Abstract
The cell surface enzyme γ-glutamyl transpeptidase (GGT) is expressed by human hepatocellular carcinomas (HCC). HCCs arise from malignant transformation of hepatocytes and are the most common form of primary liver cancer. Identification of tumor-specific, post-translational modifications of GGT may provide novel biomarkers for HCC. The HepG2 cell line, derived from a human HCC, has been used extensively in studies of liver cancer. However, the use of this cell line for studies of GGT have been stymied by reports that HepG2 cells do not process the GGT propeptide into its heterodimeric subunits. The data in this study demonstrate that HepG2 cells do, in fact, produce the mature heterodimeric form of GGT. Immunohistochemical and immunoaffinity analyses provide direct evidence that, in HepG2 cells, GGT is properly localized to the bile canaliculi. Three independent, experimental approaches demonstrate that GGT in HepG2 cells is comprised of two subunits that are more heavily N-glycosylated than GGT from normal human liver tissue. These data directly contradict the dogma in the field. These data support the use of HepG2 cells as a model system for analyzing tumor-specific changes in the post-translational modifications of GGT.
Keywords: γ-glutamyl transpeptidase, liver cancer, HepG2, post-translational modification, N-glycan, biomarker
INTRODUCTION
γ-glutamyl transpeptidase (GGT, EC 2.3.2.2) is a type II membrane glycoprotein. GGT catalyzes the first enzymatic step in the metabolism of glutathione (γ-GluCysGly), glutathione-conjugates and other γ-glutamyl-containing compounds. It plays a critical role in sustaining intracellular cysteine and glutathione levels [1; 2]. GGT activity is induced in human hepatocellular carcinomas (HCC). High levels of GGT are detected in the serum of patients with HCC and in patients with many other hepatobiliary diseases [3; 4]. However, the identification of a unique, HCC-specific GGT serum isoform in genetically distinct populations throughout the world has garnered interest in its development as a diagnostic marker for the early detection and monitoring of this disease [5].
Human tumor cell lines can recapitulate molecular phenotypes associated with primary malignancies in vivo providing important model systems for the study of potential tumor biomarkers. HepG2 cells, derived from a human HCC, exhibit features of well-differentiated HCCs, including secretion of a variety of tumor-associated proteins, including alpha fetoprotein (AFP) and GP73 [6; 7]. HepG2 cells have also been used as a model system for studying tumor-specific patterns of glycosylation [7]. However, the use of HepG2 cells for the study of GGT synthesis and post-translational modification in liver tumors has been stymied by previous reports, which concluded that HepG2 cells express enzymatically active GGT that is not cleaved into its two constituent subunits [8; 9]. These publications have been widely referenced in the literature, examples of more than 30 citations include [4; 10; 11; 12; 13]. In all other cells and tissues studied, the GGT propeptide autocleaves into a large and a small subunit, a process required for enzymatic activity [14; 15]. The conclusion that HepG2 cells do not cleave the GGT propeptide was based on the apparent molecular mass of a protein immunoprecipitated by a GGT antibody from radiolabelled HepG2 membranes. Here, we provide direct evidence that HepG2 cells do, in fact, express fully-matured, heterodimeric GGT.
MATERIAL AND METHODS
Cells
HepG2 cells (ATCC #HB-8065), a human cell line derived from a well-differentiated hepatocellular carcinoma was purchased from the American Type Culture Collection (ATCC; Manassas, VA). The cells were purchased in 2008. An additional stock of HepG2 cells, frozen in 1988, was obtained from Dr. Henry Pitot, University of Wisconsin (Madison, WI). The experiments were repeated with cells from both stocks. There were no significant differences in the data obtained from the two stocks. All data presented was obtained with the HepG2 cells from ATCC. The cells were cultured in EMEM (Eagle’s minimal essential medium containing 2mM glutamine; ATCC) supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT) and penicillin/streptomycin at 37°C in a 5% CO2.
GGT subcellular localization
HepG2 cells were grown on glass coverslips; 2 × 105 cells were plated per P35 dish. After 4 days in culture, the cells were fixed and stained. For histochemical staining, the cells were fixed for 30 min in −20°C acetone, air dried then stained in: 400μL-glutamic acid γ-(4-methoxy-β-naphthylamide) (GMNA; Sigma-Aldrich, St. Louis, MO), 30mM glycyl-glycine, 1.2mM Fast Blue, 100mM NaCl and 25mM Tris pH 7.5. The cells were rinsed in saline, the stain fixed in 0.1M copper sulfate for 2 min, rinsed in saline and mounted with glycerol. Negative controls included serine-borate in the reaction mixture to inhibit GGT activity. For antibody staining, the cells were fixed and stained as previously described [16]. GGT129, an affinity purified rabbit polyclonal antibody against a 19 amino peptide at the C-terminus of the large subunit, was used as the primary antibody [17]. The secondary antibody was Alexa Fluor 488-conjugated goat anti-rabbit IgG (Invitrogen). Nuclei were visualized with 4′-6-Diamidino-2-phenylindole (DAPI). Fluorescent images were obtained by confocal microscopy at the OUHSC Flow and Image Cytometry Core Laboratory.
Human kidney and liver microsomal fractions
Human liver and kidney were obtained from the National Disease Research Interchange (NDRI, Manassass, VA). Microsomes from normal human kidney cortex and liver tissue were prepared as described previously [18]. A 0.5% Triton X-100 microsomal extract containing GGT was used for SDS-PAGE and western blot analysis.
GGT biochemical assay
GGT activity was assayed by the method of Tate and Galbraith [8]. A unit of activity is defined as the amount of GGT that produces 1 nmol of p-nitroaniline/h from 1.0mM L-γ-glutamyl-p-nitroanilide (Sigma-Aldrich) in the presence of 20mM Glycyl-glycine at pH 8.0. Quantitation of total protein was determined by BCA assay (Thermo Scientific, Rockford, IL).
Cell lysates and western blotting
Cells were scraped off the plate in 100mM Tris, pH 7.4, 150mM NaCl, 1μM leupeptin, 1μg/mL aprotinin, pelleted at 1,000g for 10 min at 4°C. Cells and Triton-solubilized tissue extracts were heat-denatured in Laemmli sample buffer (2% SDS, 5% glycerol, 5% 2-mercaptoethanol, 0.002% bromphenol blue and 62.5 mM Tris, pH 6.8) and subjected to electrophoresis on 8 or 10% SDS-PAGE gels prior to electroblot transfer to nitrocellulose and western blotting. Primary antibodies included: GGT129 directed against the heavy subunit of GGT, GGT1/2 H-170 (Santa Cruz Biotech., Santa Cruz, CA; 1/1500 dilution) directed against the light chain of GGT and F-3165 (Sigma-Aldrich; 1/2000 dilution) directed against the FLAG epitope. HRP-conjugated secondary antibodies were visualized by chemiluminescence (ECL Plus, GE Healthcare).
Deglycosylation of GGT
HepG2 cells, harvested as described above, and Triton-solubilized tissue extracts were incubated at 95°C for 10 min in denaturing buffer [25mM Tris (pH 7.4), 25mM BME, 0.1% SDS, and 0.5% Triton X-100] and then supplemented with the protease inhibitors leupeptin (1μM) and aprotinin (1μg/ml). Samples were incubated in the presence or absence of 125 units of protein N-glycosidase F (PNGase F; New England Biolabs) for 18 h at 37°C. These samples were then denatured and resolved on 8 or 10% SDS-PAGE gels for western analysis.
Transfection of Flag-Tagged GGT into HepG2 cells
The N-terminal Flag-tagged full length human GGT (GGT-NFLAG; Flag epitope = DYKDDDDK) was constructed. The open reading frame of human GGT was PCR amplified from a pcDNA3.1(+) vector containing full-length, wild-type GGT cDNA [1]. The forward and reverse primers were 5′-TTTTTTGAATTCAATGAAGAAGAAGTTAGTGGTGCTGGG-3′ and 5′-TTTTTTGATATCTCAGTAGCCGGCAGGCTCCCCGCCTTTCCTGGAGTC-3′. The amplification product was cloned into the EcoRI and EcoRV sites of the pFLAG-CMV-4 expression vector (Sigma). HepG2 cells were transfected with 5μg of the GGT-NFLAG plasmid and 1μg of pEYFP-C1 (Clontech) with lipofectamine (Invitrogen, Carlsbad, CA) and Plus Reagent (Invitrogen), according to the manufacturer’s protocol. Control cells were transfected with the pE-YFP plasmid only. Transient transfectants were cultured for three days prior to harvesting.
All experiments and assays were performed in triplicate.
RESULTS
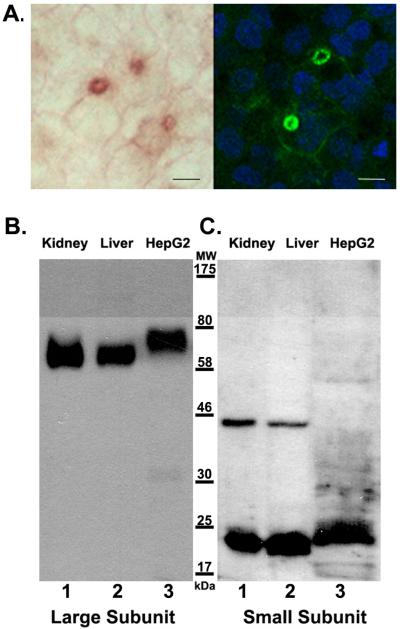
GGT is a single-pass transmembrane protein with its active site on the cell surface. In the liver, GGT is expressed by hepatocytes and is localized to the bile canaliculi [17]. As the cells become confluent, HepG2 cells form structures between adjacent cells that have been identified as bile canaliculi based on immunostaining for aminopeptidase N and dipeptidyl-peptidase IV, both of which localize to the apical bile canalicular surface of hepatocytes in vivo [19]. A histochemical stain for GGT activity localizes enzymatically-active GGT to these structures (Fig. 1A, left panel). To identify all of the GGT in the cells, the cells were fixed and incubated with the GGT129 antibody, which is directed against a peptide within the large subunit of GGT. The antibody also localized to the bile canalicular structures (Fig. 1A, right panel). Both the histochemical stain and immunolabeling also revealed very weak cell surface localization of GGT on areas of the membrane outside of the bile canalicular structures (Fig. 1A). These data confirm that all of the GGT protein co-localizes with GGT activity expressed on the cell surface.
Figure 1.
Immunodetection of GGT in HepG2 cells. (A) Localization of GGT in HepG2 cells. GGT activity was localized in acetone-fixed HepG2 cells with a histochemical stain that forms a red product at the site of GGT activity (left panel). Note the bile canalicular structures that have formed between adjacent cells and are identified by high levels of GGT activity. GGT was immunolocalized with the GGT129 antibody in cells fixed in 4% formaldehyde (right panel). Green fluorescence identifies GGT. Blue fluorescence identifies DAPI stained nuclei. The GGT129 antibody also localizes to bile canalicular structures which contain high levels of GGT activity. Scale bars are 10μm. (B and C) Western analyses against the large (B) and small (C) subunits of GGT. Extracts from normal human kidney (lanes B1, C1), liver (lanes B2, C2) and HepG2 cells (lanes B3, C3) were resolved on 10% SDS-PAGE gels and analyzed by western blotting with GGT129 antibody specific to the large subunit of GGT (lanes B1-3) or GGT1/2 H-170 antibody specific to the small subunit of GGT (lanes C1-3). For each blot, equal amounts of GGT activity were loaded per lane. Positions of molecular weight (MW) markers are indicated. Lanes C1and C2 contain a non-specific immunoreactive band at ~ 44kDa.
Western blot analysis of GGT from normal human kidney and liver (Fig. 1B, lanes 1, 2) revealed that the GGT129 antibody recognized a broad band with apparent molecular mass of approximately 63 kDa in both tissues, the size previously reported for the large subunit of human GGT [8; 20]. In HepG2 cells, the GGT129 antibody recognized a single broad band with an apparent molecular mass of 75 kDa, which is larger than the immunoreactive bands in normal human kidney and liver (Fig. 1B, lane 3). This size discrepancy could be due to a failure to cleave the propeptide in HepG2 cells and/or a difference in the post-translational processing of GGT in these cells.
To further investigate whether the GGT propeptide is cleaved into a large and small subunit in HepG2 cells, we probed human kidney, liver, and HepG2 extracts with an antibody directed against the small subunit of GGT. The small subunit of GGT in extracts from normal human kidney or liver, had an apparent molecular mass of 22 kDa (Fig. 1C, lanes 1, 2). In HepG2 extracts, the small subunit antibody recognized a larger protein with an apparent molecular mass of approximately 24 kDa (Fig. 1C, lane 3). However, the antibody directed against the small subunit did not exhibit cross-reactivity with the 75 kDa species recognized by the large subunit antibody in HepG2 extracts, as would be expected if the cells failed to cleave the propeptide into two subunits. The detection of two, independent immunoreactive species of distinct apparent molecular masses by antibodies against the large and small subunits of GGT in HepG2 extracts is consistent with the maturation of the propeptide into a heterodimer.
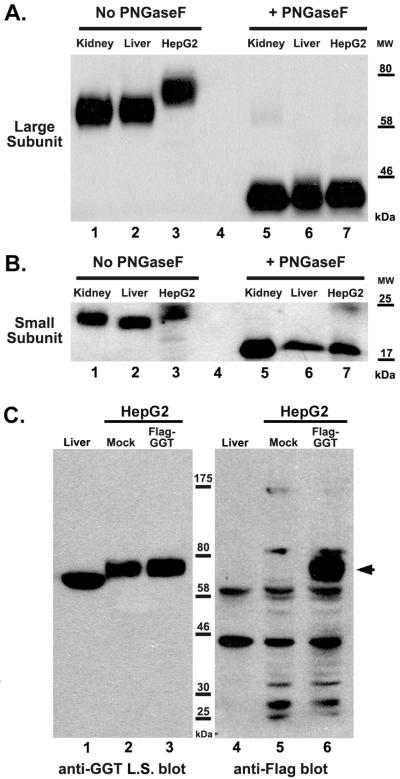
There are multiple reports of increased size and complexity of the covalently attached N-glycans on both GGT and other glycoproteins expressed by human liver tumors [21; 22; 23]. There are six potential N-glycosylation sites (sequon: NXS/T, where X≠P) on the large subunit and one on the small subunit of human GGT, all of which are glycosylated in normal human kidney tissue [20]. To determine whether the size discrepancy between the large subunit of GGT expressed in HepG2 cells and GGT from normal kidney and liver is attributable to altered glycosylation, the samples were treated with the N-glycosidase PNGase F to enzymatically remove all N-glycans. Deglycosylation resulted in a dramatic decrease in the apparent molecular mass of the large subunit of GGT from normal kidney and liver tissue from 63 kDa (Fig. 2A, lanes 1, 2) to approximately 42 kDa (Fig. 2A, lanes 5, 6), consistent with the predicted mass of the large subunit without post-translational modification. Deglycosylation of HepG2 proteins resulted in a decrease in the apparent molecular mass of the large subunit of GGT from 75 kDa to 42 kDa (Fig. 2A, lanes 3, 7), the same size as the deglycosylated GGT from kidney and liver (Fig, 2A, lanes 5, 6). These data demonstrate that the discrepancy between the apparent molecular mass of the large subunit of GGT in HepG2 cells versus the large subunit of GGT from normal kidney and liver is due to an altered pattern of glycosylation on GGT in HepG2 cells, not due to a failure to cleave the propeptide, preventing release of the small subunit from the large subunit. This conclusion was further confirmed by identifying the small subunit of GGT in the PNGFase F-treated kidney, liver, and HepG2 samples.
Figure 2.
Contribution of N-glycans to apparent size of the large and small subunits of GGT. (A and B) Extracts from normal human kidney (lanes 1, 5), liver (lanes 2, 6) and HepG2 cells (lanes 3, 7) were heat-denatured and then incubated in the absence (lanes 1-3) or presence (lanes 5-7) of the N-glycosidase, PNGaseF. The extracts were resolved on an 8% (A) or 10% (B) SDS-PAGE gel. Western analyses were carried-out against the large (A) and small (B) subunits of GGT. Positions of molecular weight (MW) markers are indicated. (C) Immunodetection of the large subunit of GGT and Flag-tagged large subunit of GGT in HepG2 cells. Extracts from human liver (lanes 1, 4), mock-transfected HepG2 cells (lanes 2, 5), and Flag-GGT transfected HepG2 cells (lanes 3, 6) were resolved on an 8% SDS-PAGE gel. Western analysis against either the large subunit (L.S) of GGT (lanes 1-3) or the Flag epitope (lanes 4-6) are shown. Arrowhead denotes position of Flag-tagged GGT. Positions of molecular weight (MW) markers are indicated.
There is one potential N-glycosylation site on the small subunit, which is quantitatively glycosylated in normal human kidney and liver tissue [20]. Treatment of kidney and liver tissue extracts with PGNase F resulted in a decrease in the apparent molecular mass of the small subunit of GGT from 22 kDa (Fig. 2B, lanes 1, 2) to 19 kDa (Fig. 2B, lanes 5, 6). Deglycosylation of the small subunit of GGT from HepG2 cells decreased the apparent molecular mass from 24 kDa to 19 kDa (Fig. 2B, lanes 3, 7). After removal of the N-glycans, the small subunit of GGT from kidney, liver and HepG2 cells all had an apparent molecular mass of 19 kDa, which is the predicted mass of the small subunit without post-translational modification. These data indicate that GGT is cleaved into a large and small subunit in kidney, liver and HepG2 cells. The increased in apparent masses of the large and small GGT subunits in HepG2 cells is due to larger N-glycans modifying the protein.
Finally, to independently confirm that the large subunit identified on western blots by the GGT129 antibody is the dominant form of GGT in HepG2 cells, we transfected HepG2 cells with an N-terminally Flag-tagged construct of human GGT. The mock-transfected HepG2 cells expressed 7,600±110 units of GGT activity/mg of cellular protein, similar to that reported by Tate and Galbraith [8]. The HepG2 cells transfected with Flag-GGT expressed 21,900±450 units GGT activity/mg of cellular protein. Western blot analysis revealed that, in both the mock-transfected and Flag-GGT transfected HepG2 cells, the GGT129 antibody recognized a single band of apparent molecular mass of 75 kDa, as was observed for endogenous GGT expressed in untransfected HepG2 cells (Fig. 2C, lanes 2, 3 and Fig. 1B, lane 3). An antibody directed against the Flag epitope identified the 75 kDa band as the only unique band in the Flag-GGT transfected cells (Fig. 2C, lane 6). A number of non-specific bands were present as is commonly observed for human tissues probed with anti-Flag-antibodies. The data obtained with the anti-Flag antibody confirm, with an epitope independent of the GGT129 epitope, that HepG2 cells cleave the propeptide into two subunits. No band corresponding to uncleaved GGT propeptide was identified by the anti-FLAG antibody (Fig. 2C, lane 6). These data demonstrate that HepG2 cells process the GGT propeptide into two subunits as observed in normal human tissues and other human cell lines.
DISCUSSION
Tate and Galbraith reported that in HepG2 cells GGT persists as an uncleaved propeptide that is enzymatically active [8]. Although this study is widely referenced there are no publications confirming their results [4; 10; 11; 12]. Tate and Galbraith identified a 120 kDa band on SDS-PAGE as the only form of GGT expressed in HepG2 cells. We show here that HepG2 cells produce enzymatically active GGT that has been cleaved into a large and small subunit. The enzyme is predominantly localized to the bile canalicular structures in polarized HepG2 cells. With immunoaffinity labeling of both the large and small subunits of endogenously-expressed GGT, as well as ectopically-expressed Flag-tagged GGT, we demonstrate that GGT expressed by HepG2 cells is composed of a 75 kDa large subunit and a 24 kDa small subunit. Therefore, the heterodimer would be approximately 99 kDa. The apparent molecular masses of the large and small subunits of GGT from normal human kidney and liver are approximately 63 and 22 kDa, respectively, giving rise to a an approximately 85 kDa heterodimeric enzyme. The size difference between GGT expressed in HepG2 cells and GGT expressed in normal human liver and kidney is eliminated by the removal of the N-linked glycans. These data are consistent with reports that there is an increase in the size and complexity of N-linked glycans on GGT originating from human liver tumors [21; 24] .
Primary human liver tumors and HepG2 cells express high levels of N-acetylglucosaminyltrasferases (GnT) IV and V relative to normal liver tissue [25; 26; 27]. These enzymes give rise to increased branching of the N-glycans (tri- and tetra-antennary structures). HepG2 cells also express high levels of alpha-(1,3)-fucosyltransferase (Fut6) and UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 (β3GnT2), which generate LewisX/sialyl LewisX structures (peripheral fucosylation) and poly-N-acetyllactosamine repeats [27; 28]. The increased expression of these enzymes in HepG2 cells is consistent with our observation of the increase in size of GGT from HepG2 cells relative to normal human liver.
There are several possible explanations for the discrepancies between our data and that of Tate and Galbraith [8]. In their study, Tate and Galbraith immunoprecipitated radioactively-labeled proteins from cells with a polyclonal antibody generated against intact, human kidney GGT. The increased glycosylation of GGT in HepG2 cells may have blocked the epitope and precluded antibody-antigen interaction, thereby limiting interaction of their antibody with the immature propeptide that is not fully glycosylated. Alternatively, they may have immunoprecipitated a complex of proteins, in which the approximately 120 kDa protein that they assigned as GGT was the most strongly labeled. The protein bands were detected by the I125 label and were never independently verified by western blotting. We did not observe a 120 kDa protein by western blotting with antibodies directed against either the large or the small subunit of human GGT in extracts of either of our two stocks of HepG2 cells.
Tate and Galbraith demonstrated that HepG2-derived GGT mRNA translated by dog pancreatic microsomes gave rise to a mature enzyme that was comprised of a large and small subunit [9]. They interpreted these data as indicating that the requisite protease was absent in HepG2 cells (but present in dog microsomes). However, evidence from both bacterial and mammalian cells indicate that the cleavage is an autocatalytic event [29; 30]. Moreover, point mutations that preserve GGT activity yet result in slow heterodimeric processing have been used to show that the transient subpopulation of GGT that remains as the uncleaved precursor does not possess enzymatic activity [14].
In light of its potential as a sensitive reporter for diagnosing and monitoring liver tumors, complete characterization of the tumor-specific modifications of GGT is of great significance. HepG2 cells exhibit high fidelity in recapitulating HCC-specific phenotypes associated with other potential tumor markers and may also serve as an important surrogate for studying tumor-specific changes on GGT [7; 31; 32].
Research Highlights.
West and Hanigan Manuscript
γ-glutamyl transpeptidase (GGT) propeptide is cleaved into two subunits in HepG2 cells
Altered N-glycosylation in this tumor cell line increases the apparent size of the subunits
These data disprove the dogma that GGT persists as a propeptide in HepG2 cells
Hepg2 Cells Provide A Model System For The Study Of Ggt Glycosylation In Liver Tumors
ACKNOWLEDGEMENTS
The authors gratefully acknowledge the assistance of the Dr. Brian Ceresa at OUHSC, Dr. Jim Henthorn, The Flow and Image Cytometry Laboratory at OUHSC; Randal May, The A.I. M. Core at OUHSC and Dr. Gary Gorbsky at the OMRF for their assistance with immunostaining and the microscopy images. This work was supported by National Institutes of Health Grant RO1 CA57530 (M.H.H.) and F32 CA128338 (M.B.W.).
Abbreviations
- EMEM
Eagle’s minimal essential medium
- GGT
γ-glutamyl transpeptidase
- HCC
hepatocellular carcinoma
- PNGase F
peptide: N-glycosidase F
- SDS
sodium dodecyl sulfate
- PAGE
polyacrylamide gel electrophoresis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Hanigan MH, Ricketts WA. Extracellular glutathione is a source of cysteine for cells that express gamma-glutamyl transpeptidase. Biochemistry. 1993;32:6302–6306. doi: 10.1021/bi00075a026. [DOI] [PubMed] [Google Scholar]
- [2].Lieberman MW, Wiseman AL, Shi ZZ, Carter BZ, Barrios R, Ou CN, Chevez-Barrios P, Wang Y, Habib GM, Goodman JC, Huang SL, Lebovitz RM, Matzuk MM. Growth retardation and cysteine deficiency in gamma-glutamyl transpeptidase-deficient mice. Proc Natl Acad Sci U S A. 1996;93:7923–7926. doi: 10.1073/pnas.93.15.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Stefaniuk P, Cianciara J, Wiercinska-Drapalo A. Present and future possibilities for early diagnosis of hepatocellular carcinoma. World J Gastroenterol. 2010;16:418–424. doi: 10.3748/wjg.v16.i4.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ikeda Y, Taniguchi N. Gene expression of gamma-glutamyltranspeptidase. Methods Enzymol. 2005;401:408–425. doi: 10.1016/S0076-6879(05)01025-6. [DOI] [PubMed] [Google Scholar]
- [5].Yao DF, Dong ZZ. Hepatoma-related gamma-glutamyl transferase in laboratory or clinical diagnosis of hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int. 2007;6:9–11. [PubMed] [Google Scholar]
- [6].Knowles BB, Howe CC, Aden DP. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science. 1980;209:497–499. doi: 10.1126/science.6248960. [DOI] [PubMed] [Google Scholar]
- [7].Norton PA, Comunale MA, Krakover J, Rodemich L, Pirog N, D’Amelio A, Philip R, Mehta AS, Block TM. N-linked glycosylation of the liver cancer biomarker GP73. J Cell Biochem. 2008;104:136–149. doi: 10.1002/jcb.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Tate SS, Galbraith RA. A human hepatoma cell line expresses a single-chain form of gamma-glutamyl transpeptidase. J Biol Chem. 1987;262:11403–11406. [PubMed] [Google Scholar]
- [9].Tate SS, Galbraith RA. In vitro translation and processing of human hepatoma cell (Hep G2) gamma-glutamyl transpeptidase. Biochem Biophys Res Commun. 1988;154:1167–1173. doi: 10.1016/0006-291x(88)90263-x. [DOI] [PubMed] [Google Scholar]
- [10].Pawlak A, Cohen EH, Octave JN, Schweickhardt R, Wu SJ, Bulle F, Chikhi N, Baik JH, Siegrist S, Guellaen G. An alternatively processed mRNA specific for gamma-glutamyl transpeptidase in human tissues. J Biol Chem. 1990;265:3256–3262. [PubMed] [Google Scholar]
- [11].Yoshida K, Arai K, Kobayashi N, Uchijima Y, Saitoh H. Purification and properties of gamma-glutamyl transpeptidase from the tissue of human benign prostatic hypertrophy. J Urol. 1991;146:895–899. doi: 10.1016/s0022-5347(17)37956-9. [DOI] [PubMed] [Google Scholar]
- [12].Joyce-Brady M, Takahashi Y, Oakes SM, Rishi AK, Levine RA, Kinlough CL, Hughey RP. Synthesis and release of amphipathic gamma-glutamyl transferase by the pulmonary alveolar type 2 cell. Its redistribution throughout the gas exchange portion of the lung indicates a new role for surfactant. J Biol Chem. 1994;269:14219–14226. [PubMed] [Google Scholar]
- [13].Arai K, Sumi SH, Yoshida K, Komoda T. A precursor form of human kidney gamma-glutamyl transferase in normal and cancerous tissues, and its possible post-translational modification. Biochim Biophys Acta. 1995;1253:33–38. doi: 10.1016/0167-4838(95)00141-g. [DOI] [PubMed] [Google Scholar]
- [14].Hashimoto W, Suzuki H, Yamamoto K, Kumagai H. Effect of site-directed mutations on processing and activity of gamma-glutamyltranspeptidase of Escherichia coli K-12. J Biochem. 1995;118:75–80. doi: 10.1093/oxfordjournals.jbchem.a124894. [DOI] [PubMed] [Google Scholar]
- [15].Tate SS. Single-chain precursor of renal gamma-glutamyl transpeptidase. FEBS Lett. 1986;194:33–38. doi: 10.1016/0014-5793(86)80046-1. [DOI] [PubMed] [Google Scholar]
- [16].Dinneen JL, Ceresa BP. Expression of dominant negative rab5 in HeLa cells regulates endocytic trafficking distal from the plasma membrane. Exp Cell Res. 2004;294:509–522. doi: 10.1016/j.yexcr.2003.12.006. [DOI] [PubMed] [Google Scholar]
- [17].Hanigan MH, Frierson HF., Jr. Immunohistochemical detection of gamma-glutamyl transpeptidase in normal human tissue. J Histochem Cytochem. 1996;44:1101–1108. doi: 10.1177/44.10.8813074. [DOI] [PubMed] [Google Scholar]
- [18].King JB, West MB, Cook PF, Hanigan MH. A novel, species-specific class of uncompetitive inhibitors of gamma-glutamyl transpeptidase. J Biol Chem. 2009;284:9059–9065. doi: 10.1074/jbc.M809608200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lian WN, Tsai JW, Yu PM, Wu TW, Yang SC, Chau YP, Lin CH. Targeting of aminopeptidase N to bile canaliculi correlates with secretory activities of the developing canalicular domain. Hepatology. 1999;30:748–760. doi: 10.1002/hep.510300302. [DOI] [PubMed] [Google Scholar]
- [20].West MB, Segu ZM, Feasley CL, Kang P, Klouckova I, Li C, Novotny MV, West CM, Machref Y, Hanigan MH. Analysis of site-specific glycosylation of renal and hepatic {gamma}-glutamyl transpeptidase from normal human tissue. J Biol Chem. 2010 doi: 10.1074/jbc.M110.145938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yamashita K, Totani K, Iwaki Y, Takamisawa I, Tateishi N, Higashi T, Sakamoto Y, Kobata A. Comparative study of the sugar chains of gamma-glutamyltranspeptidases purified from human hepatocellular carcinoma and from human liver. J Biochem (Tokyo) 1989;105:728–735. doi: 10.1093/oxfordjournals.jbchem.a122736. [DOI] [PubMed] [Google Scholar]
- [22].Kobata A, Amano J. Altered glycosylation of proteins produced by malignant cells, and application for the diagnosis and immunotherapy of tumours. Immunol Cell Biol. 2005;83:429–439. doi: 10.1111/j.1440-1711.2005.01351.x. [DOI] [PubMed] [Google Scholar]
- [23].Blomme B, Van Steenkiste C, Callewaert N, Van Vlierberghe H. Alteration of protein glycosylation in liver diseases. J Hepatol. 2009;50:592–603. doi: 10.1016/j.jhep.2008.12.010. [DOI] [PubMed] [Google Scholar]
- [24].Wang NY, Zhang D, Zhao W, Fang GX, Shi YL, Duan MH. Clinical application of an enzyme-linked immunosorbent assay detecting hepatoma-specific gamma-glutamyltransferase. Hepatol Res. 2009;39:979–987. doi: 10.1111/j.1872-034X.2009.00538.x. [DOI] [PubMed] [Google Scholar]
- [25].Takamatsu S, Inoue N, Katsumata T, Nakamura K, Fujibayashi Y, Takeuchi M. The relationship between the branch-forming glycosyltransferases and cell surface sugar chain structures. Biochemistry. 2005;44:6343–6349. doi: 10.1021/bi047606a. [DOI] [PubMed] [Google Scholar]
- [26].Ito Y, Miyoshi E, Sakon M, Takeda T, Noda K, Tsujimoto M, Ito S, Honda H, Takemura F, Wakasa K, Monden M, Matsuura N, Taniguchi N. Elevated expression of UDP-N-acetylglucosamine: alphamannoside beta1,6 N-acetylglucosaminyltransferase is an early event in hepatocarcinogenesis. Int J Cancer. 2001;91:631–637. [PubMed] [Google Scholar]
- [27].Ito H, Kuno A, Sawaki H, Sogabe M, Ozaki H, Tanaka Y, Mizokami M, Shoda J, Angata T, Sato T, Hirabayashi J, Ikehara Y, Narimatsu H. Strategy for glycoproteomics: identification of glyco-alteration using multiple glycan profiling tools. J Proteome Res. 2009;8:1358–1367. doi: 10.1021/pr800735j. [DOI] [PubMed] [Google Scholar]
- [28].Togayachi A, Akashima T, Ookubo R, Kudo T, Nishihara S, Iwasaki H, Natsume A, Mio H, Inokuchi J, Irimura T, Sasaki K, Narimatsu H. Molecular cloning and characterization of UDP-GlcNAc:lactosylceramide beta 1,3-N-acetylglucosaminyltransferase (beta 3Gn-T5), an essential enzyme for the expression of HNK-1 and Lewis X epitopes on glycolipids. J Biol Chem. 2001;276:22032–22040. doi: 10.1074/jbc.M011369200. [DOI] [PubMed] [Google Scholar]
- [29].Boanca G, Sand A, Okada T, Suzuki H, Kumagai H, Fukuyama K, Barycki JJ. Autoprocessing of Helicobacter pylori gamma-glutamyltranspeptidase leads to the formation of a threonine-threonine catalytic dyad. J Biol Chem. 2007;282:534–541. doi: 10.1074/jbc.M607694200. [DOI] [PubMed] [Google Scholar]
- [30].Kinlough CL, Poland PA, Bruns JB, Hughey RP. Gamma-glutamyltranspeptidase: disulfide bridges, propeptide cleavage, and activation in the endoplasmic reticulum. Methods Enzymol. 2005;401:426–449. doi: 10.1016/S0076-6879(05)01026-8. [DOI] [PubMed] [Google Scholar]
- [31].Ohno M, Nishikawa A, Koketsu M, Taga H, Endo Y, Hada T, Higashino K, Taniguchi N. Enzymatic basis of sugar structures of alpha-fetoprotein in hepatoma and hepatoblastoma cell lines: correlation with activities of alpha 1-6 fucosyltransferase and N-acetylglucosaminyltransferases III and V. Int J Cancer. 1992;51:315–317. doi: 10.1002/ijc.2910510223. [DOI] [PubMed] [Google Scholar]
- [32].Capurro M, Wanless IR, Sherman M, Deboer G, Shi W, Miyoshi E, Filmus J. Glypican-3: a novel serum and histochemical marker for hepatocellular carcinoma. Gastroenterology. 2003;125:89–97. doi: 10.1016/s0016-5085(03)00689-9. [DOI] [PubMed] [Google Scholar]