Abstract
Objective
Apply Dicer siRNA to study functions of Dicer and miRNA during oogenesis.
Materials and Methods
Mouse oocytes were injected with Dicer siRNA and negative control siRNA and then matured in vitro. After IVM, oocytes were examined for maturation rates, spindle and chromosomal organization, and various gene expressions.
Results
Dicer siRNA significantly reduced maturation rates, increased abnormal spindle and chromosomal organization, and reduced the transcripts of Dicer miRNAs, spindle formation proteins (plk1 and AURKA) and spindle check points (Bub1, Bublb). Depletion of bulb16 markedly prohibited the first polar body extrusion and increased the incidence of misaligned chromosomes and abnormal meiotic spindle assembly.
Conclusion
Dicer siRNA triggered a cascade reduction for gene expressions starting from Dicer to miRNAs than to spindle assembly proteins and checkpoints which led to abnormal spindle and chromosomal organization. Thus, Dicer and miRNA appeared to play an important role during oogenesis and were essential for meiotic completion.
Keywords: Dicer, microRNAS, siRNA, Oocyte meiosis, Post transcriptional gene silencing
Introduction
Gene expression occurs through the transcription of the gene to mRNA and the subsequent translation of mRNA into proteins. This process is usually positively regulated during the transcriptional and translational steps. However, the discovery of hundreds of small non-coding micro-RNA (miRNAs) in various organisms has shown that gene expression can also be negatively regulated at the post-transcriptional level [1–4]. Dorsha and Dicer are two types of ribonuclease-IIIs (RNAase-IIIs) that process miRNA gene primary transcripts (pri-miRNAs) into mature miRNA (21–25 bp) and thus regulate the biogenesis of miRNA [5–7]. miRNA regulates gene expression by assembling and base-pairing with its target mRNA, forming an effector-complex (called the RNA-induced silencing complex RISC) to induce post-transcriptional gene silencing (PTGS) either by translational repression or by cleavage of the target mRNAs [8–10]. Worms, plants, and flies use a similar biological process called RNA interference (RNAi) as a defense mechanism to protect the genome against virus infection and transposons [11–14]. In mammalian tissues, RNAi was first found in mouse oocytes and early embryos [15]. Like miRNA, RNAi also requires Dicer for the generation of small interference RNA (siRNA) and RISC effector to induce PTGS [16, 17]. Both RNAi and miRNA work in response to their exogenous triggers (namely double stranded RNAs or dsRNAs) [18].
RNAi has been utilized as a new tool for screening the loss-of-function of specific genes in an increasing number of mammalian species such as human [19], mouse [20] bovine [21] and porcine [22]. Recently, the microinjection of dsRNAs or siRNAs to knockdown specific transcripts in mouse oocytes or early embryos has been extensively adopted in the studies of some crucial genes which include spindle assembly checkpoints [23], transcription factors [24] and cell cycle regulators [25]. Therefore, RNAi efficiently induced by siRNA introduction is an excellent method for exploring the role of maternal transcripts recruited either during oocyte maturation or embryo development [26].
Early embryo development is controlled by the spatial and temporal expression of genes from either maternal or embryonic origin or from both genomes. During oogenesis, the oocyte accumulates mRNAs and proteins not only for their growth, but also for their maturation, fertilization and further embryonic development [27]. Maternal mRNAs and proteins are essential for early embryonic development before activation of embryonic genome [28]. In general, embryonic genome activation takes place at the 2-cell stage in mouse embryos and at the 8-cell stage in bovine embryos. After activation of embryonic genome, embryonic transcription initiates. Dicer, which controls miRNA genesis, is capable of negatively regulating both the maternal and embryonic gene expression through the inactivation of maternal and embryonic mRNA. Much evidences have supported the theory that Dicer may be an important player in oogenesis and embryogenesis. Indeed, evidence of the important role of Dicer can be observed in the case of Dicer-deficiency, which can either arrest embryonic development or lead to embryonic death [29, 30]. Furthermore, it has been observed that mutations in homologous dicer gene in various model organisms may lead to developmental abnormalities: a diverse range of developmental defects in C. elegans [31], abnormal embryogenesis in A. thealiana [32], developmental arrest in zebrafish [33] and depletion of stem cells in mice [34]. Transcriptions of Dicer and microRNA have also been detected and profiled in non-fertilized oocytes, zygotes, and various stages of embryos [35–37]. Dicer mRNA is one of highly abundant transcripts in mouse oocytes where the relative amount of Dicer mRNA is the highest when compared with other cell types [38]. Finally, Dicer’s importance in oogenesis was confirmed through conditional knockout experiments in which oocytes of mice lacking Dicer were found to arrest at meiosis I with multiple disorganized spindles and severe defect of chromosome congression [35, 39]. Even though we have confirmed that Dicer is essential for oogenesis, the molecular mechanism that led to the observed defects in oocytes of mice lacking Dicer remains a mystery. In this paper, RNAi to suppress Dicer1 transcripts was utilized to investigate whether miRNA and spindle assembly proteins and checkpoints implicated in meiosis dysfunction in Dicer-knockdown oocytes.
Materials and methods
Isolation of mouse cumulus free oocytes (CFOs) and cumulus-oocyte complexes (COCs)
B6D2 F1 female mice (8–12 week old) were stimulated by a single i.p. injection of 5 IU pregnant mare serum gonadotropin (PMSG, Sigma St. Louis, MO). Immature GV oocytes, enclosed by cumulus cells (COCs), were collected by puncturing the ovarian follicles at 46–48 h post injection. In general, approximately 20–25 COCs were produced by each mouse. COCs isolated from ovaries of super-ovulated mice were incubated in M2 medium (Sigma) supplemented with 0.25 mM 3-isobutyl-1-methylxanthine (IBMX, Sigma) to inhibit germinal vesicle breakdown (GVBD). Attached cumulus cells were removed from the collected COCs by repeated gentle pipetting through a narrow bore glass pipette to obtain cumulus-free-oocytes (CFOs) for study.
Microinjection of siRNA
In this study, injection of siRNA was used to knockdown a specific gene in order to study the function of the gene. Negative control siRNA, siRNAs of Dicer, Bub1 and Bub1b pre-designed to knockdown its expression in mouse (Ambion, Inc., Houston, TX) were diluted with buffer to a final concentration of 50 μM, and stored at −20°C. Approximately 10 pl of the diluted siRNA solution was injected into the germinal vesicle (GV) of COCs and CFOs using the Eppendorf FemtoJet system. Injection of siRNA was performed using previously described methods by Jaff et al. [40]. Briefly, COCs were compressed slightly between two cover slides to visualize their GVs and the diluted siRNA were then injected into the GV of COCs utilizing the Eppendorf FemtoJet system. Oocytes without injection were used as controls while oocytes injected with negative control siRNA were used as negative controls. After injection, oocytes were pre-incubated in M2 medium (Sigma) supplemented with IBMX at 37°C in a humidified atmosphere of 5% CO2 and 95% air for 4 h.
In vitro maturation
IVM medium contained α-MEM (Invitrogen, Carlsbad, CA) supplemented with 3 mg/mL of bovine serum albumin (BSA, Fraction V, Sigma), 0.23 mM sodium pyruvate, antibiotic and antimycotic solution, 10 ng/mL of mouse epithelial growth factor of culture-grade (EGF, Invitrogen), and 1.5 IU/mL of human chorionic gonadotropin (Sigma). Following injection and pre-incubation, oocytes were removed from M2 medium containing IBMX to IVM medium and cultured at 37°C in a humidified atmosphere of 5% CO2 and 95% air. At the end of IVM, oocytes were assessed for their maturation stage using a Nikon inverted microscope with a magnification of ×200.
Oocyte maturation rates
Following transfer to IVM medium, mouse GV oocytes underwent GV breakdown (GVBD) and extrusion of first polar body, representing the completion of meiosis I. Oocyte maturation rates were calculated by evaluating the proportion of oocytes at the MII stage 18 h after IVM.
Effect of Dicer siRNA on oocyte maturation
The effect of Dicer siRNA was tested with CFOs and COCs at the GV stage. Experiments involving CFOs were repeated 8 times, utilizing 116–141 CFOs extracted from five to seven mice. Similarly, experiments involving COCs were repeated 7 times, utilizing 109–125 oocytes extracted from five mice each time. For each experiment, the obtained CFOs and COCs were randomly divided into non-injected oocytes as controls, Dicer siRNAs and negative control siRNAs groups. siRNAs were injected into CFOs and COCs as previously described. After injection, oocytes were allowed to mature in-vitro. A portion of MII oocytes (265 oocytes from CFOs and 255 oocytes from COCs) were used to investigate spindle integrity by immunofluorescence staining and the other portion of MII oocytes (180 oocytes from CFOs and 112 oocytes from COCs) were used to detect gene expressions by real-time RT-PCR.
Immunofluorescence staining
The zona pellucida of the oocytes was removed with Acid Tyrode’s solution (Chemicon, Billerica, MA). Oocytes were fixed with 4% paraformaldehyde in PBS for 15 min at room temperature, permeabilized with 0.5% Triton X-100 in PBS for 15 min, washed three times in 0.1% Tween in PBS (T-PBS), and then blocked with 3% BSA in T-PBS for 1 h at 37°C. Blocked oocytes were incubated overnight at 4°C in the 1:800 diluted anti-α-tubulin monoclonal antibodies (sigma). Oocytes were then washed twice in T-PBS and incubated for 1 h at room temperature in the 1:1,000 diluted rabbit anti-mouse IgG antibody conjugated with fluorescein isothiocyanate (FITC)(Molecular probe). After rinsing with T-PBS, oocytes were mounted in Vectashield with DAPI (Vector laboratories, Burlington CA) and analyzed using the Zeiss microscope (Carl Zeiss).
Real-time RT-PCR
Following IVM, sixty MII oocytes were used to detect the expressions of Dicer and two miRNAs (let 7d and miR-30c), spindle formation proteins (plk1 and ARUKA), and spindle checkpoint (Bub1, Bub1b, Zw10 and Mad2l1) genes by real time RT-PCR using predesigned primers (Applied Biosystem). Individual oocytes were transferred to 5 μl Tri-reagent (Molecular Research) in Eppendorf tubes and subjected to total RNA isolation according to the manufacture’s guidelines. For analysis of Dicer1 and GAPDH expression, RT reaction was carried out using oligo (dT)18 in a 25 μl volume with SuperScript III RT (Invitrogen) and real-time PCR was performed using TaqMan gene expression assay kit (Assay ID Mm00521733_g1, Applied Biosystem). Analyses of let-7d and mir-30c were performed using TaqMan® MicroRNA Reverse Transcription Kit and TaqMan gene expression assay kit (Assay ID: 001178 and 000419, Applied Biosystem). Relative quantification of gene expression was analyzed using the 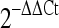 method. In all experiments, GAPDH mRNA was employed as an internal standard.
method. In all experiments, GAPDH mRNA was employed as an internal standard.
Ooplasmic replacement
The reversibility of siRNA by ooplasmic replacement was also investigated. Experiments were repeated three times. Each time, seven to eight mice were used and 126–167 CFOs were obtained. Oocytes were randomly divided into Dicer siRNA, negative control siRNA and non-injected control groups. All three groups (with or without microinjection) underwent in-vitro maturation in IVM medium for 4 h. As oocytes maturing neared the MI stage, one third of their ooplasm was replaced with that of non-injected MI oocytes. Meanwhile, a group of CFOs injected with negative controls but without ooplasmic replacement were used as controls.
For ooplasmic replacement, the studied oocytes were washed and cultured in the α-MEM medium Supplemented with 5% fetal bovine serum for 3 h. As oocytes matured to the GVBD stage, they were then cultured in M2 medium supplemented with 7.5 mg/ml cytochalasin B (Sigma) for 15 min at 30°C. A small slit was made on the zona pellucida with a sharp needle. Through the slit, one third of ooplasm were gently aspirated from the oocytes using micromanipulation (Narishige micromanipulation system), and then the same amount of ooplasm from non-injected GVBD oocytes were transferred into the perivitelline space of recipient oocytes. Couplets of recipient and donor ooplasts were briefly washed by fusion medium (300 mM mannitol, 0.1 mM MgSO4, 0.1 mg/ml Polyvinyl Alcohol, and 3 mg/ml Bovine Serum Albumin), and then transferred into a fusion chamber with two wires 0.5 mm apart and loaded with fusion medium. After applying two electric pulses (1.4 KV/cm, 26 μs each), ooplasmic replaced oocytes were incubated in M2 containing 20% FCS for recovery.
After ooplasmic replacement, oocytes were again incubated in IVM medium for final maturation. At the end of IVM, all oocytes were examined for maturation rate and for gene expressions by real-time RT-PCR.
Results
-
Oocyte maturation rates were significantly reduced by Dicer siRNA injection
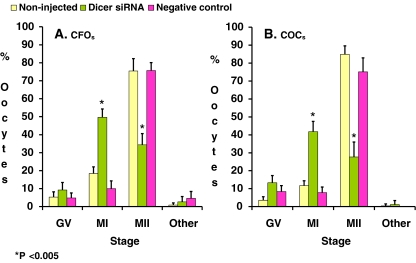
In all studied groups, the majority of oocytes matured beyond the GV stage after microinjection and in vitro maturation. However, a small portion of the oocytes either remained in GV stage or became degenerated (Fig. 1). In non-injected and negative control groups, a large portion of matured oocytes advanced to the MII stage, while in the Dicer siRNA group, more than half of the matured oocytes arrested at MI stage (Fig. 1). Oocyte maturation rates were judged by the percentages of oocyte at the MII stage. There were no significant differences between oocyte maturation rates of the negative control and the non-injected control (75.7% vs. 75.4% for CFOs, p = 0.8; and 75.1% vs. 84.9% for COCs, p = 0.2) (Fig. 1). However, injection of Dicer siRNA significantly reduced oocyte maturation rates in CFOs (34.4% vs. 75.7%, p < 0.005) as well as in COCs (27.7% vs. 75.1%, p < 0.005) (Fig. 1). Thus, injection of Dicer siRNA appears to inhibit meiotic maturation by arresting oocytes at the MI stage and circumventing the completion of meiosis I. In addition, we also found that injection of siRNAs could induce extrusion of irregular polar bodies such as double polar bodies or enlarged polar bodies in nearly 3% siRNA injected oocytes. These oocytes also exhibited abnormal patterns of chromosomal segregation (Data not shown).
-
Abnormal spindle organization and chromosome alignment rates were increased by Dicer siRNA
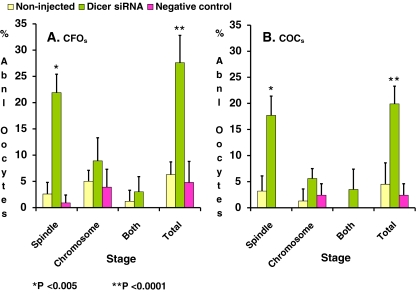
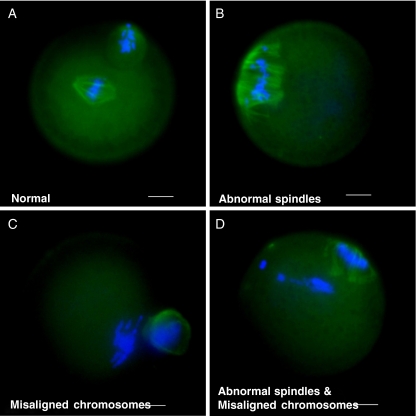
Anti-tubulin antibody immunostaining revealed that Dicer siRNA injection significantly increased abnormal spindle organization and abnormal chromosome alignment. When compared with their negative control, Dicer siRNA injection significantly increased abnormal spindle organization, abnormal chromosome alignment, or both abnormal spindle organization and abnormal chromosome alignment in both CFOs (21.9% vs. 0.9%, 8.9% vs. 3.9%, and 3.2% vs. 0%, respectively) (Fig. 2) and COCs (17.7% vs. 0%, 5.6% vs. 2.4%, and 3.5% vs. 0%, respectively) (Fig. 2). Overall, Dicer siRNA significantly increased the total abnormal spindle and chromosome organization rates in CFO (27.6% vs. 4.8%, p < 0.0001) and in COCs (19.9% vs. 2.4%, p < 0.0001) (Fig. 2). Figure 3 illustrates phenotypes of some normal and abnormal spindle and chromosome organizations. This data imply that incompletion of meiotic maturation may be due to spindle disorganization and chromosome misalignment.
-
Dicer siRNA significantly reduced the transcripts of Dicer and miRNAs in oocytes
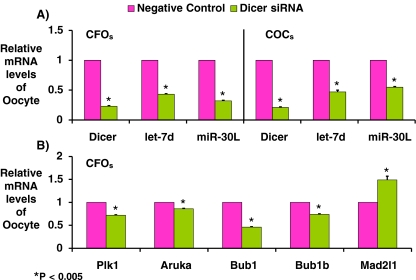
Transcripts of Dicer and the two studied miRNAs (i.e., let 7d and miR30c) in oocytes were detected quantitatively by real time RT-PCR. Dicer siRNA significantly reduced the transcripts of Dicer, let 7d and miR-30c to 23%, 43%, and 32% in CFOs, respectively. Similarly, in COCs, Dicer siRNA significantly reduced the transcripts of Dicer, let 7d and miR-30c to 21%, 47%, and 55%, respectively (Fig. 4a). Thus, Dicer siRNA can also effectively knockdown expression of miRNAs in mouse oocytes.
-
Dicer siRNA simultaneously reduced the transcriptions of genes related to spindle formation proteins and spindle checkpoint regulation
Along with reduction of Dicer and miRNA expression, Dicer siRNA simultaneously altered the expressions of Bub1, Bub1b, and Mad2l1 (spindle checkpoints) to 46%, 74%, and 149%, respectively (Fig. 4b) and reduced the expressions of Plk1 and AURKA (spindle formation proteins) to72% and 86% respectively.
-
Depletion of Bub1b by RNAi causes MI arrest, misaligned chromosomes and abnormal meiotic spindle assembly in oocytes
To further investigate the roles of Bub1 and Bub1b on the regulation of oocyte meiosis, both siRNA of Bub1 and Bub1b and negative control siRNA were microinjected to CFOs. RNAi efficiency was examined by real-time RT-PCR. As compared to its negative controls, siRNA injection significantly reduced Bub1 and Bub1b expression to 14% and 6% in the MI stage oocytes and 22% and 9% in the MII staged oocytes, respectively (data not shown). With no doubt, the expression of Bub1 and Bub1b were successfully knockdown by RNAi as well. We further found that 70% of Bub1bdepleted and 19% of Bub1depleted oocytes were arrested at the MI stage. Improper homologous chromosome segregation and lagging chromosomes were observed in 27% of Bub1b-depleted oocytes. The above experiments were conducted in triplicates.
-
The effect of Dicer siRNA can be reversed by ooplasmic replacement
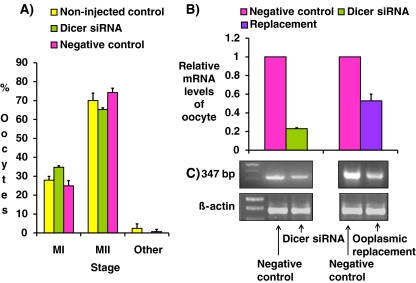
The reversibility of Dicer siRNA effects was tested by ooplasma replacement as described in Materials and Methods. After ooplasma replacement, most of Dicer1 siRNA injected oocytes could escape from MI arrest, followed by first polar bodies extrusion, and finally reached to the MII stage. Their oocyte maturation rates shown by percentages of oocyte at the MII stage were not significantly different among Dicer siRNA, negative control, and non-injected control (65.3%. 74.2% and 70.0%, p = 0.333) (Fig. 5a). The relative oocyte Dicer mRNA levels after ooplasma replacement increased from 23% to 53% of their negative controls (Fig. 5b). Meanwhile, the amplicons of RT-PCR had further been identified as 347 bp Dicer mRNA by gel electrophoresis using beta actin as an internal control (Fig. 5c). Our data showed that, ooplasmic replacement can neutralize the effect of Dicer siRNA injection.
Fig. 1.
MII formation rates. After microinjection and IVM, the distribution of GV, MI, MII and degenerated oocytes in non-injected control, Dicer siRNA and negative control of CFOs (right panel) and COCs (left panel) were recorded for comparison. Data are presented as means+S.D. of eight repeats of CFOs and seven repeats of COCs. Data were analyzed by Pearson’s Chi-square test. P < 0.05 was statistically significant. Data were analyzed by Pearson’s Chi square paired test. P < 0.05 was considered statistically significant. * indicate significant difference from its negative controls
Fig. 2.
Abnormal spindle organization rates after Dicer siRNA injection. After microinjection and IVM, oocytes were examined for their spindle integrity by immunofluorescence staining as described in text. The rates of abnormal spindle organization (shown as spindle), chromosomal misalignment (shown as chromosome), abnormal spindle organization and chromosomal alignment (shown as both), and total abnormal oocytes (shown as total) in CFOs (right panel) or in COCs (left panel) were illustrated. Data are presented as means+S.D. of eight repeats of CFOs and seven repeats of COCs. Data were analyzed by Pearson’s Chi-square test. P < 0.05 was statistically significant. Data were analyzed by Pearson’s Chi square test. P < 0.05 was considered statistically significant. * indicates significant difference from its negative controls, p < 0.005. ** indicates significant difference from its negative controls, p < 0.0001
Fig. 3.
Dicer siRNA causes a various types of spindle and chromosome disorganizations. Oocytes in negative control and Dicer siRNA groups were fixed with 4% parafirnaldehyde and stained with alphe tubulin immunofluroscence staining (green) and DNA was counterstained with DAPI (red). Representative images are shown. Bar, 25 um. a normal spindle and chromosomal organization. Oocytes exhibited single barrel-shaped MI spindles with chromosomes congregated at the metaphase plate. b abnormal spindle organization. Oocytes exhibited irregularly dispersed spindle microtubules. c chromosomal misaligned. Oocytes exhibited chromosomes scattering away from the metaphase plate. d abnormal spindle organization plus chromosomal misalignment. Oocytes were with irregularly dispersed spindle microtubules and with chromosomes scattered away from metaphase plate
Fig. 4.
Relative mRNA levels of MII oocytes were examined by real-time RT-PCR. After IVM, mRNAs of individual MII oocytes were extracted to detect various gene expressions by quantitative real-time RT-PCR. The 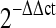 method was used to calculate the relative changes in gene expression. Data are presented as mean+S.D. Three replicates of 20 oocytes each were assayed from each group and beta actin was used as an internal standard. a Relative mRNA levels of Dicer and two miRNAs (let 7d and miRi30C) from Dicer siRNA and negative control injected groups. b Relative mRNA levels of Plk1and AURKA (spindle proteins) and Bub1, Bub1b and Mad2l1 (spindle checkpoints) from Dicer siRNA and negative control injected groups. Data were analyzed by a student t-test. P < 0.05 was considered statistically significant. * indicates significant difference from its negative control, p < 0.005
method was used to calculate the relative changes in gene expression. Data are presented as mean+S.D. Three replicates of 20 oocytes each were assayed from each group and beta actin was used as an internal standard. a Relative mRNA levels of Dicer and two miRNAs (let 7d and miRi30C) from Dicer siRNA and negative control injected groups. b Relative mRNA levels of Plk1and AURKA (spindle proteins) and Bub1, Bub1b and Mad2l1 (spindle checkpoints) from Dicer siRNA and negative control injected groups. Data were analyzed by a student t-test. P < 0.05 was considered statistically significant. * indicates significant difference from its negative control, p < 0.005
Fig. 5.
Effect of Dicer siRNA can be rescued by ooplasmic replacement. Ooplasmic replacement was performed as described in text. The experiments were performed three times and data are presented as mean+S.D. a shows the maturation rates of Dicer siRNA, negative control, and non-injected control. b shows relative mRNA of Dicer in negative control, Dicer siRNA and ooplasmic replacement. c shows the identification of amplicons of real-time RT-PCR by gel electrophoresis and beta actin is used as internal control. Data in a were analyzed by a Pearson’s Chi square test. P < 0.05 was considered statistically significant. The maturation rates (judged by percentages of oocyte at the MII stage) were not significantly different among the three studied groups. P = 0.333
Discussion
The new emerging technique of using RNAi for post-transcriptional gene silencing has been found to be a useful tool to elucidate the function of specific genes [41]. In past decades, genomic DNAs have been sequenced and the transcripts of their gene expressions had been profiled. Yet, the functions of these genes remained unknown. The advent of RNAi technology appears to be a tool to link gene sequence and gene function together. This technology requires the introduction of long dsRNA, siRNA or short hairpin RNA (shRNA) into oocytes, embryos or cells by transfection, electroporation, or microinjection [38, 42–45]. In this study, we used microinjection to introduce Dicer siRNA into mouse GV oocytes to deplete endogenous transcripts. Our data showed that the injection of Dicer siRNA: a) significantly reduced oocyte maturation rate (Fig. 1), b) increased spindle disorganization and chromosome misalignment rates (Figs. 2 and 3), and c) reduced the expression of Dicer, let 7d and miR-30c , spindle formation proteins and spindle check point genes (Fig. 4a and b). This data indicated that injection of Dicer siRNA could effectively knockdown the expression of Dicer with consequences of reducing meiotic maturation, possibly due to disorganization of spindle and chromosome misalignment. These results were in line with that obtained from knockout mouse experiment in which oocytes lacking Dicer from knockout mice were reported to be arrested in meiosis I with multiple disorganized spindles and severe chromosome congression defects [35, 39].
As expected, our experiments clearly demonstrated that the expression of Dicer was immediately and effectively reduced after Dicer siRNA injection (Fig. 4a). Since Dicer is responsible for generating miRNAs, injection of Dicer siRNA was expected to knockdown the expression of miRNAs as well [35, 39, 46]. In congruity with this prediction, the two studied miRNAs (i.e., let-7d and miR-30c) were significantly co-suppressed with Dicer after injection (Fig. 4a). Along with the reduction of Dicer and miRNAs expression, oocytes injected with Dicer siRNAs were unable to complete meiosis I. We observed that most oocytes arrested at the MI stage with significantly low percentages of oocytes advancing to the MII stage (Fig. 1a, b). Thus, the presence of Dicer appeared to be required for the completion of meiosis I. Our immunostaining further revealed that many Dicer siRNA injected oocytes contained defects in meiotic spindle organization and chromosome alignment (Fig. 2a, b). This indicated that Dicer and miRNAs are essential for meiotic spindle integrity which is necessary for normal meiotic maturation.
Oocytes with high Dicer and miRNAs activity (i.e., negative control group) were associated with high MII formation rates and low spindle disorganization and chromosome misalignment rates (Figs. 1a, b, 3a, b, and 5a). On the other hand, low activity of Dicer and miRNAs (i.e., Dicer siRNA group) were associated with low MII formation rates and high spindle disorganization and chromosome misalignment rates (Figs. 1a, b, 3a, b, and 5a). Thus, Dicer and miRNAs activities were positively correlated with spindle organization and completion of meiosis. Data also implied that high expressions of Dicer and miRNAs insured normal oocyte maturation and could be a marker for oocyte quality.
Dicer may function through its downstream miRNAs to maintain integrity of spindle and chromosome organization. As mentioned above, miRNAs were able to negatively control gene expression post-transcriptionally [1–4]. Thus, miRNAs in oocytes could control maternal mRNAs storage, translation, and degradation. In fact, miRNAs were reported to directly or indirectly control more than one third of the maternal genes expressed in oocytes [35]. A large portion of maternal transcripts were up-regulated in Dicer knockout oocytes. Previous expression analysis has identified a set of maternal mRNAs that was especially degraded during meiotic maturation [47]. Of interest to note, there was a strong correlation between these degraded mRNAs and those up-regulated transcripts upon the knockdown of Dicer [39]. Many of these up-regulated genes, such as kifl6b (kinesin family member 16B), kif4 (kinesin family member 2C), Kif4 (Kinesis family 4), Morel (microtubule-associated protein 1), Mtap2 (microtubule-associated protein 2), Mtap7 (microtubule-associated protein 7) and Tubb5 (tubulin beta 5), were microtubule-associated genes and had putative binding sites to let-7d and miR-30c [39]. In other words, some microtubule related genes might be potential target genes of let-7d and miR-30c. Therefore, reduction of let-7d and miR-30c expression in Dicer siRNA oocytes would ultimately reduce the transcription and translation of these microtubule-associated genes. Microtubule-associated proteins (constitutional proteins for spindle formation and kinesins) can generate forces that move chromosomes during meiosis. Abnormality in the regulation of these gene expressions might contribute to abnormal spindle formation, kinetochore assembly, and chromosome segregation.
Indeed, our data demonstrated that Dicer siRNA acted directly to interrupt the transcripts of some spindle proteins such as plk1 and AURKA( Fig. 4b) and altered the transcripts of some spindle assembly checkpoint (SAC) such as Bub1, Bub1b, Mad2l1 (Fig. 4b), which led to an abnormal spindle formation and assembly, and delay in spindle checkpoint. Delaying spindle checkpoint would prolong metaphase I arrest and result in arresting a good portion of oocytes at MI stage (Fig. 1a, b), allowing only a small portion proceeded to the MII stage (Fig. 1a, b). To characterize the role of Bub1 and Bub1b in oocyte maturation, we employed RNAi gene silencing approach to deplete endogenous transcripts in the GV stage of oocytes. Interestingly, RNAi not only dramatically knockdown transcripts of Bub1 and Bub1b, but also arrested majority of the depleted oocytes at the MI stage with high incidence of abnormal spindle organization and chromosomal misalignment (see section 5 of Results). This may imply that Bub1 and Bub1b might work directly on organization of SAC. Many studies have shown that Bub1 and Bub1b like other major checkpoint proteins (Bub3, Mad1-3 and Maps1), act as the controllers of prophase I arrest in response to the status of the spindle assembly [48, 49]. In most cases, SACs-depleted oocytes could abolish the spindle checkpoint arrest in response to spindle damage [48]. However, recent findings revealed that Bub1b may work to control an important anaphase-trigger, anaphase-promoting complex (APC) [49]. APC is a complex of several proteins (APCCdc 20 or APCCdh1), which is activated during meiosis to initiate chromatid separation and entrance into anaphase. The APCCdc20 protein complex has two main downstream targets, securin and cyclinB1. APCCdc20 and APCCdh1 target securin for destruction, enabling the consequent degradation of cohesin and sister chromatid separation. Using the morpholino based gene-silencing method, other labs have proven that only 6% of Bub1b-depleted oocytes could extrude first polar bodies [49]. Their study also indicated that after Bub1b depletion, Cdh1 was significantly decreased by 50% to 70% throughout prometaphase I, eventually resulting in a notably raised level of securin as high as twice the control value in Bub1b-depleted oocytes. This is suggestive that meiotic defects in Bub1b-depleted oocytes are due to reduced activity of the anaphase-promoting complex by diminishing levels of coactivator Cdh1.
Our data may reveal the molecular mechanisms of Dicer siRNA effects. Injection of Dicer siRNA may trigger a cascade reduction of gene expressions, starting from Dicer to miRNAs then to SAC proteins and checkpoints. The reduction of SAC proteins and checkpoints were key factors responsible for abnormal spindle organization and chromosomal alignments. Incomplete meiotic maturation caused by Dicer siRNA could be prevented through ooplasmic replacement (Fig. 5a), strongly indicating there was a crucial ooplasmic factor responsible in neutralizing the silencing effect of Dicer siRNA. Ultimately, we have identified this factor as 347 bp Dicer mRNA (Fig. 5b). Ooplasmic replacement did increase Dicer transcripts in recipient oocytes. This further confirmed that increasing the expressions of Dicer would improve oocyte quality and regulating Dicer expression may be a new strategy to improve oocyte quality.
In conclusion, we have detected Dicer transcripts in mouse oocytes and have demonstrated that Dicer siRNA effectively reduced Dicer and miRNAs expression. Accompanying this reduction, the injected oocytes significantly reduced their maturation rates and increased their spindle disorganization rates. Thus, Dicer and miRNA were essential for spindle organization and meiotic maturation. Dicer may prevent miRNAs from regulating the expression of microtubule related genes. Dicer may also act indirectly to reduce or alter the transcripts of spindle proteins and checkpoints. Dicer and miRNAs appeared to play important roles during oogenesis and can serve as potential markers of oocyte quality. Controlling the expression of Dicer and miRNAs may become a novel strategy in controlling the quality of oocytes.
Footnotes
Capsule
Dicer and miRNAs are essential for spindle integrity and completion of meiosis I. They appear to play important roles during oogenesis and can serve as potential markers of oocyte quality.
References
- 1.Ambros B. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/S0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Lai EC. MicroRNAs: runts of the genome assert themselves. Curr Biol. 2003;13:R925–R936. doi: 10.1016/j.cub.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 4.He L, Hannon GJ. MicroRNAs: Small RNAS with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 5.Lee Y, Ahn C, Han J, Cho H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 6.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 8.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: fenomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Bagga S, Bracht J, Hunger S, Massirer K, Holtz J, Eachus R, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 11.Aufsatz W, Mette MF, Winden J, Matzke AJ, Matzke M. RNA-directed DNA methylation in Arabidopsis. Proc Natl Acad Sci USA. 2002;99(Suppl. 4):16499–16506. doi: 10.1073/pnas.162371499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mette MF, Aufsatz W, Winden J, Matzke MA, Matzke AJ. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 2000;19:5194–5201. doi: 10.1093/emboj/19.19.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ketting RF, Haverkamp TH, Luenen HG, Plasterk RH. Mut-7 of C. Elegans, required for transposon silencing and RNA interference is a homolog of Werner syndrome helicase and RnaseD. Cell. 1999;99:133–141. doi: 10.1016/S0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- 14.Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, et al. The rde-1 gene, RNA interference and transposon silencing in C. Elegans. Cell. 1999;99:123–132. doi: 10.1016/S0092-8674(00)81644-X. [DOI] [PubMed] [Google Scholar]
- 15.Svoboda P, Stein P, Hayashi H, Schultz RM. Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development. 2000;127(19):4147–4156. doi: 10.1242/dev.127.19.4147. [DOI] [PubMed] [Google Scholar]
- 16.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAS mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 17.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAS. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 19.Brusselmans K, Schrijver E, Verhoeven G, Swinnen JV. RNA interference-mediated silencing of the acetyl-CoA-carboxylase-alpha gene induces growth inhibition and apoptosis of prostate cancer cells. Cancer Res. 2005;65(15):6719–6725. doi: 10.1158/0008-5472.CAN-05-0571. [DOI] [PubMed] [Google Scholar]
- 20.Wianny F, Zernicka-Goetz MS. Specific interference with gene function by double-stranded RNA in early mouse development. Nat Cell Biol. 2000;2(2):70–75. doi: 10.1038/35000016. [DOI] [PubMed] [Google Scholar]
- 21.Nganvongpanit K, Müller H, Rings F, Hoelker M, Jennen D, Tholen E, et al. Selective degradation of maternal and embryonic transcripts in in vitro produced bovine oocytes and embryos using sequence specific double-stranded RNA. Reproduction. 2006;131(5):861–874. doi: 10.1530/rep.1.01040. [DOI] [PubMed] [Google Scholar]
- 22.Cabot RA, Prather RS. Cleavage stage porcine embryos may have differing developmental requirements for karyopherins alpha2 and alpha3. Mol Reprod Dev. 2003;64(3):292–301. doi: 10.1002/mrd.10238. [DOI] [PubMed] [Google Scholar]
- 23.Li M, Li S, Yuan J, Wang ZB, Sun SC, Schatten H, et al. Bub3 is a spindle assembly checkpoint protein regulating chromosome segregation during mouse oocyte meiosis. PLoS One. 2009;4(11):e7701. doi: 10.1371/journal.pone.0007701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J, Tam WL, Tong GQ, Wu Q, Chan HY, Soh BS, et al. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat Cell Biol. 2006;8(10):1114–1123. doi: 10.1038/ncb1481. [DOI] [PubMed] [Google Scholar]
- 25.Schindler K, Schultz RM. CDC14B acts through FZR1 (CDH1) to prevent meiotic maturation of mouse oocytes. Biol Reprod. 2009;80(4):795–803. doi: 10.1095/biolreprod.108.074906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Svoboda P. RNA silencing in mammalian oocytes and early embryos. Curr Top Microbiol Immunol. 2008;320:225–256. doi: 10.1007/978-3-540-75157-1_11. [DOI] [PubMed] [Google Scholar]
- 27.Memili E, First NL. Zygotic and embryonic gene expression in cow: a review of timing and mechanisms of early gene expression as compared with other species. Zygote. 2000;8:87–96. doi: 10.1017/S0967199400000861. [DOI] [PubMed] [Google Scholar]
- 28.Dean J. Oocyte-specific genes regulate follicle formation, fertility and early mouse development. J Reprod Immunol. 2002;53:171–180. doi: 10.1016/S0165-0378(01)00100-0. [DOI] [PubMed] [Google Scholar]
- 29.Yang WJ, Yang DD, Na S, Sandusky GE, Zhang Q, Zhao G. Dicer is required for embryonic angiogenesis during mouse development. J Biol Chem. 2005;280:9330–9335. doi: 10.1074/jbc.M413394200. [DOI] [PubMed] [Google Scholar]
- 30.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 31.Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol. 2002;12:1484–1485. doi: 10.1016/S0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wienholds E, Koudijs MJ, Eeden FJ, Cuppen E, Plasterk RH. The microRNA-producing enzyme Dicer1 is essential for mouse development. Nature Genet. 2003;35:215–217. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- 34.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, et al. Dicer is essential for mouse development. Nature Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 35.Tang F, Kaneda M, O’Carroll D, Hajkova P, Barton SC, Sun YA, et al. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007;21:644–648. doi: 10.1101/gad.418707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amanai M, Brahmajosyula M, Perry AC. A restricted role for sperm-borne microRNAs in mammalian fertilization. Biol Reprod. 2006;75:877–884. doi: 10.1095/biolreprod.106.056499. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, et al. Identification and characterization of two novel classes of small RNAS in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAS in testes. Genes Dev. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Svoboda P, Stein P, Hayashi H, Schultz RM. Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development. 2000;127:4147–4156. doi: 10.1242/dev.127.19.4147. [DOI] [PubMed] [Google Scholar]
- 39.Murchison EP, Stein P, Xuan ZY, Pan H, Zhang MQ, Schultz RM, et al. Critical roles for dicer in the female germline. Genes Dev. 2007;21:682–693. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaffe LA, Norris RP, Freudzon M, Ratzan WJ, Mehlmann LM. Microinjection of follicle-enclosed mouse oocytes. Method Mol Biol. 2009;51:157–173. doi: 10.1007/978-1-59745-202-1_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fire A. RNA-triggered gene silencing. Trends Genet. 1999;15:358–363. doi: 10.1016/S0168-9525(99)01818-1. [DOI] [PubMed] [Google Scholar]
- 42.Wianny F, Zernicka-Goetz M. Specific interference with gene function by double-stranded RNA in early mouse development. Nat Cell Biol. 2000;2:70–75. doi: 10.1038/35000016. [DOI] [PubMed] [Google Scholar]
- 43.Grabarek JB, Plusa B, Glover DM, Zernicka-Goetz M. Efficient delivery of dsRNA into zona-enclosed mouse oocytes and preimplantation embryos by electroporation. Genesis. 2002;32:269–276. doi: 10.1002/gene.10076. [DOI] [PubMed] [Google Scholar]
- 44.Paradis F, Vigneault C, Robert C, Sirard MA. RNA interference as a tool to study gene function in bovine oocytes. Mol Reprod Dev. 2005;70:111–121. doi: 10.1002/mrd.20193. [DOI] [PubMed] [Google Scholar]
- 45.Siddall LS, Barcroft LC, Watson AJ. Targeting gene expression in the preimplantation mouse embryo using morpholino antisense oligonucleotides. Mol Reprod Dev. 2002;63:413–421. doi: 10.1002/mrd.10202. [DOI] [PubMed] [Google Scholar]
- 46.Murchison EP, Hannon GJ. MiRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr Opin Cell Biol. 2004;16:223–229. doi: 10.1016/j.ceb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 47.Su YQ, Sugiura K, Woo Y, Wigglesworth K, Kamdar S, Eppig AJ, et al. Selective degradation of transcripts during meiotic maturation of mouse oocytes. Dev Biol. 2006;302:104–117. doi: 10.1016/j.ydbio.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yin S, Wang Q, Liu JH, Ai JS, Liang CG, Hou Y, et al. Bub1 prevents chromosome misalignment and precocious anaphase during mouse oocyte meiosis. Cell Cycle. 2006;5(18):2130–2137. doi: 10.4161/cc.5.18.3170. [DOI] [PubMed] [Google Scholar]
- 49.Homer H, Gui L, Carroll J. A spindle assembly checkpoint protein functions in prophase I arrest and prometaphase progression. Science. 2009;326(5955):991–994. doi: 10.1126/science.1175326. [DOI] [PMC free article] [PubMed] [Google Scholar]