Abstract
The zebrafish mutant silent partner is characterized by a dysmorphic, non-contractile ventricle resulting in an inability to generate normal blood flow. We have identified the genetic lesion in the zebrafish homolog of the slow twitch skeletal/cardiac troponin C gene. Although human troponin C1 (TNNC1) is expressed in both cardiac and skeletal muscle, duplication of this gene in zebrafish has resulted in tissue specific partitioning of troponin C expression and function. Mutation of the zebrafish paralog tnnc1a, which is expressed predominantly in the heart, results in a loss of contractility and myofibrillar organization within ventricular cardiomyocytes, while skeletal muscle remains functional and intact. We further show that defective contractility in the developing heart results in abnormal atrial and ventricular chamber morphology. Together, our results suggest that tnnc1a is required both for the function and structural integrity of the contractile machinery in cardiomyocytes, helping to clarify potential mechanisms of troponin C mediated cardiomyopathy.
Keywords: zebrafish, troponin C, contractility, cardiovascular, heart
Introduction
Cardiovascular disease remains the leading cause of death in the developed world and cardiomyopathies, disorders that affect the heart muscle, are among the most common causes of this disease (Towbin and Bowles, 2002). Observation of cardiac defects in animal models with mutations in genes associated with familial cardiomyopathies has provided the means to study the molecular mechanisms of these disorders (Ross, 2002; Sehnert et al., 2002). In particular, the unique attribute of zebrafish embryos to survive early development without cardiac function has facilitated the analysis of several mutations that result in loss of normal cardiac muscle contraction resulting in early lethality in other animal models (Chen et al., 1996; Stainier et al., 1996).
Here, we describe the characterization of the silent partner (sil) mutation, which was identified in a large-scale zebrafish genetic screen as a recessive embryonic lethal mutation that results in defective ventricular contractility (Stainier et al., 1996). We defined the sil gene by positional cloning and show that it encodes a zebrafish homolog of the slow twitch skeletal/cardiac troponin C protein (cTnC), a subunit of the troponin (Tn) complex. The Tn complex acts as a calcium-sensitive regulator of striated muscle contraction and consists of three subunits: the calcium-binding subunit troponin C (TnC), the inhibitory subunit troponin I (TnI), and the tropomyosin-binding subunit troponin T (TnT) (Greaser and Gergely, 1973). Previously, the cardiac TnT (Tnnt2) subunit of the zebrafish Tn complex was shown to be required for contraction of both the atrium and the ventricle (Sehnert et al., 2002).
There are at least two genes for TnC found in the genomes of higher animals; one encodes the fast-twitch skeletal isoform (sTnC) and the second encodes the slow twitch skeletal muscle and the heart isoform (cTnC) (Kawasaki and Kretsinger, 1994; Tobacman, 1996; Filatov et al., 1999). Mutations in the human homolog of the cardiac Troponin C (cTnC) gene, TNNC1, have been linked to certain forms of familial cardiomyopathy (Hoffmann et al., 2001; Gomes and Potter, 2004). While the importance of TNNC1 in human disease has been documented, the phenotype resulting from a complete loss of cTnC function has yet to be described.
Human TNNC1 is expressed in both cardiac and skeletal muscle, whereas zebrafish contains two copies of the gene for cTnC due to an ancestral duplication event (Amores et al., 1998). Here, we report the characterization of these duplicated genes, annotated as tnnc1a and tnnc1b, which we find to be predominantly expressed in the heart or skeletal muscle, respectively. Furthermore, we have described the sil mutant phenotype, which we show to result from disruption of the tnnc1a gene. We have examined two sil mutant alleles, one of which results in a loss of any detectable tnnc1a expression. We find that the loss of tnnc1a function results in defective heart contractility that correlates with abnormal atrial and ventricular chamber morphology as well as a loss of normal myofibrillar structure within sarcomeres. These findings enhance our understanding of the link between TNNC1 mutation in humans and cardiomyopathy.
Results
The sil gene is essential for ventricular contractility
Large-scale genetic screens performed in zebrafish have resulted in the identification of several mutations affecting the form and function of cardiac muscle (Chen et al., 1996; Stainier et al., 1996). One such mutation, sil, was first identified as an N-ethyl-N-nitrosourea (ENU)-induced recessive embryonic lethal mutation with defective ventricular contraction (Stainier et al., 1996). As seen in other mutants with defects in cardiac contractility, homozygous mutant sil embryos develop pericardial edema and are never able to generate blood circulation (Figure 1, Supplemental Movies 1A,B). However, the initial steps of cardiac development appear to progress normally in the sil mutants, with specification of atrial and ventricular chambers detectable by 24 hours post-fertilization (hpf; Figure 2A,C,D). As development proceeds, the ventricular contractility defect becomes obvious and after 2 days post-fertilization (dpf), the ventricles of sil mutants no longer display ventricular myosin heavy chain (vmhc) expression (Figure 2E). This loss of vmhc expression from the ventricle suggests that the contractile function of the ventricular chamber may be required for the sustained expression of contractile protein genes.
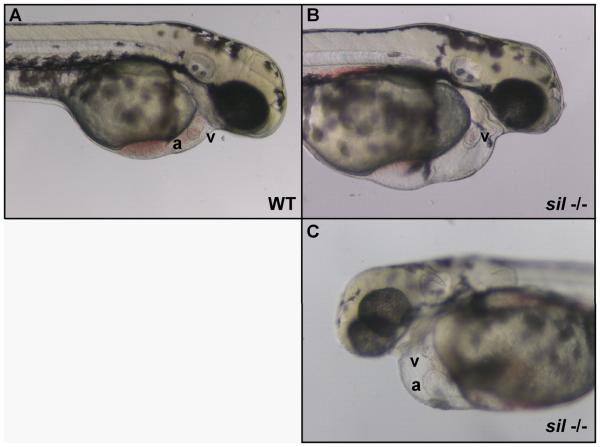
Figure 1. Characterization of the sil phenotype.
Lateral view of wild-type (WT) (A) and silm656 mutant (−/−) (B, C) embryos at 2 dpf. The ventricle (v) of the sil mutant embryo (B, C) is visibly compacted and misshapen compared to wild-type (A), while the atrium (a) of the sil mutant (C) has become enlarged. The silm656 mutant embryo (B, C) also exhibits pericardial edema and has reduced circulation as compared to wild-type (A). Mutant embryos homozygous for the silsk25 allele exhibit the same phenotype.
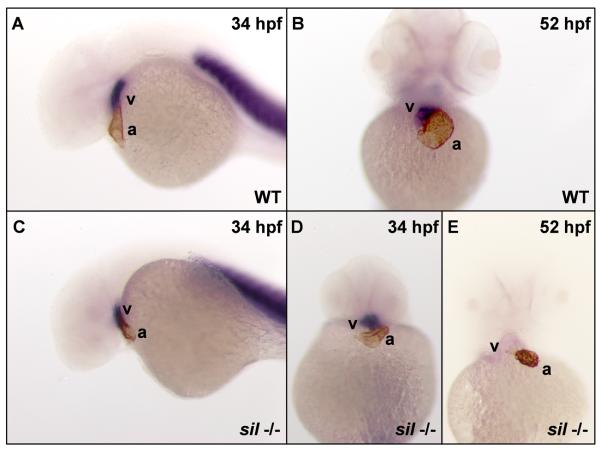
Figure 2. The sil mutant exhibits a loss of expression of ventricular contractile protein genes.
Double staining with an atrial-specific antibody (S46) and an in situ RNA probe for ventricular myosin heavy chain (vmhc) (Yelon et al., 1999) was used to distinguish the atrial and ventricular chambers in wild-type (WT) (A, B) and silm656 mutant (−/−) (C-E) embryos. The hearts of wild-type (A) and silm656 mutant (C, D) embryos at 34 hpf express markers specific for the two cardiac chambers. However, by 52 hpf, the hearts of silm656 mutant (E) embryos no longer express the ventricle specific vmhc marker while both chamber markers are still visible in wild-type embryos (B).
The onset of chamber contraction does not proceed normally in sil −/− hearts; contraction of both chambers is detectable in wild-type hearts by 1 dpf while the hearts of sil mutant embryos remain non-contractile. In the sil −/− heart, by 48 hpf the atrium has started to contract but the ventricle remains silent throughout the remainder of the embryo's development (Supplemental Movies 1A, B). At this stage, the atrium exhibits pronounced enlargement compared to wild-type and the ventricle has become compact and misshapen (Figure 1B, C), reflecting a role for cardiac function in the morphogenesis of this organ.
We more closely examined the structure of cardiomyocytes from the ventricles of wild-type and sil mutant hearts using transmission electron microscopy (TEM). By 50 hpf, myofibrillar structures are visible within the cardiomyocytes of wild-type embryos (Figure 3A, B). These highly organized contractile structures are disrupted or fail to form normally in the ventricular cardiomyocytes of sil mutants (Figure 3C, D).
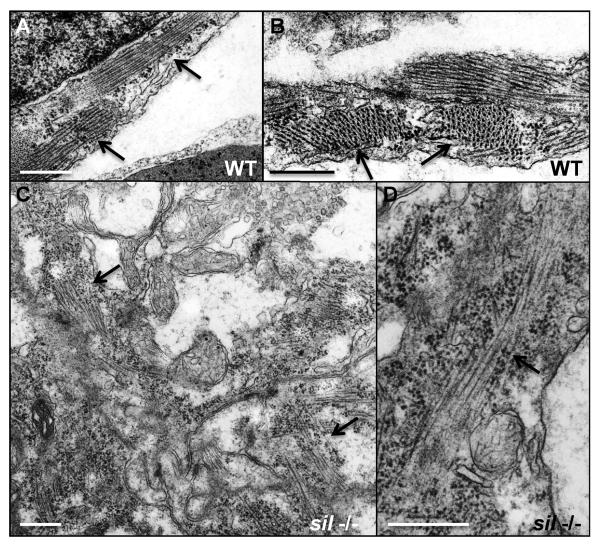
Figure 3. Myofibrillar structure is disorganized in the sil mutant.
Transmission electron microscopy was performed to study the structure of ventricular cardiomyocytes from wild-type (WT) (A, B) and silm656 mutant (−/−) (C, D) embryos at 50 hpf. (A, B) Myofibrils are easily detectable in the cardiomyocytes from ventricles of wild-type embryos. (C, D) silm656 mutant embryos exhibit very sparse and poorly organized sarcomeres. Arrows are used to indicate myofibrils. Bars, 500 nm.
The sil locus encodes zebrafish tnnc1a
By positional cloning, we identified sil as a gene homologous to the human slow twitch skeletal/cardiac cTnC gene (TNNC1, Supplemental Figure 1A) that we have annotated here as tnnc1a. Initial linkage to the marker z4003 on chromosome 23 was determined by bulk segregant analysis (data not shown, (Michelmore et al., 1991)). Other markers in the region, including z13781 which is at the same genetic map position as z4003, were used to genotype embryos and establish flanking markers. The marker z23232 (Ensembl build Zv8, Chromosome 23: 3,971,231-3,975,624) was determined to be the most closely linked genetic marker and maps near an Ensembl gene annotated as tnnc1(tnnc1, Ensembl build Zv8, Chromosome 23: 4,055,544-4,060,124).
This troponin C gene was deemed to be a strong candidate because of the biological relevance of its mammalian homologs for cardiac contractile function and its proximity to the sil genetic locus. Therefore, we amplified tnnc1a DNA from wild-type embryos and sil mutant embryos, focusing on two sil alleles: silm656 (identified as an ENU-induced mutation in the Fishman lab; (Stainier et al., 1996) and silsk25 (identified as an ENU-induced mutation in the Yelon lab). Sequencing of tnnc1a from silm656 mutant embryos predicts a splicing defect due to a splice donor mutation within the intron following exon 2 (TGgt→TGgc; Supplemental Figure 1B), which results in an in-frame insertion at the end of exon 2. Amplification of tnnc1a DNA from silsk25 mutants reveals a deletion within the locus (Supplemental Figure 1C), predicting a loss of tnnc1a expression.
Partitioning of TnC expression and function in zebrafish
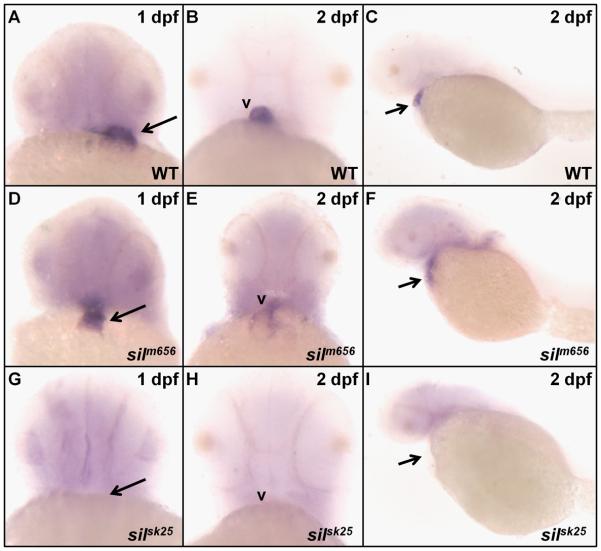
The cTnC gene is very highly conserved across vertebrate species (91.3% identity between zebrafish and human, Supplemental Figure 1D). The human genome contains a single copy of the TNNC1 gene that is expressed in both cardiac and skeletal muscle (Gahlmann et al., 1988). However, analysis of tnnc1a mRNA in wild-type embryos using whole-mount in situ hybridization reveals staining predominantly in the heart (Figure 4A-C). Expression of tnnc1a is seen throughout the heart during the first day of development (Figure 4A, B), but by 2 dpf, staining is mostly restricted to the ventricular chamber (Figure 4C). This expression pattern correlates precisely with the sil mutant phenotype; the contractility defect initially manifests as a lack of contractility throughout the heart but becomes restricted to the ventricle by 2 dpf. Appreciable expression of tnnc1a in skeletal muscle is not detectable by wholemount in situ analysis during the first 2 days of development (Figure 4D, D′).
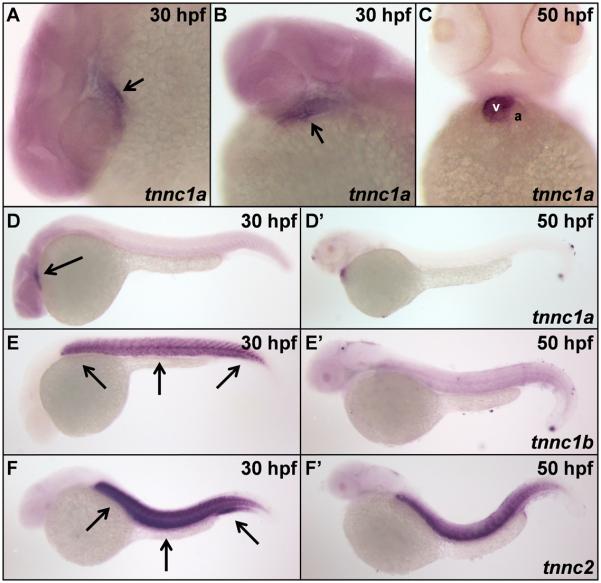
Figure 4. Expression of tnnc1a, tnnc1b, and tnnc2 mRNA in zebrafish.
(A, B) The zebrafish tnnc1a gene is expressed throughout the heart of wild-type zebrafish embryos at 30 hpf, indicated by arrows. (C) By 50 hpf, tnnc1a expression is predominantly restricted to the ventricle (v) of wild-type hearts and excluded from the atrium (a). The tnnc1a gene is expressed in cardiac tissue (indicated by the arrow) at both 30 hpf (D) and 50 hpf (D'). The second cTnC gene in zebrafish (tnnc1b) is detected predominantly in skeletal muscle (indicated by arrows) at 30 hpf (E) with decreased expression by 50 hpf (E'). The sTnC gene in zebrafish (tnnc2) is expressed in skeletal muscle (indicated by arrows) at both 30 hpf (F) and 50 hpf (F').
Our analysis of ESTs for zebrafish slow twitch skeletal/cardiac troponin C revealed two distinct populations of transcripts, indicating the possibility of two genes with distinct genomic loci. We confirmed that in addition to tnnc1a, the zebrafish genome contains a second copy of the cTnC gene, which we have annotated as tnnc1b (stnnc, Ensembl build Zv8, Chromosome 23: 19,608,998-19,611,413). The zebrafish genome contains several duplicated genes as the result of an ancestral duplication event (Amores et al., 1998). The tnnc1b gene, which varies by only 18 amino acids from the tnnc1a gene (Supplemental Figure 1D), is expressed mainly in skeletal but not cardiac muscle during the first day of development (Figure 4E); expression is substantially weaker by 2 dpf (Figure E′). The predominant expression of tnnc1a and tnnc1b in cardiac and skeletal muscle, respectively, is consistent with other duplicated zebrafish genes that have undergone partitioning of tissue expression, and subsequently, function (Serluca et al., 2001).
The zebrafish homolog of the sTnC gene, designated here as tnnc2 (tnnc, Ensembl build Zv8, Chromosome 23: 19,953,352-19,957,979) shares 61% and 63% identity to the slow twitch skeletal/cardiac isoforms tnnc1a and tnnc1b, respectively. Consistent with what is known about expression of human TNNC2, we see strong expression of tnnc2 in the zebrafish skeletal muscle during these same developmental stages (Figure 4F, F′).
Expression of tnnc1a mRNA in sil mutant embryos
We next examined the expression of tnnc1a mRNA in the sil mutant lines by in situ hybridization. In silm656 mutant embryos, expression of tnnc1a at 1 dpf can be detected in the heart as seen in wild-type embryos (Figure 5A-E). However, by 2 dpf, the expression of tnnc1a in the ventricles of silm656 mutant embryos has become diffuse (Figure 5F), similar to the loss of vmhc staining at 2 dpf in these embryos (Figure 2E). In agreement with the deletion we have detected within the tnnc1a locus of silsk25 mutant embryos, no tnnc1a expression is detected at either 1 or 2 dpf in these embryos (Figure 5G-I).
Figure 5. In situ analysis of tnnc1a mRNA expression in sil mutant embryos.
In silm656 mutant embryos at 1 dpf (D), tnnc1a expression is found throughout the heart as in wild-type (WT) embryos (A). However, by 2 dpf, the expression of tnnc1a in silm656 embryos (E, F) has become diffuse compared to wild-type (B, C). No tnnc1a expression can be detected in silsk25 mutant embryos either at 1 or 2 dpf (G-I).
Morpholino confirmation and rescue of the genetic lesions
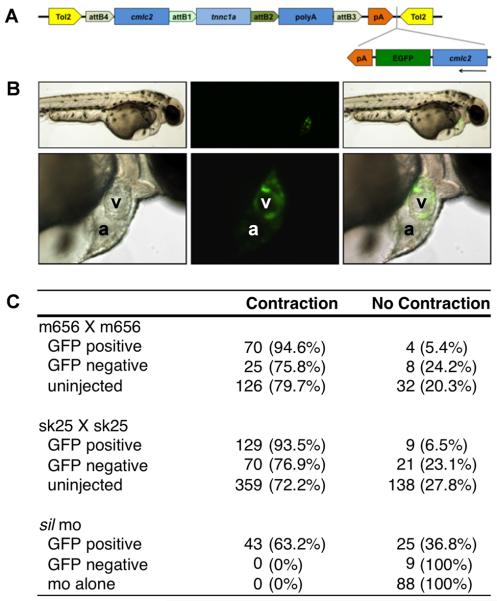
To further confirm that the phenotype of sil mutants is caused by disruption of the tnnc1a gene, we designed an antisense morpholino oligomer targeted to the splice donor site at the end of exon 3 of the tnnc1a mRNA (for RT-PCR confirmation of altered splice site usage in morpholino injected embryos, see Supplemental Figure 2). Injection of this morpholino at the 1-cell stage in wild-type embryos produced a complete phenocopy of the genetic mutants (Table 1, Supplemental Movies 2A, B). Next, we attempted to rescue the mutant sil phenotype by expression of wild-type tnnc1a. Injection of wild-type tnnc1a mRNA resulted in early developmental defects due to impaired gastrulation, which we hypothesize result from early ubiquitous expression of tnnc1a. To avoid this problem, we used the cardiac myosin light chain 2 (cmlc2) promoter (Huang et al., 2003; Mably et al., 2003) to drive myocardial-specific expression of tnnc1a. We designed a construct in which the cmlc2 promoter driving expression of tnnc1a is flanked by Tol2 elements (Kawakami et al., 2004; Kwan et al., 2007) to improve our rate of transgenesis. This construct also includes a cassette in which the cmlc2 promoter drives expression of cytoplasmic enhanced green fluorescent protein (EGFP). This reporter is cloned in the reverse orientation to the cmlc2:tnnc1a fusion to minimize the chance of interference between the two expression cassettes (construct shown in Figure 6A). Coinjection of this construct and Tol2 transposase mRNA into embryos at the 1-2 cell stage results in a mosaic pattern of EGFP expression in the heart, indicating which cardiomyocytes express the construct (Figure 6B). When we injected the construct with transposase mRNA into the progeny of a cross between either heterozygous silm656 or silsk25 zebrafish, the mosaic expression of EGFP in the cardiomyocytes correlated with the rescue of contractility in those cells (Figures 6C and Supplemental Figure 3, Supplemental Movie 3). The ability of mosaic tnnc1a expression to rescue cardiomyocyte contractility suggests that tnnc1a regulates contractility in a cell-autonomous manner. We also co-injected the tnnc1a expression construct and Tol2 transposase RNA with the morpholino targeting the exon 3-intron 3 donor site, which would only target processing of endogenous tnnc1a mRNA. As with the genetic mutants, mosaic expression of wild-type tnnc1a is able to rescue cardiomyocyte contractility (Figure 6C). Together, these data suggest that the ventricular contractility defect found in sil zebrafish as well as embryos injected with the tnnc1a morpholino is caused primarily by the disruption of tnnc1a function.
Table 1. Cardiac and skeletal muscle phenotypes at 2 dpf in embryos injected with morpholinos targeting the three zebrafish troponin C genes.
Embryos were injected at the one- to four-cell stage with tnnc1a, tnnc1b, or tnnc2 morpholinos either singly or in pair-wise fashion (see Methods). Embryos were then assayed for (A) heart or (B) skeletal muscle contraction at 2 dpf and were characterized as one of the following phenotypes; wild-type (contraction of both chambers with blood circulation); sil phenotype (contraction of the atrium, silent ventricle, and absence of blood circulation); contraction, no flow (contraction of both chambers with the absence of blood circulation); no contraction, no flow (no contraction in either chamber with the absence of blood circulation); sk. m. impaired (decreased touch-response); paralyzed (no touch response).
| Table 1A. Cardiac phenotype at 2 dpf in embryos injected with morpholinos targeting three zebrafish troponin C genes. | ||||
|---|---|---|---|---|
| wild-type | sil phenotype | contraction no flow |
no contraction no flow |
|
| uninjected | 316 (99.7%) | 0 (0%) | 1 (0.3%) | 0 (0%) |
| tnnc1a | 1 (0.7%) | 150 (98.6%) | 1 (0.7%) | 0 (0%) |
| tnnc1b | 134 (75.3%) | 0 (0%) | 43 (24.1%) | 1 (0.6%) |
| tnnc2 | 137 (99.3%) | 0 (0%) | 1 (0.7%) | 0 (0%) |
| tnnc1a + tnnc1b | 0 (0%) | 2 (2.3%) | 0 (0%) | 84 (97.7%) |
| tnnc1a + tnnc2 | 0 (0%) | 74 (100%) | 0 (0%) | 0 (0%) |
| tnnc1b + tnnc2 | 51 (77.3%) | 0 (0%) | 15 (22.7%) | 0 (0%) |
| Table 1B. Skeletal muscle phenotype at 2 dpf in embryos injected with morpholinos targeting three zebrafish troponin C genes. | |||
|---|---|---|---|
| wild-type | sk. m. impaired | paralyzed | |
| uninjected | 317 (100%) | 0 (0%) | 0 (0%) |
| tnnc1a | 152 (100%) | 0 (0%) | 0 (0%) |
| tnnc1b | 0 (0%) | 178 (100%) | 0 (0%) |
| tnnc2 | 0 (0%) | 138 (100%) | 0 (0%) |
| tnnc1a + tnnc1b | 0 (0%) | 86 (100%) | 0 (0%) |
| tnnc1a + tnnc2 | 0 (0%) | 31 (42%) | 43(58%) |
| tnnc1b + tnnc2 | 0 (0%) | 0 (0%) | 66(100%) |
Figure 6. Myocardial specific tnnc1a expression rescues the sil mutant phenotype.
(A) Depiction of the tnnc1a rescue construct used for injection of zebrafish embryos at the 1-2 cell stage. (B) 2 dpf wild-type embryos coinjected with tnnc1a morpholino (MO), the rescue construct depicted in (A), and transposase RNA. The top row shows an embryo at 4x magnification and the bottom row shows the same embryo at 20x magnification. GFP indicates expression of the construct in the cardiomyocytes of this embryo. (C) Table summarizing the results of rescue experiments in the silm656 line, the silsk25 line, and wild-type embryos injected with tnnc1a MO. Uninjected controls are compared with injected embryos that either have at least one GFP positive cell (GFP positive) or no GFP positive cells (GFP negative).
Compensatory roles of tnnc1a and tnnc1b in contractility
As previous work has demonstrated the essential role of tnnt2 in cardiomyocyte contractility throughout the entire zebrafish heart (Sehnert et al., 2002), we were surprised to find that loss of tnnc1a expression does not result in a loss of atrial contraction. We hypothesized that while tnnc1b expression appears to be restricted to the skeletal muscle, there might be low-level expression in the heart that is able to partially compensate for the loss of tnnc1a, allowing cardiomyocytes in the atrium to contract. To test this hypothesis, we designed a morpholino targeted to the splice donor site at the end of exon 1 of the tnnc1b mRNA. Injection of this morpholino alone does not appear to affect heart contraction in the majority of embryos. Approximately one quarter of injected embryos maintain contraction of both heart chambers, but are not able to generate blood circulation (Table 1A, Supplemental Movie 4A). However, when the tnnc1b and tnnc1a morpholinos are injected in combination, the heart remains completely non-contractile (Table 1A, Supplemental Movie 4B). These results suggest that in the sil mutants, weak expression of tnnc1b in the heart is sufficient for atrial contraction to commence at 2 dpf.
To confirm that tnnc1b gene expression alone is sufficient to maintain atrial contraction, we also designed a morpholino targeted to the splice donor site at the end of exon 2 of the tnnc2 gene. Injection of this morpholino alone results in a wild-type heart contraction phenotype, while injection in combination with the tnnc1a morpholino results in a sil phenotype (Table 1A, Supplemental Movies 5A, B). Therefore, tnnc1b alone is sufficient and required for atrial contraction while expression of tnnc1a at 2 dpf is dispensable for atrial function.
Interestingly, the coinjection of tnnc1a and tnnc2 morpholinos results in a complete loss of skeletal muscle function in 42% of injected embryos (Table 1B). These embryos are completely insensitive to touch. We also saw this complete loss of skeletal muscle function in all embryos coinjected with tnnc1b and tnnc2 morpholinos, however the majority of these embryos had a completely wild-type heart phenotype (Table 1B, Supplemental Movie 5C). Injection of tnnc1b or tnnc2 morpholino alone results in embryos with decreased touch-response, however, the embryos are able to swim for short distances (For summary of morpholino experiments, see Table 2).
Table 2. Co-morpholino injections reveal compensatory roles of tnnc1a and tnnc1b in contractility.
Chart summarizes the results of experiments presented in Table 1. Grey boxes represent duplicate conditions presented elsewhere in the table.
| Summary of troponin C morpholino co-injection phenotypes. | |||
|---|---|---|---|
| none | tnnc1a | tnnc1b | |
| tnnc1a | silent ventricle WT sk. muscle |
||
| tnnc1b | WT heart impaired sk. muscle |
silent heart impaired sk. muscle |
|
| tnnc2 | WT heart impaired sk. muscle |
silent ventricle paralyzed |
WT heart paralyzed |
Discussion
The sil mutant was identified in a large-scale genetic screen in zebrafish and we have shown that the associated contractility defect results from disruption of cTnC protein function. The human TNNC1 gene is expressed in both cardiac and skeletal muscle and has been found to be associated with certain forms of familial cardiomyopathy (Hoffmann et al., 2001; Gomes and Potter, 2004). While the cTnC gene is highly conserved across vertebrates, in zebrafish, both the expression and function of cTnC have been partitioned in a tissue specific manner due to an ancestral genome duplication. The zebrafish tnnc1a gene is expressed predominantly in the heart, while the tnnc1b gene is expressed in skeletal muscle. As we have demonstrated, mutations in the tnnc1a locus alone do not appear to result in defects in skeletal muscle contraction, but tnnc1a expression is required for contraction of the ventricular chamber in the heart.
We found that tnnc1a expression and function is not required for contraction of the atrial chamber following the first day post-fertilization. Previously, mutations in another zebrafish subunit of the troponin complex, Tnnt2, were shown to cause a loss of contractility in both the atrial and ventricular chambers of the heart (Sehnert et al., 2002). Based on this observation, we surmised that another troponin C gene may be expressed in the place of tnnc1a in the atrium of the zebrafish beginning at 2 dpf. Knockdown of tnnc1b in addition to tnnc1a results in a loss of contraction in both the ventricular and atrial chambers. These findings suggest that the partitioning of expression and function of the two tnnc1 genes may not be absolute and that there is a residual requirement for tnnc1b expression in the atrium after 1 dpf.
The loss of tnnc1a expression, which results in disruption of cardiac contractility, also gives rise to abnormal chamber morphology. In addition to the presence of an enlarged atrium and a compacted ventricle, sil mutants also exhibit altered expression of the ventricular specific marker vmhc by 2 dpf. The impaired morphogenesis of the cardiac chambers suggests that normal contractile function of the chambers is required for maintenance of tissue structure, consistent with previous studies on the role of chamber function on chamber morphology (Berdougo et al., 2003; Auman et al., 2007). Furthermore, the observation that vmhc expression is initially present in the ventricle of sil mutant embryos and then dissipates by 2 dpf, suggests a feedback model in which expression of contractile protein genes is necessary for the initiation of contraction and that cardiac function itself is necessary to sustain expression of cardiac contractile protein genes. This is further supported by the observation that tnnc1a expression in the silm656 mutant, while not affected during earlier stages of development, eventually also dissipates.
The silm656 allele predicts a Tnnc1a protein containing a 21 amino acid insertion at the end of exon 2. This implies the possibility that the phenotypes seen in the mutant embryos may result from a dominant negative effect; however, silm656 is a recessive mutant allele. The phenotype of the silm656 mutant embryos is also visually identical to that of embryos carrying the silsk25 allele, which is characterized by a gene deletion and loss of tnnc1a expression. Furthermore, mutant embryos resulting from a cross of heterozygous silm656 with heterozygous silsk25 zebrafish are visually identical to embryos that are homozygous for either mutant allele (data not shown). A conditional antisense knockdown has also been reported for the tnnc1a gene in zebrafish (Ho et al., 2009). Embryos displayed impaired contractility although the chambers were not reported to be silent at the developmental stages examined, consistent with an incomplete loss of tnnc1a function. Together with our results, these data suggest that the phenotypes of both the silm656 and silsk25 mutant alleles result from a loss of function rather than dominant negative effects resulting from mutations in the tnnc1a gene.
The obvious conclusion drawn from identification of TNNC1 mutations in dilated and familial cardiomyopathy has been to link the genetic changes with abnormal contractile function and thereby, disease. Our findings help to clarify this disease mechanism by demonstrating that cTnC is essential not only for cardiac contractility but also for normal morphological development. Furthermore, alteration of Ca2+ sensitivity has been shown to increase susceptibility to cardiac arrhythmia and sudden death in mice (Baudenbacher et al., 2008). Unlike other sarcomeric protein genes such as troponin T and myosin heavy chain, few mutations have been shown in the TNNC1 gene in association with human familial cardiomyopathies (Hoffmann et al., 2001; Mogensen et al., 2004). This dearth of mutations is not simply attributable to the small gene size since large numbers of troponin T gene mutations have been identified in familial hypertrophic cardiomyopathy; rather, it could illustrate that mutations in the TNNC1 gene have a pronounced affect on calcium binding affinity, more so than mutations in other troponin genes. Such mutations may be incompatible with normal cardiac development and function, thereby accounting for the few troponin C mutations detectable in cardiomyopathic patients and illustrating the key role of cTnC in both the formation and function of the heart.
A greater understanding of the way in which contractile proteins affect both the function and dysfunction of cardiomyocytes can provide insights into the underlying factors responsible for heart failure. The zebrafish mutant sil that we describe here provides a potential model for the study of human TNNC1 mutations in an in vivo system. In particular, the silsk25 allele, which completely lacks tnnc1a expression, could be used to express cDNAs of the human TNNC1 genes bearing point mutations known to be associated with familial cardiomyopathies, akin to our reported rescue experiments. This approach could provide valuable insights into the biological defects caused by these mutations.
Experimental Procedures
Positional cloning
Embryos were separated into mutant and wild-type pools based on phenotypic analysis. Genomic DNA was isolated from individual embryos by incubation in DNA isolation buffer overnight at 50°C (DNA isolation buffer: 10 mM Tris-HCl [pH 8.3], 50 mM KCl, 0.3% Tween-20, 0.3% Nonidet P40, and 0.5 mg/ml proteinase K). Proteinase K was inactivated prior to PCR setup by heating samples to 98°C for 10 minutes. PCR was performed using diluted genomic DNA as previously described (Knapik et al., 1996). Initial linkage of the silent partner locus to chromosome 23 was determined by bulk segregant analysis as previously described (Michelmore et al., 1991; Mably et al., 2003).
Whole-mount in situ hybridization and antibody staining
For whole-mount in situ hybridization and immunohistochemistry, embryos were fixed in 4% paraformaldehyde in phosphate-buffered saline, and then stored in 100% methanol at −20°C. Digoxigenin-labeled antisense RNA probes were generated by in vitro transcription (Roche), and in situ hybridization was carried out as previously described (Jowett and Lettice, 1994; Mably et al., 2006). The vmhc (ventricular myosin heavy chain) probe was derived from a PCR fragment of vmhc, performed as described previously (Joseph, 2004), in pCS2, digested with EcoRV followed by transcription with SP6. The tnnc1a probe was derived from a partial EST in pCS2 (GenBank:CO921100, IMAGE:7418798), digested with HindIII followed by transcription with T7. The tnnc1b probe was derived from a partial EST in pCS2 (GenBank:CO932630, IMAGE:7432662), digested with ClaI followed by transcription with T7. The tnnc2 probe was derived from an annotated cDNA clone for Danio rerio troponin C, fast skeletal, mRNA (MGC:77594, IMAGE:6996322). Embryos were allowed to develop in BM purple (Roche) at 28°C and were then stopped by several rinses in 1X PBT and stored at 4°C.
Antibody staining with the S46 antibody from the Developmental Studies Hybridoma Bank (DSHB) was performed as previously described (Yelon et al., 1999; Mably et al., 2003).
Transmission electron microscopy
Transmission electron microscopy of zebrafish embryos was performed as described previously (Rottbauer et al., 2001).
Morpholino analysis
The antisense morpholino oligonucleotide designed over the tnnc1a exon 3 donor site, tnnc1a_e3i3, [5′-CTGGACTTTACCATCTTCATCTACT-3′, GeneTools, LLC] was dissolved at a concentration of 200 μM in 1x Danieau's buffer (5 mM Hepes pH 7.6, 58 mM NaCl, 0.7 mM KCl, 0.6 mM Ca(NO3)2, 0.4 mM MgSO4). Approximately 1 nl of this solution (1.5-2.0 ng) was injected into each one- to four-cell staged embryo before allowing the embryos to develop at 28.5°C. Analysis with the tnnc1b exon 1 donor site morpholino, tnnc1b_e1i1, [5′-CCCATAACTTACCGCTGCTTTATAT-3′, GeneTools, LLC] and the tnnc2 exon 2 donor site morpholino, tnnc2_e2i2, [5′-GTATATAGACAACTCACCGGCAAGC-3′, GeneTools, LLC] were performed in the same manner.
DNA cloning and rescue
To drive cardiomyocyte-specific gene expression, a fragment of the zebrafish cmlc2 promoter was amplified by PCR from the plasmid pDestTol2CG2 (Kwan et al., 2007) using the following primers: 5′-CTTCGCAAAGCTTAAATC-3′ and 5′-GGTCACTGTCTGCTTTGC-3′. The PCR product was cloned into the pENTR 5′-TOPO vector (Invitrogen) and confirmed by sequencing. Amplification of tnnc1a from pC2+tnnc1a was performed using specific primers flanked with the Gateway system (Invitrogen) attB1 and attB2 sites: (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTGCCACCATGAACGACATCTACAAAGC-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTATTCCACCCCCTTCATGAACTCC-3′). The PCR product was recombined with pDONR221 (Invitrogen) by using BP Clonase II (Invitrogen) and was confirmed by sequencing.
The resulting cmlc2 5′-entry vector and tnnc1a middle entry vector were recombined with the 3′-entry vector p3E-polyA and the destination vector pDestTol2CG2 (Kwan et al., 2007) using LR Clonase II (Invitrogen) to generate the final tnnc1a rescue plasmid. One- to two-cell staged embryos were coinjected with one nl of this construct at a concentration of 25 ng/μl in addition to in vitro transcribed Tol2 transposase mRNA at a concentration of 35 ng/μl. Embryos were then allowed to develop at 28.5°C and were assayed for the presence of heart contractions at ~30 hpf.
Supplementary Material
(A) The sil gene on chromosome 23 was linked to z4003/z13781 by bulk segregant analysis (Michelmore et al., 1991) and then refined to the region between markers z33996 and z6376. The closest marker was determined to be z23232, within 0.24cM of the mutant gene. The troponin C gene was initially identified as a candidate gene based on linkage to this position of chromosome 23 and its biological function in other systems. r, number of recombinants; m, number of meioses. The asterisk indicates that z13781 is only a half-informative marker (see (Knapik et al., 1996) for explanation). (B) The lesion in the silm656 allele was determined by sequence analysis of genomic DNA. The single base pair change, which disrupted the splice donor site at the 3′ end of exon 2 of the tnnc1a gene is indicated by the vertical line. The asterisks are inserted to indicate the position of a low quality base call in certain reads. The analysis was performed using the programs phred/phrap/consed (Ewing and Green, 1998; Ewing et al., 1998; Gordon et al., 1998). (C) PCR analysis of genomic DNA from the silsk25 allele revealed a deletion within the tnnc1a locus. PCR analysis of genomic DNA was performed for each of the 6 tnnc1a exons (numbered accordingly next to each agarose gel image). Lane 1, (−) no DNA control; Lanes 2-4, DNA from 3 individual wt siblings; Lanes 5-7, DNA from 3 silsk25 mutants. (D) Alignment of zebrafish (Dr) and human (Hs) cTnC genes. Boxshade is used to represent identical residues as white letters with black shading and similar residues as white letters with gray shading. The identification of two distinct EST populations coding for proteins with near complete sequence suggested a possible gene duplication and the corresponding genes were annotated tnnc1a and tnnc1b. The tnnc1a gene, currently annotated by Entrez GeneID: 353247, tnnc1 troponin C type 1 (slow), corresponded to the EST population closely linked to marker z23232 and identified as the gene disrupted in the sil mutant.
(A) The hearts of embryos injected with a morpholino targeting the fast-twitch troponin C gene (tnnc2) contract normally. (B) Embryos injected with tnnc1a and tnnc2 morpholinos together display the sil phenotype. (C) The hearts of the majority of embryos injected with tnnc1b and tnnc2 morpholinos contract normally (representative embryo, see Table 1B), while the remaining embryos display a phenotype similar to the embryo shown in Supplemental Movie 4A.
mRNA was isolated from pooled embryos at 1 dpf and analyzed using primers flanking the morpholino targeted exon donor site. PCR was performed using the Qiagen OneStep RT-PCR kit. Lane 1, no DNA control; Lane 2, uninjected embryos; Lane 3, morpholino injected embryos. Embryos were injected with 200 μM tnnc1a, 200 μM tnnc1b, or 500 μM tnnc2 morpholino.
Overlay of DIC and FITC channel of a frame from suppl. movie 3 demonstrating expression of the cmlc2::tnnc1a/cmlc2::GFP transgene on the dorsal surface of the ventricle. The GFP expression correlates with the region of rescued myocardium.
View of heart contraction in wild-type (A) and silm656 mutant (B) embryos at 2 dpf. While contraction of the atrium and ventricle is apparent in the wild-type embryo, only contraction of the atrium is apparent in the silm656 embryo. silsk25 mutant embryos appear identical.
View of heart contraction in (A) uninjected or (B) tnnc1a morpholino injected embryos at 2 dpf. Embryos injected with 200 μM of the tnnc1a morpholino display the sil phenotype.
View of heart contraction in a 2 dpf silsk25 embryo coinjected with the tnnc1a rescue construct and Tol2 transposase RNA. GFP indicates expression of the tnnc1a rescue construct in the cardiomyocytes of this embryo. The rescue of contraction in the ventricular cardiomyocytes corresponds with the localization of GFP. Cells lacking GFP expression in the ventricle remain silent.
(A) Approximately 25% of embryos injected with a morpholino targeting the skeletal muscle slow twitch/cardiac troponin C paralog (tnnc1b) have hearts that are able to contract weakly in both chambers but lack blood circulation (representative embryo, see Table 1B). This is in contrast with the approximately 75% of embryos displaying wild-type heart contraction and circulation (not shown) (B) Embryos injected with morpholinos to both tnnc1a and tnnc1b lack contraction of both chambers.
Acknowledgments
We thank C. Fabricant for initial analysis of the sk25 allele and Amy Doherty and Jennifer Ser for zebrafish embryo collection and genotyping. We are grateful to Jonathan Rosen for construction of the cmlc2 5′-entry vector. This work was supported by a National Scientist Development Grant (0635363N) from the American Heart Association and a sponsored Research agreement with the Novartis Institutes for Biomedical Research. Work in the Yelon lab is funded by the NIH, AHA, and March of Dimes.
Grant Information: American Heart Association; Scientist Development Grant #0635363N (JDM) and Established Investigator Award #0940041N (DLY) NIH; R01 HL081911 (DLY) Novartis Institutes for Biomedical Research; Sponsored Research agreement (CAM, JDM)
References
- Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL, Westerfield M, Ekker M, Postlethwait JH. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Auman HJ, Coleman H, Riley HE, Olale F, Tsai HJ, Yelon D. Functional modulation of cardiac form through regionally confined cell shape changes. PLoS Biol. 2007;5:e53. doi: 10.1371/journal.pbio.0050053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudenbacher F, Schober T, Pinto JR, Sidorov VY, Hilliard F, Solaro RJ, Potter JD, Knollmann BC. Myofilament Ca2+ sensitization causes susceptibility to cardiac arrhythmia in mice. J Clin Invest. 2008;118:3893–3903. doi: 10.1172/JCI36642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdougo E, Coleman H, Lee DH, Stainier DY, Yelon D. Mutation of weak atrium/atrial myosin heavy chain disrupts atrial function and influences ventricular morphogenesis in zebrafish. Development. 2003;130:6121–6129. doi: 10.1242/dev.00838. [DOI] [PubMed] [Google Scholar]
- Chen JN, Haffter P, Odenthal J, Vogelsang E, Brand M, van Eeden FJ, Furutani-Seiki M, Granato M, Hammerschmidt M, Heisenberg CP, Jiang YJ, Kane DA, Kelsh RN, Mullins MC, Nusslein-Volhard C. Mutations affecting the cardiovascular system and other internal organs in zebrafish. Development. 1996;123:293–302. doi: 10.1242/dev.123.1.293. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Filatov VL, Katrukha AG, Bulargina TV, Gusev NB. Troponin: structure, properties, and mechanism of functioning. Biochemistry (Mosc) 1999;64:969–985. [PubMed] [Google Scholar]
- Gahlmann R, Wade R, Gunning P, Kedes L. Differential expression of slow and fast skeletal muscle troponin C. Slow skeletal muscle troponin C is expressed in human fibroblasts. J Mol Biol. 1988;201:379–391. doi: 10.1016/0022-2836(88)90145-3. [DOI] [PubMed] [Google Scholar]
- Gomes AV, Potter JD. Molecular and cellular aspects of troponin cardiomyopathies. Ann N Y Acad Sci. 2004;1015:214–224. doi: 10.1196/annals.1302.018. [DOI] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Greaser ML, Gergely J. Purification and properties of the components from tropinin. J Biol Chem. 1973;248:2125–2133. [PubMed] [Google Scholar]
- Ho YL, Lin YH, Tsai WY, Hsieh FJ, Tsai HJ. Conditional antisense-knockdown of zebrafish cardiac troponin C as a new animal model for dilated cardiomyopathy. Circ J. 2009;73:1691–1697. doi: 10.1253/circj.cj-09-0210. [DOI] [PubMed] [Google Scholar]
- Hoffmann B, Schmidt-Traub H, Perrot A, Osterziel KJ, Gessner R. First mutation in cardiac troponin C, L29Q, in a patient with hypertrophic cardiomyopathy. Hum Mutat. 2001;17:524. doi: 10.1002/humu.1143. [DOI] [PubMed] [Google Scholar]
- Huang CJ, Tu CT, Hsiao CD, Hsieh FJ, Tsai HJ. Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev Dyn. 2003;228:30–40. doi: 10.1002/dvdy.10356. [DOI] [PubMed] [Google Scholar]
- Joseph EM. Zebrafish IRX1b in the embryonic cardiac ventricle. Dev Dyn. 2004;231:720–726. doi: 10.1002/dvdy.20172. [DOI] [PubMed] [Google Scholar]
- Jowett T, Lettice L. Whole-mount in situ hybridizations on zebrafish embryos using a mixture of digoxigenin- and fluorescein-labelled probes. Trends Genet. 1994;10:73–74. doi: 10.1016/0168-9525(94)90220-8. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Kawasaki H, Kretsinger RH. Calcium-binding proteins. 1: EF-hands. Protein Profile. 1994;1:343–517. [PubMed] [Google Scholar]
- Knapik EW, Goodman A, Atkinson OS, Roberts CT, Shiozawa M, Sim CU, Weksler-Zangen S, Trolliet MR, Futrell C, Innes BA, Koike G, McLaughlin MG, Pierre L, Simon JS, Vilallonga E, Roy M, Chiang PW, Fishman MC, Driever W, Jacob HJ. A reference cross DNA panel for zebrafish (Danio rerio) anchored with simple sequence length polymorphisms. Development. 1996;123:451–460. doi: 10.1242/dev.123.1.451. [DOI] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Mably JD, Chuang LP, Serluca FC, Mohideen MA, Chen JN, Fishman MC. santa and valentine pattern concentric growth of cardiac myocardium in the zebrafish. Development. 2006;133:3139–3146. doi: 10.1242/dev.02469. [DOI] [PubMed] [Google Scholar]
- Mably JD, Mohideen MA, Burns CG, Chen JN, Fishman MC. heart of glass regulates the concentric growth of the heart in zebrafish. Curr Biol. 2003;13:2138–2147. doi: 10.1016/j.cub.2003.11.055. [DOI] [PubMed] [Google Scholar]
- Michelmore RW, Paran I, Kesseli RV. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci U S A. 1991;88:9828–9832. doi: 10.1073/pnas.88.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen J, Murphy RT, Shaw T, Bahl A, Redwood C, Watkins H, Burke M, Elliott PM, McKenna WJ. Severe disease expression of cardiac troponin C and T mutations in patients with idiopathic dilated cardiomyopathy. J Am Coll Cardiol. 2004;44:2033–2040. doi: 10.1016/j.jacc.2004.08.027. [DOI] [PubMed] [Google Scholar]
- Ross J., Jr Dilated cardiomyopathy: concepts derived from gene deficient and transgenic animal models. Circ J. 2002;66:219–224. doi: 10.1253/circj.66.219. [DOI] [PubMed] [Google Scholar]
- Rottbauer W, Baker K, Wo ZG, Mohideen MA, Cantiello HF, Fishman MC. Growth and function of the embryonic heart depend upon the cardiac-specific L-type calcium channel alpha1 subunit. Dev Cell. 2001;1:265–275. doi: 10.1016/s1534-5807(01)00023-5. [DOI] [PubMed] [Google Scholar]
- Sehnert AJ, Huq A, Weinstein BM, Walker C, Fishman M, Stainier DY. Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nat Genet. 2002;31:106–110. doi: 10.1038/ng875. [DOI] [PubMed] [Google Scholar]
- Serluca FC, Sidow A, Mably JD, Fishman MC. Partitioning of tissue expression accompanies multiple duplications of the Na+/K+ ATPase alpha subunit gene. Genome Res. 2001;11:1625–1631. doi: 10.1101/gr.192001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stainier DY, Fouquet B, Chen JN, Warren KS, Weinstein BM, Meiler SE, Mohideen MA, Neuhauss SC, Solnica-Krezel L, Schier AF, Zwartkruis F, Stemple DL, Malicki J, Driever W, Fishman MC. Mutations affecting the formation and function of the cardiovascular system in the zebrafish embryo. Development. 1996;123:285–292. doi: 10.1242/dev.123.1.285. [DOI] [PubMed] [Google Scholar]
- Tobacman LS. Thin filament-mediated regulation of cardiac contraction. Annu Rev Physiol. 1996;58:447–481. doi: 10.1146/annurev.ph.58.030196.002311. [DOI] [PubMed] [Google Scholar]
- Towbin JA, Bowles NE. The failing heart. Nature. 2002;415:227–233. doi: 10.1038/415227a. [DOI] [PubMed] [Google Scholar]
- Yelon D, Horne SA, Stainier DY. Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev Biol. 1999;214:23–37. doi: 10.1006/dbio.1999.9406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) The sil gene on chromosome 23 was linked to z4003/z13781 by bulk segregant analysis (Michelmore et al., 1991) and then refined to the region between markers z33996 and z6376. The closest marker was determined to be z23232, within 0.24cM of the mutant gene. The troponin C gene was initially identified as a candidate gene based on linkage to this position of chromosome 23 and its biological function in other systems. r, number of recombinants; m, number of meioses. The asterisk indicates that z13781 is only a half-informative marker (see (Knapik et al., 1996) for explanation). (B) The lesion in the silm656 allele was determined by sequence analysis of genomic DNA. The single base pair change, which disrupted the splice donor site at the 3′ end of exon 2 of the tnnc1a gene is indicated by the vertical line. The asterisks are inserted to indicate the position of a low quality base call in certain reads. The analysis was performed using the programs phred/phrap/consed (Ewing and Green, 1998; Ewing et al., 1998; Gordon et al., 1998). (C) PCR analysis of genomic DNA from the silsk25 allele revealed a deletion within the tnnc1a locus. PCR analysis of genomic DNA was performed for each of the 6 tnnc1a exons (numbered accordingly next to each agarose gel image). Lane 1, (−) no DNA control; Lanes 2-4, DNA from 3 individual wt siblings; Lanes 5-7, DNA from 3 silsk25 mutants. (D) Alignment of zebrafish (Dr) and human (Hs) cTnC genes. Boxshade is used to represent identical residues as white letters with black shading and similar residues as white letters with gray shading. The identification of two distinct EST populations coding for proteins with near complete sequence suggested a possible gene duplication and the corresponding genes were annotated tnnc1a and tnnc1b. The tnnc1a gene, currently annotated by Entrez GeneID: 353247, tnnc1 troponin C type 1 (slow), corresponded to the EST population closely linked to marker z23232 and identified as the gene disrupted in the sil mutant.
(A) The hearts of embryos injected with a morpholino targeting the fast-twitch troponin C gene (tnnc2) contract normally. (B) Embryos injected with tnnc1a and tnnc2 morpholinos together display the sil phenotype. (C) The hearts of the majority of embryos injected with tnnc1b and tnnc2 morpholinos contract normally (representative embryo, see Table 1B), while the remaining embryos display a phenotype similar to the embryo shown in Supplemental Movie 4A.
mRNA was isolated from pooled embryos at 1 dpf and analyzed using primers flanking the morpholino targeted exon donor site. PCR was performed using the Qiagen OneStep RT-PCR kit. Lane 1, no DNA control; Lane 2, uninjected embryos; Lane 3, morpholino injected embryos. Embryos were injected with 200 μM tnnc1a, 200 μM tnnc1b, or 500 μM tnnc2 morpholino.
Overlay of DIC and FITC channel of a frame from suppl. movie 3 demonstrating expression of the cmlc2::tnnc1a/cmlc2::GFP transgene on the dorsal surface of the ventricle. The GFP expression correlates with the region of rescued myocardium.
View of heart contraction in wild-type (A) and silm656 mutant (B) embryos at 2 dpf. While contraction of the atrium and ventricle is apparent in the wild-type embryo, only contraction of the atrium is apparent in the silm656 embryo. silsk25 mutant embryos appear identical.
View of heart contraction in (A) uninjected or (B) tnnc1a morpholino injected embryos at 2 dpf. Embryos injected with 200 μM of the tnnc1a morpholino display the sil phenotype.
View of heart contraction in a 2 dpf silsk25 embryo coinjected with the tnnc1a rescue construct and Tol2 transposase RNA. GFP indicates expression of the tnnc1a rescue construct in the cardiomyocytes of this embryo. The rescue of contraction in the ventricular cardiomyocytes corresponds with the localization of GFP. Cells lacking GFP expression in the ventricle remain silent.
(A) Approximately 25% of embryos injected with a morpholino targeting the skeletal muscle slow twitch/cardiac troponin C paralog (tnnc1b) have hearts that are able to contract weakly in both chambers but lack blood circulation (representative embryo, see Table 1B). This is in contrast with the approximately 75% of embryos displaying wild-type heart contraction and circulation (not shown) (B) Embryos injected with morpholinos to both tnnc1a and tnnc1b lack contraction of both chambers.