Abstract
Mitochondria are the major source of reactive oxygen species. Both complex I and complex II mediate O2•− production in mitochondria and host reactive protein thiols. To explore the functions of the specific domains involved in the redox modifications of complexes I and II, various peptide-based antibodies were generated against these complexes, and their inhibitory effects were subsequently measured. The redox domains involved in S-glutathionylation and nitration, as well as the binding motif of the iron-sulfur cluster (N1a) of the complexes I and II were utilized to design B cell epitopes for generating antibodies. The effect of antibody binding on enzyme-mediated O2•− generation was measured by EPR spin trapping. Binding of either antibody AbGSCA206 or AbGSCB367 against glutathione (GS)-binding domain to complex I inhibits its O2•− generation, but does not affect electron transfer efficiency. Binding of antibody (Ab24N1a) against the binding motif of N1a to complex I modestly suppresses both O2•− generation and electron transfer efficiency. Binding of either antibody Ab75 or Ab24 against non-redox domain decreases electron leakage for O2•− production. In complex II, binding of antibody AbGSC90 against GS-binding domain to complex II marginally decreases both O2•− generation and electron transfer activity. Binding of antibody AbY142 to complex II against the nitrated domain modestly inhibits electron leakage, but does not affect the electron transfer activity of complex II. In conclusion, mediation of O2•− generation by complexes I and II can be regulated by specific redox and non-redox domains.
Keywords: Peptide, Antibody, Superoxide Generation, Complex I, Complex II
INTRODUCTION
Superoxide anion radical (O2•−) production in biological systems plays an important role in oxidative cellular injury. In the cardiovascular system, the mechanism of O2•− production is mainly controlled by several enzymatic systems. Under disease conditions such as ischemia and reperfusion, O2•− production via endothelial NO synthase (eNOS) and xanthine oxidase plays a critical role in endothelial dysfunction. Furthermore, an inflammatory response can trigger O2•− overproduction via leukocyte-derived NADPH oxidase. In the myocyte, mitochondrial electron transport chain (ETC) is the major source of O2•− production under disease conditions. In mitochondria, the generation of O2•− and the oxidants derived from it can act as a redox signal in triggering cellular events such as apoptosis, proliferation, and senescence.
The major components of the ETC are located in the inner membrane of mitochondria, hosting four major electron transfer complexes (complexes I-IV), cytochrome c, and ubiquinone. O2•− production via electron leakage can be mediated by complex I,1–4 complex II,5 and complex III.6–9 In addition, phosphorylation of complex IV under ischemic conditions also results in electron leakage for O2•− generation.10 The redox cofactors that directly participate in the catalysis of the electron transfer reaction are normally responsible for the electron leakage for O2•− generation. As documented in the literature, the ETC redox centers involved in O2•− generation include flavin mononucleotide (FMN) of complex I,1,2 iron-sulfur cluster of complex I,3 ubisemiquinone of complexes I-III, flavin adenine dinucleotide (FAD) of complex II, and heme b of complex III.9 Therefore, targeting of the structural environment surrounding redox cofactors might serve as an effective approach in reducing enzyme-mediated O2•− generation.
A specific redox modification on the protein matrix of ETC has been reported to alter its electron transfer activity and subsequent O2•− generation activity.5,11–14 In complex I, protein S-glutathionylation involved in the 51 kDa FMN-binding subunit and 75 kDa iron-sulfur protein has been extensively characterized in vitro and in vivo.11–13,15,16 In complex II, specific S-glutathionylation has been identified to be at the 70 kDa FAD-binding subunit.5 Based on an EPR spin-trapping study, GSSG (oxidized glutathione)-induced glutathionylation of either complex I or complex II affects their O2•− generation activity by marginally decreasing electron leakage and increasing electron transfer efficiency.5,12,16 High dosage of GSSG or diamide-induced glutathionylation of complex I tends to decrease the catalytic function of complex I and increase enzyme-mediated O2•− generation.11,13 Nevertheless, the catalytic role of each identified GS-binding domain remains unknown.
In addition to redox modification with S-glutathionylation, we have previously reported that in oxidative damage to complex I, the C206 moiety of the 51 kDa subunit plays a unique role as a reactive thiol, based on the evidence of immuno spin-trapping with 5, 5-dimethyl pyrroline N-oxide (DMPO) and mass spectrometry.17 With a proteomic approach, we further determined that the C206 of the 51 kDa subunit is involved in site-specific S-glutathionylation.5 The peptide identified as DMPO-binding and GS-binding is 200GAGAYIC206GEETALIESIEGK219, which is highly conserved in the bacterial, fungal, and mammalian enzymes (90% sequence identity in the bacterial enzyme, 176GAGAYIC182GEETALMNSLEGLR196). In complex II, the 70 kDa FAD-binding subunit undergoes protein tyrosine nitration following deglutathionylation under the conditions of ischemia and reperfusion.14 In vitro studies indicate protein nitration of complex II impairs electron transfer activity and increases electron leakage for O2•− generation.14
The specific functional domains involved in GS binding, protein radical formation, and nitration is thus proposed as targets for suppressing enzyme- mediated O2•− generation using an antibody-based approach. Moreover, probing the functional roles of each domain may be accomplished with antibodies against specific target domains in either complex I or complex II. Peptide-based immunochemistry is an ideal approach to generate high affinity antibodies against specific functional domains present in the electron transfer complex. The development of antibodies with high titer and high specificity would also facilitate the detection of complex I/II-derived redox modifications ex vivo and in vivo.
In this study, we generated several peptide-based antibodies against specific domains of complexes I and II. The effect of antibody binding on enzyme-mediated O2•− generation was analyzed by EPR spin-trapping with 5-diethoxylphosphoryl-5-methyl-1-pyrroline N-oxide (DEPMPO). The use of a peptide-based antibody may represent an application to reduce oxidant stress. Practically, the peptide-based antibody has potential use in developing a peptide-based vaccine. Therefore, this pilot study could be valuable for therapeutic applications.
MATERIALS AND METHODS
Reagents
Ammonium sulfate, diethylenetriaminepentaacetic acid (DTPA), ubiquinone-1 (Q1), sodium cholate, deoxycholic acid, rotenone, and β-nicotinamide adenine dinucleotide (reduced form, NADH) were purchased from Sigma Chemical Company (St. Louis, MO) and used as received. The spin trap of DEPMPO was purchased from ALEXIS Biochemicals (San Diego, CA).
Model Building
The amino acids sequences of complex I (NQR) from Thermus thermophilus and from Bos taurus were retrieved from NCBI. The NCBI reference sequence IDs are YP_143354.1, YP_143355.1, YP_143356.1 (T. thermophilus 24, 51, 75 kDa subunits) and XP_001250335.2, NP_777233.1, NP_777245.1 (B. taurus 24, 51, and 75 kDa subunits), respectively. The amino acid sequences were aligned by ClustalW218 to evaluate the sequence identity and homology.
Several homology models were generated by SWISS-MODEL19 based on either automatically or manually selected templates and these models were subsequently assessed by ANOLEA20 and PROCHECK21 tools. The best model was chosen based on these assessment results. The model and the template structures were superimposed by DaliLite22 and visualized by PyMOL23. The predicted amino acid sequences (p24, p51, and p75 in Table 1) in the exposed surface area of the model structures were manually identified as a B cell epitope for the subsequent synthesis of designed MVF fusion peptides.
Table 1.
Amino Acid Sequence of Designed Peptides and Their Corresponding MVF Fusion Peptides Used as Immunogens.
| No | Amino Acid Sequence | Seq. Code | #AA |
|---|---|---|---|
| 1 | GAGAYICGEETALIESIEGK | pGSCA206 | 20 |
| 2 | KLLSLIKGVIVHRLEGVEGPSLGAGAYICGEETALIESIEGK | pMVFGSCA206 | 42 |
| 3 | SGDTTAPKKTSFGSLKDFDR | p51 | 20 |
| 4 | KLLSLIKGVIVHRLEGVEGPSLSGDTTAPKKTSFGSLKDEDR | pMVF51 | 42 |
| 5 | VDSDTLCTEEVFPTAGAGTDLR | pGSCB367 | 22 |
| 6 | KLLSLIKGVIVHRLEGVEGPSLVDSDTLCTEEVFPTAGAGTDLR | pMVFGSCB367 | 44 |
| 7 | WNILTNSEKTKKAREGVMEFL | p75 | 21 |
| 8 | KLLSLIKGVIVHRLEGVEGPSLWNILTNSEKTKKAREGVMEFL | pMVF75 | 43 |
| 9 | KYHIQVCTTTPCMLRNSDSILE | p24N1a | 22 |
| 10 | KLLSLIKGVIVHRLEGVEGPSLKYHIQVCTTTPCMLRNSDSILE | pMVF24N1a | 44 |
| 11 | EDLTPKDIEEIIDELKAGK | p24 | 19 |
| 12 | KLLSLIKGVIVHRLEGVEGPSLEDLTPKDIEEIIDELKAGK | pMVF24 | 41 |
| 13 | GAGLRAAFGLSEAGFNTACVTK | pGSC90 | 22 |
| 14 | KLLSLIKGVIVHRLEGVEGPSLGAGLRAAFGLSEAGFNTACVTK | pMVFGSC90 | 44 |
| 15 | DQDAIHYMTEQAPASVVEL | pY142 | 19 |
| 16 | KLLSLIKGVIVHRLEGVEGPSLDQDAIHYMTEQAPASVVEL | pMVFY142 | 41 |
The Construction of Designer MVF Fusion Peptides
An MVF fusion peptide was a chimeric construct of three distinct sequence motives: a T cell epitope, a linker, and a B cell epitope. The T cell epitope sequence (KLLSLIKGVIVHRLEGVE, Table 1) was a motif of 18 residues derived from a promiscuous T-helper measles virus fusion protein. Following the T cell epitope was a four-residue linker (GPSL). The linker was incorporated into the fusion peptide to help in independent folding of both the T cell epitope and the B cell epitope. The sequences of B cell epitopes (Table 1) were specific functional domains of the complex I or complex II subunits. These B cell epitope sequences were determined based on either experimental MS data or theoretical homology models.
Peptide Synthesis and Purification
Peptide synthesis was performed on a Milligen/Biosearch 9600 solid-phase peptide synthesizer (Bedford MA) using Fmoc chemistry. Regioselective side chain protection on cys residues were Trt or Acm as described by Kaumaya et. al.24,25 Preloaded Fmoc-amino acids on CLEAR ACID resin (0.36 meq/g) (Peptides International, Louisville, KY) were used for the peptide synthesis using the PyBop/HOBt coupling method. All peptides were cleaved from the resin using global deprotection reagent B [Trifluoroacetic acid (TFA): Phenol: Water: Triisopropylsilane (TIS), 90:4:4:2]. The protecting group from cys (Trt) comes off in the global cleavage reaction. Crude peptides were purified by preparative RP-HPLC using C-4 Vydac column in a water (0.1% TFA): acetonitrile (0.1% TFA) gradient system. Pure fractions were analyzed using HPLC, pooled together and lyophilized in 10% acetic acid. The purified peptide was hydrolyzed dry and kept at 20°C to prevent oxidation of free sulfhydryl groups of cys residues.
Peptide Immunization and Antibody Purification
Two New Zealand white rabbits (6–8 weeks old, female outbreed, Harlan, Indianapolis, IN) were immunized with each purified MVF fusion peptide (1 mg) dissolved in H2O (500 μl) with 100 mg of a muramyl dipeptide adjuvant, nor-MDP (N-acetylglucosamine-3-yl-acetyl-L-alanyl-D-isoglutamine). Peptides were emulsified (50:50) in a Montanide ISA 720 vehicle (Seppic, France). 2 ml of blood was drawn for pre-immunization sera. All rabbits were immunized subcutaneously at four spots on the back. After the first immunization, the same dose of booster injections was administered at 3, 6, and 9 weeks. Sera were collected by bleeding from the rabbit’s ear after each immunization for determination of antibody titers. Antibody titers were determined by ELISA. High titered sera were purified on a protein A/G-agarose column (Pierce, Rockford, IL). Eluted antibodies were concentrated and exchanged in PBS using 100 kDa cut-off centrifuge filter units (Millipore, Bedford, MA).
Preparations of Mitochondrial Complex I
Bovine heart mitochondrial complex I was prepared under non-reducing conditions according to the published method with modifications.26 Submitochondrial particles (SMP) were prepared as described and used as the starting material,27 beginning with 2.5 lb of trimmed bovine hearts with fat and connective tissues removed. The SMP preparation was suspended in 50 mM Tris-Cl buffer, pH 8.0, containing 1 mM histidine and 0.66 M sucrose (TSH), and then subjected to KCl fractionation (72 gm KCl was added per liter of SMP) in the presence of deoxycholate (0.3 mg/mg protein). The supernatant thus obtained was mixed with an appropriate amount of cold water to precipitate trace amounts of cytochrome c oxidase, and then dialyzed against 10 mM Tris-Cl, pH 8.0, containing 1 mM EDTA for 6 h with one change of the buffer. The dialysate was subjected to centrifugation (96,000 × g for 75 min). The pellet containing complexes I, II, and III was homogenized in TSH buffer, and then subjected to repeated ammonium acetate fractionation in the presence of deoxycholate (0.5 mg/mg protein). Complex I was finally resolved (39% saturation of ammonium sulfate) and separated using ammonium sulfate precipitation (35.9% saturation) in the presence of potassium cholate (0.4 mg/mg protein).
The flavin subcomplex of complex I containing NADH dehydrogenase (NDH) was isolated from SMP under non-reducing conditions by following the established method described in a previous publication.17
Preparations of Mitochondrial Complex II
Bovine heart mitochondrial supercomplex hosting complexes II and III, succinate-cytochrome c reductase (SCR), was prepared and assayed according to the published method developed by Yu et. al.28 The purified SCR contained ~ 4.0–4.2 nmol heme b per mg protein and exhibited an activity of approximately 8.5μmol cytochrome c reduced min−1 mg−1 protein. Complex II was isolated from SCR by calcium phosphate-cellulose chromatography under non-reducing conditions according to the published method described by Yu et. al.28,29 Complex II-containing fractions obtained from the second calcium phosphate-cellulose column were concentrated by 43% ammonium sulfate saturation and centrifuged at 48,000 × g for 20 minutes.29 The precipitate obtained was dissolved in 50 mM Na/K phosphate, pH 7.8, containing 0.2% sodium cholate and 10% glycerol. The specific activity of the purified SQR was ~ 15.2 μmol succinate oxidized or DCPIP (dichlorophenol indophenols) reduced min−1 mg−1 protein.
Analytical Methods
Optical spectra were measured on a Shimadzu 2401 UV/VIS recording spectrophotometer. The protein concentrations of complex I and complex II were determined by the Lowry method using BSA as standard. The concentration of Q1 or Q2 was determined by absorbance spectra from NaBH4 reduction using the extinction coefficient ε(275 nm – 290 nm) = 12.25 mM−1 cm−1.30 To measure the electron transfer activity of complex I, an appropriate amount of complex I was added to an assay mixture (1.00 ml) containing 20 mM potassium phosphate buffer, pH 8.0, 2mM NaN3, 0.1 mM Q1, and 0.15 mM NADH as described by Hatefi et al.31 The complex I activity was determined by measuring the decrease in absorbance at 340 nm. The specific activity of complex I was calculated using the extinction coefficient ε340nm = 6.22 mM−1 cm−1. The purified complex I exhibited a specific activity of ~ 1.0 μmol NADH oxidized min−1 mg−1.
The enzyme activity of complex II was assayed by measuring Q2-stimulated DCPIP reduction by succinate as described in the literature.29 To measure the electron transfer activity of complex II, an appropriate amount of complex II was added to an assay mixture (1.00 ml) containing 50 mM phosphate buffer, pH 7.4, 0.1 mM EDTA, 75 μM DCPIP, 50 μM Q2, and 20 mM succinate as developed by Hatefi et. al.31 The complex II activity was determined by measuring the decrease in absorbance at 600 nm. The specific activity of complex II [nmol DCPIP reduced (or succinate oxidized) min−1 mg−1] was calculated using the extinction coefficient ε600 nm = 21 mM−1cm−1.
Electron Paramagnetic Resonance Experiments
EPR measurements were carried out on a Bruker EMX spectrometer operating at 9.86 GHz with 100 kHz modulation frequency at room temperature (RT). The reaction mixture was transferred to a 50 μl capillary, which was then positioned into the HS cavity (Bruker Instrument, Billerica, MA). The sample was scanned using the following parameters: center field, 3510 G; sweep width, 140 G; power, 20 mW; receiver gain, 2×105; modulation amplitude, 1 G; time of conversion, 163.84 ms; time constant, 163.84 ms; number of scans, 3. The spectral simulations were performed using the WinSim program developed at NIEHS by Duling.32 The hyperfine coupling constants used to simulate the spin adduct of DEPMPO/•OOH were isomer 1: aN = 13.14 G, aHß= 11.04 G, aHγ = 0.96 G, aP = 49.96 G (80% relative concentration); isomer 2: aN = 13.18 G, aHß = 12.59 G, aHγ = 3.46 G, aP = 48.2 G (20% relative concentration).33 The correlation coefficient of simulated spectrum was typically more than 0.950. Therefore, the simulated spectrum was suitable for spin quantitation.17,34
Immunoblotting Analysis
The reaction mixture was mixed with Laemmli sample buffer at a 4:1 (v/v) ratio, incubated at 70 °C for 10 min, and then immediately loaded onto a 4–12% Bis-Tris polyacrylamide gradient gel. Samples were run at RT for 55 min at 190 V. Protein bands were electrophoretically transferred to nitrocellulose membrane in 25 mM Bis-Tris, 25 mM Bicine, 0.029% (w/v) EDTA, and 10% methanol. Membranes were blocked for 1 h at RT in Tris-buffered saline containing 0.1% Tween-20 (TTBS) and 5% dry milk. The blots were then incubated overnight with generated antibodies at 4 °C. Blots were then washed 3 times in TTBS, and incubated for 1 h with horseradish peroxidase-conjugated anti-rabbit IgG in TTBS at RT. The blots were washed twice in TTBS and twice in TBS and then visualized by ECL Western Blotting Detection Reagents (GE Healthcare, Fairfield, CT).
RESULTS AND DISCUSSION
O2•− Generation Mediated by Complex I as Measured by EPR Spin-Trapping with DEPMPO
To obtain direct evidence for O2•− production mediated by complex I, we employed the EPR spin-trapping technique to measure O2•− generation. Of theavailable spin traps, DEPMPO is ideal for measuring O2•− production by complex I based on the following reasons: a) DEPMPO assay is 40-fold more sensitive than the cyt c assay for the detection of O2•− 35, b) DEPMPO traps O2•− with an efficiency of 60–70%,35 and c) the O2•− adduct of DEPMPO/•OOH is more stable than that of DMPO/•OOH 33.
When isolated complex I (0.1 mg/ml) was incubated with DEPMPO (20 mM) in PBS and the reaction initiated by the addition of NADH (0.5 mM), a multi-line EPR spectrum was produced that was characteristic of DEPMPO/•OOH (Fig. 1A, solid line) based on the hyperfine coupling constants [isomer 1: aN = 13.14 G, aHß = 11.04 G, aHγ = 0.96 G, aP = 49.96 G (80% relative concentration); isomer 2: aN = 13.18 G, aH ß= 12.59 G, aHγ = 3.46 G, aP = 48.20 G (20% relative concentration)17,33] obtained from computer simulation (Fig. 1A, dashed line). Trapping of complex I-mediated O2•− by DEPMPO was thus highly specific and suitable for quantitative analysis.
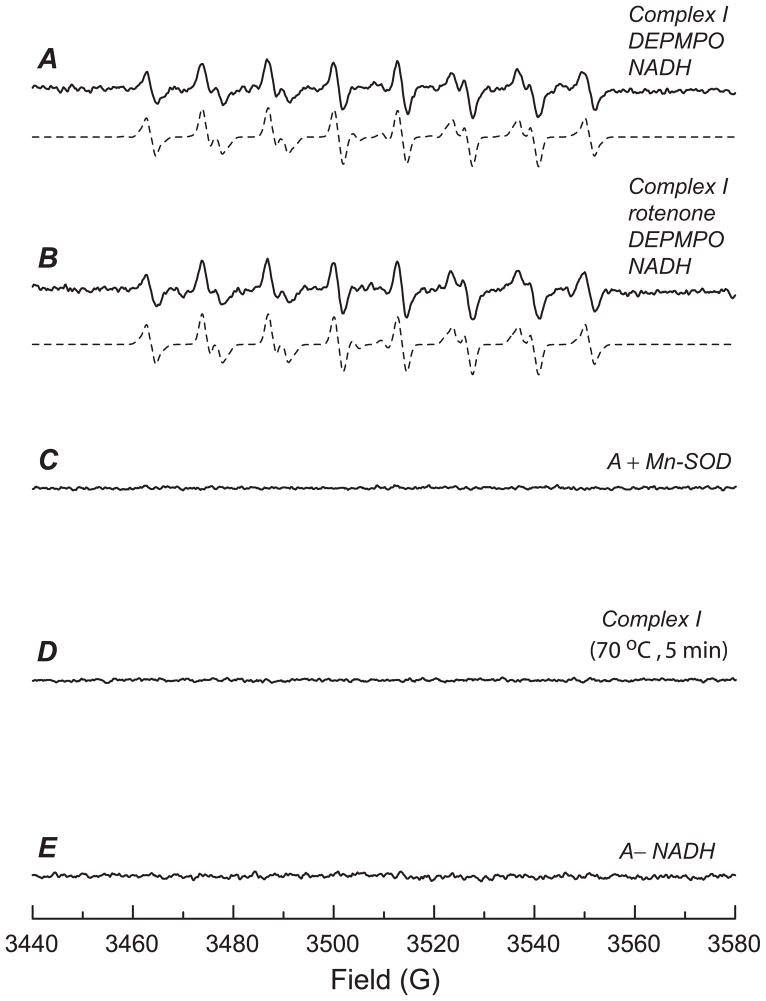
Fig. 1. EPR spin-trapping of O2•− generated from complex I in the presence of DEPMPO.
A, The computer simulation (dashed line) superimposed on the experimental spectrum (solid line) obtained using complex I (0.1 mg/ml), DEPMPO (20 mM), diethylenetriaminopentaacetic acid (1 mM), and NADH (0.5 mM) in PBS. The experimental spectrum was recorded after signal averaging 3 scans at RT. B, the same as A, except that rotenone (25 μM) was added to the mixture before the reaction was initiated by NADH. C, the same as A, except that Mn-SOD (0.2 unit/μl) was added to the mixture before the reaction was initiated by NADH. D, the same as A, except that complex I was heated at 70 °C for 5min. E, the same as A, except that the NADH was omitted from the system.
Rotenone, a known inhibitor of complex I, is proposed to inhibit electron transfer activity through blocking the quinone-binding site. Pretreatment of complex I with rotenone (25 μM) inhibited ~ 80% of the electron transfer activity from NADH to Q1, but failed to inhibit the O2•− production mediated by complex I (97.8 ± 2.2% superoxide-generation activity (SGA) remained, Fig. 1B), implying that the flavin subcomplex (Fp) controls complex I-mediated O2•− production in the absence of Q1.
Upon addition of manganese-superoxide dismutase (Mn-SOD, 200 units/ml), the EPR signal of DEPMPO/•OOH adduct derived from the trapping of O2•− was quenched (Fig. 1C). In the absence of complex I, no DEPMPO/•OOH adduct was detected (data not shown), indicating the enzymatic dependence of the DEPMPO/•OOH adduct formation. When native complex I was replaced with heat-denatured (70°C, 5 min) complex I, the formation of DEPMPO/•OOH was inhibited (Fig. 1D), suggesting that electron transfer activity in the enzyme is required for O2•− generation. The amount of DEPMPO/•OOH was greatly decreased when NADH was omitted from the system (Fig. 1E), supporting the role of NADH as the direct electron donor for complex I-mediated O2•− production.
Homology Models of the Complex I 51, 75 and 24 kDa Subunits
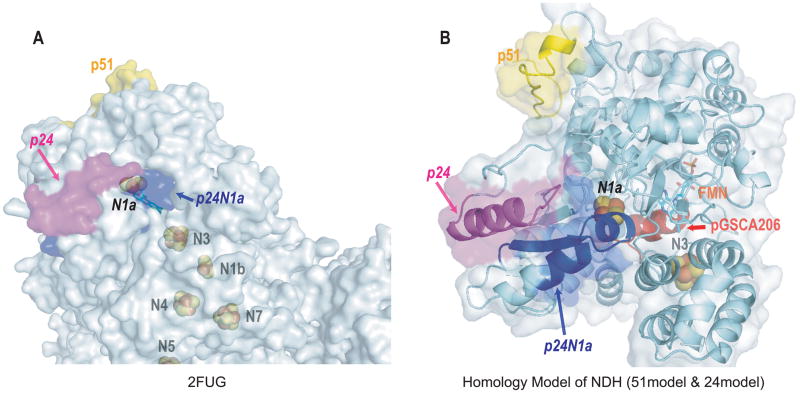
The three homology models of the complex I 51, 75 and 24 kDa subunits generated from SWISS-MODEL were named 51model (Fig. 2A), 75model (Fig. 3A), and 24model (Fig. 5A) respectively. The ClustalW2 sequences alignment of complex I (NQR) from T. thermophilus and from B. taurus indicated all residues involved in the coordination of iron-sulfur clusters were identical (Fig. S1 for the sequence alignment result of 51, 75 and 24 kDa subunits). The conserved residues were C379, C382, C385 and C425 for cluster N3 in the 51model (in Fig. 2A). The 75model (in Fig. 3A) consisted of three iron-sulfur clusters, N1b (C64, C75, C78 and C92), N4 (C176, C179, C182 and C226) and N5 (H124, C128, C131 and C137) while the 24model (in Fig. 5A) contained cluster N1a (C140, C145, C181 and C185). Since these iron-sulfur clusters play essential roles in the electron transfer chain activities, these residues were expected to be highly conserved across different organisms. The SWISS-MODEL details and the Ramachandran plot results of the three models are summarized in Table 2.
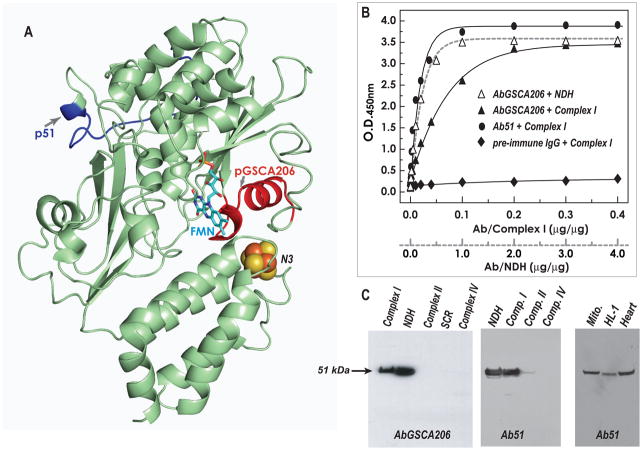
Fig. 2. Immunological specificity of the antibodies AbGSCA206 and Ab51 as analyzed by ELISA and Western blotting.
A, Homolog model (51model in Table 2, aa: 16–432 in bovine mature protein) of the 51 kDa subunit using the crystal structure of T. thermophilus (2FUG) as a template. Arrows show the domains of pGSCA206 and p51, denoted by red and blue ribbons. The peptides of pGSCA206 and p51 are designed to be the B cell epitopes of immunogens. B, the antigens, 400 μg of complex I and 40 μg of NDH, were coated on the ELISA plate and then reacted with various amounts of purified AbGSCA206 (Δ and ▲), Ab51 (●), and pre-immune IgG (◆). A goat anti-rabbit IgG-horseradish peroxidase conjugate was then added. The binding of AbGSCA206 and Ab51 to the antigens was quantified by measuring the optical density at 450 nm of the diimine product resulting from two-electron oxidation of 3,3′,5,5′-tetramethylbenzidine (TMB) using a TMB Peroxidase ELISA Substrate Kit (Bio-RAD Laboratories, Hercules, CA). C, SDS-PAGE was carried out according to the procedure described under “Materials and Methods.” The isolated enzymes, complex I (40 μg), NDH (4 μg), complex II (40 μg), SCR (40 μg), and complex IV (40 μg) were immunoblotted with AbGSCA206 in left panel and Ab51 in middle panel respectively. Right panel: isolated mitochondria (Mito. 100 μg) from rat heart, cardiac myocytes (HL-1, 100 μg), and tissue homogenates of rat heart (Heart, 100 μg), were probed with Ab51.
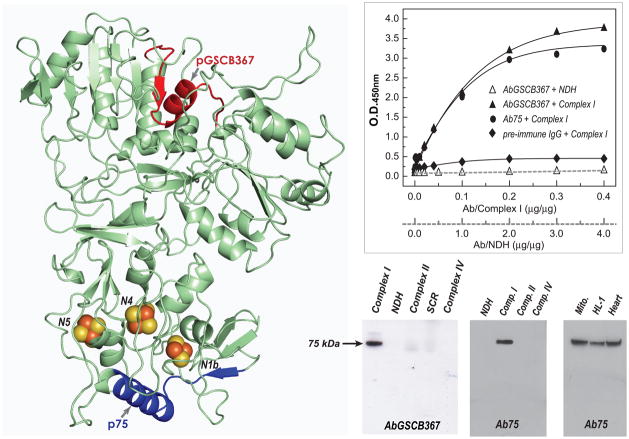
Fig. 3. Immunological analysis of the antibodies AbGSCB367 and Ab75.
A, Homolog model (75model in Table 2, aa: 8–687 in bovine mature protein) of the 75 kDa subunit using the crystal structure of T. thermophilus (2FUG) as a template. Arrows show the domains of pGSCB367 and p75, denoted by red and blue ribbons. Both pGSCB367 and p75, containing the conformation of β-sheet-α-turn-helix based on the homology model in A, were designed to be the B cell epitopes for antibodies generation. B, the antigens, 400 μg of complex I was reacted with various amounts of purified AbGSCB367 ( Δ and ▲), Ab75 (●), and pre-immune IgG (◆), and then subjected to ELISA analysis. C, The antigens, 4 μg of NDH, 40 μg of complex I, 40 μg of complex II, 40 μg of SCR, and 40 μg of complex IV were probed with AbGSCB367 (left panel) and p75 (middle panel). Right panel: isolated mitochondria (Mito. 100 μg) from rat heart, cardiac myocytes (HL-1, 100 μg), and tissue homogenates of rat heart (Heart, 100 μg), were probed with antibodies Ab75.
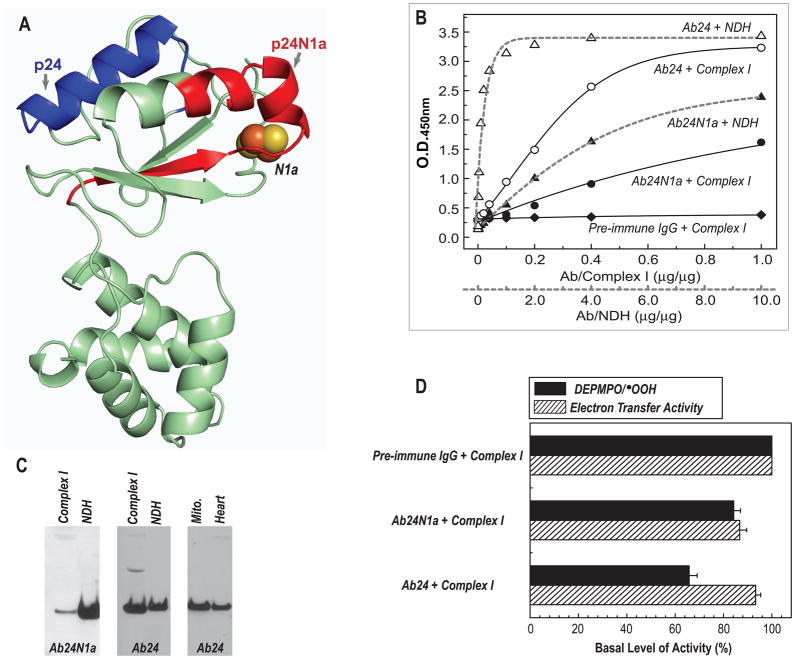
Fig. 5. Immunological analysis of antibodies Ab24N1a and Ab24.
A, Homolog model (24model in Table 2, aa: 38–201 in bovine mature protein) of the 24 kDa subunit using the crystal structure of T. thermophilus (2FUG) as a template. Arrows show the domains of p24N1a and p24, denoted by red and blue ribbons. The peptides of p24N1a and p24 were designed to be the B-cell epitopes for antibodies generation. B, ELISA analysis of Ab24N1a and Ab24 using the antigens of complex I (solid line, 114 μg) and NDH (dashed line, 11 μg). C, immunoblotting analysis of Ab24N1a and Ab24 using the complex I (40 μg), NDH (4 μg), isolated mitochondria (Mito., 100 μg) from rat heart, and tissue homogenates of rat heart (100 μg). D, effect of antibodies Ab24N1a and Ab24 on the O2•− generation (black bars) and electron transfer activity (dashed bars) mediated by complex I.
Table 2.
Model Generation (SWISS-MODEL) and Evaluation (Ramachandran Plot) Statistics.
| 24model | 51model | 75model | |
|---|---|---|---|
| SWISS-MODEL details | |||
| Modelled residue range | 60 to 233 | 36 to 452 | 31 to 710 |
| Based on template | 3i9v2 (3.10 Å) | 2fugA (3.30 Å) | 3i9vC (3.10 Å) |
| Sequence Identity [%] | 34.286 | 45 | 25.546 |
| Evalue: | 0.00e–1 | 0.00e–1 | 0.00e–1 |
| Ramachandran Plot | |||
| Residues in most favoured regions [A,B,L] | 124 | 266 | 455 |
| Residues in additional allowed regions [a,b,l,p] | 21 | 65 | 127 |
| Residues in generously allowed regions [~a,~b,~l,~p] | 2 | 8 | 8 |
| Residues in disallowed regions | 1 | 5 | 6 |
| Number of non-glycine and non-proline residues | 148 | 344 | 596 |
| Number of end-residues (excl. Gly and Pro) | 2 | 2 | 2 |
| Number of glycine residues | 9 | 50 | 50 |
| Number of proline residues | 15 | 21 | 32 |
| Total number of residues | 174 | 417 | 680 |
Generation of Antibodies against the 51 kDa Subunit of Complex I
We had previously reported a specific tryptic 20-residue peptide, 200GAGAYIC206GEETALIESIEGK219, (pGSCA206, Table 1 and 51model in Fig. 2A) involved in the S-glutathionylation of the 51 kDa subunit of complex I.12 The C206 site was also known to be the specific site of protein thiyl radical formation resulting from oxygen free-radical attack.17 To gain deeper insight into the functional role of this identified GS-binding/DMPO-binding domain, a high titer antibody against the pGSCA206 sequence was generated. A 42-residue peptide, KLLSLIKGVIVHRLEGVEGPSLGAGAYICGEETALIESIEGK, (pMVFGSC206, Table 1) was synthesized. The peptide pMVFGSC206 consists of a T cell epitope (KLLSLIKGVIVHRLEGVE), a linker (GPSL), and a B cell epitope (pGSCA206, GAGAYICGEETALIESIEGK). The crude peptide was purified to homogeneity by reverse phase HPLC and characterized by matrix assisted laser desorption ionization mass spectroscopy (MALDI), which gave the exact mass unit [(M + H)+ 4359.41].
To provide an extra experimental control in addition to using pre-immune IgG, the N terminal amino acid residues 1–20 of the 51 kDa mature protein (p51, 21SGDTTAPKKTSFGSLKDEDR40, Table I and 51model in Fig. 2A) was chosen as the other B cell epitope. The corresponding chimeric construct of the fusion peptide used as an immunogen wasKLLSLIKGVIVHRLEGVEGPSLSGDTTAPKKTSFGSLKDEDR (pMVF51, mass unit = 4479.61). Rabbits were immunized with the immunogen pMVFGSCA206 or pMVF51. The antibodies generated and purified were named AbGSCA206 and Ab51 respectively.
Immunological Specificity of Antibodies of AbGSCA206 and Ab51
The immunological cross-reactivity was analyzed by ELISA using intact complex I and its Fp subcomplex (NDH) as an antigen. As shown in Fig. 2B, both AbGSCA206 and Ab51 showed high titer and bound with antigens contained in the 51 kDa subunit in intact complex I and NDH. The binding affinity of Ab51 to complex I is marginally higher than that of AbGSCA206. Furthermore, the binding affinity of Fp subcomplex is higher than that of intact complex I, suggesting that detaching Fp from the intact complex I renders the domain of pGSCA206 more surface-exposed. There was virtually undetectable binding between pre-immune IgG and the antigen of complex I (Fig. 2B) or NDH (data not shown). The protein concentration of complex I used for ELISA was 11 times higher than that of NDH, because the amount of Fp subcomplex present in complex I is about 8.3%.
The immunological specificity of antibodies was further characterized by Western blot (Fig. 2C) against complex I, NDH, complex II, SCR, and complex IV. Both Ab51 (left panel in Fig. 2C) and AbGSCA206 (middle panel in Fig. 2C) exhibited specific binding to the 51 kDa subunit of complex I and NDH. As expected, there was no detectable binding of Ab51 or AbGSCA206 to complex II, SCR, or complex IV. Further immunological specificity of Ab51 was analyzed by immunoblotting protein samples from isolated mitochondria of the rat heart, rat myocardial tissue, and the mouse cardiac myocyte cell line HL-1 (right panel in Fig. 2C). Ab51 bound specifically to the 51 kDa subunit of complex I, demonstrating that Ab51 is suitable for in vivo, ex vivo, or in vitro studies.
Antibodies against the 75 kDa Subunit of Complex I
A tryptic peptide 361VDSDTLC367TEEVFPTAGAGTDLR382 (pGSCB367, Table 1 and 75model in Fig. 3A) involved in the S-glutathionylation of the 75 kDa subunit of complex I was previously identified12. To generate antibodies against pGSCB367, a 44-residue chimeric MVF fusion peptide KLLSLIKGVIVHRLEGVEGPSLVDSDTLCTEEVFPTAGAGTDLR (pMVFGSCB367, Table 1) was synthesized. The purified pMVFGSCB367 was characterized by MS, which gave the observed mass [(M + H)+ 4519.61] as calculated [(M + H)+ 4519.49].
To provide an experimental control, residues 77–97 of the 75 kDa mature protein, 100WNILTNSEKTKKAREGVMEFL120 (p75, Table 1 and 75model in Fig. 3A) was designed as a B cell epitope. The purified 43-residue MVF fusion peptide, KLLSLIKGVI VHRLEGVEGPSLWNILTNSEKTKKAREGVMEFL (pMVF75, mass unit = 4834.79), was used as an immunogen. The polyclonal antibodies generated against pMVFGSC367 and pMVF75 were named AbGSCB367 and Ab75, respectively.
Immunological Specificity of Antibodies against the 75 kDa Subunit of Complex I
Immunological specificity of AbGSCB367 and Ab75 was characterized by ELISA (Fig. 3B) and Western blotting (Fig. 3C). As indicated in Fig. 3B, both AbGSCB367 and Ab75 antibodies showed high titer and bound with antigen of complex I but not with the antigen of NDH due to the absence of the 75 kDa polypeptide in the Fp subcomplex. The immunological specificity was further probed with Western blotting using complex I, NDH, complex II, SCR, and complex IV. As indicated in Fig. 3C (left-middle panels), both antibodies only bound specifically to the 75 kDa subunit of complex I. Ab75 was further tested for specificity against the 75 kDa subunit of complex I in isolated mitochondria of the rat heart, rat myocardial tissue, and mouse myocyte cell line HL-1, where Ab75 bound specifically to the 75 kDa subunit (right-side panel in the Fig. 3C). Taken together, A75 is both highly specific and highly sensitive, thus suitable for in vivo, ex vivo, in vitro studies.
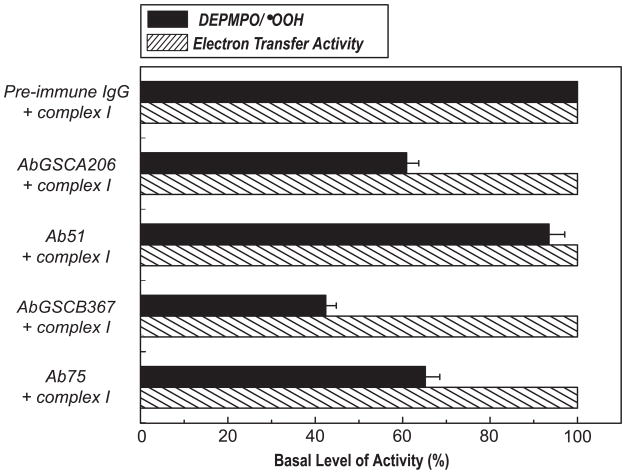
Immunoinhibition of Complex I-mediated O2•− Generation by AbGSCA206, Ab51, AbGSCB367, and Ab75
To test the role of the GS-binding domain from the 51 kDa and 75 kDa subunits in the complex I-derived SGA, the effect of antibodies on O2•− generation by complex I was assayed by EPR spin-trapping with DEPMPO. Mediation of O2•− generation by complex I was induced by NADH in the absence of Q1. The production of DEPMPO/•OOH adduct was thus mainly controlled by the Fp subcomplex or the hydrophilic domain of complex I under the above assay conditions. When the isolated intact complex I was incubated with antibodies of AbGSCA206 or AbGSCB367, the O2•− generation activity of complex I decreased by ~ 37 % and ~ 57%, respectively (Fig. 4), indicating that the binding of AbGSCA206 or AbGSCB367 to the GS-binding domains of complex I significantly decreased the electron leakage mediated by the Fp subcomplex or hydrophilic domain. The binding of the antibodies AbGSCA206 or AbGSCB367 did not affect the electron transfer activity from NADH to Q1 catalyzed by complex I (Fig. 4), indicating that the identified GS-binding domain from the 51 kDa or 75 kDa subunit was not essential for the catalytic function of complex I.
Fig. 4. Immunoinhibition of Complex I-mediated O2•− generation activity by AbGSCA206, Ab51, AbGSCB367, and Ab75 as assayed by EPR spin-trapping with DEPMPO.
Isolated complex I (0.91 mg/ml) was incubated with pre-immune IgG or AbGSCA206 or Ab51 or AbGSCB367 or Ab75 (0.36 mg/ml) at 4°C for 12 h. The O2•− generation mediated by complex I was initiated by NADH (0.5 mM) in the presence of DEPMPO (20 mM), and subsequently measured by EPR as described in “Materials and Methods”. The O2•− generation activity (black bars) was analyzed based on the spin quantitation obtained by double integration of the simulation spectrum. The electron transfer activity (dashed bars) was assayed by Q1-stimilated NADH oxidation at RT.
The use of pre-immune IgG has provided a good experimental control for the study (Fig. 4). Nevertheless, pre-immune IgG does not bind to complex I. To provide an additional experimental control, the effects of antibody binding on the enzymatic function of complex I was determined using Ab51 and Ab75. Binding of Ab51 or Ab75 to complex I did not significantly alter its electron transfer activity (Fig. 4). However, binding of Ab51 to complex I slightly reduced the mediation of O2•− generation (by ~7% vs. ~37% reduction by AbGSCA206 binding) as shown by EPR spin trapping (Fig. 4). Binding of Ab75 to complex I inhibited O2•− generation by ~35% (vs. ~57% inhibition by AbGSCB367 binding) as indicated in Fig. 4.
The above results support the concept of C206 as the redox sensitive thiol.17,36 The redox GS-binding domain hosting C206 is highly conserved among the enzymes from bacteria, fungi, and mammals. Therefore, the redox domain of the 51 kDa polypeptide is likely responsible for modulating electron leakage for O2•− production. An X-ray crystal structure (protein data bank entry 2FUG) of the hydrophilic domain of respiratory complex I from T. thermophilus indicates that the conserved cysteine (Cys182 in T. thermophilus) is only 6Å from FMN36. FMN serves as a source of O2•−, thus the binding of AbGSCA206 may trigger a conformational change in this domain and decrease the electron leakage from FMNH2. Based on the homolog model (Fig. 2A), binding of AbGSCA206 may induce a shielding effect on FMN, thus preventing FMN from accessing molecular oxygen and thus decreases electron leakage for O2•− production.
The B cell epitope of Ab51 antibody is located on the 20 amino acid residues at the N-terminus of the 51 kDa subunit of complex I. The sequence is not conserved (15% sequence similarity) in complex I of T. thermophilus. The binding of Ab51 has little or no effect on the electron transfer or O2•− generation activities of complex I, indicating that the N-terminal domain of the 51 kDa subunit is not essential for the catalytic functions of complex I (Fig 4).
The binding of AbGSCB367 to complex I inhibited O2•− generation by up to 57%. Furthermore, the binding of Ab75 antibody to complex I resulted in the inhibition of O2•− generation by 35% (Fig. 4). These results suggest that both glutathionylated and non-glutathionylated domains of the 75 kDa subunit can regulate electron leakage for O2•− production by the hydrophilic domain of complex I, which may further imply an essential role for the 75 kDa subunit in the catalytic function of complex I-mediated O2•− generation.
Based on the 75model, p75 may exhibit a β-sheet-α-turn-helix (blue region in Fig. 3A). A similar structure (91MVVDTLSDVVREAQAGMVEFT111 in the Nqo3 of 2FUG) is observed in T. thermophilus. Together with the X-ray structure, the homology model may explain the prevention of electron leakage for O2•− generation by Ab75. The nearest distances between the major α-helix of p75 and the iron sulfur clusters are about 11.2 Å (from M117 (Cα) to N5), 11.6Å (to N1b), and 14.3 Å (to N4) based on the homolog model (Fig. 3A and Fig. S2). Three possible mechanisms are proposed. (i) The binding of Ab75 may stabilize the conformation of the protein matrix surrounding the iron sulfur clusters, thus enhancing electron transfer efficiency. (ii) The binding of Ab75 may protect the protein matrix from molecular oxygen’s accessing the iron sulfur clusters including N1b, N4 and N5 in bovine protein, thereby minimizing the electron leakage from iron sulfur clusters. The above mechanism is proposed based on the hypothesis that electron leakage can occur at the iron-sulfur clusters.3 (iii) The binding of Ab75 could induce a conformational change at the flavin subcomplexes of complex I and prevent electron leakage from FMNH2.
According to 75model (Fig. 3A), the domain of pGSCB367 is surface-exposed and its conformation is mostly conserved in the Nqo3 of T. thermophilus. The closest distance to the iron sulfur clusters is about 23Å (from the E370 (Cα) of helix to N4 center). The distance is too long to support a direct mechanism. Therefore, the binding of AbGSCB367 to the 75 kDa subunit of complex I may trigger a long-range conformational change, and thus hinder the approach of O2. The significantly inhibitory result of AbGSCB367 binding strongly supports the notion that the GS-binding domain of complex I is closely related to the regulation of enzyme-mediated O2•− generation.12
Generation of Antibodies against the 24 kDa Subunit of Complex I
The 24 kDa subunit is part of NDH, and houses one [2Fe-2S] cluster which is responsible for EPR signal N1a with very low Em,7 ~ −370 mV. The binding motif for [2Fe-2S] in the bovine protein is CXXXXCX35CXXXC. To understand the role of [2Fe-2S] in the complex I-mediated O2•− generation, the partial sequence of [2Fe-2S]-binding motif, 97KYHIQVCTTTPCMLRNSDSILE118 (p24N1a in Table 1 and 24model in Fig. 5A), was synthesized as a chimeric construct of the fusion peptide (pMVF24N1a in Table 1). The reduced form of the synthetic peptide was used as an antigen to raise antibodies in rabbits. The antibodies were named Ab24N1a.
To provide a parallel control to Ab24N1a, we have designed B cell epitope of the 24 kDa subunit. The 19-residue sequence 162EDLTPKDIEEIIDELKAGK180 (p24 in Table 1 and 24model in Fig. 5A) corresponds to the amino acid residues 162–180 of the 24 kDa mature protein. The polyclonal antibody against the immunogen of chimeric fusion peptide (pMVF24 in Table 1) was named Ab24.
Immunoinhibition of Complex I-mediated O2•− Generation by Ab24N1a and Ab24
Effects of Ab24N1a- and Ab24-binding on complex I and NDH was evaluated by ELISA and immunoblotting. As indicated in Fig. 5B (ELISA) and 5C (Western blot), both Ab24N1a and Ab24 bound specifically to the 24 kDa subunit of complex I and NDH. Based on the result of ELISA (Fig. 5B), the binding affinity of Ab24N1a to complex I was about 50% of the binding affinity to NDH. Whereas the binding affinity of Ab24 to complex I was about 30% of the binding affinity to NDH. Furthermore, the binding affinity of Ab24N1 to complex I was about 50 % of the binding affinity of Ab24 to complex I.
As measured by EPR spin-trapping with DEPMPO, the binding of Ab24N1a to complex I modestly reduces the mediation of O2•− generation by 15.8 %, and inhibits the electron transfer activity of complex I by 13.3% (Fig. 5D). Whereas binding of Ab24 to complex I inhibits complex I-mediated O2•− generation by 34.1% and its electron transfer activity by 6.9% (Fig. 5D).
Based on the homology model of the bovine 24 kDa subunit (24model in Fig. 6B) and the X-ray structure of the bacterial Nqo2 (2FUG, Fig. 6A), the 1st helix of p24N1a is buried inside the protein coordinating to the 2Fe-2S center (N1a, Fig. 5A and Fig. 6A from 2FUG). In addition, the 2nd helix of p24N1a is only partially surface-exposed (Fig. 5A and Fig. 6A from 2FUG). The reduced surface-exposure area of p24N1a may explain the fact that a relatively weak binding of Ab24N1a to the intact complex I was observed (Fig. 5B). The reduced binding affinity of Ab24N1a to complex I may also result in weak inhibition of complex I-derived electron transfer and O2•− generation activities. In the 3D model of NDH hosting 51 kDa and 24 kDa subunits (Fig. 6B),37 the surface-exposed nature of 1st and 2nd helical domains is increased, thus enhancing the overall binding affinity of Ab24N1a to NDH.
Fig. 6. Surface diagrams of 2FUG Nqo1, Nqo2 and Nqo15 (A) and homology model of NDH hosting 51model and 24model (B).
In the T. thermophilus complex I X-ray structure, there is an additional subunit Nqo15 associating with the Nqo1 and Nqo2.36 The function of Nqo15 is not clear, and it is proposed to stabilize the complex I structure and/or to provide the storage of iron. The Nqo15 further blocks the accessible surface of the iron-sulfur cluster N1a. Ab24N1a is designed to target the p24N1a region of the 24model. Compared to the better surface-exposed p24 (magenta area) and p51 (yellow area), p24N1a (blue area) is partially buried in the pocket and less favorable for antibody binding. A reduced binding affinity of Ab24N1a to NDH was also observed (Fig. 5B, ▲). The binding of Ab24N1a to complex I was further reduced (Fig. 5B, ●), suggesting other subunits in bovine protein may associate with NDH and further block the p24N1a accessible surface.
The major α-helical domain of B cell epitope, p24, is highly surface-exposed based on the structure of 2FUG (Fig. 6A). Therefore, Ab24 showed a high titer and high specificity for intact complex I (Fig. 5B), isolated mitochondria, and the tissue homogenates of rat heart (Fig. 5C).
Binding of Ab24N1a to intact complex I only modestly inhibits its SGA and ETA (<15% inhibition, Fig. 5D), presumably due to low affinity and weak binding between the antigen and the antibody. Binding of Ab24 to intact complex I significantly decreases O2•− generation by 34 % (Fig. 5D), but only slightly affects the electron transfer efficiency. Presumably, a conformational change was induced by Ab24-binding to protect the protein matrix from O2 accessing the N1a center in the bovine protein, therefore reducing the electron leakage from the 2Fe-2S cluster. The closest distance of p24 domain to the N1a center is ~ 18.5 Å (from the K167 residue of helix domain to the cluster N1a). Therefore, the conformational change induced by Ab24 binding could have decreased electron leakage from FMNH2 since the 24 kDa subunit is structurally associated with 51 kDa subunit in intact complex I (see the homology structure of NDH in the Fig. 6B).
Generation of Antibodies against the 70 kDa Subunit of Complex II
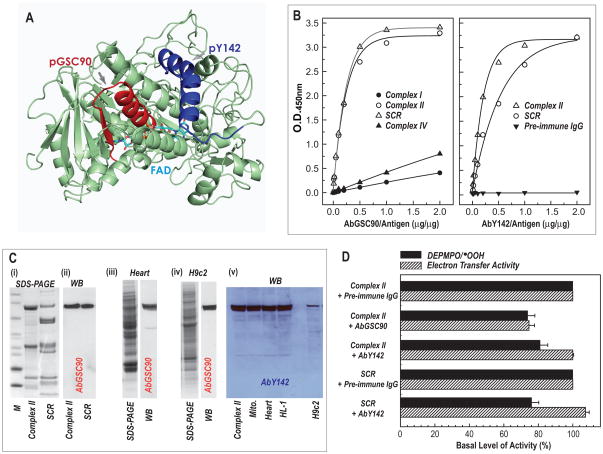
In complex II, a specific tryptic peptide, 77AAFGLSEAGFNTACVTK93, was identified to be involved in the GS-binding domain of the 70 kDa FAD-binding subunit.5 The peptide sequence 72GAGLRAAFGLSEAGFNTACVTK93 (pGSC90, Table 1) was designed with additional residues at the N-terminus as a B cell epitope. This 22-residue sequence contains an α-helix-β-turn-sheet structure38 as indicated in the Fig. 7A (pGSC90, red ribbon in Fig. 7A). The peptide pMVFGSC90 (in Table 1) was synthesized as a chimeric construct. The crude peptide was purified to homogeneity by reverse phase HPLC, and used as an immunogen to raise antibody AbGSC90.
Fig. 7. Immunological analysis of antibodies AbGSC90 and AbY142.
A, The peptides of pGSC90 (a 22-residue with an α-helix-β-turn-sheet conformation denoted by red ribbon based on x-ray structure of mammalian complex II (1ZOY)) and pY142 (a 19-residue with a major α-helical conformation denoted by blue ribbon) were designed to the B-cell epitopes. B, ELISA analysis of AbGSC90 (in left panel) and (in right panel) using the antigens of complex I (400 μg), complex II (40 μg), SCR (120 μg), complex IV (80 μg). C, Left panel (i–iv): immunoblotting analysis of AbGSC90 using the antigens of complex II (10 μg), SCR (30 μg), tissue homogenates of rat heart (100 μg), and cell lysate of H9c2 (100 μg). Right panel (v): immunoblotting analysis of AbY142 using the antigens of complex II (10 μg), isolated mitochondria (100 μg), tissue homogenates of rat heart (100 μg), cell lysates of HL-1 and H9c2 (100 μg). D, effect of antibody AbGSC90 and AbY142 on the O2•− generation and electron transfer activity mediated by complex II. O2•− generation mediated by complex II and SCR was measured by EPR spin-trapping with DEPMPO according to a published method. 5,14
The identified tryptic peptide involved in the protein nitration of complex II 70 kDa subunit is 130GSDWLGDQDAIHYMTEQAPASVVELENYGMPFSR163. The peptide of pY142 containing 19 residues (aa 136–154) exhibits the conformation of α-helix (blue ribbon in the Fig. 7A). The chimeric construct (pMVFY142 in Table I) of pY142 was used as the immunogen to generate antibody AbY142.
Immunological Specificity of AbGSC90 and AbY142
The immunological cross-reactivity of AbGSC90 and AbY142 was analyzed by ELISA (Fig. 7B) using isolated complex I, complex II and complex IV as antigens. AbGSC90 showed high titer and bound with the 70 kDa FAD binding subunit in complex II or SCR (a supercomplex hosting complex II and complex III); however, there was little binding detected between AbGSC90 and either complex I or complex IV (Fig. 7B). The immunological cross-reactivity of AbY142 showed a high titer and bound to complex II and, with decreased affinity to SCR.(Fig. 7B, right panel). These results suggest that the specific domain of Y142 in complex II may be structurally associated with complex III in the supercomplex.
The immunological specificity of AbGSC90 and AbY142 was further characterized by a Western blot; both antibodies bound specifically to the 70 kDa subunit of complex II and SCR (Fig. 7C). The antibodies also bound specifically to the 70 kDa subunit of complex II in myocardial tissue homogenates of the rat heart and cell lysates from the rat cardiac myoblast cell line H9c2 or mouse myocyte cell line HL-1 (Fig. 7C). These results demonstrate that both AbGSC90 and AbY142 are highly specific with high sensitivity, and thus are suitable for in vivo or in vitro studies.
Immunoinhibition of Complex II by AbGSC90 and AbY142
The effect of AbGSC90 binding on the electron transfer and O2•− generation activities of complex II was measured (Fig. 7D). When the isolated complex II was incubated with various amounts of the antibodies, the electron transfer activity (ETA) of complex II decreased as the amount of the antibodies increased (data not shown). A maximum inhibition of 25% was observed with 400 μg of the antibody per nmol (based on heme b) of complex II. The binding of antibodies also moderately decreased the O2•− generation mediated by complex II by 26% as measured by EPR spin trapping with DEPMPO (Fig. 7D). The results suggest that the binding of AbGSC90 with the epitope that is involved in GS-binding (aa 77–93) moderately decreases the electron transfer activity required for the electron leakage for O2•− production.9
In the previous studies,12 it was reported that a modest increase of the ETA resulted from in vitro S-glutathionylation of complex II. However, the binding of AbGSC90 leads to a moderate reduction of ETA in complex II. These results suggest that the GS-binding domain is involved, directly or indirectly, in the catalytic activity of complex II. The binding of AbGSC90 also moderately decreased, by 26%, the O2•− generation mediated by complex II as measured by EPR spin trapping with DEPMPO (Fig. 7D) or the acetylated cytochrome c reduction assay (data not shown). The decreased O2•− generation can be explained by the ETA decrease resulting from AbGSC90 binding.
Binding of AbY142 to complex II results in the modest inhibition (19%) of SGA, but does notaffect its ETA (Fig. 7D). Binding of AbY142 to the supercomplex of SCR inhibits its SGA by 24%, but slightly increases its electron transfer efficiency (Fig. 7D). The peptide of pY142 is related to the specific domain of protein tyrosine nitration (Y142 is nitrated) mediated by peroxynitrite.14 X-ray structure of mammalian complex II (protein data bank entry 1ZOY) reveals that the major α-helical domain of pY142 is highly surface-exposed and situated in the hydrophilic environment. Binding affinity of AbY142 to the isolated complex II is significantly higher than the binding affinity to the SCR (the right panel of Fig. 7B). Presumably, structural association with complex III limits the access of antibodies to the pY142 domain on the surface of the SCR supercomplex. Binding of AbY142 modestly inhibits complex II-mediated O2•− generation, but does not affect the electron transfer efficiency of complex II (Fig. 7D). A similar result was also observed in the binding of AbY142 to the SCR, indicating the regulatory function of pY142 domain in complex II-mediated O2•− generation.
Conclusion
The novel findings resulting from this investigation include (i) defining the functional role of a specific redox domain in complexes I and II in mediating O2•− generation and (ii) exploring the functional non-redox domain in regulating O2•− generation based on the homology model of complex I. With the use of unique peptide-based antibodies that block O2•− generation, we have established that several redox domains are involved in O2•− production by mitochondrial complex I and complex II, including GS-binding domains, a binding motif of iron-sulfur cluster (N1a), and the domain involved in protein tyrosine nitration. With the aid of the homology model of complex I, the antibodies with high affinity and specificity were also obtained, including Ab24/Ab51/Ab75 for complex I and AbGSC90/AbY142 for complex II. Binding of Ab24 and Ab75 impacts the O2•− production by complex I, suggesting the involvement of the non-redox domain in the regulation of its O2•− generation. The information obtained from this study may also pave the way for the development of therapeutic approaches using drugs to suppress oxidative stress.
Supplementary Material
Acknowledgments
This work was supported by RO1 grant from National Institutes of Health [HL083237 to Y-R C].
Abbreviations
- Ab
antibody
- MVF
measles virus fusion
- Fmoc
N-(9-fluorenyl)methoxycarbonyl
- NQR
NADH ubiquinone reductase or complex I
- NDH
NADH dehydrogenase or flavin protein subcomplex of complex I
- SCR
succinate cytochrome c reductase or mitochondrial supercomplex hosting complex II and complex III
- GS
glutathione
- O2•−
superoxide anion radical
- ETC
electron transport chain
- SMP
submitochondrial particles
- Q1
ubiquinone-1
- DEPMPO
5-diethoxylphosphoryl-5-methyl-1-pyrroline N-oxide
- FMN
flavin mononucleotide
- SDS-PAGE
SDS polyacrylamide gel electrophoresis
- EPR
electron paramagnetic resonance
- MS
mass spectrometry
- PBS
phosphate buffered saline
- ETA
electron transfer activity
- SGA
superoxide generation activity
References
- 1.Kudin AP, Bimpong-Buta NY, Vielhaber S, Elger CE, Kunz WS. J Biol Chem. 2004;279:4127–4135. doi: 10.1074/jbc.M310341200. [DOI] [PubMed] [Google Scholar]
- 2.Galkin A, Brandt U. J Biol Chem. 2005;280:30129–30135. doi: 10.1074/jbc.M504709200. Epub 32005 Jun 30128. [DOI] [PubMed] [Google Scholar]
- 3.Ohnishi ST, Ohnishi T, Muranaka S, Fujita H, Kimura H, Uemura K, Yoshida K, Utsumi K. J Bioenerg Biomembr. 2005;37:1–15. doi: 10.1007/s10863-005-4117-y. [DOI] [PubMed] [Google Scholar]
- 4.Kussmaul L, Hirst J. Proc Natl Acad Sci U S A. 2006;103:7607–7612. doi: 10.1073/pnas.0510977103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen YR, Chen CL, Pfeiffer DR, Zweier JL. J Biol Chem. 2007;282:32640–32654. doi: 10.1074/jbc.M702294200. [DOI] [PubMed] [Google Scholar]
- 6.Turrens JF, Alexandre A, Lehninger AL. Arch Biochem Biophys. 1985;237:408–414. doi: 10.1016/0003-9861(85)90293-0. [DOI] [PubMed] [Google Scholar]
- 7.Zhang L, Yu L, Yu CA. J Biol Chem. 1998;273:33972–33976. doi: 10.1074/jbc.273.51.33972. [DOI] [PubMed] [Google Scholar]
- 8.Muller F, Crofts AR, Kramer DM. Biochemistry. 2002;41:7866–7874. doi: 10.1021/bi025581e. [DOI] [PubMed] [Google Scholar]
- 9.Chen YR, Chen CL, Yeh A, Liu X, Zweier JL. J Biol Chem. 2006;281:13159–13168. doi: 10.1074/jbc.M513627200. [DOI] [PubMed] [Google Scholar]
- 10.Prabu SK, Anandatheerthavarada HK, Raza H, Srinivasan S, Spear JF, Avadhani NG. J Biol Chem. 2006;281:2061–2070. doi: 10.1074/jbc.M507741200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hurd TR, Requejo R, Filipovska A, Brown S, Prime TA, Robinson AJ, Fearnley IM, Murphy MP. J Biol Chem. 2008;283:24801–24815. doi: 10.1074/jbc.M803432200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen CL, Zhang L, Yeh A, Chen CA, Green-Church KB, Zweier JL, Chen YR. Biochemistry. 2007;46:5754–5765. doi: 10.1021/bi602580c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor ER, Hurrell F, Shannon RJ, Lin TK, Hirst J, Murphy MP. J Biol Chem. 2003;278:19603–19610. doi: 10.1074/jbc.M209359200. [DOI] [PubMed] [Google Scholar]
- 14.Chen CL, Chen J, Rawale S, Varadharaj S, Kaumaya PP, Zweier JL, Chen YR. J Biol Chem. 2008;283:27991–28003. doi: 10.1074/jbc.M802691200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beer SM, Taylor ER, Brown SE, Dahm CC, Costa NJ, Runswick MJ, Murphy MP. J Biol Chem. 2004;279:47939–47951. doi: 10.1074/jbc.M408011200. Epub 42004 Aug 47930. [DOI] [PubMed] [Google Scholar]
- 16.Chen J, Chen CL, Rawale S, Chen CA, Zweier JL, Kaumaya PP, Chen YR. J Biol Chem. 2010;285:3168–3180. doi: 10.1074/jbc.M109.056846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen YR, Chen CL, Zhang L, Green-Church KB, Zweier JL. J Biol Chem. 2005;280:37339–37348. doi: 10.1074/jbc.M503936200. [DOI] [PubMed] [Google Scholar]
- 18.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 19.Arnold K, Bordoli L, Kopp J, TS Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 20.Melo F, Feytmans E. J Mol Biol. 1998;277:1141–1152. doi: 10.1006/jmbi.1998.1665. [DOI] [PubMed] [Google Scholar]
- 21.Laskowski RA, MacArthur MW, Moss D, Thornton JM. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 22.Holm L, Park J. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 23.DeLano WL. 2002 [Google Scholar]
- 24.Dakappagari NK, Lute KD, Rawale S, Steele JT, Allen SD, Phillips G, Reilly RT, Kaumaya PT. J Biol Chem. 2005;280:54–63. doi: 10.1074/jbc.M411020200. [DOI] [PubMed] [Google Scholar]
- 25.Sundaram R, Lynch MP, Rawale SV, Sun Y, Kazanji M, Kaumaya PT. J Biol Chem. 2004;279:24141–24151. doi: 10.1074/jbc.M313210200. [DOI] [PubMed] [Google Scholar]
- 26.Galante YM, Hatefi Y. Arch Biochem Biophys. 1979;192:559–568. doi: 10.1016/0003-9861(79)90126-7. [DOI] [PubMed] [Google Scholar]
- 27.Vinogradov AD, King TE. Methods Enzymol. 1979;55:118–127. doi: 10.1016/0076-6879(79)55017-4. [DOI] [PubMed] [Google Scholar]
- 28.Yu L, Yu CA. J Biol Chem. 1982;257:2016–2021. [PubMed] [Google Scholar]
- 29.Lee GY, He DY, Yu L, Yu CA. J of Biol Chem. 1995;270:6193–6198. doi: 10.1074/jbc.270.11.6193. [DOI] [PubMed] [Google Scholar]
- 30.Redfearn ER, Whittaker PA. Biochim Biophys Acta. 1966;118:413–418. doi: 10.1016/s0926-6593(66)80050-4. [DOI] [PubMed] [Google Scholar]
- 31.Hatefi Y. Methods Enzymol. 1978;53:11–14. doi: 10.1016/s0076-6879(78)53006-1. [DOI] [PubMed] [Google Scholar]
- 32.Duling DR. J Magn Reson B. 1994;104:105–110. doi: 10.1006/jmrb.1994.1062. [DOI] [PubMed] [Google Scholar]
- 33.Frejaville C, Karoui H, Tuccio B, Le Moigne F, Culcasi M, Pietri S, Lauricella R, Tordo P. J Med Chem. 1995;38:258–265. doi: 10.1021/jm00002a007. [DOI] [PubMed] [Google Scholar]
- 34.Vasquez-Vivar J, Kalyanaraman B, Martasek P, Hogg N, Masters BS, Karoui H, Tordo P, Pritchard KA., Jr Proc Natl Acad Sci U S A. 1998;95:9220–9225. doi: 10.1073/pnas.95.16.9220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roubaud V, Sankarapandi S, Kuppusamy P, Tordo P, Zweier JL. Anal Biochem. 1997;247:404–411. doi: 10.1006/abio.1997.2067. [DOI] [PubMed] [Google Scholar]
- 36.Sazanov LA, Hinchliffe P. Science. 2006;311:1430–1436. doi: 10.1126/science.1123809. [DOI] [PubMed] [Google Scholar]
- 37.Barker CD, Reda T, Hirst J. Biochemistry. 2007;46:3454–3464. doi: 10.1021/bi061988y. [DOI] [PubMed] [Google Scholar]
- 38.Kobs-Conrad S, Lee H, DiGeorge AM, Kaumaya PT. J Biol Chem. 1993;268:25285–25295. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.