Abstract
Experiences during early development profoundly affect development of the CNS to impart either risk for or resilience to later psychopathology. Work in the developmental neuroscience field is providing compelling data that epigenetic marking of the genome may underlie aspects of this process. Experiments in rodents continue to show that experiences during sensitive periods of development influence DNA methylation patterns of several genes. These experience-induced DNA methylation patterns represent stable epigenetic modifications that alter gene transcription throughout the lifespan and promote specific behavioral outcomes. We discuss the relevance of these findings to humans, and also briefly discuss these findings in the broader contexts of cognition and psychiatric disorder. We conclude by discussing the implications of these observations for future research.
Keywords: early-life experience, maternal care, epigenetic, DNA methylation, histone modification
Introduction
It is clear that there are periods during postnatal development in which the developing brain has heightened sensitivity to environmental influences. These sensitive periods represent heightened epochs of brain plasticity, where the early environment is able to shape neural circuits and thus determine structural and functional aspects of brain and behavior for the lifespan. It is through this so-called programming effect on the CNS that early-life experiences are thought to confer either risk or resilience regarding later psychopathology.
Recent work in the developmental neuroscience field continues to advance our understanding of the mechanisms whereby early-life experiences and environmental influences interact directly with genes in the developing brain. Studies continue to show that epigenetic mechanisms, or the chemical markings of the DNA and the surrounding histone proteins that regulate gene activity in the CNS, are modified by experiences, particularly those occurring within the context of caregiving. Thus, an epigenetic hypothesis for environmental contributions to cognitive health and risk for psychopathology continues to gain traction (e.g. Costa et al., 2009; Duman & Monteggia, 2006; McGowan & Szyf, 2010; Roth, Lubin, Sodhi, & Kleinman, 2009; Rutter, Moffitt, & Caspi, 2006; Tsankova, Renthal, Kumar, & Nestler, 2007). Here we discuss evidence of epigenetic marking of the genome by several types of early-life experiences, and the hypothesized role this plays in mediating the long-term influence of these experiences on brain and behavioral outcome.
Epigenetic modifications
Epigenetic mechanisms that help regulate gene activity in the CNS have historically been recognized for their role in processes involved in cellular differentiation, genomic imprinting, and disease (namely cancer). However, developmental and behavioral neuroscience research continue to highlight that the epigenome remains exquisitely sensitive to environmental influences and that epigenetic mechanisms mediate gene-environment interplay throughout the lifespan. Post-translational modifications of histones and DNA methylation have been the most intensely studied epigenetic mechanisms in this regard, and a growing body of evidence indicates that changes in gene activity as a result of a variety of environmental factors, including toxins, diet, stress, and other behaviourally-relevant stimuli, occur through epigenetic mechanisms (e.g. Fagiolini, Jensen, & Champagne, 2009; Franklin & Mansuy, 2009; Gräff & Mansuy, 2008; Jirtle & Skinner, 2007; Liu, Van Groen, Kadish, & Tollefsbol, 2009; Roth & Sweatt, 2009; Zhang & Meaney, 2010).
Histones are proteins that help organize and condense DNA within the nucleus. One can imagine chromatin as a core of eight histone proteins (histones 2A, 2B, 3, and 4, with two copies of each molecule) with DNA wrapped around much like thread is on a spool. The interaction between histones and DNA is mediated in part by the N-terminal tail of histone proteins, and covalent modifications to these tails help determine whether DNA is accessible for gene transcription (Strahl & Allis, 2000). For example, acetylation at lysine residues, via enzymes called histone acetyltransferases (HATs), decreases the affinity between the protein tail and DNA (Marmorstein & Trievel, 2009). This relaxes chromatin structure and provides access for transcriptional machinery. Thus, histone acetylation is generally associated with transcriptional activation and is widely regarded as one of the epigenetic marks associated with active chromatin, or what is commonly referred to as euchromatin (Figure 1). Histone acetylation is a reversible process, and the enzymes that catalyze the reversal of histone acetylation are known as histone deacetylases (HDACs) (Haberland, Montgomery, & Olson, 2009).
Figure 1.
Epigenetic modifications in the CNS. Reversible and site-specific histone modifications occur at amino-acid residues of histone tails via histone modifying enzymes, some of which are depicted on the diagram. DNA methylation occurs at cytosine residues at the 5-position of the pyrimidine ring in a reaction catalyzed by DNA methyltransferases (DNMTs). DNA methylation is a reversible process, whereby several rounds of cell division without DNMT-mediated remethylation are necessary to erase epigenetic marks. Several active DNA demethylation pathways have also been proposed for post-mitotic cells, including the direct removal of methyl groups by a DNA demethylase (Bhattacharya, Ramchandani, Cervoni, & Szyf, 1999) or a Gadd45-coupled DNA repair-like process (Barreto et al., 2007; Ma et al., 2009). Together, these modifications provide a unique epigenetic signature that governs chromatin structure (active vs. closed) and thus gene transcription.
As it is currently best understood, euchromatin is also characterized by mostly unmethylated cytosines, whereas heterochromatin, or transcriptionally inactive chromatin, is characterized by methylated cytosines and both histone deacetylation and histone methylation at specific sites (Figure 1). The addition of methyl groups to cytosines, a process referred to as DNA methylation, is catalyzed by the enzymatic activity of DNA methyltransferases (DNMTs). Methylated cytosines in turn bind repressor proteins, including the methyl-binding domain protein MeCP2 and HDACs (Bird, 2002; Miranda & Jones, 2007). This promotes a higher-affinity interaction between DNA and the histone core through localized regulation of the three-dimensional structure of DNA and its associated histone proteins, which ultimately suppresses gene transcription. However, it is important to point out that recent studies do suggest that MeCP2 can also be associated with active genes (Chahrour et al., 2008; Cohen, Zhou, & Greenberg, 2008; Yasui et al., 2007), thus our understanding of DNA methylation in the regulation of genes appears incomplete.
Lasting epigenetic modifications by early-life experiences –rodent evidence
Over the past half-century, it has become increasingly clear that environmental influences early in development remain pervasive across the lifespan. Though it has been appreciated for some time that the long-term consequences of early-life experiences represent epigenetic influences, it has not been until recently that we have begun to collect empirical data to support this view. DNA methylation, with its stable and even heritable nature regarding gene regulation, offers an ideal substrate for long-lasting cellular changes, and thus has been the most commonly studied epigenetic modification regarding experience-induced changes in the postnatal environment. Below we review data from studies that have capitalized on the utility of animal models to exploit sensitive periods of development to help support the view that early environmental influences and infant-caregiver experiences produce lasting epigenetic modifications, stable changes in CNS gene activity, and behavior. The majority of these studies have focused on environmental influences during the first ten days of life, which represents a sensitive period in rat development known to facilitate early learning and infant-caregiver attachment (e.g. Barr et al., 2009; Moriceau, Shionoya, Jakubs, & Sullivan, 2009; Sullivan, Landers, Yeaman, & Wilson, 2000). For each study, we have made a point to specify whether the observations are for males, females, or both sexes, as sex-specific epigenetic effects continue to emerge as an interesting phenomenon (McCarthy et al., 2009).
Epigenetic programming by variations in maternal care
Michael Meaney’s group provided the first demonstration that maternal behavior during early postnatal development directly modifies chromatin (Weaver et al., 2004). Prior to this study, their previous work had indicated that the maternal behavior of Long-Evans hooded rats during the first ten days after birth was variable, and that based upon this variability mothers could be categorized as either mothers that displayed high levels of pup licking/grooming and arched back nursing (high LG-ABN) or mothers that displayed low levels of pup licking/grooming and nursing position (low LG-ABN) (Caldji et al., 1998; Champagne, Francis, Mar, & Meaney, 2003; Liu et al., 1997). Adult offspring that were raised by high-LG mothers were found to have a more moderate stress response (decreased HPA activation) and to exhibit less fear during exploration of a novel environment in comparison to offspring that were raised by low LG-ABN mothers (Caldji et al., 1998; Liu et al., 1997). They had also previously shown that these adult phenotypes were attributable to several molecular changes, including increased hippocampal glucocorticoid receptor (GR) expression, (receptors that help to moderate neural and behavioral responses to stress), increased expression of a transcription factor (NGFI-A), decreased hypothalamic corticotrophin releasing factor (CRF) expression, and enhanced glucocorticoid feedback sensitivity (Francis, Caldji, Champagne, Plotsky, & Meaney, 1999; Liu et al., 1997; Meaney et al., 2000).
In an effort to provide an explanation as to how maternal care is able to influence transcriptional regulation and behavior into adulthood, Ian Weaver led a study where they examined whether maternal care during the first week of life produced epigenetic modifications (Weaver et al., 2004). Indeed, they found that patterns of DNA methylation of the GR gene (exon 17, within the NGF1-A consensus sequence) in the hippocampus were the direct result of the mother’s caregiving behaviors. Specifically, adult males raised by high LG-ABN mothers were found to have significantly less methylation of GR DNA and greater overall histone acetylation within their hippocampus in comparison to adults who had been raised by low LG-ABN mothers. Through a series of cross-fostering studies, they were able to demonstrate that the levels of GR promoter methylation were determined by the mother’s behavior during the postnatal period and were not a product of the biological mother’s behavioral predilection. These data were key in providing an association between the levels of caregiving behavior and DNA methylation of the GR gene promoter. Finally, in an effort to help establish a causal link between the observed epigenetic modifications, gene expression patterns, and adult behavior, they demonstrated that administration of an HDAC inhibitor was capable of removing the group differences in DNA methylation, histone acetylation, gene expression, and behavior.
A later study indicated that LG-ABN behavior is also capable of altering DNA methylation of the estrogen receptor alpha (ER-alpha) promoter, a gene whose activation and downstream effects on oxytocin receptor binding support the expression of maternal behavior (Champagne et al., 2006). Variations in LG-ABN had been shown to influence both the expression of the ER-alpha gene in the medial preoptic area and the type of maternal behavior the offspring would eventually display (Champagne, Diorio, Sharma, & Meaney, 2001). In a 2006 study led by Frances Champagne, the levels of cytosine methylation of the ER-alpha promoter were shown to likewise be directly influenced by the mother’s care (Champagne et al., 2006). Adult female offspring of high LB-ABN mothers were found to have lower levels of ER-alpha DNA methylation and increased binding of a transcription factor (Stat5) at a binding site within the promoter. Taken together, these data indicate that higher levels of LG-ABN during the first week of life stably modifies DNA methylation of the GR and ER-alpha genes such that it alters their gene transcription throughout the lifespan and promotes adult behavior that is characterized by stress resilience and increased maternal care.
Epigenetic programming by caregiver maltreatment
Abusive and neglectful experiences from the caregiver are known to leave a child particularly susceptible to cognitive and mental dysfunction. Indeed, there is a significant association of reported childhood maltreatment and the later diagnosis of adolescent and adulthood schizophrenia, borderline personality disorder, posttraumatic stress disorder, and major depression (e.g. Bremner, 2003; Heim & Nemeroff, 2001; Kaufman, Plotsky, Nemeroff, & Charney, 2000; Schore, 2002). Imaging studies on adults with a history of childhood maltreatment indicate that the frontal cortex, corpus callosum, amygdala, locus coeruleus, hippocampus, HPA axis, and cerebellum are particularly impacted by these experiences (e.g. De Bellis, 2005; Gunnar & Quevedo, 2007; Perry, Pollard, Blakely, Baker, & Vigilante, 1995; Teicher et al., 2003).
Akin to the clinical data, rodent and non-human primate models of early-life adversity show that early trauma produces lasting changes in neural function and behavior (e.g. Gunnar & Quevedo, 2007; Kaffman & Meaney, 2007; Korosi & Baram, 2009; Pryce & Feldon, 2003; Sanchez, 2006). Though these models indicate that changes in function and responsiveness of the HPA axis represent the most consistent outcome of exposure to early-life adversity, dysregulation of mediators of neural function and plasticity in brain regions such as the prefrontal cortex has also emerged as a leading candidate responsible for the lasting changes in cognition induced by early-life stress (Fumagali, Molteni, Racagni, & Riva, 2007; McEwen, 2007). One mediator of neural function and plasticity that has received much attention is the brain-derived neurotrophic factor (Bdnf) protein, and indeed changes in Bdnf gene activity continues to be hypothesized as a candidate molecular mechanism through which early-life experiences persistently modifies brain structure and function (Branchi, Francia, & Alleva, 2004; Fumagali et al., 2007, Casey et al., 2009). This led us to hypothesize that epigenetic modification to the Bdnf gene could provide an avenue for lasting alterations in Bdnf gene activity, and published a study in 2009 addressing this (Roth, Lubin, Funk, & Sweatt, 2009).
To examine whether early-life adversity alters Bdnf gene expression through epigenetic modifications, we exposed male and female infant rats to a stressed, “abusive” caretaker for 30 min daily during the first seven days of life (maltreatment condition). We generated abusive caretakers by placing them in an unfamiliar environment with limited nesting resources. This treatment was sufficient at potentiating caretakers to frequently display behaviors that both elicited distress responses and are deemed abusive and potentially harmful in non-human primates and humans (behaviors included frequent stepping on, dropping, dragging, actively rejecting, and roughly handling infants). We used two control conditions, where we exposed littermates to either a non-stressed, positive caretaker (cross-fostered care condition) or simply weighed and marked the remaining littermates and immediately returned them to the homecage (normal maternal care control condition). We found that infant maltreatment resulted in significant methylation of Bdnf DNA in the prefrontal cortex of both males and females, and that the hypermethylated DNA persisted through development and into adulthood (Figure 2). We also found that the DNA hypermethylation paralleled a lasting deficit in expression of the gene (both sexes). The observed decrease in Bdnf gene expression was in line with previous findings where early-life experiences are known to having a lasting impact on this gene; however, our results provided novel evidence regarding an epigenetic basis for such lasting effects.
Figure 2.
Effect of infant experience on DNA methylation of the Bdnf gene. Adult animals (at least 3 months old) that were maltreated as infants were found to have significant methylation of a regulatory region of the Bdnf gene (exon IV) in their prefrontal cortex in comparison to adults that had experienced either cross-foster care or remained with the biological mother (portrayed as the normal infancy group on the graph). Figure adapted from Roth et al., 2009a.
To address whether there was a causal relationship between the observed epigenetic markings and deficits in gene expression, we exposed adult male and female rats that had experienced maltreatment to a 7-day drug treatment regimen of intracranial infusions of a DNA methylation inhibitor called zebularine. We found that this drug treatment regimen was sufficient to rescue both the aberrant DNA methylation and gene expression patterns incited by early-life adversity. These data helped establish an association between the levels of DNA methylation and mRNA levels of the gene. Interestingly, the drug-treatment effect was specific to the maltreatment group. We found no effects of the treatment in control animals, indicating a specific interaction with early-life experience.
Our data indicate that experiences with an abusive caregiver during the first week of life can stably modify DNA methylation and gene expression, and intriguingly, that the effects of early-life adversity are potentially modifiable. These latter results dovetail those from Meaney and his colleagues (Weaver et al., 2004, 2005; Weaver, Meaney, & Szyf, 2006), and together, these data provide further argument that DNA methylation and its enzymatic machinery remains labile following cellular differentiation and in post-mitotic cells, which offers promising therapeutic prospects.
As experiences in the nest provide a powerful learning environment that serves to program later maternal behavior, we have explored whether our early-life experience regimen affects maternal behavior of females, and whether the observed epigenetic modifications might be transmittable across a generation. We found that females with a history of maltreatment (maltreated-females) showed the same types of abusive behaviors toward offspring that they had experienced themselves as infants (Figure 3). In contrast, females with a history of normal infancy rarely showed these behaviors (step on, 9.2% occurrence; drop, 3.3%; drag, 0.0%; actively avoid, 0.0%; roughly handle, 12.4%). We also found that eight-day-old offspring (both males and females) derived from the maltreated-females had significant methylation of Bdnf DNA in their prefrontal cortex and hippocampus in comparison to offspring derived from normal-treated females (Figure 4). Of course these results then raised the question: Was the transgenerational epigenetic inheritance due to the mother’s caregiving behaviors (mostly abusive), or was it due to a biological component, i.e. the patterns were not completely erased between generations?
Figure 3.
Effect of mother’s history on her maternal behavior. Left panel – Qualitative assessment of maternal behaviors directed toward infants in our maltreatment condition show that infants had overall experienced an adverse caregiving environment that was characterized by mainly abusive behaviors. Right panel – Qualitative assessment of maternal behaviors of females that had experienced our infant maltreatment regimen indicate that they grow up to display considerable abusive behaviors toward their own offspring. Please note that observation conditions in this experiment (homecage situated at eyelevel) also enabled us to determine whether females were displaying low vs. high nursing postures. Figure adapted from Roth et al., 2009a.
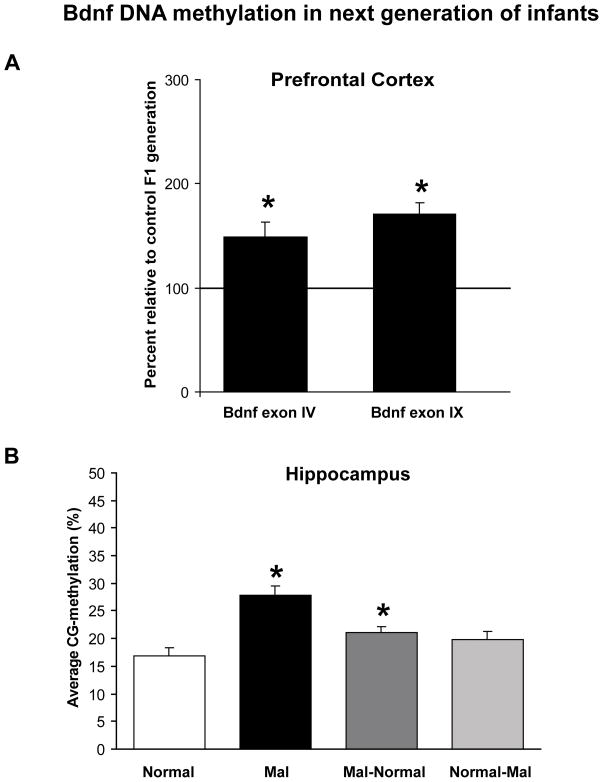
Figure 4.
Effect of mother’s history on offspring DNA methylation patterns. Offspring of females that had been maltreated were found to have significantly higher levels of methylated Bdnf DNA in their prefrontal cortex (A) and hippocampus (B) in comparison to offspring from females that had a normal infancy. Data collected in panel A were from methylation-specific real-time PCR (2 Bdnf loci examined), and data collected in panel B were from direct bisulfite sequencing (Bdnf exon IV). (B) Cross-fostering of these offspring (Mal-Normal) did not completely rescue CNS DNA methylation, nor did it induce significant methylation in controls (Normal-Mal). Figure adapted from Roth et al., 2009a.
To address this, we performed a series of cross-fostering experiments, where offspring born to maltreated-females were cross-fostered to normal females (maltreated-normal) and vice versa. To our surprise, cross-fostering did not completely reverse the patterns of DNA methylation in second-generation offspring, nor did it induce significant methylation (Figure 4B). These data suggest that the perpetuation of maltreatment-induced DNA methylation patterns was not simply a product of the postnatal experience, but likely reflected some prenatal component. Perhaps these results reflect a prenatal stress effect, as the maltreated-females were shown to have increased anxiety-like behaviors during the prepartum period. On the other hand, these results may reflect the intriguing possibility that the germ line was altered as well. Regardless of the mechanism, this is an example of transgenerational inheritance of acquired epigenetic marks in the CNS, and suggestive of how adverse experiences in one generation can potentially affect another (see Franklin & Mansuy, 2009 for a review of additional transgenerational studies).
Epigenetic programming by mother-infant separation
There is now evidence that separation of an infant from a caregiver can stably modify DNA methylation and gene expression. In another demonstration of the amazing plasticity of the HPA axis during early postnatal development, Chris Murgatroyd and Dietmar Spengler have shown that periodic separation of an infant from the caregiver (3 hr daily; ELS) during a sensitive period of development is sufficient to induce epigenetic modifications that alter HPA activity (Murgatroyd et al., 2009). In line with prior research, they first demonstrated in their study that infant separation during the first 10 days of life resulted in hyperactivity of the HPA axis, which was characterized by increased corticosterone secretion both at basal conditions and in response to stress. ELS mice as adults were also shown to have significant deficits in memory of an inhibitory avoidance task and increased immobility in the forced swim task.
Due to its role in mediating mood and cognition and its stimulatory effect on HPA activity, the authors examined whether there were any epigenetic modifications to the arginine vasopressin (Avp) gene. AVP is a hypothalamic secretagogue that induces the synthesis and release of adrenocorticotropin from the pituitary. When they examined methylation of the Avp gene in the paraventricular nucleus (PVN), they found that ELS mice (males) as far as 1 year out from the manipulation had significantly less methylation of several CpG sites in comparison to controls (non-separated mice), and that levels of DNA methylation were inversely correlated with Avp expression. They also found that there was reduced binding of MeCP2 to the Avp gene in ELS mice. Together, their results provide a nice demonstration that separation from the caregiver leads to epigenetic marking of a regulatory region of the Avp gene, which may be responsible for the sustained increase in Avp expression and HPA hyperactivity.
Epigenetic programming by prenatal stress
Prenatal experiences are recognized too for their profound effects on neurobehavioral development, and have been hypothesized to do so through excess glucorticoids (Cottrell & Seckl, 2009; Goel & Bale, 2009; Kinsella & Monk, 2009; Lupien, McEwen, Gunnar, & Heim, 2009). In 2008, Tracy Bale’s group published a study in which they examined whether the maladaptive effects of prenatal stress on adult HPA responsivity and behavior might be mediated though epigenetic mechanisms (Mueller & Bale, 2008). They administered a chronic, variable daily stress regimen to pregnant mice for seven days either during early (days 1–7), mid (days 8–14), or late gestation (days 15–21). Adult males who had been exposed to stress during early gestation were found to have altered tail suspension immobility and altered forced swimming behavior, suggestive of a depressive-like phenotype. They also exhibited changes in CRF and GR expression, and increased corticosterone release after a restraint stress challenge (indicative of altered HPA-axis responsivity). Examination of the CRF gene in these adults indicated that the prenatal stress regimen had significantly reduced methylation of specific cytosines with the regulatory regions of the CRF gene in both the hypothalamus and amygdala. Their results provide important evidence that a mother’s experience can program the brain and behavior during in utero fetal development, via epigenetic mechanisms. Further evidence for this comes from a study in which maternal cocaine administration during the second and third trimesters of gestation was shown (in males) not only to elicit global changes in hippocampal DNA methylation that were apparent in infancy, but changes that later emerged in adolescence (Novikova et al., 2008).
Evidence of epigenetic marking in humans
The studies reviewed above utilize animal models to demonstrate the lasting effects of early-life experiences and environmental influences on the epigenome. An important question to ask is: Is there any evidence of this occurring in humans? In an effort to translate some of their rodent work to human relevance, Michael Meaney and Moshe Szyf have examined expression and DNA methylation of the human glucocorticoid receptor (Nr3c1) gene in hippocampal samples derived from male suicide victims with a history of childhood maltreatment (abuse and/or neglect). They found decreased levels of GR mRNA that were correlated with increased cytosine methylation of the Nr3c1 promoter (McGowan et al., 2009). These results highlight that human caregiver experiences may likewise program genes through epigenetic modifications.
Other evidence of early epigenetic marking in humans comes from studies examining whether there is epigenetic fetal programming by environmental influences. For example, Tim Oberlander’s group has shown that human infants (both sexes) of mothers with high levels of depression and anxiety during the third trimester have increased methylation of the Nr3c1 gene promoter in cord blood cells (Oberlander, Weinberg, Papsdorf, Grunau, & Am, 2008). Infants (both sexes) delivered by C-section have been shown to have higher levels of global DNA methylation in leucocytes compared to those delivered vaginally (Schlinzig, Johansson, Gunnar, Ekström, & Norman, 2009). Remarkably, these studies indicate that the epigenome of a prenatally developing infant is sensitive to the mother’s experiences, the prenatal environment, and even the experience of birth. While there is no clear consensus yet on whether peripheral measures of DNA methylation accurately reflect CNS methylation, the potential for its clinical utility as a biomarker of psychiatric disorder continues to garner interest (e.g. Gavin & Sharma, 2009; Peedicayil, 2008).
Epigenetic modifications in cognition and mental disorder
In parallel to the developmental neuroscience studies, studies in the behavioural neuroscience field indicate that adult neurons respond to environmental signals via epigenetic mechanisms, and that epigenetic regulation of genes is necessary for experience-induced changes in adult brain function and behavior. We will briefly mention a few of these studies here, and direct readers to several recent reviews for more complete coverage of this topic (Borrelli, Nestler, Allis, & Sassone-Corsi, 2008; Gräff & Mansuy, 2008; Roth & Sweatt, 2009).
Investigators continue to show that learning and memory processes evoke alteration of epigenetic marks in the adult CNS (male rodents were used in the studies highlighted here). For example, in some of our own studies we have used the contextual-fear conditioning paradigm in rodents to demonstrate that epigenetic marking of hippocampal genes is associated with fear memory formation. We have shown that following fear conditioning and during a period when the fear memory is being formed, adult rats have a decrease in methylation (demethylation) and transcriptional activation of the memory enhancing gene Reelin, and an increase in methylation and transcriptional silencing of the memory suppressor gene Protein Phosphatase 1 (Miller & Sweatt, 2007). More recently, we have shown that fear memory formation also evokes both DNA methylation and demethylation of the Bdnf gene, with localized histone modifications occurring at specific Bdnf promoters that support upregulation of Bdnf transcription (Lubin, Roth, & Sweatt, 2008). Localized histone modifications also occur at specific Bdnf promoters following chronic social defeat (Tsankova et al., 2006). Finally, Li-Huei Tsai’s group has shown that the beneficial effects of environmental enrichment on restoring learning and memory capacity in cognitively-impaired mice involve increased hippocampal and cortical H3 acetylation (Fischer, Sananbenesi, Wang, Dobbin, & Tsai, 2007). These are exemplary studies indicating that epigenetic regulation of genes appears to play an active process in regulating an animal’s ability to respond to and form memories of its environment and experiences. These data then suggest that epigenetic modifications made to genes early in development would certainly have the capacity to subsequently affect cognition.
There is also growing evidence that aberrant epigenetic marking of genes has a role in psychiatric disorders, and the most studied disorder in this regard has been schizophrenia. Reductions in expression of Reelin and GAD1 genes (which support synaptic function and memory) in the hippocampus and cortex of schizophrenic patients (both sexes) have been one the most consistent findings in postmortem studies (Costa et al., 2009; Grayson et al., 2009). Data generated using a variety of approaches suggest that the down-regulation of these transcripts and their respective proteins is most likely due to hypermethylation of their promoters (Abdomaleky et al., 2005; Costa et al., 2009; Grayson et al., 2009). More recently investigators have found there to be as many as 100 loci with altered CpG methylation in schizophrenia (Connor & Akbarian, 2008; Mill et al., 2008). Altogether, evidence suggests there is some type of relationship between schizophrenia and cytosine methylation (see Roth et al., 2009b for a comprehensive review of epigenetic alterations in schizophrenia). These findings lead to the highly speculative notion that, to the extent that an increased incidence of schizophrenia is associated with early-life adversity, epigenetic changes triggered in early prenatal or postnatal development might predispose the development of schizophrenia later in life.
Summary
Overall, there is a growing body of work from the developmental neuroscience field that supports the view that epigenetic changes underlie the long-term impact of early-life experiences. At the same time, data illustrate that epigenetic alterations are potentially reversible, and that the epigenome can be modified via pharmacological and behavioral routes. Thus, understanding the role of the epigenome in behavioral modifications driven by early-life experiences will clearly be important for and relevant to the field of child psychiatry.
While the data discussed in this review are certainly intriguing, much more study is needed to fully appreciate and understand the discovered epigenetic phenomena. We conclude by discussing a few key issues that are sure to guide future research.
To date, developmental studies have used a candidate-gene approach to determine whether early-life experiences produce epigenetic modifications at specific gene loci. An important question that future research will need to address is: Are chromatin modifications occurring across the genome, or are only a handful of genes subject to such environmental programming? Though several studies have provided intriguing evidence that the epigenetic marks induced by early-life experience can be pharmacologically reversed (Roth et al., 2009a; Weaver et al., 2004, 2005, 2006), the data are incomplete and straightforward interpretation of their clinical relevance is difficult. For example, the ability of these pharmacological strategies to alter DNA methylation and gene expression patterns on a long-term scale has not been adequately addressed. Furthermore, there is the caveat that these types of chromatin-modifying drugs would possibly alter epigenetic states on a global scale.
Though there is convincing evidence that early-life experiences produce lasting changes in DNA methylation, the effect of these experiences on histone modifications remains largely undetermined. There is an overall emerging hypothesis in the behavioral neuroscience field that a complex pattern of specific post-translational histone modification occurs at memory-linked gene promoters to alter chromatin structure and subsequently control transcription of the genes in response to environmental cues (Roth & Sweatt, 2009; Strahl & Allis, 2000). As discussed earlier in this review, the amino acid residues of histone tails are subject to covalent modifications that include: lysine acetylation, methylation, sumoylation, and ubiquitinylation; arginine methylation; and, serine phosphorylation (Berger, 2007). Depending on the amino acid residue modified and the nature of the modification (dimethylation vs. trimethylation for example) there can be different affects on gene transcription. It is likely the case that in combination with DNA methylation modifications, a complex histone code is involved in mediating the long-term effects of early environmental influences. This aspect of early-experience induced epigenetic modification in the CNS will also need to be addressed in future studies.
Most of what we know regarding epigenetics in mediating the long-term effects of early-life experiences has come from studies using animal models where the behavioral outcome is fairly uniform and that exclude any confounds of genetic variability. It is important to consider that in humans, however, genetic polymorphisms exist and it is often the case that experiences that produce a particular outcome in some people do not in others. Thus, functional polymorphisms are predicted to add an extra layer of complexity to understanding how behavioral outcome is moderated by early-life experiences, and there is evidence to support this view.
For example, maltreated children with a genotype conferring low levels of monoamine oxidase A (MAOA) expression have been shown to be more likely to develop antisocial problems, as measured by the likelihood to develop conduct disorder, be convicted of a violent crime, and self-report antisocial behavior (Caspi et al., 2002). Though there has been some debate regarding the role of serotonin transporter genotype × environmental interaction in mood disorders (reviewed in Brown & Harris, 2008), there is evidence indicating that emotional problems are more likely in individuals who experienced stressful life-events and have one or two copies of the short allele of the serotonin transporter gene (e.g. Caspi et al., 2003; Kumsta et al., 2010). Finally, more recent work indicates that there is an interaction between early-life stress, Bdnf genotype, and neurobehavioral outcome. Specifically, individuals that were institutionalized and Met allele carriers were found to have increased amygdala volume and be more anxious in comparison to individuals that were institutionalized and homozygous for the Val allele (reviewed in Casey et al., 2009). The interactions of these genetic variables with experience-driven epigenetic changes will be an important area of future investigation.
As a final point, the data reviewed here then argue that to be in a position to fully understand how early experiences impart resilience or psychopathology, both epigenetic modifications and genetic variation need to be examined. One of the key challenges for future research will be to establish animal models to facilitate such study.
Key Points.
Over the past half-century, it has become increasingly clear that environmental influences and experiences early in development are able to shape neural circuits and thus determine structural and functional aspects of brain and behavior for the lifespan.
Recent work in the development neuroscience field has begun to highlight the molecular mechanisms whereby early-life experiences interact directly with genes in the developing brain.
Studies have shown that experiences during sensitive periods of development can program genes through DNA methylation. To date, such epigenetic alterations have been identified for several genes known to play prominent roles in cognition and psychiatric disorders.
Studies have also demonstrated that acquired epigenetic alterations can be inherited and may be potentially reversible.
An understanding of the role of the epigenome is not only relevant to helping us better understand how early-life experiences confer either risk or resilience regarding later psychopathology, but will be important and relevant for future therapeutics.
Acknowledgments
This work has been supported by grants from the National Institutes of Health, the National Alliance for Research on Schizophrenia and Depression, Civitan International, the Rotary Clubs CART fund, and the Evelyn F. McKnight Brain Research Foundation.
References
- Abdolmaleky HM, Cheng KH, Russo A, Smith CL, Faraone SV, Wilcox M, Shafa R, Glatt SJ, Nguyen G, Ponte JF, Thiagalingam S, Tsuang MT. Hypermethylation of the reelin (RELN) promoter in the brain of schizophrenic patients: a preliminary report. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2005;134:60–66. doi: 10.1002/ajmg.b.30140. [DOI] [PubMed] [Google Scholar]
- Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, Doderlein G, Maltry N, Wu W, Lyko F, Niehrs C. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- Barr GA, Moriceau S, Shionoya K, Muzny K, Gao P, Wang S, Sullivan RM. Transitions in infant learning are modulated by dopamine in the amygdala. Nature Neuroscience. 2009;12:1367–1369. doi: 10.1038/nn.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes & development. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Borrelli E, Nestler EJ, Allis CD, Sassone-Corsi P. Decoding the epigenetic language of neuronal plasticity. Neuron. 2008;60:961–974. doi: 10.1016/j.neuron.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branchi I, Francia N, Alleva E. Epigenetic control of neurobehavioural plasticity: the role of neurotrophins. Behavioural Pharmacology. 2004;15:353–362. doi: 10.1097/00008877-200409000-00006. [DOI] [PubMed] [Google Scholar]
- Bremner JD. Long-term effects of childhood abuse on brain and neurobiology. Child & Adolescent Psychiatric Clinics of North America. 2003;12:271–292. doi: 10.1016/s1056-4993(02)00098-6. [DOI] [PubMed] [Google Scholar]
- Brown GW, Harris TO. Depression and the serotonin transporter 5-HTTLPR polymorphism: A review and a hypothesis concerning gene-environment interaction. Journal of Affective Disorders. 2008;111:1–12. doi: 10.1016/j.jad.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Caldji C, Tannenbaum B, Sharma S, Francis D, Plotsky PM, Meaney MJ. Maternal care during infant regulates the development of neural systems mediating the expression of fearfulness in the rat. Proceedings of the National Academy of Sciences. 1998;95:5335–5340. doi: 10.1073/pnas.95.9.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey BJ, Glatt CE, Tottenham N, Soliman F, Bath K, Amso D, Altemus M, Pattwell S, Jones R, Levita L, McEwen B, Magariños AM, Gunnar M, Thomas KM, Mezey J, Clark AG, Hempstead BL, Lee FS. Brain-derived neurotrophic factor as a model system for examining gene by environment interactions across development. Neuroscience. 2009;164:108–120. doi: 10.1016/j.neuroscience.2009.03.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi A, McClay J, Moffitt TE, Mill J, Martin J, Craig IW, Taylor A, Poulton R. Role of genotype in the cycle of violence in maltreated children. Science. 2002;297:851–854. doi: 10.1126/science.1072290. [DOI] [PubMed] [Google Scholar]
- Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, Mcclay J, Mill J, Martin J, Braithwaite A, Poulton R. Influence of life stress on depression: moderation by a polymorphism in the 5-HT gene. Science. 2003;301:386–400. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong STC, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne F, Diorio J, Sharma S, Meaney MJ. Naturally occurring variations in maternal behavior in the rat are associated with differences in estrogen-inducible central oxytocin receptors. Proceedings of the National Academy of Sciences. 2001;98:12736–12741. doi: 10.1073/pnas.221224598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne F, Weaver I, Diorio J, Dymov S, Szyf M, Meaney MJ. Maternal care associated with methylation of the estrogen receptor-alpha1b promoter and estrogen receptor-alpha expression in the medial preoptic area of female offspring. Endocrinology. 2006;147:2909–2915. doi: 10.1210/en.2005-1119. [DOI] [PubMed] [Google Scholar]
- Champagne FA, Francis DD, Mar A, Meaney MJ. Variations in maternal care in the rat as a mediating influence for the effects of environment on development. Physiology & Behavior. 2003;79:359–371. doi: 10.1016/s0031-9384(03)00149-5. [DOI] [PubMed] [Google Scholar]
- Cohen S, Zhou Z, Greenberg ME. Activating a repressor. Science. 2008;320:1172–1173. doi: 10.1126/science.1159146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor CM, Akbarian S. DNA methylation changes in schizophrenia and bipolar disorder. Epigenetics. 2008;3:55–58. doi: 10.4161/epi.3.2.5938. [DOI] [PubMed] [Google Scholar]
- Costa E, Chen Y, Dong E, Grayson DR, Kundakovic M, Maloku E, Ruzicka W, Satta R, Veldic M, Zhubi A, Guidotti A. GABAergic promoter hypermethylation as a model to study the neurochemistry of schizophrenia vulnerability. Expert Review of Neurotherapeutics. 2009;9:87–98. doi: 10.1586/14737175.9.1.87. [DOI] [PubMed] [Google Scholar]
- Cottrell EC, Seckl J. Prenatal stress, glucocorticoids and the programming of adult disease. Frontiers in Behavioral Neuroscience. 2009:3. doi: 10.3389/neuro.08.019.2009. Epub ahead of print Sep 7 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bellis MD. The psychobiology of neglect. Child Maltreatment. 2005;10:150–172. doi: 10.1177/1077559505275116. [DOI] [PubMed] [Google Scholar]
- Duman RS, Monteggia LM. A neurotrophic model for stress-related mood disorders. Biological Psychiatry. 2006;59:1116–1127. doi: 10.1016/j.biopsych.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Fagiolini M, Jensen CL, Champagne FA. Epigenetic influences on brain development and plasticity. Current Opinion in Neurobiology. 2009;19:207–212. doi: 10.1016/j.conb.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodeling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- Francis DD, Caldji C, Champagne F, Plotsky PM, Meaney MJ. The role of corticotropin-releasing factor-norepinephrine systems in mediating the effects of early experience on the development of behavioral and endocrine responses to stress. Biological Psychiatry. 1999;46:1153–1166. doi: 10.1016/s0006-3223(99)00237-1. [DOI] [PubMed] [Google Scholar]
- Franklin TB, Mansuy IM. Epigenetic inheritance in mammals: Evidence for the impact of adverse environmental effects. Neurobiology of Disease. 2009 doi: 10.1016/j.nbd.2009.11.012. Epub ahead of print Nov 27 2009. [DOI] [PubMed] [Google Scholar]
- Fumagalli F, Molteni R, Racagni G, Riva MA. Stress during development: Impact on neuroplasticity and relevance to psychopathology. Progress in Neurobiology. 2007;81:197–217. doi: 10.1016/j.pneurobio.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Gavin DP, Sharma RP. Chromatin from peripheral blood mononuclear cells as biomarkers for epigenetic abnormalities in schizophrenia. Cardiovascular Psychiatry and Neurology. 2009 doi: 10.1155/2009/409562. 2009, Article ID 409562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel N, Bale TL. Examining the intersection of sex and stress in modeling neuropsychiatric disorders. Journal of Neuroendocrinology. 2009;21:415–420. doi: 10.1111/j.1365-2826.2009.01843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff J, Mansuy IM. Epigenetic codes in cognition and behaviour. Behavioural Brain Research. 2008;192:70–87. doi: 10.1016/j.bbr.2008.01.021. [DOI] [PubMed] [Google Scholar]
- Grayson D, Chen Y, Dong E, Kundakovic M, Guidotti A. From trans-methylation to cytosine methylation: evolution of the methylation hypothesis of schizophrenia. Epigenetics. 2009;4:144–149. doi: 10.4161/epi.4.3.8534. [DOI] [PubMed] [Google Scholar]
- Gunnar M, Quevedo K. The neurobiology of stress and development. Annual Review of Psychology. 2007;58:145–173. doi: 10.1146/annurev.psych.58.110405.085605. [DOI] [PubMed] [Google Scholar]
- Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nature Reviews Genetics. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim C, Nemeroff CB. The role of childhood trauma in the neurobiology of mood and anxiety disorders: preclinical and clinical studies. Biological Psychiatry. 2001;49:1023–1039. doi: 10.1016/s0006-3223(01)01157-x. [DOI] [PubMed] [Google Scholar]
- Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nature Reviews Genetics. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffman A, Meaney MJ. Neurodevelopmental sequelae of postnatal maternal care in rodents: clinical and research implications of molecular insights. Journal of Child Psychology and Psychiatry. 2007;48:224–244. doi: 10.1111/j.1469-7610.2007.01730.x. [DOI] [PubMed] [Google Scholar]
- Kaufman J, Plotsky PM, Nemeroff CB, Charney DS. Effects of early adverse experiences on brain structure and function: clinical implications. Biological Psychiatry. 2000;48:778–790. doi: 10.1016/s0006-3223(00)00998-7. [DOI] [PubMed] [Google Scholar]
- Kinsella MT, Monk C. Impact of maternal stress, depression and anxiety on fetal neurobehavioral development. Clinical Obstetrics and Gynecology. 2009;52:425–440. doi: 10.1097/GRF.0b013e3181b52df1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korosi A, Baram TZ. The pathways from mother's love to baby's future. Frontiers in Behavioral Neuroscience. 2009:3. doi: 10.3389/neuro.08.027.2009. Epub ahead of print Sep 24 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumsta R, Stevens S, Brookes K, Schlotz W, Castle J, Beckett C, Kreppner J, Rutter M, Sonuga-Barke E. 5HTT genotype moderates the influence of early institutional deprivation on emotional problems in adolescence: evidence from the English and Romanian Adoptee (ERA) study. Journal of Child Psychology and Psychiatry. 2010 doi: 10.1111/j.1469-7610.2010.02249.x. Epub ahead of print Mar 25 2010. [DOI] [PubMed] [Google Scholar]
- Liu D, Diorio J, Tannenbaum B, Caldji C, Francis D, Freedman A, Sharma S, Pearson D, Plotsky PM, Meaney MJ. Maternal care, hippocampal glucocorticoid receptors, and hypothalamic-pituitary-adrenal responses to stress. Science. 1997;277:1659–1662. doi: 10.1126/science.277.5332.1659. [DOI] [PubMed] [Google Scholar]
- Liu L, Van Groen T, Kadish I, Tollefsbol TO. DNA methylation impacts on learning and memory in aging. Neurobiology of Aging. 2009;30:549–560. doi: 10.1016/j.neurobiolaging.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of bdnf gene transcription in the consolidation of fear memory. Journal of Neuroscience. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien SJ, McEwen BS, Gunnar MR, Heim C. Effects of stress throughout the lifespan on the brain, behaviour and cognition. Nature Reviews Neuroscience. 2009;10:434–445. doi: 10.1038/nrn2639. [DOI] [PubMed] [Google Scholar]
- Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, Flavell RA, Lu B, Ming GL, Song H. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein R, Trievel RC. Histone modifying enzymes: Structures, mechanisms, and specificities. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2009;1789:58–68. doi: 10.1016/j.bbagrm.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MM, Auger AP, Bale TL, De Vries GJ, Dunn GA, Forger NG, Murray EK, Nugent BM, Schwarz JM, Wilson ME. The Epigenetics of Sex Differences in the Brain. Journal of Neuroscience. 2009;29:12815–12823. doi: 10.1523/JNEUROSCI.3331-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BS. Physiology and neurobiology of stress and adaptation: central role of the brain. Physiological Reviews. 2007;87:873–904. doi: 10.1152/physrev.00041.2006. [DOI] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, D'alessio AC, Dymov S, Labonte B, Szyf M, Turecki G, Meaney MJ. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nature Neuroscience. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan PO, Szyf M. The epigenetics of social adversity in early life: Implications for mental health outcomes. Neurobiology of Disease. 2010 doi: 10.1016/j.nbd.2009.12.026. Epub ahead of print Jan 4 2010. [DOI] [PubMed] [Google Scholar]
- Meaney MJ, Diorio J, Francis D, Weaver S, Yau J, Chapman K, Seckl JR. Postnatal handling increases the expression of cAMP-inducible transcription factors in the rat hippocampus: The effects of thyroid hormones and serotonin. Journal of Neuroscience. 2000;20:3926–3935. doi: 10.1523/JNEUROSCI.20-10-03926.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L, Jia P, Assadzadeh A, Flanagan J, Schumacher A, Wang SC, Petronis A. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. The American Journal of Human Genetics. 2008;82:696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Miranda TB, Jones PA. DNA methylation: The nuts and bolts of repression. Journal of Cellular Physiology. 2007;213:384–390. doi: 10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]
- Moriceau S, Shionoya K, Jakubs K, Sullivan RM. Early-Life stress disrupts attachment learning: The role of amygdala corticosterone, locus ceruleus corticotropin releasing hormone, and olfactory bulb norepinephrine. Journal of Neuroscience. 2009;29:15745–15755. doi: 10.1523/JNEUROSCI.4106-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller BR, Bale TL. Sex-specific programming of offspring emotionality after stress early in pregnancy. Journal of Neuroscience. 2008;28:9055–9065. doi: 10.1523/JNEUROSCI.1424-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmuhl Y, Fischer D, Holsboer F, Wotjak CT, Almeida OFX, Spengler D. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nature Neuroscience. 2009;12:1559–1566. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- Novikova SI, He F, Bai J, Cutrufello NJ, Lidow MS, Undieh AS. Maternal cocaine administration in mice alters DNA methylation and gene expression in hippocampal neurons of neonatal and prepubertal offspring. PLoS ONE. 2008;3:e1919. doi: 10.1371/journal.pone.0001919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberlander T, Weinberg J, Papsdorf M, Grunau R, SM, AmD Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics. 2008;3:97–106. doi: 10.4161/epi.3.2.6034. [DOI] [PubMed] [Google Scholar]
- Peedicayil J. Epigenetic biomarkers in psychiatric disorders. British Journal of Pharmacology. 2008;155:795–796. doi: 10.1038/bjp.2008.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry BD, Pollard R, Blakely T, Baker W, Vigilante D. Childhood trauma, the neurobiology of adaptation and 'use-dependent' development of the brain: How “states” become “traits”. Infant Mental Health Journal. 1995;16:271–291. [Google Scholar]
- Pryce CR, Feldon J. Long-term neurobehavioral impact of postnatal environment in rats: manipulations, effects and mediating mechanisms. Neuroscience & Biobehavioral Reviews. 2003;27:57–71. doi: 10.1016/s0149-7634(03)00009-5. [DOI] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biological Psychiatry. 2009a;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Sodhi M, Kleinman JE. Epigenetic mechanisms in schizophrenia. Biochimica et Biophysica Acta (BBA) - General Subjects. 2009b;1790:869–877. doi: 10.1016/j.bbagen.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Sweatt JD. Regulation of chromatin structure in memory formation. Current Opinion in Neurobiology. 2009;19:336–342. doi: 10.1016/j.conb.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter M, Moffitt TE, Caspi A. Gene-environment interplay and psychopathology: multiple varieties but real effects. Journal of Child Psychology and Psychiatry. 2006;47:226–261. doi: 10.1111/j.1469-7610.2005.01557.x. [DOI] [PubMed] [Google Scholar]
- Sanchez MM. The impact of early adverse care on HPA axis development: Nonhuman primate models. Hormones and Behavior. 2006;50:623–631. doi: 10.1016/j.yhbeh.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Schlinzig T, Johansson S, Gunnar A, Ekström TJ, Norman M. Epigenetic modulation at birth - altered DNA-methylation in white blood cells after Caesarean section. Acta Pædiatrica. 2009;98:1096–1099. doi: 10.1111/j.1651-2227.2009.01371.x. [DOI] [PubMed] [Google Scholar]
- Schore AN. Dysregulation of the right brain: a fundamental mechanism of traumatic attachment and the psychopathogenesis of posttraumatic stress disorder. Australian and New Zealand Journal of Psychiatry. 2002;36:9–30. doi: 10.1046/j.1440-1614.2002.00996.x. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Sullivan RM, Landers M, Yeaman B, Wilson DA. Good memories of bad events in infancy. Nature. 2000;407:38–39. doi: 10.1038/35024156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teicher MH, Andersen SL, Polcari A, Anderson CM, Navalta CP, Kim DM. The neurobiological consequences of early stress and childhood maltreatment. Neuroscience & Biobehavioral Reviews. 2003;27:33–44. doi: 10.1016/s0149-7634(03)00007-1. [DOI] [PubMed] [Google Scholar]
- Tsankova N, Berton O, Renthal W, Kumar A, Neve R, Nestler E. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nature Neuroscience. 2006;9:519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nature Reviews Neuroscience. 2007;8:355–367. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- Weaver I, Champagne F, Brown S, Dymov S, Sharma S, Meaney MJ, Szyf M. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. Journal of Neuroscience. 2005;25:11045–11054. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver I, Meaney MJ, Szyf M. Maternal care effects on the hippocampal transcriptome and anxiety-mediated behaviors in the offspring that are reversible in adulthood. Proceedings of the National Academy of Sciences. 2006;103:3480–3485. doi: 10.1073/pnas.0507526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver ICG, Cervoni N, Champagne FA, D'alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nature Neuroscience. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Yasui DH, Peddada S, Bieda MC, Vallero RO, Hogart A, Nagarajan RP, Thatcher KN, Farnham PJ, Lasalle JM. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proceedings of the National Academy of Sciences. 2007;104:19416–19421. doi: 10.1073/pnas.0707442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang TY, Meaney MJ. Epigenetics and the environmental regulation of the genome and Its function. Annual Review of Psychology. 2010;61:439–466. doi: 10.1146/annurev.psych.60.110707.163625. [DOI] [PubMed] [Google Scholar]