with these proteases as well as with another target protease, thrombin, by a ternary complex bridging mechanism in which the binding of the protease to the glycosaminoglycan alongside antithrombin promotes the serpin-protease interaction [16–20].
Rapid kinetic studies of heparin pentasaccharide binding to antithrombin have shown that allosteric activation is a two-step process in which the negatively charged pentasaccharide recognizes and binds to a positively charged site on the serpin and then induces the protein into an activated conformational state that is reported by CD, UV and fluorescence changes [9, 21]. Allosteric activation is driven by an induced-fit mechanism that shifts the protein into a higher energy activated state because of the increased pentasaccharide binding energy in this state [22, 23]. X-ray structures of free and pentasaccharide-complexed antithrombin have shown the nature of the allosteric activating changes produced by pentasaccharide binding and revealed that such changes involve both the heparin binding site and the protease binding site with an allosteric core mediating communication between these sites [24, 25].
More recent studies have suggested that the allosteric activating changes that are initiated in the heparin binding site and propagated to the protease binding site may be resolvable into distinct steps. X-ray structures of an intermediate activated state of native antithrombin have thus been reported in which the conformational changes in the heparin binding site have occurred without the changes in the protease binding site [26, 27]. Our recent studies support such a two-stage allosteric activation mechanism in solution from our studies of conformationally altered latent and cleaved forms of antithrombin that have lost the ability to be activated as protease inhibitors but retain the ability to bind the heparin pentasaccharide [28]. These conformationally altered antithrombins thus bind the pentasaccharide through a two-step induced conformational change mechanism similar to that shown for the native antithrombin-pentasaccharide interaction, although with reduced affinity. Notably, an X-ray structure of latent antithrombin complexed with the pentasaccharide shows a remarkable resemblance to the intermediate native antithrombin-pentasaccharide complex in showing the same conformational changes in the heparin binding site without the global changes of fully-activated native antithrombin [25]. Such findings suggest that pentasaccharide binding to the conformationally altered antithrombins may reflect an intermediate stage in the allosteric activation of native antithrombin. The present study was undertaken to assess whether the kinetics of pentasaccharide binding to native and latent antithrombins could be reconciled with a 3-step mechanism for allosteric activation of native antithrombin and allow dissection of the structural, spectroscopic and activating changes associated with each step of the 3-step mechanism.
Experimental
Simulations of pentasaccharide binding to native and latent forms of antithrombin by the 2-step mechanism of scheme 1 or the 3-step mechanism of scheme 2 were done by numerical integration of the differential equations for these mechanisms using Scientist software (Micromath) and the indicated values for K1, k+2, k−2, k+3, and k−3 in Table 3. These values were chosen based on apparent kinetic parameters obtained in kinetic studies of the binding interactions. For the initial binding step, the forward rate constant, k+1, was set at the diffusion-limited value of 100 μM−1s−1 and the reverse rate constant, k−1, was set at 1000 s−1 to provide a K1 of 10 μM close to the measured value and rapid equilibrium binding (k−1≫k+2). The predicted value for KD was calculated from the kinetic parameters as K1K2/(1+K2) for the 2-step mechanism and K1K2K3/(1+K2K3+K3) for the 3-step mechanism where K1, K2 and K3 equal k−1/k+1, k−2/k+2, and k−3/k+3, respectively [29]. Progress curves for pentasaccharide binding to antithrombin were generated for pentasaccharide concentrations ranging from 0.25–15 μM and fixed antithrombin concentrations of 0.025 μM. The curves for formation of the final complex were fit well by a single exponential function to yield kobs. kobs values were then plotted against the pentasaccharide concentration and fitted by the rectangular hyperbolic equation in the text to generate values for K1,app, k+lim.app, k−lim,app= koff and k+lim,app/K1,app = kon.
Scheme 1.
Scheme 2.
Table 3.
Kinetic constants used to simulate two- and three-step mechanisms of pentasaccharide binding to native and latent antithrombinsa
| K1 (μM) | k+1 (μM−1 s−1) | k−1 (s−1) | k+2 (s−1) | k−2 (s−1) | k+3 (s−1) | k−3 (s−1) | KD (nM) | |
|---|---|---|---|---|---|---|---|---|
| Latent 2-step | 10 | 100 | 1000 | 200 | 2 | -- | -- | 100 |
| Native 2-step | 10 | 100 | 1000 | 200 | 0.1 | -- | -- | 5.0 |
| Native 3-step | 10 | 100 | 1000 | 200 | 2 | 2000 | 100 | 4.8 |
Kinetic constants were chosen to approximate measured values in Table 1 for 2-step mechanisms. For the 3-step native antithrombin binding mechanism, kinetic constants for the first two steps were assumed to be identical to those for the latent antithrombin interaction. Kinetic constants for the third step were chosen to make k+3 ≫ k+2 and to constrain k−3/k+3 to a value that yielded an overall KD equal to that for the 2-step native antithrombin mechanism. KD was calculated from individual rate constants based on the equations given in Methods. Since the assumed rate constants conform to the conditions, k−1 ≫ k+2, k+3 ≫ k+2, and k+2 ≫ k−2, kon is approximated by k+2/K1 for both 2- and 3-step mechanisms and koff is approximated by k−2 for the 2-step mechanism and k−2 x k−3/k+3 for the 3-step mechanism [31].
Overlays of X-ray structures of antithrombin-heparin pentasaccharide complexes available from the Protein Data Bank and RMSD calculations were done using Swiss PDB viewer software.
Results and Discussion
Kinetics of pentasaccharide binding to native and conformationally altered antithrombins
The kinetics of heparin pentasaccharide binding to native antithrombin monitored by intrinsic tryptophan or bound TNS fluorescence changes that report the binding show a saturable dependence of the observed pseudo-first order binding rate constant on heparin concentration. This dependence is indicative of a two-step induced conformational change binding mechanism in which the pentasaccharide binds to antithrombin in an initial rapid equilibrium step characterized by a dissociation constant, K1, and then induces a conformational change in the protein with forward and reverse rate constants, k+2 and k−2 (Scheme 1) [21]. This latter step is reported by protein or TNS fluorescence changes. That the first binding step is in rapid equilibrium is supported by the observation that progress curves for heparin pentasaccharide binding to native antithrombin show no lags over a wide range of saccharide concentrations [21]. Surprisingly, the kinetics of pentasaccharide binding to conformationally altered latent and cleaved forms of antithrombin show a strikingly similar saturable dependence of the binding rate constant on heparin concentration. Tables 1 and 2 compare the kinetic and equilibrium binding parameters we recently measured for pentasaccharide binding to native and latent antithrombins and the fluorescence changes associated with this binding under identical experimental conditions [28]. The values determined for the cleaved antithrombin interaction are not shown, but are similar to those for the latent antithrombin interaction. Notably, pentasaccharide binding to both forms of antithrombin is reported by comparable 24–28% fluorescence changes of the extrinsic probe, TNS, but markedly different tryptophan fluorescence changes of 40% for the native antithrombin interaction and 6% for the latent antithrombin interaction. Most striking, the kinetic parameters are experimentally indistinguishable for the initial pentasaccharide binding step (K1) as well as for the rate constant, k+2, for the subsequent conformational change step. Since the overall on-rate constant (kon) for pentasaccharide binding is equal to the ratio of these parameters, k+2/K1, kon is also the same for the two interactions. The kinetic parameters differ only in the reverse rate constant for the conformational change step, this being equal to the off-rate constant for pentasaccharide dissociation (koff) when the initial binding step is in rapid equilibrium. The latter difference therefore accounts for the 20–30-fold greater binding affinity of the pentasaccharide for native antithrombin as compared to the latent form. These findings suggest that native and latent antithrombins bind the pentasaccharide and undergo induced conformational changes that are highly similar but not identical.
Table 1.
Kinetic and equilibrium parameters for heparin pentasaccharide binding to native and latent antithrombins at I 0.15, pH 7.4, 10°Ca
| Antithrombin form | K1 (μM) | k+2 (s−1) | kon = k+2/K1 (μM−1 s−1)b | k−2 = koff (s−1)b | KD (nM) |
|---|---|---|---|---|---|
| Native | 6±0.4 | 200±5 | 33±1 | 0.3±0.9 | 4±2 |
| Latent | 9±1 | 200±15 | 30±4 | 3±1 | 150±50 |
Kinetic parameters, taken from Schedin-Weiss et al. [28], were measured by rapid kinetic analyses of pentasaccharide binding to native and latent antithrombins monitored by tryptophan or TNS fluorescence changes. Parameters for the two-step mechanism of scheme 1 were obtained by fitting the dependence of the observed pseudo-first order binding rate constant on pentasaccharide concentration to a hyperbolic saturation function. Dissociation constants were obtained by equilibrium binding titrations of antithrombin with pentasaccharide monitored by tryptophan or TNS fluorescence changes.
Values measured from the slope and intercept of the initial linear dependence of kobs versus pentasaccharide concentration. Deviations between the kon obtained from the slope and that calculated from the ratio, k+2/K1, reflect the greater experimental error in the latter.
Table 2.
Equilibrium binding constants, fluorescence changes and ionic and nonionic contributions to binding for the interactions of the heparin pentasaccharide with native and latent antithrombins at I 0.15, pH 7.4, 25°Ca
| Antithrombin form | KD (nM) | % Trp fluorescence change | % TNS fluorescence change | Number of ionic interactions | Non-ionic KD (μM) |
|---|---|---|---|---|---|
| Native | 45±7 | 40 | 28 | 3.8±0.9 | 15±17 |
| Latent | 1200±200 | 6 | 24 | 1.7±0.2 | 18±7 |
Equilibrium binding parameters for heparin pentasaccharide interactions with antithrombin, taken from Schedin-Weiss et al. [28], were measured by fluorescence titrations performed as a function of ionic strength.
X-ray structures of native and latent antithrombin-pentasaccharide complexes
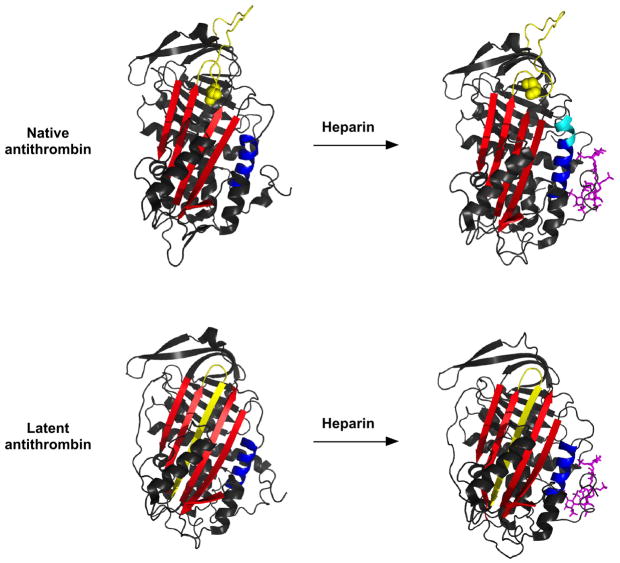
The above interpretation is borne out by comparing the X-ray structures of native and latent antithrombin both free and complexed with the pentasaccharide (Fig. 1) [24, 25]. These structures are derived from the same crystal because native and latent antithrombins were crystallized together as a dimer. A closeup of the heparin binding sites in the native and latent antithrombin-pentasaccharide complex structures in fact show that the pentasaccharide binds to the same site in latent as in native antithrombin and induces similar conformational changes in the site (Fig. 2). These changes involve movements of the three regions that make up the heparin binding site, namely, the N-terminal region, helix A, and helix D, as well as the formation of a new P helix in the loop preceding helix D. Together, these changes create a complementary binding site for the pentasaccharide in which basic residues of the protein are positioned to interact with negatively charged sulfates and carboxylates of the pentasaccharide [25]. A notable difference between the two antithrombin-heparin pentasaccharide complex structures within the heparin binding site is that the pentasaccharide induces an extension of helix D at its C-terminal end in native antithrombin but not in the latent form. This change appears to be linked to other conformational changes induced in the protease binding site that are unique to native antithrombin and that cause full activation of antithrombin reactivity with proteases. The latter include the closing of a gap in sheet A in which the RCL hinge is buried and the expulsion of the RCL from the gap and its extension away from the serpin body (Fig. 1) [30].
Figure 1. X-ray structures of native and latent antithrombins with and without bound pentasaccharide.
-Ribbon models of native (upper panel) and latent (lower panel) antithrombins are shown in the free state (left-hand side, pdb 1E05) and complexed with pentasaccharide (right-hand side, pdb 1E03). The RCL is colored yellow, sheet A is red, helix D is blue and the heparin pentasaccharide is shown in stick and colored magenta. The extension of helix D that accompanies pentasaccharide binding and activation of native antithrombin is colored in cyan. The P14 RCL hinge residue that is buried in sheet A in free native antithrombin and expelled from the sheet in pentasaccharide-activated native antithrombin is shown in space-filling representation.
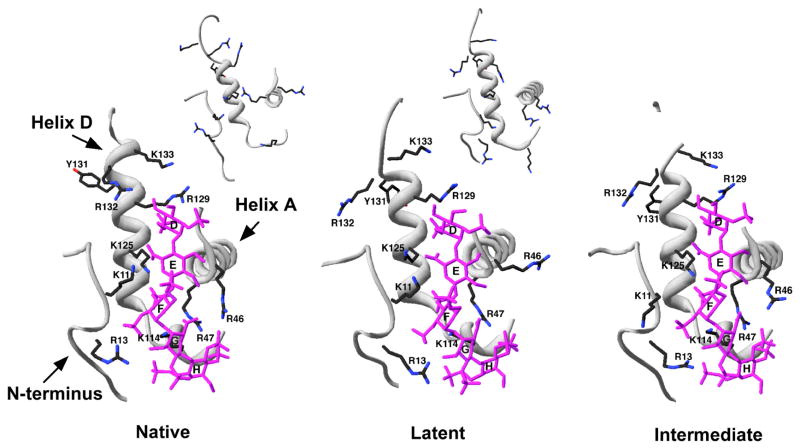
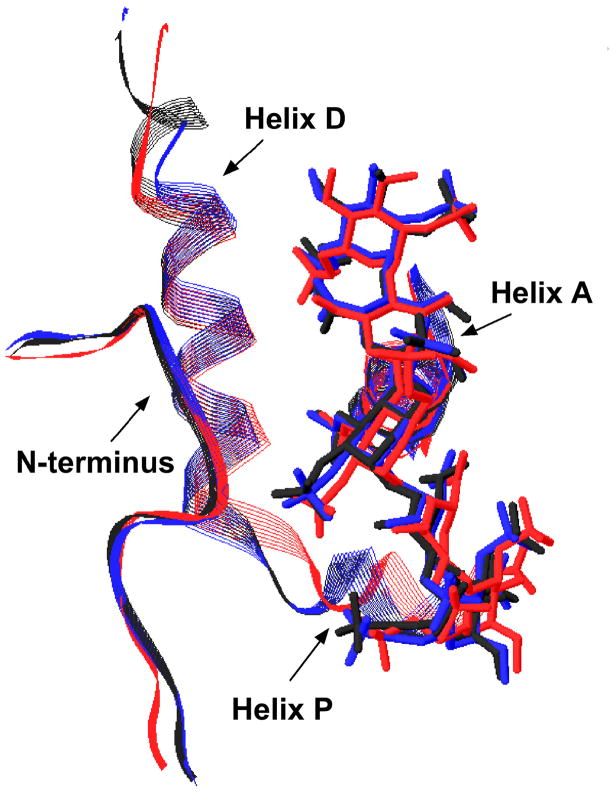
Figure 2. Close-up of the heparin binding site in native, latent and intermediate antithrombin-pentasaccharide complex structures.
The N-terminus, helix A and helix D regions of the heparin binding site from X-ray structures of pentasaccharide complexes with native (pdb 1E03), latent (pdb 1E03) and native intermediate (pdb 1NQ9) forms of antithrombin are shown in light gray ribbon representation. Basic sidechains that interact with the pentasaccharide are shown in stick representation with carbons in black, nitrogens in blue and oxygens in red. The pentasaccharide is shown in stick representation with all atoms colored magenta. The insets for the native and latent antithrombin structures show the uncomplexed forms of the protein (pdb 1E05) and reveal the significant changes in side-chain orientation involved in the induced-fit binding of the pentasaccharide.
In view of our findings that native and latent antithrombin interactions with the pentasaccharide show similar binding kinetics and a common set of structural changes induced in the heparin binding site by binding, the recent reports of two structures of an intermediate form of native antithrombin complexed with the pentasaccharide is particularly noteworthy [26, 27]. Remarkably, the intermediate complex bears a striking resemblance to the latent antithrombin-pentasaccharide complex within the heparin binding site (Fig. 2). Thus, in both structures the conformational changes in the heparin binding site have occurred but helix D extension and the activating changes in the protease binding site have not. Such findings suggest that the latent antithrombin-pentasaccharide complex may be a good model for the intermediate complex.
Three-step mechanism for pentasaccharide binding and activation of antithrombin
Together, the similarity in the kinetics of pentasaccharide binding to native and latent antithrombins and in the structures of native, latent and intermediate antithrombin-pentasaccharide complexes imply a three-step mechanism for pentasaccharide binding and allosteric activation of native antithrombin in which the activating conformational changes occur in two stages (Scheme 2). In steps one and two of the mechanism, the pentasaccharide initially binds with dissociation constant, K1, and then induces the first stage conformational changes within the heparin binding site with forward and reverse rate constants, k+2 and k−2, respectively. In step three, the second stage conformational changes are induced with forward and reverse rate constants, k+3 and k−3, respectively. These latter changes involve the extension of helix D and the activating changes in the protease binding site.
While such a mechanism was previously suggested based on the observed intermediate antithrombin-pentasaccharide complex crystal structures [27], we wished to determine whether the 3-step mechanism could be reconciled with the observed kinetics of pentasaccharide binding to native antithrombin in solution. Assuming that the kinetics of pentasaccharide binding to latent antithrombin is a reasonable model for the two steps leading to the intermediate in the 3-step binding to native antithrombin, we performed kinetic simulations of the 3-step mechanism. The kinetic parameters, K1, k+2 and k−2, were thus fixed at values of 10 μM, 200 s−1 and 2 s−1, respectively, that were similar to those measured for the latent antithrombin interaction. In keeping with the rapid equilibrium of the first binding step, we assumed a diffusion-limited on-rate constant (k+1) of 108 M−1s−1 for binding that required the off-rate constant (k−1) to have a value of 1000 s−1. Importantly, this set of rate constants satisfied the rapid equilibrium constraint, k−1 ≫ k2. We then reasoned that to account for the similar limiting rate constants measured for latent and native antithrombin interactions at saturation, k+2 must represent a common rate-limiting conformational change step for the two interactions. We therefore set k+3 for the third step stage II conformational changes to a value much greater (10-fold) than k+2. We finally constrained k−3 to have a value that would result in a calculated overall equilibrium dissociation constant that corresponded to the measured KD for the native antithrombin interaction given the assumed values for K1, k+2, k−2 and k+3 [29]. It should be noted that the simulations do not depend on the exact values chosen for k+3 and k−3 as long as k+3 ≫ k+2 and the ratio of k+3/k−3 is fixed at a value that yields the observed overall dissociation constant for binding. The simulations are also independent of the chosen values for k+1 and k−1 as long as the ratio k−1/k+1 is fixed at the measured value for K1 and k−1≫k+2 so that rapid equilibrium conditions prevail. Table 3 summarizes the rate constants used for the simulations.
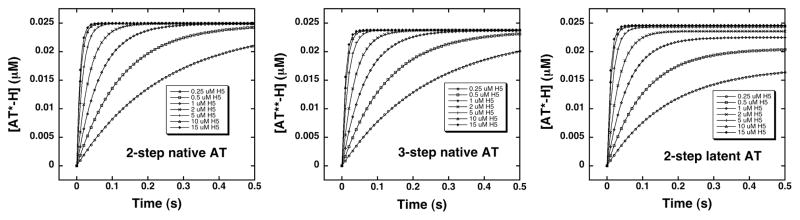
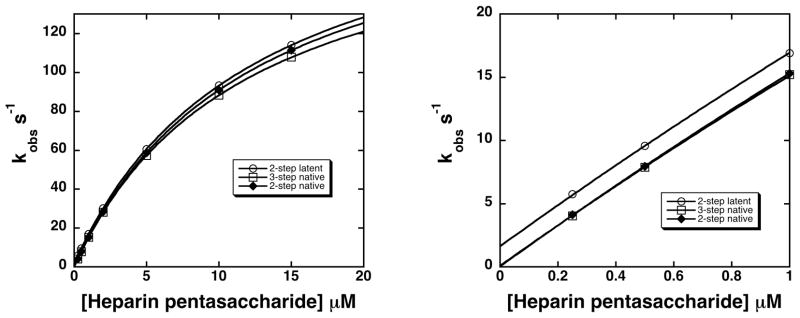
Progress curves for pentasaccharide binding to native and latent antithrombins were simulated for two and three-step binding mechanisms by numerical integrations of the differential equations for the mechanisms under the pseudo-first order conditions used experimentally. Figure 3 shows the simulated progress curves over the range of pentasaccharide concentrations used experimentally, concentrations that partially saturate the initial binding interaction (0.25–15 μM), and employing an antithrombin concentration of 0.025 μM to yield pseudo-first order conditions and exponential binding progress curves. The simulated progress curves were fit by an exponential function as was done with the experimental progress curves and observed pseudo-first order rate constants (kobs) for pentasaccharide binding plotted as a function of pentasaccharide concentration and computer fit by the hyperbolic function for a two-step saturable binding process:
Figure 3. Simulated progress curves for pentasaccharide binding to native and latent antithrombins by two-step and three-step induced-fit binding mechanisms.
Simulations of pentasaccharide binding to native and latent antithrombins by the two-step mechanism of scheme 1 or the three-step mechanism of scheme 2 were generated by numerical integration of the differential rate equations for the binding models using the kinetic constants in table 3. A series of binding progress curves were calculated for an antithrombin concentration of 0.025 μM and pentasaccharide concentrations ranging from 0.25–15 μM (circles) to duplicate the pseudo-first order experimental conditions and pentasaccharide concentrations used to measure the binding kinetics. Progress curves were fit by a single exponential function (solid lines) to obtain the observed pseudo-first order binding rate constant.
In this equation, K1,app is the apparent dissociation constant for the initial binding step and k+lim,app and k−lim,app are apparent forward and reverse rate constants for one or more subsequent rate-limiting conformational change steps. Figure 4 shows that the saturable dependence of kobs on pentasaccharide concentration obtained for the simulations for the native antithrombin interaction resulted in similar fits by the kinetic equation whether the two-step or three-step mechanisms were used. The parameters obtained from these fits shown in Table 4 differ insignificantly (within the experimental error of actual kinetic measurements), indicating that the kinetic data previously interpreted in terms of a two-step mechanism are also fully consistent with the 3-step mechanism.
Figure 4. Dependence of kobs for pentasaccharide binding to native and latent antithrombins on pentasaccharide concentration for two and three-step binding mechanisms.
kobs values were obtained from fits of progress curves in figure 3 for two-step (diamonds) and three-step (squares) binding mechanisms of the native antithrombin-pentasaccharide interaction and for the two-step (circles) mechanism of the latent antithrombin-pentasaccharide interaction and plotted as a function of pentasaccharide concentration. Data were fit by the hyperbolic saturation curve given in the text to obtain the kinetic parameters, K1,app, k+lim,app and k−lim,app, shown in table 4. The left-hand panel shows the full range of pentasaccharide concentrations examined and the right-hand panel shows the low pentasaccharide concentration range that reveals the different ordinate intercepts corresponding to k−lim,app.
Table 4.
Apparent kinetic parameters obtained from analyses of simulated binding progress curves by a two-step saturable binding mechanisma
| K1,app (μM) | k+lim,app (s−1) | kon (μM−1s−1) | k−lim,app = koff (s−1) | |
|---|---|---|---|---|
| Latent 2-step | 12.4±0.1 | 206±1 | 16.5±0.1 | 1.65±0.04 |
| Native 2-step | 12.2±0.1 | 202±1 | 16.5±0.1 | 0.05±0.03 |
| Native 3-step | 11.8±0.1 | 192±1 | 16.3±0.2 | 0.10±0.07 |
Kinetic constants were determined by fitting the dependence of kobs values obtained from simulated progress curves on pentasaccharide concentration to the hyperbolic saturation function in the text.
The simulations of 3-step and 2-step mechanisms for pentasaccharide binding to native and latent antithrombin, respectively, also predict the observed similarity in values for K1,app and k+lim,app, but differences in values for k−lim,app when these interactions are assumed to have the first two steps in common. k−lim,app, obtained from the intercept of the plots of kobs vs. pentasaccharide concentration (fig. 4), represents the overall off-rate constant and is equal to k−2 for the two-step latent antithrombin binding mechanism but is approximated by k−2 x k−3 /k+3 for the 3-step native antithrombin mechanism [31]. The slower off-rate constant for the native as compared to the latent antithrombin interaction thus suggests that the pentasaccharide interaction with the intermediate is tightened following the stage II conformational changes that lead to full activation. This is reflected by the equilibrium constants for the two successive conformational change steps, K2 and K3 (equal to k−2/k+2 and k−3/k+3, respectively), which indicate that pentasaccharide binding is enhanced 100-fold by the stage I conformational changes leading to the intermediate and a further 20-fold by the stage II conformational changes resulting in full activation. The two stages of conformational changes are thus both driven by increases in pentasaccharide binding energy that result from progressively better fits of the pentasaccharide in the binding site.
Spectroscopic reporters of antithrombin conformational changes
The 3-step mechanism for pentasaccharide binding and allosteric activation of antithrombin has additional implications for understanding the differential fluorescence changes associated with pentasaccharide binding to native and latent antithrombins. The observation that TNS fluorescence changes accompanying pentasaccharide interactions with the two antithrombins are similar suggests that these changes report the stage I conformational changes in the heparin binding site. This would be in keeping with the mapping of a TNS binding site in the serpin, PAI-1, between sheet A and helices D and E [32]. TNS bound to a comparable site in antithrombin might thus be expected to sense the stage I conformational changes in the adjacent heparin binding site. By contrast, tryptophan fluorescence changes reporting the two interactions differ significantly, with a 6% change for the latent antithrombin interaction and a 40% change for the native antithrombin interaction. These changes have been deconvoluted for the native antithrombin interaction by time-resolved fluorescence measurements of single Phe mutants of each of the four tryptophans of antithrombin [33]. Such studies suggest that Trp49 and Trp189, the residues closest to the heparin binding site, account for the 6% change observed for the latent antithrombin interaction whereas Trp225 and Trp307 account for the larger changes reporting the native antithrombin interaction. The Trp fluorescence changes reporting the stage I conformational changes that produce the intermediate are thus small relative to the fluorescence changes reporting the stage II conformational changes. The latter reflect the expulsion of the RCL hinge from sheet A and closing of the gap in sheet A in the protease binding region.
Activating effects of antithrombin conformational changes
The 3-step mechanism is supported by the effects of mutations in antithrombin that uncouple the conformational changes in the heparin binding site from those in the protease binding site. A notable example is a Lys133Pro mutant that blocks the extension of helix D and the Trp fluorescence changes that report RCL expulsion from sheet A [34]. A deletion of residues in the C-terminal loop of helix D produced a similar loss of Trp fluorescence changes but retained the TNS fluorescence changes that report the stage I conformational changes in the heparin binding site [35]. Importantly, the Lys133Pro mutation resulted in a ~20-fold decrease in heparin binding affinity, comparable to the heparin affinity differences between native and latent antithrombin. Moreover, this decrease in affinity was solely due to an increase in the apparent off-rate constant, the initial binding interaction and the limiting conformational change rate constant that determine the on-rate not being affected at all. This once again parallels the differences in pentasaccharide interactions with native and latent antithrombins and is consistent with the mutant antithrombin undergoing only the stage I conformational changes characteristic of the intermediate.
While the latent antithrombin-pentasaccharide complex appears to be a good model for the intermediate native antithrombin-pentasaccharide complex, the latent serpin is inactive as a protease inhibitor and therefore the effects of the stage I conformational changes on the activation of the intermediate complex cannot be assessed with latent antithrombin. These effects can be surmised, however, in the case of the Lys133Pro variant which provides the best model for the intermediate native antithrombin-pentasaccharide complex. Significantly, pentasaccharide binding to the variant causes a partial activation of antithrombin reactivity with factor Xa, as evidenced by a 30–40-fold pentasaccharide enhancement of the second order inhibition rate constant for the variant antithrombin-factor Xa reaction as compared to a 100–200-fold enhancement of the wild-type serpin reaction. This would suggest that the two conformational changes we have resolved in the allosteric activation pathway both contribute to activation. This would be in keeping with a recently proposed model of activation in which two distinct structural changes in the protease binding region, one that relieves repulsive interactions in an exosite outside the RCL and the other that extends the RCL away from the serpin body, contribute to enhancing antithrombin reactivity with factor Xa and factor IXa [14]. It would further imply that the induced-fit changes in the heparin binding site that occur in the stage I conformational changes are transmitted to the protease binding region through an allosteric core to partially activate antithrombin without the RCL activating conformational changes in stage II.
Increases in pentasaccharide interactions drive antithrombin conformational changes
Comparison of the salt dependence of pentasaccharide interactions with native and latent antithrombins indicates that the differences in binding affinity result from two fewer ion pair interactions made in the latent antithrombin-pentasaccharide complex than in the native antithrombin complex (Table 2) [28]. The contribution of other hydrogen bonding and van der Waals interactions as well as hydrophobic attractions that account for more than half of the binding energy of the native inhibitor interaction are similar for the two complexes [9]. Assuming that the latent antithrombin interaction is representative of that in the intermediate complex, the stage II conformational changes would thus appear to enhance pentasaccharide binding by the formation of two additional ion pair interactions. Clues to the origin of these additional interactions comes from comparing the structures of native, latent and intermediate antithrombin-pentasaccharide complexes within the heparin binding site (Fig. 5). Clear differences exist between native and latent structures from overlays of main chain atoms of the structures as verified by RMSD calculations using four native and three latent structures. These differences are most pronounced in the C-terminal extension of helix D, as expected, but are also significant in the P helix region and involve the bound pentasaccharide. Such differences suggest that rearrangements of the pentasaccharide occur in the second stage conformational changes to produce the net increase in ion pair interactions. Because of the cooperativity between antithrombin basic residues in binding the pentasaccharide, however, it is difficult to ascribe the gain in ionic interactions to specific residues solely from comparisons of these structures. Surprisingly, the overlays of two intermediate complex structures reveals a resemblance more to the native than the latent complex except for the C-terminal extension of helix D, implying that the so-called intermediate structures may be “frozen” in a state in which pentasaccharide binding interactions are approaching the fully-activated native structure before helix D extension and RCL expulsion have occurred.
Figure 5. Overlay of the heparin binding site for native, latent and intermediate antithrombin-pentasaccharide complex structures.
Residues 6–20 of the N-terminus, 45–57 of the A-helix and 112–137 of the P- and D-helix regions are shown in ribbon presentation for crystal structures of pentasaccharide complexes of native (pdb 1E03), latent (pdb 1E03) and intermediate (pdb 1NQ9) antithrombin variants in black, red and blue, respectively. The pentasaccharide is shown in stick in the same colors as the respective antithrombin form. The figure was prepared in swiss PDB viewer after structural alignment based on the main chain residues of the pentasaccharide-binding region including residues 7–25, 44–50 and 112–133. Native antithrombin (pdb 1E03-I) was used as a reference. Similar differences between structures were observed when overlays of the 1E03 native structure were done with 1TB6, 2GD4, 3KCG native structures, 1NQ9, 3EVJ intermediate structures and 1E03, 1NQ9, 3EVJ latent structures as assessed from RMSD calculations.
Contrasting our proposal that the stage II conformational changes are driven by improved binding interactions of the pentasaccharide in the fully-activated state, an alternative proposal has suggested that these conformational changes are driven instead by the greater thermodynamic stability of the fully-activated state when the pentasaccharide is bound [26, 27]. However, it is difficult to rationalize this proposal with the fact that the native antithrombin conformation is considerably more thermodynamically stable than the fully-activated conformation, based on the ~5 degree higher melting temperature of native versus activated forms of antithrombin [36, 37] and the equilibrium between these forms that favors the native over the activated state by at least 100:1[38]. This differential stability reflects the greater stability of antithrombin forms in which the RCL is inserted into sheet A, implying that the intermediate state of antithrombin in which the RCL is buried in sheet A and the fully-activated state in which the RCL is expelled are likely to have similar differential stabilities. That pentasaccharide binding does not reverse the relative stabilities of these states is indicated by the observation that mutations of positively charged heparin binding site residues to neutral or negatively charged residues which could be considered to mimic pentasaccharide binding fail to activate antithrombin [39]. It thus seems more likely that the stage II conformational changes are intrinsically unfavorable and are driven by the coupling of these changes to changes in the heparin binding site, i.e., helix D extension, that result in an improved fit of the pentasaccharide in the site. That there is an energetic cost to inducing the second stage conformational changes that is paid for by increases in pentasaccharide binding energy is supported by the observation that changes in antithrombin structure that reduce this energetic cost enhance heparin binding affinity [38, 40].
Conclusions
In summary, kinetic data comparing pentasaccharide binding to native and latent antithrombin together with kinetic simulations and X-ray structures of native, latent and intermediate antithrombin-pentasaccharide complexes support a three-step mechanism for heparin pentasaccharide binding and conformational activation of antithrombin. In this mechanism an initial weak binding of the pentasaccharide induces two sequential conformational change steps that are driven by progressive increases in pentasaccharide binding energy and that result in progressive activation of the serpin. The first set of conformational changes involve an induced-fit of the pentasaccharide with the heparin binding site that cause partially activating changes in the protease binding site transmitted through an allosteric communication network. These changes are reported by TNS fluorescence changes and changes in Trp49 and Trp189 fluorescence. The changes are common to native, latent and Lys133Pro variant antithrombins and account for the similar on-rate constants for pentasaccharide binding to these antithrombin forms. The second set of conformational changes involve further improvements in the fit of the pentasaccharide with the heparin binding site due to the extension of helix D that are linked to sheet A closure and RCL extension away from the sheet in the protease binding site that cause full allosteric activation. These changes are reported by changes in Trp225 and Trp307 fluorescence. This second set of conformational changes are unique to native antithrombin and account for the reduced off-rate constant for pentasaccharide dissociation from native as compared to latent and Lys133Pro variant antithrombins.
Research Highlights.
Heparin interacts with native and latent forms of antithrombin by similar induced-fit mechanisms
Kinetic evidence supports a 2-stage mechanism of allosteric activation of antithrombin by heparin
The first stage involves heparin binding and induced-fit steps common to latent antithrombin
The second stage involves activating conformational changes unique to native antithrombin
Each stage partially activates antithrombin and is driven by an induced-fit binding of heparin
Acknowledgments
We thank Dr. Peter Gettins of the University of Illinois at Chicago for his valued comments on the manuscript. This work was funded in part by NIH R01 grant HL-39888 to STO and by American Heart Association Fellowship 0920118G to BR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Björk I, Olson ST. Antithrombin, a bloody important serpin. In: Church FC, Cunningham DD, Ginsburg D, Hoffman M, Stone SR, Tollefsen DM, editors. Chemistry and Biology of Serpins. Plenum Press; New York: 1997. pp. 17–33. [PubMed] [Google Scholar]
- 2.Church FC, Pike RN, Tollefsen DM, Buckle DM, Ciaccia AV, Olson ST. Regulation of hemostasis by heparin binding serpins. In: Silverman GA, Lomas DA, editors. Molecular and Cellular Aspects of the Serpinopathies and Disorders of Serpin Activity. World Scientific Publishing; Singapore: 2007. pp. 509–554. [Google Scholar]
- 3.Gettins P. Serpin Structure, Mechanism, and Function. Chem Rev. 2002;102:4751–4803. doi: 10.1021/cr010170+. [DOI] [PubMed] [Google Scholar]
- 4.Stratikos E, Gettins PGW. Formation of the covalent serpin-proteinase complex involves translocation of the proteinase by more than 70 Å and full insertion of the reactive center loop into β-sheet A. Proc Natl Acad Sci, USA. 1999;96:4808–4813. doi: 10.1073/pnas.96.9.4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huntington JA, Read RJ, Carrell RW. Structure of a serpin-protease complex shows inhibition by deformation. Nature. 2000;407:923–926. doi: 10.1038/35038119. [DOI] [PubMed] [Google Scholar]
- 6.Marcum JA, Atha DH, Fritze LMS, Nawroth PP, Stern DM, Rosenberg RD. Cloned Bovine Aortic Endothelial Cells Synthesize Anticoagulantly Active Heparin Sulfate Proteoglycan. J Biol Chem. 1986;261:7507–7517. [PubMed] [Google Scholar]
- 7.De Agostini AI, Watkins SC, Slayter HS, Youssoufian H, Rosenberg RD. Localization of anticoagulantly active heparan sulfate proteoglycans in vascular endothelium: antithrombin binding on cultured endothelial cells and perfused rat aorta. J Cell Biol. 1990;111:1293–1304. doi: 10.1083/jcb.111.3.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petitou M, Casu B, Lindahl U. 1976–1983, A Critical Period in the History of Heparin: The Discovery of the Antithrombin Binding Site. Biochimie. 2003;85:83–89. doi: 10.1016/s0300-9084(03)00078-6. [DOI] [PubMed] [Google Scholar]
- 9.Olson ST, Björk I, Sheffer R, Craig PA, Shore JD, Choay J. Role of the antithrombin-binding pentasaccharide in heparin acceleration of antithrombinproteinase reactions. Resolution of the antithrombin conformational change contribution to heparin rate enhancement. J Biol Chem. 1992;267:12528–12538. [PubMed] [Google Scholar]
- 10.Chuang YJ, Swanson R, Raja SM, Olson ST. Heparin Enhances the Specificity of Antithrombin for Thrombin and Factor Xa Independent of the Reactive Center Loop Sequence. J Biol Chem. 2001;276:14961–14971. doi: 10.1074/jbc.M011550200. [DOI] [PubMed] [Google Scholar]
- 11.Izaguirre G, Zhang W, Swanson R, Bedsted T, Olson ST. Localization of an antithrombin exosite that promotes rapid inhibition of factors Xa and IXa dependent on heparin activation of the serpin. J Biol Chem. 2003;278:51433–51440. doi: 10.1074/jbc.M309266200. [DOI] [PubMed] [Google Scholar]
- 12.Izaguirre G, Olson ST. Residues Tyr253 and Glu255 in strand 3 of β-sheet C of antithrombin are key determinants of an exosite made accessible by heparin activation to promote rapid inhibition of factors Xa and IXa. J Biol Chem. 2006;281:13424–13432. doi: 10.1074/jbc.M600415200. [DOI] [PubMed] [Google Scholar]
- 13.Johnson DJ, Li W, Adams TE, Huntington JA. Antithrombin-S195A factor Xa-heparin structure reveals the mechanism of antithrombin activation. EMBO J. 2006;25:2029–2037. doi: 10.1038/sj.emboj.7601089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gettins PGW, Olson ST. Activation of antithrombin as a factor IXa and Xa inhibitor involves mitigation of repression rather than positive enhancement. FEBS Lett. 2009;583:3397–3400. doi: 10.1016/j.febslet.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson DJD, Langdown J, Huntington JA. Molecular basis of factor IXa recognition by heparin-activated antithrombin revealed by a 1.7 Å structure of the ternary complex. Proc Natl Acad Sci, USA. 2010;107:645–650. doi: 10.1073/pnas.0910144107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olson ST, Björk I. Predominant contribution of surface approximation to the mechanism of heparin acceleration of the antithrombin-thrombin reaction. Elucidation from salt concentration effects. J Biol Chem. 1991;266:6353–6364. [PubMed] [Google Scholar]
- 17.Dementiev A, Petitou M, Herbert JM, Gettins PGW. The ternary complex of antithrombin-anhydrothrombin-heparin reveals the basis of inhibitor specificity. Nat Struc Mol Biol. 2004;11:863–867. doi: 10.1038/nsmb810. [DOI] [PubMed] [Google Scholar]
- 18.Li W, Johnson DJD, Esmon CT, Huntington JA. Structure of the antithrombin-thrombin-heparin ternary complex reveals the antithrombotic mechanism of heparin. Nat Struc Mol Biol. 2004;11:857–862. doi: 10.1038/nsmb811. [DOI] [PubMed] [Google Scholar]
- 19.Rezaie AR. Calcium Enhances Heparin Catalysis of the Antithrombin-Factor Xa Reaction by a Template Mechanism: Evidence that calcium alleviates Gla domain antagonism of heparin binding to factor Xa. J Biol Chem. 1998;273:16824–16827. doi: 10.1074/jbc.273.27.16824. [DOI] [PubMed] [Google Scholar]
- 20.Bedsted T, Swanson R, Chuang YJ, Bock PE, Björk I, Olson ST. Heparin and Calcium Ions Dramatically Enhance Antithrombin Reactivity with Factor IXa by Generating New Interaction Exosites. Biochemistry. 2003;42:8143–8152. doi: 10.1021/bi034363y. [DOI] [PubMed] [Google Scholar]
- 21.Olson ST, Srinivasan KR, Björk I, Shore JD. Binding of high affinity heparin to antithrombin III. Stopped flow kinetic studies of the binding interaction. J Biol Chem. 1981;256:11073–11079. [PubMed] [Google Scholar]
- 22.Desai UR, Petitou M, Björk I, Olson ST. Mechanism of Heparin Activation of Antithrombin. Role of Individual Residues of the Pentasaccharide Activating Sequence in the Recognition of Native and Activated States of Antithrombin. J Biol Chem. 1998;273:7478–7487. doi: 10.1074/jbc.273.13.7478. [DOI] [PubMed] [Google Scholar]
- 23.Olson ST, Björk I, Bock SC. Identification of critical molecular interactions mediating heparin activation of antithrombin. Trends Cardiovas Med. 2002;12:198–205. doi: 10.1016/s1050-1738(02)00160-3. [DOI] [PubMed] [Google Scholar]
- 24.Carrell RW, Stein PE, Fermi G, Wardell MR. Biological implications of a 3Å structure of dimeric antithrombin. Structure. 1994;2:257–270. doi: 10.1016/s0969-2126(00)00028-9. [DOI] [PubMed] [Google Scholar]
- 25.Jin L, Abrahams JP, Skinner R, Petitou M, Pike RN, Carrell RW. The anticoagulant activation of antithrombin by heparin. Proc Natl Acad Sci, USA. 1997;94:14683–14688. doi: 10.1073/pnas.94.26.14683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson DJD, Huntington JA. Crystal structure of antithrombin in a heparin-bound intermediate state. Biochemistry. 2003;42:8712–8719. doi: 10.1021/bi034524y. [DOI] [PubMed] [Google Scholar]
- 27.Langdown J, Belzar KJ, Savory WJ, Baglin TP, Huntington JA. The critical role of hinge-region expulsion in the induced-fit heparin binding mechanism of antithrombin. J Mol Biol. 2009;386:1278–1289. doi: 10.1016/j.jmb.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schedin-Weiss S, Richard B, Hjelm R, Olson ST. Antiangiogenic forms of antithrombin specifically bind to the anticoagulant heparin sequence. Biochemistry. 2008;47:13610–13619. doi: 10.1021/bi801656u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verhamme IM, Bock PE. Rapid-reaction kinetic characterization of the pathway of streptokinase-plasmin catalytic complex formation. J Biol Chem. 2008;283:26137–26147. doi: 10.1074/jbc.M804038200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huntington JA, Olson ST, Fan B, Gettins PGW. Mechanism of Heparin Activation of Antithrombin. Evidence for Reactive Center Loop Preinsertion with Expulsion upon Heparin Binding. Biochemistry. 1996;35:8495–8503. doi: 10.1021/bi9604643. [DOI] [PubMed] [Google Scholar]
- 31.Tsodikov OV, Record MT., Jr General method of analysis of kinetic equations for multistep reversible mechanisms in the single exponential regime: Application to kinetics of open complex formation between Es70 RNA polymerase and lambdaPr promoter DNA. Biophys J. 1999;76:1320–1329. doi: 10.1016/S0006-3495(99)77294-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Egelund R, Einholm AP, Pedersen KE, Nielsen RW, Christensen A, Deinum J, Andreasen PA. A regulatory hydrophobic area in the flexible joint region of plasminogen activator inhibitor-1, defined with fluorescent activity-neutralizing ligands. J Biol Chem. 2001;276:13077–13086. doi: 10.1074/jbc.M009024200. [DOI] [PubMed] [Google Scholar]
- 33.Meagher JL, Beecham JM, Olson ST, Gettins PGW. Deconvolution of the Fluorescence Emission Spectrum of Human Antithrombin and Identification of the Tryptophan Residues That Are Responsive to Heparin Binding. J Biol Chem. 1998;273:23283–23289. doi: 10.1074/jbc.273.36.23283. [DOI] [PubMed] [Google Scholar]
- 34.Belzar KJ, Zhou A, Carrell RW, Gettins PGW, Huntington JA. Helix D Elongation and Allosteric Activation of Antithrombin. J Biol Chem. 2002;277:8551–8558. doi: 10.1074/jbc.M110807200. [DOI] [PubMed] [Google Scholar]
- 35.Meagher JL, Huntington JA, Fan B, Gettins PGW. Role of Arginine 132 and Lysine 133 in Heparin Binding to and Activation of Antithrombin. J Biol Chem. 1996;271:29353–29358. doi: 10.1074/jbc.271.46.29353. [DOI] [PubMed] [Google Scholar]
- 36.Futamura A, Gettins PGW. Serine 380 (P14)→Glutamate Mutation Activates Antithrombin as an Inhibitor of Factor Xa. J Biol Chem. 2000;275:4092–4098. doi: 10.1074/jbc.275.6.4092. [DOI] [PubMed] [Google Scholar]
- 37.Huntington JA, Gettins PGW. Conformational Conversion of Antithrombin to a Fully Activated Substrate of Factor Xa without Need for Heparin. Biochemistry. 1998;37:3272–3277. doi: 10.1021/bi972182o. [DOI] [PubMed] [Google Scholar]
- 38.Raja SM, Chhablani N, Swanson R, Thompson E, Laffan M, Lane DA, Olson ST. Deletion of P1 arginine in a novel antithrombin variant (antithrombin London) abolishes inhibitory activity but enhances heparin affinity and is associated with early onset thrombosis. J Biol Chem. 2003;278:13688–13695. doi: 10.1074/jbc.M300062200. [DOI] [PubMed] [Google Scholar]
- 39.Langdown J, Carter WJ, Baglin TP, Huntington JA. Allosteric activation of antithrombin is independent of charge neutralization or reversal in the heparin binding site. FEBS Lett. 2006;580:4709–4712. doi: 10.1016/j.febslet.2006.07.057. [DOI] [PubMed] [Google Scholar]
- 40.Turk B, Brieditis I, Bock SC, Olson ST, Björk I. The Oligosaccharide Side Chain on Asn-135 of α-Antithrombin, Absent in β-Antithrombin, Decreases the Heparin Affinity of the Inhibitor by Affecting the Heparin-Induced Conformational Change. Biochemistry. 1997;36:6682–6691. doi: 10.1021/bi9702492. [DOI] [PubMed] [Google Scholar]