In this issue of Cell Cycle, Straza et al.1 report therapeutic targeting of the transcriptional compressor C-terminal binding protein (CtBP) with a small molecule to specifically induce apoptosis in transformed cells and in cancer cells. Although CtBP has been associated with tumor promotion as a negative regulator of several tumor suppressors,2 the present study is significant since it is the first report on exploiting CtBP as an anti-cancer target.
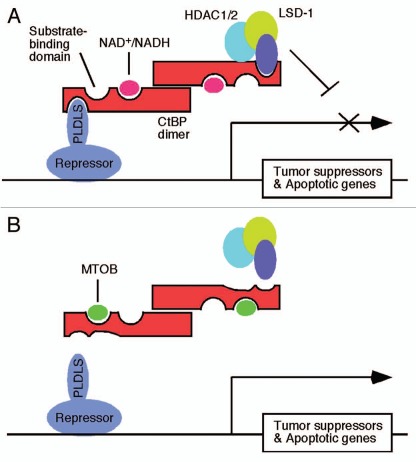
The vertebrate CtBP family proteins consist of two major related proteins, CtBP1 and CtBP2 (collectively referred as CtBP) that function as transcriptional corepressors for a large number of repressors. Both proteins exhibit overall sequence and structural similarity to the D-isomer specific 2-hydroxy acid dehydrogenases (D2-HDH).3 The role of the dehydrogenase activity in CtBP-mediated transcriptional repression remains controversial. Although CtBP1 was shown to possess a slow dehydrogenase activity,4,5 the cognate “substrate” for the CtBP dehydrogenase activity remained obscure until an intermediate of the methionine salvage pathway, 2-keto-4-methylthio-2-oxo butyrate (MTOB) was identified as a good substrate for CtBP1 dehydrogenase.6 The analysis of the constituents of the CtBP1 protein complex has helped to establish a general mechanism of transcriptional repression by CtBP7 (Fig.1). A DNA-binding transcriptional repressor recruits CtBP to the target promoter through a PLDLS-like motif first identified in adenovirus E1A. Using the individual subunits in the CtBP dimer, CtBP simultaneously interacts with a transcriptional repressor and a chromatin-modifying protein complex that consists of enzymes such as histone deacetylases (HDAC) 1/2 and lysine specific demethylase-1 (LSD-1)4 that suppress gene transcription. By structural and biochemical studies, an NAD(H)-binding domain in CtBP has been found to be a redox sensor of the cellular NAD+/NADH ratio, altering CtBP conformation and/or dimerization to regulate CtBP interaction with repressors/chromatin modifying enzymes.
Figure 1.
(A) Transcriptional repression of tumor suppressors and apoptotic genes by the CtBP dimer under normal conditions. CtBP is recruited to the target promoter through interaction with the PLDLS-like motif in the repressor which is anchored to the target promoter. The second subunit in the CtBP dimer interacts with DNA-modifying enzymes including HDACs and LSD-1. Epigenetic modifications of the chromatin by the CtBP complex result in gene silencing. NAD+/NADH binding domain and substrate (MTOB)-binding domain are illustrated. (B) Transcriptional activation of tumor suppressors and apoptotic genes after binding of MTOB to CtBP. MTOB binding to the substrate-binding domain of CtBP might induce conformational changes in the PLDLS-binding cleft and cause dissociation of the CtBP complex from the repressor, resulting in derepression of the target genes.
The present study by Straza et al. has revealed an important clue to the role of the dehydrogenase “substrate”-binding domain in CtBP function. In the CtBP structure, the substrate-binding domain and the PLDLS-binding cleft are physically very close to the dehydrogenase catalytic center.8,9 Although PLDLS-binding does not seem to compete with substrate binding in vitro,8 data by Straza et al. suggest that substrate loading can exert a significant impact on CtBP recruitment to target promoters. The CtBP substrate MTOB was shown to relieve CtBP2-mediated repression of a pro-apoptotic gene Bik at high concentrations, resulting in enhanced apoptosis of transformed cells and cancer cells in contrast to the relative resistance of normal cells to MTOB. This effect was correlated with reduced recruitment of CtBPs, especially CtBP2, to the Bik promoter. Although CtBP1/2 were previously reported to repress several pro-apoptotic genes including Noxa, Perp, Bax and Bik,10,11 Bik appears to be most critical for MTOB-induced apoptosis since shRNA-mediated depletion of Bik relieved most of MTOB-induced apoptosis in colorectal cancer cells. This result suggests that molecules that maximize the cell killing activity of Bik may constitute attractive anticancer agents. Although no detailed mechanism of MTOB-suppression of CtBP recruitment to target promoters has been provided by the authors, one possibility might be that MTOB loading to the CtBP substrate-binding domain causes conformational changes in the PLDLS-binding cleft, resulting in reduced CtBP interaction with promoter-bound transcriptional repressors (Fig.1). Should this be the case, then experiments with mutants in the substrate-binding dozmain of CtBP could be performed to show resistance of the mutants to inhibition by MTOB. It is of high significance that this study also showed that CtBP is over-expressed in many primary colon tumors and CtBP expression appears to be inversely correlated with expression of tumor suppressor ARF. More detailed expression profiling of CtBP, tumor suppressors, and apoptosis-regulatory genes including Bik in various tumor samples may help uncover new correlations between these genes and help better understand the mutual regulations and roles of these genes during tumorigenesis.
The authors demonstrated the utility of MTOB as an anti-cancer agent in cell culture as well as in the mouse xenograft model. Since MTOB appears to inhibit the transcriptional activity of CtBP at relatively high concentrations, identification of new generation CtBP substrate molecules that function at relatively low concentrations might constitute more effective anti-cancer drugs. Should the biochemical mechanism of MTOB-mediated inhibition of CtBP be clarified further, there might be MTOB derivatives that could bind to the CtBP substrate-binding domain and inhibit CtBP function more specifically and more efficiently.
References
- 1.Straza MW, et al. Cell Cycle. 2010:3740–3750. doi: 10.4161/cc.9.18.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chinnadurai G, et al. Cancer Res. 2009;69:731–734. doi: 10.1158/0008-5472.CAN-08-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chinnadurai G, et al. Mol Cell. 2002;9:213–224. doi: 10.1016/s1097-2765(02)00443-4. [DOI] [PubMed] [Google Scholar]
- 4.Balasubramanian P, et al. FEBS Lett. 2003;537:157–160. doi: 10.1016/s0014-5793(03)00119-4. [DOI] [PubMed] [Google Scholar]
- 5.Shi Y, et al. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- 6.Achouri Y, et al. Biochem Biophys Res Commun. 2007;352:903–906. doi: 10.1016/j.bbrc.2006.11.111. [DOI] [PubMed] [Google Scholar]
- 7.Chinnadurai G, et al. Int J Biochem Cell Biol. 2007;39:1593–1607. doi: 10.1016/j.biocel.2007.01.025. [DOI] [PubMed] [Google Scholar]
- 8.Kumar V, et al. Mol Cell. 2002;10:857–869. doi: 10.1016/s1097-2765(02)00650-0. [DOI] [PubMed] [Google Scholar]
- 9.Nardini M, et al. EMBO J. 2003;22:3122–3130. doi: 10.1093/emboj/cdg283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grooteclaes M, et al. Proc Natl Acad Sci USA. 2003;100:4568–4573. doi: 10.1073/pnas.0830998100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kovi RC, et al. Cell Death Differ. 2010;17:513–521. doi: 10.1038/cdd.2009.140. [DOI] [PMC free article] [PubMed] [Google Scholar]